

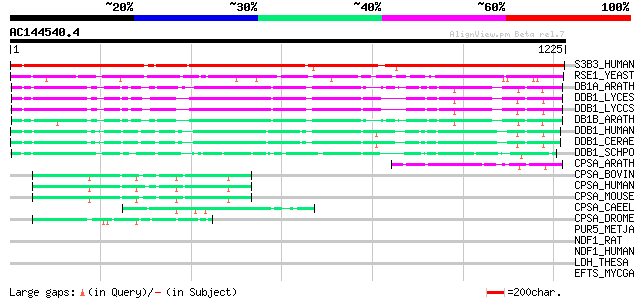
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144540.4 - phase: 0
(1225 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
S3B3_HUMAN (Q15393) Splicing factor 3B subunit 3 (Spliceosome as... 1363 0.0
RSE1_YEAST (Q04693) Pre-mRNA splicing factor RSE1 305 5e-82
DB1A_ARATH (Q9M0V3) DNA damage binding protein 1a (UV-damaged DN... 223 2e-57
DDB1_LYCES (Q6QNU4) DNA damage binding protein 1 (UV-damaged DNA... 219 4e-56
DDB1_LYCCS (Q6E7D1) DNA damage binding protein 1 (UV-damaged DNA... 218 6e-56
DB1B_ARATH (O49552) DNA damage binding protein 1b (UV-damaged DN... 210 2e-53
DDB1_HUMAN (Q16531) DNA damage binding protein 1 (Damage-specifi... 206 4e-52
DDB1_CERAE (P33194) DNA damage binding protein 1 (Damage-specifi... 203 2e-51
DDB1_SCHPO (O13807) DNA damage binding protein 1 (Damage-specifi... 67 2e-10
CPSA_ARATH (Q9FGR0) Probable cleavage and polyadenylation specif... 67 3e-10
CPSA_BOVIN (Q10569) Cleavage and polyadenylation specificity fac... 55 1e-06
CPSA_HUMAN (Q10570) Cleavage and polyadenylation specificity fac... 55 2e-06
CPSA_MOUSE (Q9EPU4) Cleavage and polyadenylation specificity fac... 54 2e-06
CPSA_CAEEL (Q9N4C2) Probable cleavage and polyadenylation specif... 54 2e-06
CPSA_DROME (Q9V726) Cleavage and polyadenylation specificity fac... 51 2e-05
PUR5_METJA (Q57656) Phosphoribosylformylglycinamidine cyclo-liga... 35 0.96
NDF1_RAT (Q64289) Neurogenic differentiation factor 1 (NeuroD1) ... 35 1.2
NDF1_HUMAN (Q13562) Neurogenic differentiation factor 1 (NeuroD1... 35 1.2
LDH_THESA (Q7X5C9) L-lactate dehydrogenase (EC 1.1.1.27) (L-LDH) 35 1.2
EFTS_MYCGA (Q7NC21) Elongation factor Ts (EF-Ts) 35 1.2
>S3B3_HUMAN (Q15393) Splicing factor 3B subunit 3 (Spliceosome
associated protein 130) (SAP 130) (SF3b130) (Pre-mRNA
splicing factor SF3b 130 kDa subunit) (STAF130)
Length = 1217
Score = 1363 bits (3528), Expect = 0.0
Identities = 691/1244 (55%), Positives = 914/1244 (72%), Gaps = 51/1244 (4%)
Query: 1 MYLYNLTLQRPTGIVCAINGNFSGSDDGITQEIVVARGKVLELLRPD-KFGRIQSILSVQ 59
M+LYNLTLQR TGI AI+GNFSG+ QEIVV+RGK+LELLRPD G++ ++L+V+
Sbjct: 1 MFLYNLTLQRATGISFAIHGNFSGTKQ---QEIVVSRGKILELLRPDPNTGKVHTLLTVE 57
Query: 60 VFGTIRSLSQFRLTGAQKDFIVVGSDSGRIVILDYNKQKNVFDKIHQETFGKSGCRRIVP 119
VFG IRSL FRLTG KD+IVVGSDSGRIVIL+Y KN+F+KIHQETFGKSGCRRIVP
Sbjct: 58 VFGVIRSLMAFRLTGGTKDYIVVGSDSGRIVILEYQPSKNMFEKIHQETFGKSGCRRIVP 117
Query: 120 GQYLAIDPKGRAVMIAACEKKKLVYVLNRDSLARLTISSPLEANKSHTIVFSICAVDCGF 179
GQ+LA+DPKGRAVMI+A EK+KLVY+LNRD+ ARLTISSPLEA+K++T+V+ + VD GF
Sbjct: 118 GQFLAVDPKGRAVMISAIEKQKLVYILNRDAAARLTISSPLEAHKANTLVYHVVGVDVGF 177
Query: 180 ENPIFAAIELDCSDADQDATGVAASQAQKHLIFYELDLGLNHVSRKWSDQVDNGANMLVT 239
ENP+FA +E+D +A D TG AA+ Q+ L FYELDLGLNHV RK+S+ ++ N L+T
Sbjct: 178 ENPMFACLEMDYEEAGNDPTGEAAANTQQTLTFYELDLGLNHVVRKYSEPLEEHGNFLIT 237
Query: 240 VPGGADGPSGVLVCAENFVIYKNQG-HPDVRAVIPRRA-DLP-AERGVLIVSAAMHKTKN 296
VPGG+DGPSGVL+C+EN++ YKN G PD+R IPRR DL ERG++ V +A HKTK+
Sbjct: 238 VPGGSDGPSGVLICSENYITYKNFGDQPDIRCPIPRRRNDLDDPERGMIFVCSATHKTKS 297
Query: 297 LKPEEFKLFFLLQTEYGDIFKVTLTDGGGDRVSELNIKYFDTIAVAVSICVLKSGFLFAA 356
+ FF QTE GDIFK+TL + D V+E+ +KYFDT+ VA ++CVLK+GFLF A
Sbjct: 298 M------FFFWAQTEQGDIFKITL-ETDEDMVTEIRLKYFDTVPVAAAMCVLKTGFLFVA 350
Query: 357 SEFGNHALYQFKGIGDDDNDVGASSASLMETEEGFQPVFFQPRRLKNLVRIDQVESLMPV 416
SEFGNH LYQ +GDDD + SSA +E + F FFQPR LKNLV +D+++SL P+
Sbjct: 351 SEFGNHYLYQIAHLGDDDEEPEFSSAMPLEEGDTF---FFQPRPLKNLVLVDELDSLSPI 407
Query: 417 MDMKVSNLFEEETPQIFTLCGRGPRSSLRIMRTGLAVSEMAVSKLPGIPSAVWTVKKNVM 476
+ ++++L E+TPQ++ CGRGPRSSLR++R GL VSEMAVS+LPG P+AVWTV++++
Sbjct: 408 LFCQIADLANEDTPQLYVACGRGPRSSLRVLRHGLEVSEMAVSELPGNPNAVWTVRRHIE 467
Query: 477 DEFDAYIVVSFTNATLVLSIGETADEVSDSGFLDTAPSLAVSLIGDDSLMQVHPNGIRHI 536
DEFDAYI+VSF NATLVLSIGET +EV+DSGFL T P+L+ SL+GDD+L+QV+P+GIRHI
Sbjct: 468 DEFDAYIIVSFVNATLVLSIGETVEEVTDSGFLGTTPTLSCSLLGDDALVQVYPDGIRHI 527
Query: 537 REDGRTNEWQTSGKRTIAKVGSNRLQVVIALNGGELIYFEVDVTGQLME-VERHEMSGDV 595
R D R NEW+T GK+TI K N+ QVVIAL GGEL+YFE+D +GQL E ER EMS DV
Sbjct: 528 RADKRVNEWKTPGKKTIVKCAVNQRQVVIALTGGELVYFEMDPSGQLNEYTERKEMSADV 587
Query: 596 ACLDIAPVPKGRLRSRFLAVGSYDKTIRILSLDPDDCMQTLGIQSLSSAPESLLFLEVQA 655
C+ +A VP G RSRFLAVG D T+RI+SLDP DC+Q L +Q+L + PESL +E+
Sbjct: 588 VCMSLANVPPGEQRSRFLAVGLVDNTVRIISLDPSDCLQPLSMQALPAQPESLCIVEM-- 645
Query: 656 SVGGEDGADHPAS------LFLNAGLQNGVLSRTVVDMVTGLLSDTRSRFLGLKAPKLFP 709
GG + D L+LN GLQNGVL RTV+D VTG LSDTR+R+LG + KLF
Sbjct: 646 --GGTEKQDELGERGSIGFLYLNIGLQNGVLLRTVLDPVTGDLSDTRTRYLGSRPVKLFR 703
Query: 710 IIVRGKRAMLCLSSRPWLGYIHQGHFLLTPLSYETLEYAASFSSDQCFEGVVSVASEALR 769
+ ++G+ A+L +SSR WL Y +Q F LTPLSYETLE+A+ F+S+QC EG+V++++ LR
Sbjct: 704 VRMQGQEAVLAMSSRSWLSYSYQSRFHLTPLSYETLEFASGFASEQCPEGIVAISTNTLR 763
Query: 770 IFTVERLGETFNQNVIPLRYTPRKFVLQPKRKLLVVIESDQGALTAEEREAARKECFEAA 829
I +E+LG FNQ PL+YTPRKFV+ P+ L++IE+D A T EA
Sbjct: 764 ILALEKLGAVFNQVAFPLQYTPRKFVIHPESNNLIIIETDHNAYT------------EAT 811
Query: 830 HAGENKTGSEDQMENGGEDEDN----------DDSLSDEHYGYPKSESDKWVSCIRVLDP 879
A + +E+ +E GEDE +++L + +G PK+ + +W S IRV++P
Sbjct: 812 KAQRKQQMAEEMVEAAGEDERELAAEMAAAFLNENLPESIFGAPKAGNGQWASVIRVMNP 871
Query: 880 RTGNTTCLLELQENEAAFSICTVNFHDKEYGTLLAVGTAKGLQFTPKRSLTAGFIHIYRF 939
GNT L++L++NEAAFS+ F + + VG AK L P RS+ GF++ Y+
Sbjct: 872 IQGNTLDLVQLEQNEAAFSVAVCRFSNTGEDWYVLVGVAKDLILNP-RSVAGGFVYTYKL 930
Query: 940 LDDGRSLELLHKTQVEGVPLALCQFQGRLLAGIGPVLRLYDLGKRRLLRKCENKSFPSSI 999
+++G LE LHKT VE VP A+ FQGR+L G+G +LR+YDLGK++LLRKCENK + I
Sbjct: 931 VNNGEKLEFLHKTPVEEVPAAIAPFQGRVLIGVGKLLRVYDLGKKKLLRKCENKHIANYI 990
Query: 1000 VSIHAYRDRIYVGGIQESFHYCKYRRDENQLYIFADDSVPRWLTSSYHIDFDTMAGADKF 1059
I R+ V +QESF + +Y+R+ENQL IFADD+ PRW+T++ +D+DT+AGADKF
Sbjct: 991 SGIQTIGHRVIVSDVQESFIWVRYKRNENQLIIFADDTYPRWVTTASLLDYDTVAGADKF 1050
Query: 1060 GNIFFARLPQDVSDEIEEDPTGGKIKWEQGKLNGAPNKVEEIVQFHVGDVITSLQKASLV 1119
GNI RLP + +DE++EDPTG K W++G LNGA K E I+ +HVG+ + SLQK +L+
Sbjct: 1051 GNICVVRLPPNTNDEVDEDPTGNKALWDRGLLNGASQKAEVIMNYHVGETVLSLQKTTLI 1110
Query: 1120 PGGGECIVYGTVMGSVGALHAFTSRDDVDFFSHLEMHMRQDNPPLCGRDHMAYRSAYFPV 1179
PGG E +VY T+ G +G L FTS +D DFF H+EMH+R ++PPLCGRDH+++RS YFPV
Sbjct: 1111 PGGSESLVYTTLSGGIGILVPFTSHEDHDFFQHVEMHLRSEHPPLCGRDHLSFRSYYFPV 1170
Query: 1180 KDVIDGDLCEQFPTLPMDLQRKIADELDRTPGEILKKLEEVRNK 1223
K+VIDGDLCEQF ++ + Q+ +++ELDRTP E+ KKLE++R +
Sbjct: 1171 KNVIDGDLCEQFNSMEPNKQKNVSEELDRTPPEVSKKLEDIRTR 1214
>RSE1_YEAST (Q04693) Pre-mRNA splicing factor RSE1
Length = 1361
Score = 305 bits (781), Expect = 5e-82
Identities = 349/1384 (25%), Positives = 602/1384 (43%), Gaps = 247/1384 (17%)
Query: 1 MYLYNLTLQRPTGIVCAINGNFSGSDDGITQE---IVVARGKVLELLRPDKFGRIQSILS 57
+YLY+LTL++ T V + G+F + G +E + VA LEL G ++ I
Sbjct: 58 LYLYHLTLKKQTNFVHSCIGHFVDLEAGSKREQSQLCVATETHLELYDTAD-GELKLIAK 116
Query: 58 VQ-VFGTIRSLSQFRL--TGAQKD------FIVVGSDSGRIVILDYNKQKNVF--DKIHQ 106
Q +F TI S+ L +G++ F+ + SDSG + I+ +
Sbjct: 117 FQNLFATITSMKSLDLPHSGSRAKASNWPTFLALTSDSGNLSIVQIIMHAGALRLKTLVN 176
Query: 107 ETFGKSGCRRIVPGQYLAIDPKGRAVMIAACEKKKLVYVLNRDSLARLTISSPLEANKSH 166
+ ++ RR+ P Y+ IDP GR +++++ E+ KL +++ D +L ISSPLE + H
Sbjct: 177 QPLTRTTLRRVSPISYMEIDPNGRCIILSSVEQNKLCFLV--DYAQKLRISSPLEIIRPH 234
Query: 167 TIVFSICAVDCGFENPIFAAIELDCSDADQDATGVAASQAQKHLIFYELDLGLNHVSRKW 226
+ + VD F NP F +E+D AA+Q HLIFY L+LGLNH+ +K
Sbjct: 235 MVTLDMAVVDVNFNNPCFVTLEIDN----------AATQLSVHLIFYVLELGLNHIVKKA 284
Query: 227 SDQVDNGANMLVTVPG---------------GADGPS----GVLVCAENFVIYKN-QGHP 266
V+ AN ++++P AD + V++ EN ++ K+ G
Sbjct: 285 DYLVNPSANFVLSLPDLSRYNITTSLSDNNYDADYDTLFNPFVVIGFENHILVKDMNGFF 344
Query: 267 DVRAVIPRRADLPA-ERGVLIVSAAMHKTKNLKPEEFKLFFLLQTEYGDIFKVTLTDGGG 325
++ IP+R+ + + V I+S + K KN F LLQ+ +GD+FK+T++
Sbjct: 345 SLKVEIPKRSITNSRHKNVTIISGIVQKLKN------DFFVLLQSNHGDLFKLTVSPDTN 398
Query: 326 DRVSEL-NIKYFDTIAVAVSICVLKSGFLFAASEFGNHALYQFKGIGDDDNDVGASSASL 384
DR L + YFDTI + + + K+G+LFA SE N+ L+QF+ +G + ND +++
Sbjct: 399 DRNRPLVQLSYFDTIQNSHQLHIFKNGYLFALSEMNNNFLFQFEKLGVEKNDF----SNV 454
Query: 385 METEEGFQPVFFQPR-RLKNLVRIDQVESLMPVMDMKVSNLFEEETPQIFTLCGRGPRSS 443
+ +++ + + F+P +L+NL + Q +L P + S + + I T +
Sbjct: 455 LTSKDPNKSLVFEPSIKLQNLSILSQQLNLNPSIK---SQIVSDSPLSIAT--KHFTNNK 509
Query: 444 LRIMRTGLAVSEMAVSKLPGIPSAVWTV-KKNVMDEFDAYIVVSFTNATLVLSIGE---- 498
+ + + S + + LP + +W + + + + ++F T++L I
Sbjct: 510 IITLTNAVNYSNLISTSLPPNATKLWLIPDPATTGDNNTLLFITFPKKTMILQIDNESME 569
Query: 499 --TADEVSDSGF-LDTAPSLAVSLIGDDSLMQVHPNGIRHIREDGRTN-----EWQTSGK 550
T DE + S F L ++ L+G S++QV +RHI G++ W
Sbjct: 570 ELTPDEATRSAFKLSQDTTIHTCLMGSHSIIQVCTAELRHIVPTGKSRYSNKLTWVPPAG 629
Query: 551 RTIAKVGSNRLQVVIALNGGELIYFEVDVTG-QLMEVERHEMSGDVACLDIAP----VPK 605
I S++ Q++I+L+ EL+YF++DV+ L+E+ H LD P + +
Sbjct: 630 IRIVCATSSKTQLIISLSNYELVYFKIDVSSDSLIELTTHPE------LDTMPSKVAIVQ 683
Query: 606 GRLRSRFLAVGSYDKTIRILSL--DPDDCMQTLGIQSLSSAPESLLFLEVQASVGGEDGA 663
+ LA+ + I+I+SL +D + + +Q +S ++ + +S+G
Sbjct: 684 DTQHADLLAIADNEGMIKIMSLKDQKEDFLTVISLQLVSEKISDMIMVR-DSSIG----- 737
Query: 664 DHPASLFLNAGLQNGVLSRTVVDMVTGLLSDTRSRFLGLKAPKLF--------------- 708
L L+ GL+NGV + + V G +D + RFLGLK L
Sbjct: 738 ----QLNLHVGLENGVYMKFHIGDVDGSFTDIKRRFLGLKPVSLSYLREISVSLNNEEEE 793
Query: 709 -------------PIIVRGKRAMLCL---SSRPWLGYIHQGHFLLTPLSYETLEYAASF- 751
I G + M C+ SS W+ Y + + + L + + + F
Sbjct: 794 EEEEDDDDEKEEEEINSSGAKWMSCVVCHSSSTWVSYTWKNVWTIRQLKDQNMLSCSKFV 853
Query: 752 SSDQCFEGVVSVASEALRIFTVERLGETFNQNVIPLRYTPRKFVLQPKRKLLVVIESDQG 811
++D GV S++S +G N + + + + + E+ G
Sbjct: 854 NADVAINGVCSISSSGRL-----NIGRVSNFPTLDNWFHVHESSVNKQ-------ENGGG 901
Query: 812 ALTAEEREAARKECFEAAHAGENKTGSEDQMENGGEDEDNDDSL--SDEHYGYPKSESDK 869
+ EE E +E E + + N N S+ D H G +
Sbjct: 902 DESNEEEEDEMEEEMEMLQISTFRPRTILSFPN------NPKSILFIDNHSGKKQCRISL 955
Query: 870 WV--SCIRVLDPRTGNTTCLLELQENEAAFSICTVNFHDKEYGTLLAVGTAKGLQFTPKR 927
+ C++ G++ L ++ ++ S ++F + ++ G KR
Sbjct: 956 QIDGECLKF-----GSSDHLYKILDDIDCVSAAIIDFTRQADHLIICAG--------DKR 1002
Query: 928 SLTAGFIHIYRFLDDGRSL----ELLHKTQVEGVPLALCQFQGRLLAGIGPVLRLYDLGK 983
LT Y+ L + L ELLH+T++ A+ +F+ LL +G + LY LGK
Sbjct: 1003 LLT------YKILVNKDKLSFDIELLHQTEIISPIHAMLKFKNFLLTAMGSTIVLYGLGK 1056
Query: 984 RRLLRKCENKSFPS--SIVSIHAYR-DRIYVGGIQESFHYCKYRRDENQLYIFADDSVPR 1040
++LLR+ ++ S IVS+H + +R+ VG I ES + N + DDSV R
Sbjct: 1057 KQLLRRSVTQTPVSITKIVSMHQWNYERLAVGDIHESVTLFIWDPAGNVFIPYVDDSVKR 1116
Query: 1041 WLTSSYHIDFDTMAGADKFGNIFFARLPQD----VSDEIEEDPTGGKIKWE------QGK 1090
+T +D T+ GAD++GN + R P + +S+ + + G IK+ Q K
Sbjct: 1117 HVTVLKFLDEATVIGADRYGNAWTLRSPPECEKIMSNHDPSELSNGAIKYPLDVITLQQK 1176
Query: 1091 LNGAPN------KVEEIVQFHVGDVITSLQ-KASLVPGGGECIVYGTVMGSVGALHAFTS 1143
L PN K + + F V D+IT SL +Y + G+VG S
Sbjct: 1177 L---PNTYDCKFKFQLLNHFFVNDIITDFHILDSLSNSDRPGCIYMGLQGTVGCFIPLLS 1233
Query: 1144 RDDVDFFSHLE--------------------MHMRQDNP--------------------- 1162
+ +V ++E +MR+++
Sbjct: 1234 KGNVFMMGNIENIMAEADDTFYLDYESRKKNNNMRKEDDEEESGSVVLQGRHGIEDEIIC 1293
Query: 1163 ----PLCGRDHMAYRSAYFPVKDVIDGDLCEQFPTLPMDLQRKIADELDRTPGE-ILKKL 1217
+ GRDH YRS Y PV+ VIDGDLCE F L ++ Q +A L E I++ +
Sbjct: 1294 EGSCSILGRDHQEYRSYYAPVRKVIDGDLCENFLRLSLNEQEFLAKNLKSVQVEDIIQTI 1353
Query: 1218 EEVR 1221
EVR
Sbjct: 1354 NEVR 1357
>DB1A_ARATH (Q9M0V3) DNA damage binding protein 1a (UV-damaged
DNA-binding protein 1a) (DDB1a)
Length = 1088
Score = 223 bits (569), Expect = 2e-57
Identities = 285/1245 (22%), Positives = 513/1245 (40%), Gaps = 194/1245 (15%)
Query: 4 YNLTLQRPTGIVCAINGNFSGSDDGITQEIVVARGKVLE--LLRPDKFGRIQSILSVQVF 61
Y +T +PT + + GNF+ + ++VA+ +E LL P +Q +L V ++
Sbjct: 6 YVVTAHKPTSVTHSCVGNFTSPQE---LNLIVAKCTRIEIHLLTPQG---LQPMLDVPIY 59
Query: 62 GTIRSLSQFRLTGAQKDFIVVGSDSGRIVILDYNKQKN-VFDKIHQETFGKSGCRRIVPG 120
G I +L FR G +DF+ + ++ + +L ++ + + + + + + G R G
Sbjct: 60 GRIATLELFRPHGEAQDFLFIATERYKFCVLQWDPESSELITRAMGDVSDRIG-RPTDNG 118
Query: 121 QYLAIDPKGRAVMIAACEKKKLVYVLNRDSLARLTISSPLEANKSHTIVFSICAVDCGFE 180
Q IDP R + + + V + + + LE + I F G
Sbjct: 119 QIGIIDPDCRLIGLHLYDGLFKVIPFDNKGQLKEAFNIRLEELQVLDIKFLF-----GCA 173
Query: 181 NPIFAAIELDCSDADQDATGVAASQAQKHLIFYELDL-GLNHVSRKWS-DQVDNGANMLV 238
P A + D DA +H+ YE+ L + V WS + +DNGA++L+
Sbjct: 174 KPTIAVLYQDNKDA-------------RHVKTYEVSLKDKDFVEGPWSQNSLDNGADLLI 220
Query: 239 TVPGGADGPSGVLVCAENFVIYKNQGHPDVRAVIPRRADLPAERGVLIVSAAMHKTKNLK 298
VP GVL+ E ++Y + IP R + G + V +
Sbjct: 221 PVPPPL---CGVLIIGEETIVYCS---ASAFKAIPIRPSITKAYGRVDVDGSR------- 267
Query: 299 PEEFKLFFLLQTEYGDIFKVTLTDGGGDRVSELNIKYFDTIAVAVSICVLKSGFLFAASE 358
+LL G I + +T ++V+ L I+ ++A +I L + +F S
Sbjct: 268 -------YLLGDHAGMIHLLVITH-EKEKVTGLKIELLGETSIASTISYLDNAVVFVGSS 319
Query: 359 FGNHALYQFKGIGDDDNDVGASSASLMETEEGFQPVFFQPRRLKNLVRIDQVESLMPVMD 418
+G+ L + D + S +E +++ +L P++D
Sbjct: 320 YGDSQLVKLNLHPD-------AKGSYVEV-------------------LERYINLGPIVD 353
Query: 419 MKVSNLFEEETPQIFTLCGRGPRSSLRIMRTGLAVSEMAVSKLPGIPSAVWTVKKNVMDE 478
V +L + Q+ T G SLR++R G+ ++E A +L GI +W++K ++ +
Sbjct: 354 FCVVDLERQGQGQVVTCSGAFKDGSLRVVRNGIGINEQASVELQGI-KGMWSLKSSIDEA 412
Query: 479 FDAYIVVSFTNAT--LVLSIGETADEVSDSGFLDTAPSLAVSLIGDDSLMQVHPNGIRHI 536
FD ++VVSF + T L +++ + +E GFL +L + L+QV N +R +
Sbjct: 413 FDTFLVVSFISETRILAMNLEDELEETEIEGFLSQVQTLFCHDAVYNQLVQVTSNSVRLV 472
Query: 537 REDGR--TNEWQTSGKRTIAKVGSNRLQVVIALNGGELIYFEVDVTGQLMEVERHEMSGD 594
R +EW T+ +N QV++A GG L+Y E+ G+L EV+ + +
Sbjct: 473 SSTTRELRDEWHAPAGFTVNVATANASQVLLATGGGHLVYLEIG-DGKLTEVQHALLEYE 531
Query: 595 VACLDIAPVPKGRLRSRFLAVGSY-DKTIRILSLDPDDCMQTLGIQSLSSAPESLLFLEV 653
V+CLDI P+ S+ AVG + D ++RI SL P+ + T P S+L
Sbjct: 532 VSCLDINPIGDNPNYSQLAAVGMWTDISVRIFSL-PELTLITKEQLGGEIIPRSVLLCAF 590
Query: 654 QASVGGEDGADHPASLFLNAGLQNGVLSRTVVDMVTGLLSDTRSRFLGLKAPKLFPIIVR 713
+ +L L +G L +D TG L D + LG + L +
Sbjct: 591 E------------GISYLLCALGDGHLLNFQMDTTTGQLKDRKKVSLGTQPITLRTFSSK 638
Query: 714 GKRAMLCLSSRPWLGYIHQGHFLLTPLSYETLEYAASFSSDQCFEGVVSVASEA-LRIFT 772
+ S RP + Y L + ++ + + + F+S F +++A E L I T
Sbjct: 639 SATHVFAASDRPTVIYSSNKKLLYSNVNLKEVSHMCPFNS-AAFPDSLAIAREGELTIGT 697
Query: 773 VERLGETFNQNVIPLRYTPRKFVLQPKRKLLVVIESDQGALTAEEREAARKECFEAAHAG 832
++ + + + IPL E AR+ C H
Sbjct: 698 IDDI-QKLHIRTIPL------------------------------GEHARRIC----HQE 722
Query: 833 ENKTGSEDQMENGGEDEDNDDSLSDEHYGYPKSESDKWVSCIRVLDPRTGNTTCLLELQE 892
+ +T + N E+ S+ H+ +R+LD +T L
Sbjct: 723 QTRTFGICSLGNQSNSEE-----SEMHF-------------VRLLDDQTFEFMSTYPLDS 764
Query: 893 NEAAFSICTVNFHDKEYGTLLAVGTAKGLQFTPKRSLTAGFIHIYRFLDDGRSLELLHKT 952
E SI + +F ++ VGTA L + T G I ++ ++DGR L+L+ +
Sbjct: 765 FEYGCSILSCSF-TEDKNVYYCVGTAYVL--PEENEPTKGRILVF-IVEDGR-LQLIAEK 819
Query: 953 QVEGVPLALCQFQGRLLAGIGPVLRLY-----DLGKRRLLRKCENKSFPSSIVSIHAYRD 1007
+ +G +L F G+LLA I ++LY D G R L +C + ++ + D
Sbjct: 820 ETKGAVYSLNAFNGKLLAAINQKIQLYKWMLRDDGTRELQSECGHHGHILALY-VQTRGD 878
Query: 1008 RIYVGGIQESFHYCKYRRDENQLYIFADDSVPRWLTSSYHIDFDTMAGADKFGNIFFARL 1067
I VG + +S Y+ +E + A D W+++ +D D GA+ N+ +
Sbjct: 879 FIVVGDLMKSISLLLYKHEEGAIEERARDYNANWMSAVEILDDDIYLGAENNFNLLTVK- 937
Query: 1068 PQDVSDEIEEDPTGGKIKWEQGKLNGAPNKVEEIVQFHVGDVITSLQKASLVP------- 1120
K +G + ++E + ++H+G+ + + SLV
Sbjct: 938 -----------------KNSEGATDEERGRLEVVGEYHLGEFVNRFRHGSLVMRLPDSEI 980
Query: 1121 GGGECIVYGTVMGSVGALHAFTSRDDVDFFSHLEMHMRQDNPPLCGRDHMAYRS-----A 1175
G +++GTV G +G + A ++ F L+ +R+ + G H +RS
Sbjct: 981 GQIPTVIFGTVNGVIGVI-ASLPQEQYTFLEKLQSSLRKVIKGVGGLSHEQWRSFNNEKR 1039
Query: 1176 YFPVKDVIDGDLCEQFPTLPMDLQRKIADELDRTPGEILKKLEEV 1220
++ +DGDL E F L + I+ ++ E+ K++EE+
Sbjct: 1040 TAEARNFLDGDLIESFLDLSRNKMEDISKSMNVQVEELCKRVEEL 1084
>DDB1_LYCES (Q6QNU4) DNA damage binding protein 1 (UV-damaged
DNA-binding protein 1) (High pigmentation protein 1)
Length = 1090
Score = 219 bits (558), Expect = 4e-56
Identities = 276/1247 (22%), Positives = 506/1247 (40%), Gaps = 196/1247 (15%)
Query: 4 YNLTLQRPTGIVCAINGNFSGSDDGITQEIVVARGKVLE--LLRPDKFGRIQSILSVQVF 61
Y +T +PT + + GNF+G + +++A+ +E LL P +Q +L V ++
Sbjct: 6 YVVTAHKPTNVTHSCVGNFTGPQE---LNLIIAKCTRIEIHLLTPQ---GLQPMLDVPIY 59
Query: 62 GTIRSLSQFRLTGAQKDFIVVGSDSGRIVILDYNKQ-KNVFDKIHQETFGKSGCRRIVPG 120
G I +L FR G +D + + ++ + +L ++ + V + + + G R G
Sbjct: 60 GRIATLELFRPHGETQDLLFIATERYKFCVLQWDTEASEVITRAMGDVSDRIG-RPTDNG 118
Query: 121 QYLAIDPKGRAVMIAACEKKKLVYVLNRDSLARLTISSPLEANKSHTIVFSICAVDCGFE 180
Q IDP R + + + V + + + LE + I F G
Sbjct: 119 QIGIIDPDCRLIGLHLYDGLFKVIPFDNKGQLKEAFNIRLEELQVLDIKFLY-----GCP 173
Query: 181 NPIFAAIELDCSDADQDATGVAASQAQKHLIFYELDL-GLNHVSRKWS-DQVDNGANMLV 238
P + D DA +H+ YE+ L + + W+ + +DNGA++L+
Sbjct: 174 KPTIVVLYQDNKDA-------------RHVKTYEVSLKDKDFIEGPWAQNNLDNGASLLI 220
Query: 239 TVPGGADGPSGVLVCAENFVIYKNQGHPDVRAVIPRRADLPAERGVLIVSAAMHKTKNLK 298
VP GVL+ E ++Y + IP R + G + + +
Sbjct: 221 PVPPPL---CGVLIIGEETIVYCS---ASAFKAIPIRPSITRAYGRVDADGSRY------ 268
Query: 299 PEEFKLFFLLQTEYGDIFKVTLTDGGGDRVSELNIKYFDTIAVAVSICVLKSGFLFAASE 358
L ++ + + + ++V+ L I+ ++A +I L + F+F S
Sbjct: 269 ---------LLGDHNGLLHLLVITHEKEKVTGLKIELLGETSIASTISYLDNAFVFIGSS 319
Query: 359 FGNHALYQFKGIGDDDNDVGASSASLMETEEGFQPVFFQPRRLKNLVRI-DQVESLMPVM 417
+G+ L + QP + V + ++ +L P++
Sbjct: 320 YGDSQLVKLN---------------------------LQPDTKGSYVEVLERYVNLGPIV 352
Query: 418 DMKVSNLFEEETPQIFTLCGRGPRSSLRIMRTGLAVSEMAVSKLPGIPSAVWTVKKNVMD 477
D V +L + Q+ T G SLRI+R G+ ++E A +L GI +W+++ D
Sbjct: 353 DFCVVDLERQGQGQVVTCSGAYKDGSLRIVRNGIGINEQASVELQGI-KGMWSLRSATDD 411
Query: 478 EFDAYIVVSFTNATLVLSIG--ETADEVSDSGFLDTAPSLAVSLIGDDSLMQVHPNGIRH 535
+D ++VVSF + T VL++ + +E GF +L + L+QV N +R
Sbjct: 412 PYDTFLVVSFISETRVLAMNLEDELEETEIEGFNSQVQTLFCHDAVYNQLVQVTSNSVRL 471
Query: 536 IREDGR--TNEWQTSGKRTIAKVGSNRLQVVIALNGGELIYFEVDVTGQLMEVERHEMSG 593
+ R NEW ++ +N QV++A GG L+Y E+ G L EV+ ++
Sbjct: 472 VSSTSRDLKNEWFAPVGYSVNVATANATQVLLATGGGHLVYLEIG-DGVLNEVKYAKLDY 530
Query: 594 DVACLDIAPVPKGRLRSRFLAVGSY-DKTIRILSLDPDDCMQTLGIQSLSSAPESLLFLE 652
D++CLDI P+ + S AVG + D ++RI SL PD + T P S+L
Sbjct: 531 DISCLDINPIGENPNYSNIAAVGMWTDISVRIYSL-PDLNLITKEQLGGEIIPRSVLMCS 589
Query: 653 VQASVGGEDGADHPASLFLNAGLQNGVLSRTVVDMVTGLLSDTRSRFLGLKAPKLFPIIV 712
+ +L L +G L V+ M TG L+D + LG + L
Sbjct: 590 FE------------GISYLLCALGDGHLLNFVLSMSTGELTDRKKVSLGTQPITLRTFSS 637
Query: 713 RGKRAMLCLSSRPWLGYIHQGHFLLTPLSYETLEYAASFSSDQCFEGVVSVASEA-LRIF 771
+ + S RP + Y L + ++ + + + F+ F +++A E L I
Sbjct: 638 KDTTHVFAASDRPTVIYSSNKKLLYSNVNLKEVSHMCPFNV-AAFPDSLAIAKEGELTIG 696
Query: 772 TVERLGETFNQNVIPLRYTPRKFVLQPKRKLLVVIESDQGALTAEEREAARKECFEAAHA 831
T++ + + + IPL R+ Q + + + A++ E
Sbjct: 697 TIDEI-QKLHIRSIPLGEHARRISHQEQTRTFALCSVKYTQSNADDPEM----------- 744
Query: 832 GENKTGSEDQMENGGEDEDNDDSLSDEHYGYPKSESDKWVSCIRVLDPRTGNTTCLLELQ 891
H+ +R+LD +T L
Sbjct: 745 ---------------------------HF-------------VRLLDDQTFEFISTYPLD 764
Query: 892 ENEAAFSICTVNFHDKEYGTLLAVGTAKGLQFTPKRSLTAGFIHIYRFLDDGRSLELLHK 951
+ E SI + +F D + +GTA + T G I ++ ++DG+ L+L+ +
Sbjct: 765 QFEYGCSILSCSFSD-DSNVYYCIGTA--YVMPEENEPTKGRILVF-IVEDGK-LQLIAE 819
Query: 952 TQVEGVPLALCQFQGRLLAGIGPVLRLY------DLGKRRLLRKCENKSFPSSIVSIHAY 1005
+ +G +L F G+LLA I ++LY D G R L +C + ++ +
Sbjct: 820 KETKGAVYSLNAFNGKLLAAINQKIQLYKWASREDGGSRELQTECGHHGHILALY-VQTR 878
Query: 1006 RDRIYVGGIQESFHYCKYRRDENQLYIFADDSVPRWLTSSYHIDFDTMAGADKFGNIFFA 1065
D I VG + +S ++ +E + A D W+++ +D D GA+ N+F
Sbjct: 879 GDFIVVGDLMKSISLLIFKHEEGAIEERARDYNANWMSAVEILDDDIYLGAENNFNLFTV 938
Query: 1066 RLPQDVSDEIEEDPTGGKIKWEQGKLNGAPNKVEEIVQFHVGDVITSLQKASLV------ 1119
R K +G + +++E + ++H+G+ + + SLV
Sbjct: 939 R------------------KNSEGATDEERSRLEVVGEYHLGEFVNRFRHGSLVMRLPDS 980
Query: 1120 -PGGGECIVYGTVMGSVGALHAFTSRDDVDFFSHLEMHMRQDNPPLCGRDHMAYRSAY-- 1176
G +++GTV G +G + A D F L+ ++R+ + G H +RS Y
Sbjct: 981 DVGQIPTVIFGTVNGVIGVI-ASLPHDQYLFLEKLQTNLRKVIKGVGGLSHEQWRSFYNE 1039
Query: 1177 ---FPVKDVIDGDLCEQFPTLPMDLQRKIADELDRTPGEILKKLEEV 1220
K+ +DGDL E F L + +I+ + E++K++EE+
Sbjct: 1040 KKTVDAKNFLDGDLIESFLDLSRNRMEEISKAMSVPVEELMKRVEEL 1086
>DDB1_LYCCS (Q6E7D1) DNA damage binding protein 1 (UV-damaged
DNA-binding protein 1)
Length = 1095
Score = 218 bits (556), Expect = 6e-56
Identities = 277/1249 (22%), Positives = 506/1249 (40%), Gaps = 195/1249 (15%)
Query: 4 YNLTLQRPTGIVCAINGNFSGSDDGITQEIVVARGKVLE--LLRPDKFGRI--QSILSVQ 59
Y +T +PT + + GNF+G + +++A+ +E LL P I Q +L V
Sbjct: 6 YVVTAHKPTNVTHSCVGNFTGPQE---LNLIIAKCTRIEIHLLTPQGLQCICLQPMLDVP 62
Query: 60 VFGTIRSLSQFRLTGAQKDFIVVGSDSGRIVILDYNKQ-KNVFDKIHQETFGKSGCRRIV 118
++G I +L FR G +D + + ++ + +L ++ + V + + + G R
Sbjct: 63 IYGRIATLELFRPHGETQDLLFIATERYKFCVLQWDTEASEVITRAMGDVSDRIG-RPTD 121
Query: 119 PGQYLAIDPKGRAVMIAACEKKKLVYVLNRDSLARLTISSPLEANKSHTIVFSICAVDCG 178
GQ IDP R + + + V + + + LE + I F G
Sbjct: 122 NGQIGIIDPDCRLIGLHLYDGLFKVIPFDNKGQLKEAFNIRLEELQVLDIKFLY-----G 176
Query: 179 FENPIFAAIELDCSDADQDATGVAASQAQKHLIFYELDL-GLNHVSRKWS-DQVDNGANM 236
P + D DA +H+ YE+ L + + W+ + +DNGA++
Sbjct: 177 CPKPTIVVLYQDNKDA-------------RHVKTYEVSLKDKDFIEGPWAQNNLDNGASL 223
Query: 237 LVTVPGGADGPSGVLVCAENFVIYKNQGHPDVRAVIPRRADLPAERGVLIVSAAMHKTKN 296
L+ VP GVL+ E ++Y + IP R + G + + +
Sbjct: 224 LIPVPPPL---CGVLIIGEETIVYCS---ASAFKAIPIRPSITRAYGRVDADGSRY---- 273
Query: 297 LKPEEFKLFFLLQTEYGDIFKVTLTDGGGDRVSELNIKYFDTIAVAVSICVLKSGFLFAA 356
L ++ + + + ++V+ L I+ ++A +I L + F+F
Sbjct: 274 -----------LLGDHNGLLHLLVITHEKEKVTGLKIELLGETSIASTISYLDNAFVFIG 322
Query: 357 SEFGNHALYQFKGIGDDDNDVGASSASLMETEEGFQPVFFQPRRLKNLVRI-DQVESLMP 415
S +G+ L + QP + V + ++ +L P
Sbjct: 323 SSYGDSQLVKLN---------------------------LQPDTKGSYVEVLERYVNLGP 355
Query: 416 VMDMKVSNLFEEETPQIFTLCGRGPRSSLRIMRTGLAVSEMAVSKLPGIPSAVWTVKKNV 475
++D V +L + Q+ T G SLRI+R G+ ++E A +L GI +W+++
Sbjct: 356 IVDFCVVDLERQGQGQVVTCSGAYKDGSLRIVRNGIGINEQASVELQGI-KGMWSLRSAT 414
Query: 476 MDEFDAYIVVSFTNATLVLSIG--ETADEVSDSGFLDTAPSLAVSLIGDDSLMQVHPNGI 533
D +D ++VVSF + T VL++ + +E GF +L + L+QV N +
Sbjct: 415 DDPYDTFLVVSFISETRVLAMNLEDELEETEIEGFNSQVQTLFCHDAVYNQLVQVTSNSV 474
Query: 534 RHIREDGR--TNEWQTSGKRTIAKVGSNRLQVVIALNGGELIYFEVDVTGQLMEVERHEM 591
R + R NEW ++ +N QV++A GG L+Y E+ G L EV+ ++
Sbjct: 475 RLVSSTSRDLKNEWFAPVGYSVNVATANATQVLLATGGGHLVYLEIG-DGVLNEVKYAKL 533
Query: 592 SGDVACLDIAPVPKGRLRSRFLAVGSY-DKTIRILSLDPDDCMQTLGIQSLSSAPESLLF 650
D++CLDI P+ + S AVG + D ++RI SL PD + T P S+L
Sbjct: 534 DYDISCLDINPIGENPNYSNIAAVGMWTDISVRIYSL-PDLNLITKEQLGGEIIPRSVLM 592
Query: 651 LEVQASVGGEDGADHPASLFLNAGLQNGVLSRTVVDMVTGLLSDTRSRFLGLKAPKLFPI 710
+ +L L +G L V+ M TG L+D + LG + L
Sbjct: 593 CSFE------------GISYLLCALGDGHLLNFVLSMSTGELTDRKKVSLGTQPITLRTF 640
Query: 711 IVRGKRAMLCLSSRPWLGYIHQGHFLLTPLSYETLEYAASFSSDQCFEGVVSVASEA-LR 769
+ + S RP + Y L + ++ + + + F+ F +++A E L
Sbjct: 641 SSKDTTHVFAASDRPTVIYSSNKKLLYSNVNLKEVSHMCPFNV-AAFPDSLAIAKEGELT 699
Query: 770 IFTVERLGETFNQNVIPLRYTPRKFVLQPKRKLLVVIESDQGALTAEEREAARKECFEAA 829
I T++ + + + IPL R+ Q + + + A++ E
Sbjct: 700 IGTIDEI-QKLHIRSIPLGEHARRISHQEQTRTFALCSVKYTQSNADDPEM--------- 749
Query: 830 HAGENKTGSEDQMENGGEDEDNDDSLSDEHYGYPKSESDKWVSCIRVLDPRTGNTTCLLE 889
H+ +R+LD +T
Sbjct: 750 -----------------------------HF-------------VRLLDDQTFEFISTYP 767
Query: 890 LQENEAAFSICTVNFHDKEYGTLLAVGTAKGLQFTPKRSLTAGFIHIYRFLDDGRSLELL 949
L + E SI + +F D + +GTA + T G I ++ ++DG+ L+L+
Sbjct: 768 LDQFEYGCSILSCSFSD-DSNVYYCIGTA--YVMPEENEPTKGRILVF-IVEDGK-LQLI 822
Query: 950 HKTQVEGVPLALCQFQGRLLAGIGPVLRLY------DLGKRRLLRKCENKSFPSSIVSIH 1003
+ + +G +L F G+LLA I ++LY D G R L +C + ++ +
Sbjct: 823 AEKETKGAVYSLNAFNGKLLAAINQKIQLYKWASREDGGSRELQTECGHHGHILALY-VQ 881
Query: 1004 AYRDRIYVGGIQESFHYCKYRRDENQLYIFADDSVPRWLTSSYHIDFDTMAGADKFGNIF 1063
D I VG + +S ++ +E + A D W+++ +D D GA+ N+F
Sbjct: 882 TRGDFIVVGDLMKSISLLIFKHEEGAIEERARDYNANWMSAVEILDDDIYLGAENNFNLF 941
Query: 1064 FARLPQDVSDEIEEDPTGGKIKWEQGKLNGAPNKVEEIVQFHVGDVITSLQKASLV---- 1119
R K +G + +++E + ++H+G+ + + SLV
Sbjct: 942 TVR------------------KNSEGATDEERSRLEVVGEYHLGEFVNRFRHGSLVMRLP 983
Query: 1120 ---PGGGECIVYGTVMGSVGALHAFTSRDDVDFFSHLEMHMRQDNPPLCGRDHMAYRSAY 1176
G +++GTV G +G + A D F L+ ++R+ + G H +RS Y
Sbjct: 984 DSDVGQIPTVIFGTVNGVIGVI-ASLPHDQYLFLEKLQTNLRKVIKGVGGLSHEQWRSFY 1042
Query: 1177 -----FPVKDVIDGDLCEQFPTLPMDLQRKIADELDRTPGEILKKLEEV 1220
K+ +DGDL E F L + +I+ + E++K++EE+
Sbjct: 1043 NEKKTVDAKNFLDGDLIESFLDLSRNRMEEISKAMSVPVEELMKRVEEL 1091
>DB1B_ARATH (O49552) DNA damage binding protein 1b (UV-damaged
DNA-binding protein 1b) (DDB1b)
Length = 1102
Score = 210 bits (535), Expect = 2e-53
Identities = 292/1259 (23%), Positives = 506/1259 (39%), Gaps = 208/1259 (16%)
Query: 4 YNLTLQRPTGIVCAINGNFSGSDDGITQEIVVARGKVLE--LLRPDKFGRIQSILSVQVF 61
Y +T Q+PT + + GNF+ + ++VA+ +E LL P +Q+IL V ++
Sbjct: 6 YAVTAQKPTCVTHSCVGNFTSPQE---LNLIVAKSTRIEIHLLSPQG---LQTILDVPLY 59
Query: 62 GTIRSLSQFR----LTGAQKDFIVVGSDSGRIVILDYNKQKNVFDK-------IHQETFG 110
G I ++ FR + F+ + +L +VF + Q G
Sbjct: 60 GRIATMELFRPHVSIVFLLNTFLCLRVKHKTFCLLQLKDINSVFFNGIMSPLSLLQGAMG 119
Query: 111 KSGCRRIVP---GQYLAIDPKGRAVMIAACEKKKLVYVLNRDSLARLTISSPLEANKSHT 167
R P GQ IDP R + + + V + + + LE +
Sbjct: 120 DVSDRIGRPTDNGQIGIIDPDCRVIGLHLYDGLFKVIPFDNKGQLKEAFNIRLEELQVLD 179
Query: 168 IVFSICAVDCGFENPIFAAIELDCSDADQDATGVAASQAQKHLIFYELDL-GLNHVSRKW 226
I F G P A + D DA +H+ YE+ L N V W
Sbjct: 180 IKFLY-----GCTKPTIAVLYQDNKDA-------------RHVKTYEVSLKDKNFVEGPW 221
Query: 227 S-DQVDNGANMLVTVPGGADGPSGVLVCAENFVIYKNQGHPDVRAVIPRRADLPAERGVL 285
S + +DNGA++L+ VP GVL+ E ++Y + + IP R + G +
Sbjct: 222 SQNNLDNGADLLIPVPSPL---CGVLIIGEETIVYCS---ANAFKAIPIRPSITKAYGRV 275
Query: 286 IVSAAMHKTKNLKPEEFKLFFLLQTEYGDIFKVTLTDGGGDRVSELNIKYFDTIAVAVSI 345
+ + +LL G I + +T ++V+ L I+ ++A SI
Sbjct: 276 DLDGSR--------------YLLGDHAGLIHLLVITH-EKEKVTGLKIELLGETSIASSI 320
Query: 346 CVLKSGFLFAASEFGNHALYQFKGIGDDDNDVGASSASLMETEEGFQPVFFQPRRLKNLV 405
L + +F S +G+ L + QP + V
Sbjct: 321 SYLDNAVVFVGSSYGDSQLIKLN---------------------------LQPDAKGSYV 353
Query: 406 RI-DQVESLMPVMDMKVSNLFEEETPQIFTLCGRGPRSSLRIMRTGLAVSEMAVSKLPGI 464
I ++ +L P++D V +L + Q+ T G SLRI+R G+ ++E A +L GI
Sbjct: 354 EILEKYVNLGPIVDFCVVDLERQGQGQVVTCSGAYKDGSLRIVRNGIGINEQASVELQGI 413
Query: 465 PSAVWTVKKNVMDEFDAYIVVSFTNAT--LVLSIGETADEVSDSGFLDTAPSLAVSLIGD 522
+W++K ++ + FD ++VVSF + T L ++I + +E GFL +L
Sbjct: 414 -KGMWSLKSSIDEAFDTFLVVSFISETRILAMNIEDELEETEIEGFLSEVQTLFCHDAVY 472
Query: 523 DSLMQVHPNGIRHIREDGR--TNEWQTSGKRTIAKVGSNRLQVVIALNGGELIYFEVDVT 580
+ L+QV N +R + R N+W ++ +N QV++A GG L+Y E+
Sbjct: 473 NQLVQVTSNSVRLVSSTTRELRNKWDAPAGFSVNVATANASQVLLATGGGHLVYLEIG-D 531
Query: 581 GQLMEVERHEMSGDVACLDIAPVPKGRLRSRFLAVGSY-DKTIRILSLDPDDCMQTLGIQ 639
G L EV+ + +V+CLDI P+ S+ AVG + D ++RI L PD + T
Sbjct: 532 GTLTEVKHVLLEYEVSCLDINPIGDNPNYSQLAAVGMWTDISVRIFVL-PDLTLITKEEL 590
Query: 640 SLSSAPESLLFLEVQASVGGEDGADHPASLFLNAGLQNGVLSRTVVDMVTGLLSDTRSRF 699
P S+L + +L L +G L +D G L D +
Sbjct: 591 GGEIIPRSVLLCAFE------------GISYLLCALGDGHLLNFQLDTSCGKLRDRKKVS 638
Query: 700 LGLKAPKLFPIIVRGKRAMLCLSSRPWLGYIHQGHFLLTPLSYETLEYAASFSSDQCFEG 759
LG + L + + S RP + Y + L + ++ + + + F+S F
Sbjct: 639 LGTRPITLRTFSSKSATHVFAASDRPAVIYSNNKKLLYSNVNLKEVSHMCPFNS-AAFPD 697
Query: 760 VVSVASEA-LRIFTVERLGETFNQNVIPLRYTPRKFVLQPKRKLLVVIESDQGALTAEER 818
+++A E L I T++ + + + IP+
Sbjct: 698 SLAIAREGELTIGTIDDI-QKLHIRTIPI------------------------------G 726
Query: 819 EAARKECFEAAHAGENKTGSEDQMENGGEDEDNDDSLSDEHYGYPKSESDKWVSCIRVLD 878
E AR+ C H + +T + + N E+ S+ H+ +R+LD
Sbjct: 727 EHARRIC----HQEQTRTFAISCLRNEPSAEE-----SESHF-------------VRLLD 764
Query: 879 PRTGNTTCLLELQENEAAFSICTVNFHDKEYGTLLAVGTAKGLQFTPKRSLTAGFIHIYR 938
++ L E SI + +F D + VGTA L + T G I ++
Sbjct: 765 AQSFEFLSSYPLDAFECGCSILSCSFTD-DKNVYYCVGTAYVL--PEENEPTKGRILVF- 820
Query: 939 FLDDGRSLELLHKTQVEGVPLALCQFQGRLLAGIGPVLRLY-----DLGKRRLLRKCENK 993
+++GR L+L+ + + +G +L F G+LLA I ++LY D G R L +C +
Sbjct: 821 IVEEGR-LQLITEKETKGAVYSLNAFNGKLLASINQKIQLYKWMLRDDGTRELQSECGHH 879
Query: 994 SFPSSIVSIHAYRDRIYVGGIQESFHYCKYRRDENQLYIFADDSVPRWLTSSYHIDFDTM 1053
++ + D I VG + +S Y+ +E + A D W+T+ ++ D
Sbjct: 880 GHILALY-VQTRGDFIAVGDLMKSISLLIYKHEEGAIEERARDYNANWMTAVEILNDDIY 938
Query: 1054 AGADKFGNIFFARLPQDVSDEIEEDPTGGKIKWEQGKLNGAPNKVEEIVQFHVGDVITSL 1113
G D NIF + K +G + ++E + ++H+G+ +
Sbjct: 939 LGTDNCFNIFTVK------------------KNNEGATDEERARMEVVGEYHIGEFVNRF 980
Query: 1114 QKASLVP-------GGGECIVYGTVMGSVGALHAFTSRDDVDFFSHLEMHMRQDNPPLCG 1166
+ SLV G +++GTV G +G + A ++ F L+ +R+ + G
Sbjct: 981 RHGSLVMKLPDSDIGQIPTVIFGTVSGMIGVI-ASLPQEQYAFLEKLQTSLRKVIKGVGG 1039
Query: 1167 RDHMAYRS-----AYFPVKDVIDGDLCEQFPTLPMDLQRKIADELDRTPGEILKKLEEV 1220
H +RS K +DGDL E F L +I+ +D E+ K++EE+
Sbjct: 1040 LSHEQWRSFNNEKRTAEAKGYLDGDLIESFLDLSRGKMEEISKGMDVQVEELCKRVEEL 1098
>DDB1_HUMAN (Q16531) DNA damage binding protein 1 (Damage-specific DNA
binding protein 1) (DDB p127 subunit) (DDBa) (UV-damaged
DNA-binding protein 1) (UV-DDB 1) (Xeroderma pigmentosum
group E complementing protein) (XPCe) (X-associated
protein 1) (XAP-1)
Length = 1140
Score = 206 bits (523), Expect = 4e-52
Identities = 272/1249 (21%), Positives = 504/1249 (39%), Gaps = 163/1249 (13%)
Query: 2 YLYNLTLQRPTGIVCAINGNFSGSDDGITQEIVVARGKVLELLRPDKFGRIQSILSVQVF 61
Y Y +T Q+PT + + G+F+ ++D +++A+ LE+ G ++ + V ++
Sbjct: 3 YNYVVTAQKPTAVNGCVTGHFTSAED---LNLLIAKNTRLEIYVVTAEG-LRPVKEVGMY 58
Query: 62 GTIRSLSQFRLTGAQKDFIVVGSDSGRIVILDYNKQKNVFDKI---HQETFGKSGCRRIV 118
G I + FR G KD + + + IL+Y + D I H + G R
Sbjct: 59 GKIAVMELFRPKGESKDLLFILTAKYNACILEYKQSGESIDIITRAHGNVQDRIG-RPSE 117
Query: 119 PGQYLAIDPKGRAVMIAACEKKKLVYVLNRDSLARLTISSPLEANKSHTIVFSICAVDCG 178
G IDP+ R + + + V L+RD+ + LE + F G
Sbjct: 118 TGIIGIIDPECRMIGLRLYDGLFKVIPLDRDNKELKAFNIRLEELHVIDVKFLY-----G 172
Query: 179 FENPIFAAIELDCSDADQDATGVAASQAQKHLIFYELDLGLNHVSR-KWSDQ-VDNGANM 236
+ P + QD G +H+ YE+ L ++ W + V+ A+M
Sbjct: 173 CQAPTICFVY-------QDPQG-------RHVKTYEVSLREKEFNKGPWKQENVEAEASM 218
Query: 237 LVTVPGGADGPSGVLVCAENFVIYKNQGHPDVRAVIPRRADLPAERGVLIVSAAMHKTKN 296
++ VP G +++ E+ + + + I +++ + V + ++
Sbjct: 219 VIAVPEPFGG--AIIIGQESITYHNGDKYLAIAPPIIKQSTIVCHNRV-----DPNGSRY 271
Query: 297 LKPEEFKLFFLLQTEYGDIFKVTLTDGGGDRVSELNIKYFDTIAVAVSICVLKSGFLFAA 356
L + F+L E + T+T + +L ++ ++A + L +G +F
Sbjct: 272 LLGDMEGRLFMLLLEKEEQMDGTVT------LKDLRVELLGETSIAECLTYLDNGVVFVG 325
Query: 357 SEFGNHALYQFKGIGDDDNDVGASSASLMETEEGFQPVFFQPRRLKNLVRIDQVESLMPV 416
S G+ L + + D N+ G+ +V ++ +L P+
Sbjct: 326 SRLGDSQLVK---LNVDSNEQGSY-----------------------VVAMETFTNLGPI 359
Query: 417 MDMKVSNLFEEETPQIFTLCGRGPRSSLRIMRTGLAVSEMAVSKLPGIPSAVWTVKKNVM 476
+DM V +L + Q+ T G SLRI+R G+ + E A LPGI +W ++ +
Sbjct: 360 VDMCVVDLERQGQGQLVTCSGAFKEGSLRIIRNGIGIHEHASIDLPGI-KGLWPLRSDPN 418
Query: 477 DEFDAYIVVSFTNATLVLSI-GETADEVSDSGFLDTAPSLAVSLIGDDSLMQVHPNGIRH 535
E D +V+SF T VL + GE +E GF+D + + L+Q+ +R
Sbjct: 419 RETDDTLVLSFVGQTRVLMLNGEEVEETELMGFVDDQQTFFCGNVAHQQLIQITSASVRL 478
Query: 536 IREDGR--TNEWQTSGKRTIAKVGSNRLQVVIALNGGELIYFEVDVTGQLMEVERHEMSG 593
+ ++ + +EW+ + I+ N QVV+A+ G +Y+ +L ++ EM
Sbjct: 479 VSQEPKALVSEWKEPQAKNISVASCNSSQVVVAV--GRALYYLQIHPQELRQISHTEMEH 536
Query: 594 DVACLDIAPVPKGRLRSRFLAVGSY-DKTIRILSLDPDDCM--QTLGIQSLSSAPESLLF 650
+VACLDI P+ S A+G + D + RIL L + + + LG + + P S+L
Sbjct: 537 EVACLDITPLGDSNGLSPLCAIGLWTDISARILKLPSFELLHKEMLGGEII---PRSILM 593
Query: 651 LEVQASVGGEDGADHPASLFLNAGLQNGVLSRTVVDMVTGLLSDTRSRFLGLKAPKLFPI 710
+ +S +L L +G L +++ TGLLSD + LG + L
Sbjct: 594 TTFE------------SSHYLLCALGDGALFYFGLNIETGLLSDRKKVTLGTQPTVLRTF 641
Query: 711 IVRGKRAMLCLSSRPWLGYIHQGHFLLTPLSYETLEYAASFSSDQCFEGVVSVASEALRI 770
+ S RP + Y + + ++ + + Y +SD + + + L I
Sbjct: 642 RSLSTTNVFACSDRPTVIYSSNHKLVFSNVNLKEVNYMCPLNSDGYPDSLALANNSTLTI 701
Query: 771 FTVERLGETFNQNVIPLRYTPRKFVLQPKRKLLVVIES------DQGALTAEEREAARKE 824
T++ + + + +PL +PRK Q + V+ S G TA A+ +
Sbjct: 702 GTIDEI-QKLHIRTVPLYESPRKICYQEVSQCFGVLSSRIEVQDTSGGTTALRPSASTQA 760
Query: 825 CFEAAHAGENKTGSEDQMENG-GEDEDNDDSLSDEHYGYPKSESDKWVSCIRVLDPRTGN 883
+ + + + S E GE+ + V + ++D T
Sbjct: 761 LSSSVSSSKLFSSSTAPHETSFGEEVE--------------------VHNLLIIDQHTFE 800
Query: 884 TTCLLELQENEAAFSICTVNFHDKEYGTLLAVGTAKGLQFTPKRSLTAGFIHIYRFLDDG 943
+ +NE A S+ + K+ T VGTA + + + G I ++++ D
Sbjct: 801 VLHAHQFLQNEYALSLVSCKL-GKDPNTYFIVGTA--MVYPEEAEPKQGRIVVFQYSDG- 856
Query: 944 RSLELLHKTQVEGVPLALCQFQGRLLAGIGPVLRLYDLGKRRLLRKCENKSFPSSIVSIH 1003
L+ + + +V+G ++ +F G+LLA I +RLY+ + LR N + +
Sbjct: 857 -KLQTVAEKEVKGAVYSMVEFNGKLLASINSTVRLYEWTTEKELRTECNHYNNIMALYLK 915
Query: 1004 AYRDRIYVGGIQESFHYCKYRRDENQLYIFADDSVPRWLTSSYHIDFDTMAGADKFGNIF 1063
D I VG + S Y+ E A D P W+++ +D D GA+ N+F
Sbjct: 916 TKGDFILVGDLMRSVLLLAYKPMEGNFEEIARDFNPNWMSAVEILDDDNFLGAENAFNLF 975
Query: 1064 FARLPQDVSDEIEEDPTGGKIKWEQGKLNGAPNKVEEIVQFHVGDVITSLQKASLV---- 1119
+ + + E ++E+ FH+G+ + SLV
Sbjct: 976 VCQKDSAATTDEER------------------QHLQEVGLFHLGEFVNVFCHGSLVMQNL 1017
Query: 1120 -----PGGGECIVYGTVMGSVGALHAFTSRDDVDFFSHLEMHMRQDN--PPLCGRDHMAY 1172
P G +++GTV G +G + TS + + L+M R + + +H +
Sbjct: 1018 GETSTPTQGS-VLFGTVNGMIGLV---TSLSESWYNLLLDMQNRLNKVIKSVGKIEHSFW 1073
Query: 1173 RSAYF-----PVKDVIDGDLCEQFPTLPMDLQRKIADELDRTPGEILKK 1216
RS + P IDGDL E F + +++ L G +K+
Sbjct: 1074 RSFHTERKTEPATGFIDGDLIESFLDISRPKMQEVVANLQYDDGSGMKR 1122
>DDB1_CERAE (P33194) DNA damage binding protein 1 (Damage-specific DNA
binding protein 1) (DDB p127 subunit) (DDBa) (UV-damaged
DNA-binding protein 1) (UV-DDB 1)
Length = 1140
Score = 203 bits (517), Expect = 2e-51
Identities = 271/1249 (21%), Positives = 503/1249 (39%), Gaps = 163/1249 (13%)
Query: 2 YLYNLTLQRPTGIVCAINGNFSGSDDGITQEIVVARGKVLELLRPDKFGRIQSILSVQVF 61
Y Y +T Q+PT + + +F+ ++D +++A+ LE+ G ++ + V ++
Sbjct: 3 YNYVVTAQKPTAVNGCVTAHFTSAED---LNLLIAKNTRLEIYVVTAEG-LRPVKEVGMY 58
Query: 62 GTIRSLSQFRLTGAQKDFIVVGSDSGRIVILDYNKQKNVFDKI---HQETFGKSGCRRIV 118
G I + FR G KD + + + IL+Y + D I H + G R
Sbjct: 59 GKIAVMELFRPKGESKDLLFILTAKYNACILEYKQSGESIDIITRAHGNVQDRIG-RPSE 117
Query: 119 PGQYLAIDPKGRAVMIAACEKKKLVYVLNRDSLARLTISSPLEANKSHTIVFSICAVDCG 178
G IDP+ R + + + V L+RD+ + LE + F G
Sbjct: 118 TGIIGIIDPECRMIGLRLYDGLFKVIPLDRDNKELKAFNIRLEELHVIDVKFLY-----G 172
Query: 179 FENPIFAAIELDCSDADQDATGVAASQAQKHLIFYELDLGLNHVSR-KWSDQ-VDNGANM 236
+ P + QD G +H+ YE+ L ++ W + V+ A+M
Sbjct: 173 CQAPTICFVY-------QDPQG-------RHVKTYEVSLREKEFNKGPWKQENVEAEASM 218
Query: 237 LVTVPGGADGPSGVLVCAENFVIYKNQGHPDVRAVIPRRADLPAERGVLIVSAAMHKTKN 296
++ VP G +++ E+ + + + I +++ + V + ++
Sbjct: 219 VIAVPEPFGG--AIIIGQESITYHNGDKYLAIAPPIIKQSTIVCHNRV-----DPNGSRY 271
Query: 297 LKPEEFKLFFLLQTEYGDIFKVTLTDGGGDRVSELNIKYFDTIAVAVSICVLKSGFLFAA 356
L + F+L E + T+T + +L ++ ++A + L +G +F
Sbjct: 272 LLGDMEGRLFMLLLEKEEQMDGTVT------LKDLRVELLGETSIAECLTYLDNGVVFVG 325
Query: 357 SEFGNHALYQFKGIGDDDNDVGASSASLMETEEGFQPVFFQPRRLKNLVRIDQVESLMPV 416
S G+ L + + D N+ G+ +V ++ +L P+
Sbjct: 326 SRLGDSQLVK---LNVDSNEQGSY-----------------------VVAMETFTNLGPI 359
Query: 417 MDMKVSNLFEEETPQIFTLCGRGPRSSLRIMRTGLAVSEMAVSKLPGIPSAVWTVKKNVM 476
+DM V +L + Q+ T G SLRI+R G+ + E A LPGI +W ++ +
Sbjct: 360 VDMCVVDLERQGQGQLVTCSGAFKEGSLRIIRNGIGIHEHASIDLPGI-KGLWPLRSDPN 418
Query: 477 DEFDAYIVVSFTNATLVLSI-GETADEVSDSGFLDTAPSLAVSLIGDDSLMQVHPNGIRH 535
E D +V+SF T VL + GE +E GF+D + + L+Q+ +R
Sbjct: 419 RETDDTLVLSFVGQTRVLMLNGEEVEETELMGFVDDQQTFFCGNVAHQQLIQITSASVRL 478
Query: 536 IREDGR--TNEWQTSGKRTIAKVGSNRLQVVIALNGGELIYFEVDVTGQLMEVERHEMSG 593
+ ++ + +EW+ + I+ N QVV+A+ G +Y+ +L ++ EM
Sbjct: 479 VSQEPKALVSEWKEPQAKNISVASCNSSQVVVAV--GRALYYLQIHPQELRQISHTEMEH 536
Query: 594 DVACLDIAPVPKGRLRSRFLAVGSY-DKTIRILSLDPDDCM--QTLGIQSLSSAPESLLF 650
+VACLDI P+ S A+G + D + RIL L + + + LG + + P S+L
Sbjct: 537 EVACLDITPLGDSNGLSPLCAIGLWTDISARILKLPSFELLHKEMLGGEII---PRSILM 593
Query: 651 LEVQASVGGEDGADHPASLFLNAGLQNGVLSRTVVDMVTGLLSDTRSRFLGLKAPKLFPI 710
+ +S +L L +G L +++ TGLLSD + LG + L
Sbjct: 594 TTFE------------SSHYLLCALGDGALFYFGLNIETGLLSDRKKVTLGTQPTVLRTF 641
Query: 711 IVRGKRAMLCLSSRPWLGYIHQGHFLLTPLSYETLEYAASFSSDQCFEGVVSVASEALRI 770
+ S RP + Y + + ++ + + Y +SD + + + L I
Sbjct: 642 RSLSTTNVFACSDRPTVIYSSNHKLVFSNVNLKEVNYMCPLNSDGYPDSLALANNSTLTI 701
Query: 771 FTVERLGETFNQNVIPLRYTPRKFVLQPKRKLLVVIES------DQGALTAEEREAARKE 824
T++ + + + +PL +PRK Q + V+ S G TA A+ +
Sbjct: 702 GTIDEI-QKLHIRTVPLYESPRKICYQEVSQCFGVLSSRIEVQDTSGGTTALRPSASTQA 760
Query: 825 CFEAAHAGENKTGSEDQMENG-GEDEDNDDSLSDEHYGYPKSESDKWVSCIRVLDPRTGN 883
+ + + + S E GE+ + V + ++D T
Sbjct: 761 LSSSVSSSKLFSSSTAPHETSFGEEVE--------------------VHNLLIIDQHTFE 800
Query: 884 TTCLLELQENEAAFSICTVNFHDKEYGTLLAVGTAKGLQFTPKRSLTAGFIHIYRFLDDG 943
+ +NE A S+ + K+ T VGTA + + + G I ++++ D
Sbjct: 801 VLHAHQFLQNEYALSLVSCKL-GKDPNTYFIVGTA--MVYPEEAEPKQGRIVVFQYSDG- 856
Query: 944 RSLELLHKTQVEGVPLALCQFQGRLLAGIGPVLRLYDLGKRRLLRKCENKSFPSSIVSIH 1003
L+ + + +V+G ++ +F G+LLA I +RLY+ + LR N + +
Sbjct: 857 -KLQTVAEKEVKGAVYSMVEFNGKLLASINSTVRLYEWTTEKELRTECNHYNNIMALYLK 915
Query: 1004 AYRDRIYVGGIQESFHYCKYRRDENQLYIFADDSVPRWLTSSYHIDFDTMAGADKFGNIF 1063
D I VG + S Y+ E A D P W+++ +D D GA+ N+F
Sbjct: 916 TKGDFILVGDLMRSVLLLAYKPMEGNFEEIARDFNPNWMSAVEILDDDNFLGAENAFNLF 975
Query: 1064 FARLPQDVSDEIEEDPTGGKIKWEQGKLNGAPNKVEEIVQFHVGDVITSLQKASLV---- 1119
+ + + E ++E+ FH+G+ + SLV
Sbjct: 976 VCQKDSAATTDEER------------------QHLQEVGLFHLGEFVNVFCHGSLVMQNL 1017
Query: 1120 -----PGGGECIVYGTVMGSVGALHAFTSRDDVDFFSHLEMHMRQDN--PPLCGRDHMAY 1172
P G +++GTV G +G + TS + + L+M R + + +H +
Sbjct: 1018 GETSTPTQGS-VLFGTVNGMIGLV---TSLSESWYNLLLDMQNRLNKVIKSVGKIEHSFW 1073
Query: 1173 RSAYF-----PVKDVIDGDLCEQFPTLPMDLQRKIADELDRTPGEILKK 1216
RS + P IDGDL E F + +++ L G +K+
Sbjct: 1074 RSFHTERKTEPATGFIDGDLIESFLDISRPKMQEVVANLQYDDGSGMKR 1122
>DDB1_SCHPO (O13807) DNA damage binding protein 1 (Damage-specific DNA
binding protein 1)
Length = 1072
Score = 67.4 bits (163), Expect = 2e-10
Identities = 216/1224 (17%), Positives = 439/1224 (35%), Gaps = 204/1224 (16%)
Query: 4 YNLTLQRPTGIVCAINGNFSGSDDGITQEIVVARGKVLELLRPDKFGRIQSILSVQVFGT 63
Y L +P+ I A+ F + + ++VA+ LE+ + R+ I S +F
Sbjct: 3 YVTYLHKPSSIRNAVFCKFVNAS---SWNVIVAKVNCLEVYSYEN-NRLCLITSANIFAK 58
Query: 64 IRSLSQFRLTGAQKDFIVVGSDSGRIVILDYNKQKN-VFDKIHQETFGKSGCRRIVPGQY 122
I ++ F+ + D I+V +DS R L ++ N V + I + + R G
Sbjct: 59 IVNVKAFKPVSSPTDHIIVATDSFRYFTLFWDANDNTVSNGIKIQDCSERSLRESQSGPL 118
Query: 123 LAIDPKGRAVMIAACEKKKLVYVLNRDSLARLTISSPLEANKSHTI-VFSICAVDCGF-- 179
L +DP R + + + + + + +T + + + ++ + + VD
Sbjct: 119 LLVDPFQRVICLHVYQGLLTIIPIFKSKKRFMTSHNNPSLHDNFSVRIQELNVVDIAMLY 178
Query: 180 --ENPIFAAIELDCSDADQDATGVAASQAQKHLIFYELDLGLNHVSRKWSDQVDNGANML 237
P A + D S++ HL Y++++ + D L
Sbjct: 179 NSSRPSLAVLYKD-------------SKSIVHLSTYKINVREQEIDEDDVVCHDIEEGKL 225
Query: 238 VTVPGGADGPSGVLVCAENFVIYKNQGHPDVRAVIPRRADLPAERGVLIVSAAMHKTKNL 297
+ G GV V E +V Y ++ D+ + +L
Sbjct: 226 IPSENG-----GVFVFGEMYVYYISK-------------DIQVSKLLLTYPITAFSPSIS 267
Query: 298 KPEEFKL---FFLLQTEYGDI--FKVTLTDGGGDRVSELNIKYFDTIAVAVSICVLKSGF 352
E L +++ E G + FK TD + ++ ++A + L
Sbjct: 268 NDPETGLDSSIYIVADESGMLYKFKALFTD----ETVSMELEKLGESSIASCLIALPDNH 323
Query: 353 LFAASEFGNHALYQFKGIGDDDNDVGASSASLMETEEGFQPVFFQPRRLKNLVRIDQVES 412
LF S F N L Q I +++ + L+N V I
Sbjct: 324 LFVGSHFNNSVLLQLPSITKNNHKLEI---------------------LQNFVNI----- 357
Query: 413 LMPVMDMKVSNLFEEETPQIFTLCGRGPRSSLRIMRTGLAVSEMAVSKLPGIPSAVWTVK 472
P+ D + + ++ I T G +LRI+R + + +A+ ++ GI
Sbjct: 358 -APISDFIIDD--DQTGSSIITCSGAYKDGTLRIIRNSINIENVALIEMEGIKDFF---S 411
Query: 473 KNVMDEFDAYIVVSFTNATLVLSIGETADEVSDSGFLDTAPSLAVSLI-GDDSLMQVHPN 531
+ +D YI +S T + + ++ ++ VS I G+ ++Q+
Sbjct: 412 VSFRANYDNYIFLSLICETRAIIVSPEGVFSANHDLSCEESTIFVSTIYGNSQILQITTK 471
Query: 532 GIRHIREDGRTNEWQTSGKRTIAKVGSNRLQVVIALNGGELIYFEVDVTGQLMEVERHEM 591
IR + + + + W + T ++ + V +A GG +++FE + EV R++
Sbjct: 472 EIR-LFDGKKLHSWISPMSITCGSSFADNVCVAVA--GGLILFFE-----GITEVGRYQC 523
Query: 592 SGDVACLDIAPVPKGRLRSRFLAVGSYDKTIRILSLDPDDCMQTLGIQSLSSAPESLLFL 651
+V+ L + VG + I +L+ D T ++ L+ P S+++
Sbjct: 524 DTEVSSLCFT-------EENVVYVGLWSADIIMLTYCQDGISLTHSLK-LTDIPRSIVYS 575
Query: 652 EVQASVGGEDGADHPASLFLNAGLQNGVLSRTVVDMVTGLLSDTRSRFLGLKAPKLFPII 711
+ GG L QNG + + R LG+ L
Sbjct: 576 QKYGDDGGTLYVSTNNGYVLMFNFQNGQV----------IEHSLRRNQLGVAPIILKHFD 625
Query: 712 VRGKRAMLCLSSRPWLGYIHQGHFLLTPLSYETLEYAASFSSDQCFEGVVSVASEALRIF 771
+ K A+ L +P L Y ++TPLS + +S+ + ++ + + +
Sbjct: 626 SKEKNAIFALGEKPQLMYYESDKLVITPLSCTEMLNISSYVNPSLGVNMLYCTNSYISLA 685
Query: 772 TVERLGETFNQNVIPLRYTPRKFVLQPKRKLLVVIESDQGALTAEEREAARKECFEAAHA 831
+ + + N + ++ PR+ ++ ++ ++ T E+R
Sbjct: 686 KMSEI-RSLNVQTVSVKGFPRRICSNSLFYFVLCMQLEESIGTQEQR------------- 731
Query: 832 GENKTGSEDQMENGGEDEDNDDSLSDEHYGYPKSESDKWVSCIRVLDPRTGNTTCLLELQ 891
+S +RV + T + +
Sbjct: 732 --------------------------------------LLSFLRVYEKNTLSEIAHHKFN 753
Query: 892 ENEAAFSICTVNFHDKEYGTLLAVGTAKGLQFTPKRSLTAGFIHIYRFLDDGRSLELLHK 951
E E SI +N DK + VGT G F + + +G + ++ D ++E+ +
Sbjct: 754 EYEMVESIILMN-DDKR----VVVGT--GFNFPDQDAPDSGRLMVFEMTSD-NNIEMQAE 805
Query: 952 TQVEGVPLALCQFQGRLLAGIGPVLRLYDLGKRRLLRKCENKSFPSSIVSIHAYRDRIYV 1011
+V+G L ++ ++AGI + +++ + + + P+ + I +D I
Sbjct: 806 HKVQGSVNTLVLYKHLIVAGINASVCIFEY-EHGTMHVRNSIRTPTYTIDISVNQDEIIA 864
Query: 1012 GGIQESFHYCKYRRDENQLYIFADDSVPRWLTSSYHIDFDTMAGADKFGNIFFARLPQDV 1071
+ +S ++ D QL A D P W TS + + GN L +V
Sbjct: 865 ADLMKSITVLQFIDD--QLIEVARDYHPLWATSVEILSERKYFVTEADGNAVIL-LRDNV 921
Query: 1072 SDEIEEDPTGGKIKWEQGKLNGAPNKVEEIVQFHVGDVITSLQKASLVPGGGECIV---- 1127
S ++ + K++W + +F++G++I + + + + +V
Sbjct: 922 SPQLSDRK---KLRWYK--------------KFYLGELINKTRHCTFIEPQDKSLVTPQL 964
Query: 1128 -YGTVMGSVGALHAFTSRDDVDFFSHLEMHMRQDNPPLCGRDHMAYR----SAYFPVKDV 1182
TV GS+ + + L+ ++R+ P G H ++ D+
Sbjct: 965 LCATVDGSL-MIVGDAGMSNTPLLLQLQDNIRKVIPSFGGLSHKEWKEYRGENETSPSDL 1023
Query: 1183 IDGDLCEQFPTLPMDLQRKIADEL 1206
IDG L E + L+ I +E+
Sbjct: 1024 IDGSLIESI----LGLREPILNEI 1043
>CPSA_ARATH (Q9FGR0) Probable cleavage and polyadenylation specificity
factor, 160 kDa subunit (CPSF 160 kDa subunit)
Length = 1448
Score = 67.0 bits (162), Expect = 3e-10
Identities = 95/410 (23%), Positives = 179/410 (43%), Gaps = 64/410 (15%)
Query: 843 ENGGEDEDNDDSLSDE-HYGYPKSESDKWVSCIRVLDP-RTGN---TTCLLELQENEAAF 897
+ G+ DN + SD+ Y E + I++L+P R+G T + +Q +E A
Sbjct: 1064 QEAGQQLDNHNMSSDDLQRTYTVEEFE-----IQILEPERSGGPWETKAKIPMQTSEHAL 1118
Query: 898 SICTVNFHDKEYG---TLLAVGTAKGLQFTPKRSLTA-GFIHIYRFLDDGRSLELL---- 949
++ V + G TLLAVGTA + + A G + ++ F +G + + +
Sbjct: 1119 TVRVVTLLNASTGENETLLAVGTA----YVQGEDVAARGRVLLFSFGKNGDNSQNVVTEV 1174
Query: 950 HKTQVEGVPLALCQFQGRLLAGIGPVLRLYDLGKRRLLRKCENKSFPSSIVSIHAYRDRI 1009
+ +++G A+ QG LL GP + L+ L + P +VS++ + I
Sbjct: 1175 YSRELKGAISAVASIQGHLLISSGPKIILHKWNGTELNGVAFFDAPPLYVVSMNVVKSFI 1234
Query: 1010 YVGGIQESFHYCKYRRDENQLYIFADD--SVPRWLTSSYHIDFDTM--AGADKFGNI-FF 1064
+G + +S ++ ++ +QL + A D S+ + T + ID T+ A +D+ NI F
Sbjct: 1235 LLGDVHKSIYFLSWKEQGSQLSLLAKDFESLDCFATE-FLIDGSTLSLAVSDEQKNIQVF 1293
Query: 1065 ARLPQDVSDEIEEDPTGGKIKWEQGKLNGAPNKVEEIVQFHVGDVITSLQKASLVPGGGE 1124
P+ + W+ KL +FHVG ++ + +V G +
Sbjct: 1294 YYAPKMIES------------WKGLKLLSR-------AEFHVGAHVSKFLRLQMVSSGAD 1334
Query: 1125 -----CIVYGTVMGSVGALHAFTSRDDVDF--FSHLEMHMRQDNPPLCGRDHMAYRSAYF 1177
+++GT+ GS G + D+V F L+ + P + G + +A+R
Sbjct: 1335 KINRFALLFGTLDGSFGCIAPL---DEVTFRRLQSLQKKLVDAVPHVAGLNPLAFRQFRS 1391
Query: 1178 PVK-------DVIDGDLCEQFPTLPMDLQRKIADELDRTPGEILKKLEEV 1220
K ++D +L + LP++ Q ++A ++ T ILK L ++
Sbjct: 1392 SGKARRSGPDSIVDCELLCHYEMLPLEEQLELAHQIGTTRYSILKDLVDL 1441
Score = 45.1 bits (105), Expect = 0.001
Identities = 60/288 (20%), Positives = 105/288 (35%), Gaps = 62/288 (21%)
Query: 307 LLQTEYGDIFKVTLTDGGGDRVSELNIKYFDTIAVAVSICVLKSGFLFAASEFGNHALYQ 366
LL T+ G++ +TL G V L++ +A I + + F S G+ L Q
Sbjct: 381 LLSTKSGELLLLTLIYDGR-AVQRLDLSKSKASVLASDITSVGNSLFFLGSRLGDSLLVQ 439
Query: 367 FK----------GIGDDDNDV---GASSASLMETEEGFQPVF------------------ 395
F G+ D+D D+ G + L T + FQ
Sbjct: 440 FSCRSGPAASLPGLRDEDEDIEGEGHQAKRLRMTSDTFQDTIGNEELSLFGSTPNNSDSA 499
Query: 396 ---------FQPRRLKNLVRIDQVES----LMPVMDMKVSNLFEEETPQIFTLCGRGPRS 442
F +LV + V+ L D + + ++ ++ G G
Sbjct: 500 QVTSSVLKSFSFAVRDSLVNVGPVKDFAYGLRINADANATGVSKQSNYELVCCSGHGKNG 559
Query: 443 SLRIMRTGLAVSEMAVSKLPGIPSAVWTVKKNVM--------------DEFDAYIVVSFT 488
+L ++R + + +LPG +WTV DE+ AY+++S
Sbjct: 560 ALCVLRQSIRPEMITEVELPGC-KGIWTVYHKSSRGHNADSSKMAADEDEYHAYLIISLE 618
Query: 489 NATLVLSIGETADEVSDS--GFLDTAPSLAVSLIGDDSLMQVHPNGIR 534
T+VL + EV++S ++ A +L G ++QV +G R
Sbjct: 619 ARTMVLETADLLTEVTESVDYYVQGRTIAAGNLFGRRRVIQVFEHGAR 666
>CPSA_BOVIN (Q10569) Cleavage and polyadenylation specificity
factor, 160 kDa subunit (CPSF 160 kDa subunit)
Length = 1444
Score = 55.1 bits (131), Expect = 1e-06
Identities = 112/578 (19%), Positives = 212/578 (36%), Gaps = 111/578 (19%)
Query: 51 RIQSILSVQVFGTIRSLSQFRLTGAQKDFIVVGSDSGRIVILDYNKQKNVFDKIHQETFG 110
+++ + S FG + S++ +L GA++D +++ ++ +++Y+ + + F
Sbjct: 68 KLELVASFSFFGNVMSMASVQLAGAKRDALLLSFKDAKLSVVEYDPGTHDLKTLSLHYFE 127
Query: 111 ----KSGCRRIVPGQYLAIDPKGRAVMIAACEKKKLVYVLNRDSLARLTISSPLEANKSH 166
+ G + V + +DP GR + + +V R+SLA E +S
Sbjct: 128 EPELRDGFVQNVHTPRVRVDPDGRCAAMLIYGTRLVVLPFRRESLAEEHEGLVGEGQRSS 187
Query: 167 TIVFSIC-----------AVDCGFENPIFAAIELDCSDADQDATG-VAASQAQKHLIFYE 214
+ I VD F + + L + +Q G VA Q ++
Sbjct: 188 FLPSYIIDVRALDEKLLNIVDLQFLHGYYEPTLLILFEPNQTWPGRVAVRQDTCSIVAIS 247
Query: 215 LDLGLNHVSRKWS-DQVDNGANMLVTVPGGADGPSGVLVCAENFVIYKNQGHPDVRAVIP 273
L++ WS + + VP GV++ A N ++Y NQ P +
Sbjct: 248 LNITQKVHPVIWSLTSLPFDCTQALAVPKPI---GGVVIFAVNSLLYLNQSVPPYGVALN 304
Query: 274 RRAD------LPAERGVLIVSAAMHKTKNLKPEEFKLF--FLLQTEYGDIFKVTL-TDGG 324
L + GV I T + F + ++ + G+I+ +TL TDG
Sbjct: 305 SLTTGTTAFPLRTQEGVRI-------TLDCAQAAFISYDKMVISLKGGEIYVLTLITDG- 356
Query: 325 GDRVSELNIKYFDTIAVAV---SICVLKSGFLFAASEFGNHALYQF-------------- 367
+ + +FD A +V S+ ++ G+LF S GN L ++
Sbjct: 357 ---MRSVRAFHFDKAAASVLTTSMVTMEPGYLFLGSRLGNSLLLKYTEKLQEPPASTARE 413
Query: 368 -----------------------KGIGDDDNDVGASSASLMETEEGFQPVFFQPRRLKNL 404
K + D+ D S E + G Q + ++
Sbjct: 414 AADKEEPPSKKKRVDATTGWSGSKSVPQDEVDEIEVYGS--EAQSGTQLATYSFEVCDSI 471
Query: 405 VRIDQVESLMPVMDMKVSNLFE---EETPQIFTLCGRGPRSSLRIMRTGLAVSEMAVSKL 461
+ I + +S F+ E +I G G +L +++ + + +L
Sbjct: 472 LNIGPCANAAMGEPAFLSEEFQNSPEPDLEIVVCSGYGKNGALSVLQKSIRPQVVTTFEL 531
Query: 462 PGIPSAVWTVKKNVMDEFD------------------------AYIVVSFTNATLVLSIG 497
PG +WTV V E + ++++S ++T++L G
Sbjct: 532 PGCYD-MWTVIAPVRKEQEETLKGEGTEPEPGAPEAEDDGRRHGFLILSREDSTMILQTG 590
Query: 498 ETADEVSDSGFLDTAPSLAVSLIGDDS-LMQVHPNGIR 534
+ E+ SGF P++ IGD+ ++QV P GIR
Sbjct: 591 QEIMELDASGFATQGPTVFAGNIGDNRYIVQVSPLGIR 628
>CPSA_HUMAN (Q10570) Cleavage and polyadenylation specificity
factor, 160 kDa subunit (CPSF 160 kDa subunit)
Length = 1443
Score = 54.7 bits (130), Expect = 2e-06
Identities = 112/578 (19%), Positives = 209/578 (35%), Gaps = 109/578 (18%)
Query: 51 RIQSILSVQVFGTIRSLSQFRLTGAQKDFIVVGSDSGRIVILDYNKQKNVFDKIHQETFG 110
+++ S FG + S++ +L GA++D +++ ++ +++Y+ + + F
Sbjct: 65 KLELAASFSFFGNVMSMASVQLAGAKRDALLLSFKDAKLSVVEYDPGTHDLKTLSLHYFE 124
Query: 111 ----KSGCRRIVPGQYLAIDPKGRAVMIAACEKKKLVYVLNRDSLARLTISSPLEANKSH 166
+ G + V + +DP GR + + +V R+SLA E +S
Sbjct: 125 EPELRDGFVQNVHTPRVRVDPDGRCAAMLVYGTRLVVLPFRRESLAEEHEGLVGEGQRSS 184
Query: 167 TIVFSICAV-----------DCGFENPIFAAIELDCSDADQDATG-VAASQAQKHLIFYE 214
+ I V D F + + L + +Q G VA Q ++
Sbjct: 185 FLPSYIIDVRALDEKLLNIIDLQFLHGYYEPTLLILFEPNQTWPGRVAVRQDTCSIVAIS 244
Query: 215 LDLGLNHVSRKWS-DQVDNGANMLVTVPGGADGPSGVLVCAENFVIYKNQGHPDVRAVIP 273
L++ WS + + VP GV+V A N ++Y NQ P +
Sbjct: 245 LNITQKVHPVIWSLTSLPFDCTQALAVPKPI---GGVVVFAVNSLLYLNQSVPPYGVALN 301
Query: 274 RRAD------LPAERGVLIVSAAMHKTKNLKPEEFKLFFLLQTEYGDIFKVTL-TDGGGD 326
L + GV I T + ++ + G+I+ +TL TDG
Sbjct: 302 SLTTGTTAFPLRTQEGVRITLDCAQATFISYDK-----MVISLKGGEIYVLTLITDG--- 353
Query: 327 RVSELNIKYFDTIAVAV---SICVLKSGFLFAASEFGNHALYQF---------------- 367
+ + +FD A +V S+ ++ G+LF S GN L ++
Sbjct: 354 -MRSVRAFHFDKAAASVLTTSMVTMEPGYLFLGSRLGNSLLLKYTEKLQEPPASAVREAA 412
Query: 368 ----------------------KGIGDDDNDVGASSASLMETEEGFQPVFFQPRRLKNLV 405
K + D+ D S E + G Q + +++
Sbjct: 413 DKEEPPSKKKRVDATAGWSAAGKSVPQDEVDEIEVYGS--EAQSGTQLATYSFEVCDSIL 470
Query: 406 RIDQVESLMPVMDMKVSNLFE---EETPQIFTLCGRGPRSSLRIMRTGLAVSEMAVSKLP 462
I + +S F+ E +I G G +L +++ + + +LP
Sbjct: 471 NIGPCANAAVGEPAFLSEEFQNSPEPDLEIVVCSGHGKNGALSVLQKSIRPQVVTTFELP 530
Query: 463 GIPSAVWTVKKNVMDEFD-------------------------AYIVVSFTNATLVLSIG 497
G +WTV V E + ++++S ++T++L G
Sbjct: 531 GCYD-MWTVIAPVRKEEEDNPKGEGTEQEPSTTPEADDDGRRHGFLILSREDSTMILQTG 589
Query: 498 ETADEVSDSGFLDTAPSLAVSLIGDDS-LMQVHPNGIR 534
+ E+ SGF P++ IGD+ ++QV P GIR
Sbjct: 590 QEIMELDTSGFATQGPTVFAGNIGDNRYIVQVSPLGIR 627
>CPSA_MOUSE (Q9EPU4) Cleavage and polyadenylation specificity
factor, 160 kDa subunit (CPSF 160 kDa subunit)
Length = 1441
Score = 54.3 bits (129), Expect = 2e-06
Identities = 112/578 (19%), Positives = 212/578 (36%), Gaps = 111/578 (19%)
Query: 51 RIQSILSVQVFGTIRSLSQFRLTGAQKDFIVVGSDSGRIVILDYNKQKNVFDKIHQETFG 110
+++ + S FG + S++ +L GA++D +++ ++ +++Y+ + + F
Sbjct: 65 KLELVASFSFFGNVMSMASVQLAGAKRDALLLSFKDAKLSVVEYDPGTHDLKTLSLHYFE 124
Query: 111 ----KSGCRRIVPGQYLAIDPKGRAVMIAACEKKKLVYVLNRDSLARLTISSPLEANKSH 166
+ G + V + +DP GR + + +V R+SLA E +S
Sbjct: 125 EPELRDGFVQNVHTPRVRVDPDGRCAAMLIYGTRLVVLPFRRESLAEEHEGLMGEGQRSS 184
Query: 167 TIVFSICAV-----------DCGFENPIFAAIELDCSDADQDATG-VAASQAQKHLIFYE 214
+ I V D F + + L + +Q G VA Q ++
Sbjct: 185 FLPSYIIDVRALDEKLLNIIDLQFLHGYYEPTLLILFEPNQTWPGRVAVRQDTCSIVAIS 244
Query: 215 LDLGLNHVSRKWS-DQVDNGANMLVTVPGGADGPSGVLVCAENFVIYKNQGHPDVRAVIP 273
L++ WS + + VP GV++ A N ++Y NQ P +
Sbjct: 245 LNITQKVHPVIWSLTSLPFDCTQALAVPKPI---GGVVIFAVNSLLYLNQSVPPYGVALN 301
Query: 274 RRAD------LPAERGVLIVSAAMHKTKNLKPEEFKLF--FLLQTEYGDIFKVTL-TDGG 324
L + GV I T + F + ++ + G+I+ +TL TDG
Sbjct: 302 SLTTGTTAFPLRTQEGVRI-------TLDCAQAAFISYDKMVISLKGGEIYVLTLITDG- 353
Query: 325 GDRVSELNIKYFDTIAVAV---SICVLKSGFLFAASEFGNHALYQF-------------- 367
+ + +FD A +V S+ ++ G+LF S GN L ++
Sbjct: 354 ---MRSVRAFHFDKAAASVLTTSMVTMEPGYLFLGSRLGNSLLLKYTEKLQEPPASSVRE 410
Query: 368 -----------------------KGIGDDDNDVGASSASLMETEEGFQPVFFQPRRLKNL 404
K + D+ D S E + G Q + ++
Sbjct: 411 AADKEEPPSKKKRVEPAVGWTGGKTVPQDEVDEIEVYGS--EAQSGTQLATYSFEVCDSM 468
Query: 405 VRIDQVESLMPVMDMKVSNLFE---EETPQIFTLCGRGPRSSLRIMRTGLAVSEMAVSKL 461
+ I + +S F+ E +I G G +L +++ + + +L
Sbjct: 469 LNIGPCANAAVGEPAFLSEEFQNSPEPDLEIVVCSGYGKNGALSVLQKSIRPQVVTTFEL 528
Query: 462 PGIPSAVWTVKKNVMDEFD------------------------AYIVVSFTNATLVLSIG 497
PG +WTV V E + ++++S ++T++L G
Sbjct: 529 PGCYD-MWTVIAPVRKEEEETPKAESTEQEPSAPKAEEDGRRHGFLILSREDSTMILQTG 587
Query: 498 ETADEVSDSGFLDTAPSLAVSLIGDDS-LMQVHPNGIR 534
+ E+ SGF P++ IGD+ ++QV P GIR
Sbjct: 588 QEIMELDTSGFATQGPTVFAGNIGDNRYIVQVSPLGIR 625
>CPSA_CAEEL (Q9N4C2) Probable cleavage and polyadenylation
specificity factor, 160 kDa subunit (CPSF 160 kDa
subunit)
Length = 1454
Score = 54.3 bits (129), Expect = 2e-06
Identities = 94/461 (20%), Positives = 172/461 (36%), Gaps = 67/461 (14%)
Query: 249 GVLVCAENFVIYKNQGHPDVRAVIPRRADLPAE---RGVLIVSAAMHKTKNLKPEEFKLF 305
G LV N V+Y NQ P V+ D + + + + + + ++ E+ ++
Sbjct: 263 GALVFGSNTVVYLNQAVPPCGLVLNSCYDGFTKFPLKDLKHLKMTLDCSTSVYMEDGRI- 321
Query: 306 FLLQTEYGDIFKVTL-TDGGGDRVSELNIKYFDTIAVAVSICVLKSGFLFAASEFGNHAL 364
+ + GD+F + L T GG V L ++A S+ V G LF S G+ L
Sbjct: 322 -AVGSRDGDLFLLRLMTSSGGGTVKSLEFSKVYETSIAYSLTVCAPGHLFVGSRLGDSQL 380
Query: 365 YQF-----------KGIGDDDNDVGASSASLMETEEGFQPVFFQPRRLKNLVRIDQ---- 409
++ K + D+ D A+ L E + + ++ + +ID+
Sbjct: 381 LEYTLLKTTRDCAVKRLKIDNKDPAAAEIELDEDDMELYGGAIEEQQNDDDEQIDESLQF 440
Query: 410 -----VESLMPVMDMKVS--NLFEEETPQ---------IFTLCGRGPRSSLRIMRTGLAV 453
+ ++ PV M V N + + T G G +L + + L
Sbjct: 441 RELDRLRNVGPVKSMCVGRPNYMSNDLVDAKRRDPVFDLVTASGHGKNGALCVHQRSLRP 500
Query: 454 SEMAVSKLPGIPSAVWTVKKNVMDEFDAYIVVSFTNATLVLSIGETADEVSDSGFLDTAP 513
+ S L G +W V + +E Y++VS +TL+L +GE E+ + F+ P
Sbjct: 501 EIITSSLLEGAEQ-LWAVGRKE-NESHKYLIVSRVRSTLILELGEELVELEEQLFVTGEP 558
Query: 514 SLAVSLIGDDSL-MQVHPNGIRHIREDGRTNEWQTSGKRTIAKVGSNRLQVVIALNGGEL 572
++A + +L +QV I + + + E + + V + G L
Sbjct: 559 TVAAGELSQGALAVQVTSTCIALVTDGQQMQEVHIDSNFPVIQASIVDPYVALLTQNGRL 618
Query: 573 IYFEVDVTGQLMEVERHEMSGDVACLDIAPVPKGRLRSRFLAVGSYDKTIRILSLDPDDC 632
+ +E+ +ME D++ A ++ T + L+
Sbjct: 619 LLYEL-----VMEPYVQLREVDISATSFA---------------TWHATAQNLT------ 652
Query: 633 MQTLGIQSLSSAPESLLFLEVQASVGGEDGADHPASLFLNA 673
Q I + A E + F + S+GG G D S NA
Sbjct: 653 -QLTSISIYADASEIMKFAAAEKSMGGGGGGDGEVSTAENA 692
>CPSA_DROME (Q9V726) Cleavage and polyadenylation specificity
factor, 160 kDa subunit (CPSF 160 kDa subunit)
(CPSF-160) (dCPSF)
Length = 1455
Score = 50.8 bits (120), Expect = 2e-05
Identities = 92/437 (21%), Positives = 166/437 (37%), Gaps = 62/437 (14%)
Query: 51 RIQSILSVQVFGTIRSLSQFRLTGAQKDFIVVGSDSGRIVILDYNKQKNVFDKIHQETFG 110
R++ + + ++G + SL L GA +D +++ ++ +L ++ + F
Sbjct: 67 RLECLATYTLYGNVMSLQCVSLAGAMRDALLISFKDAKLSVLQHDPDTFALKTLSLHYFE 126
Query: 111 KSGCRRIVPGQYLA----IDPKGRAVMIAACEKKKLVYVLNRD-SLARLTISSPLEANKS 165
+ R G+Y +DP R ++ K+ +V +D SL + ++ K+
Sbjct: 127 EDDIRGGWTGRYFVPTVRVDPDSRCAVMLVYGKRLVVLPFRKDNSLDEIELADVKPIKKA 186
Query: 166 HTIVFSICAVDCGFENPIFAAIELDCSDADQDATGVAASQ------AQKHLIFYE----- 214
T + S PI A+ + D D+ V Q LI YE
Sbjct: 187 PTAMVS--------RTPIMASYLIALRDLDEKIDNVLDIQFLHGYYEPTLLILYEPVRTC 238
Query: 215 -------------LDLGLNHVSRK----WSDQVDNGANMLVTVPGGADGPSGVLVCAENF 257
+ + LN R W+ V++ + V G LV N
Sbjct: 239 PGRIKVRSDTCVLVAISLNIQQRVHPIIWT--VNSLPFDCLQVYPIQKPIGGCLVMTVNA 296
Query: 258 VIYKNQGHPDVRAVIPRRAD------LPAERGVLIVSAAMHKTKNLKPEEFKLFFLLQTE 311
VIY NQ P + AD L + GV I ++ + KL L+T
Sbjct: 297 VIYLNQSVPPYGVSLNSSADNSTAFPLKPQDGVRI---SLDCANFAFIDVDKLVISLRT- 352
Query: 312 YGDIFKVTLTDGGGDRVSELNIKYFDTIAVAVSICVLKSGFLFAASEFGNHALYQFKGIG 371
GD++ +TL V + + ICVL S ++F S GN L F
Sbjct: 353 -GDLYVLTLCVDSMRTVRNFHFHKAAASVLTSCICVLHSEYIFLGSRLGNSLLLHF--TE 409
Query: 372 DDDNDVGASSASLMETEEGFQPVFFQPRRLKNLVRIDQVESLMPVMDMKVSNLFEEETPQ 431
+D + V ++E+ + + + + L+ + +DQ+E + P S E+E +
Sbjct: 410 EDQSTVITLDEVEQQSEQQQRNLQDEDQNLEEIFDVDQLE-MAPT--QAKSRRIEDEELE 466
Query: 432 IFTLCGRGPRSSLRIMR 448
++ G G ++S+ +R
Sbjct: 467 VY---GSGAKASVLQLR 480
Score = 43.9 bits (102), Expect = 0.003
Identities = 91/479 (18%), Positives = 179/479 (36%), Gaps = 102/479 (21%)
Query: 785 IPLRYTPRKFVLQPKRKLLVVIESDQGALTAEEREAARKECFEAAHAGENKTGSEDQMEN 844
+PLR TPR+ V + ++ +I + +T R
Sbjct: 1029 VPLRCTPRQLVYHRENRVYCLITQTEEPMTKYYR-------------------------F 1063
Query: 845 GGEDEDNDDSLSDEHYGYPKSESDKWVSCIRVLDPRT----GNTTCLLELQENEAAFSIC 900
GED++ + E + YP + V ++ P T + + E E+ AF I
Sbjct: 1064 NGEDKELSEESRGERFIYPIGSQFEMV----LISPETWEIVPDASITFEPWEHVTAFKIV 1119
Query: 901 TVNFHDKEYG--TLLAVGTAKGLQFTPKRSLTA-GFIHIYRFL----DDGR-----SLEL 948
+++ G L +GT F +T+ G IHIY + + G+ ++
Sbjct: 1120 KLSYEGTRSGLKEYLCIGT----NFNYSEDITSRGNIHIYDIIEVVPEPGKPMTKFKIKE 1175
Query: 949 LHKTQVEGVPLALCQFQGRLLAGIGPVLRLYDLGKRRLLRKCENKSFPSSIVSIH---AY 1005
+ K + +G A+ G L+ G+G + ++ L L+ +F + + +H
Sbjct: 1176 IFKKEQKGPVSAISDVLGFLVTGLGQKIYIWQLRDGDLI----GVAFIDTNIYVHQIITV 1231
Query: 1006 RDRIYVGGIQESFHYCKYRRDENQLYIFADDSVPRWLTSSYHIDF---DTMAG---ADKF 1059
+ I++ + +S +++ + L + + D P Y I+F ++ G D
Sbjct: 1232 KSLIFIADVYKSISLLRFQEEYRTLSLASRDFNP---LEVYGIEFMVDNSNLGFLVTDAE 1288
Query: 1060 GNIFFARLPQDVSDEIEEDPTGGKIKWEQGKLNGAPNKVEEIVQFHVGDVITSL------ 1113
NI + + + GG+ K+ +H+G V+ ++
Sbjct: 1289 RNIIVYMYQPEARESL-----GGQ-------------KLLRKADYHLGQVVNTMFRVQCH 1330
Query: 1114 -----QKASLVPGGGECIVYGTVMGSVGALHAFTSRDDVDFFSHLEMHMRQDNPPLCGRD 1168
Q+ + +VYGT+ G++G + F L+ + LCG +
Sbjct: 1331 QKGLHQRQPFLYENKHFVVYGTLDGALGYCLPLPEKVYRRFLM-LQNVLLSYQEHLCGLN 1389
Query: 1169 HMAYRS-------AYFPVKDVIDGDLCEQFPTLPMDLQRKIADELDRTPGEILKKLEEV 1220
YR+ P + +IDGDL + + + ++A ++ EIL L E+
Sbjct: 1390 PKEYRTLKSSKKQGINPSRCIIDGDLIWSYRLMANSERNEVAKKIGTRTEEILGDLLEI 1448
Score = 37.7 bits (86), Expect = 0.19
Identities = 24/76 (31%), Positives = 43/76 (56%), Gaps = 9/76 (11%)
Query: 468 VWTV-----KKNVMDEFDAYIVVSFTNATLVLSIGETADEVSDSGFLDTAPSLAVSLIGD 522
VWTV KK+ ++ ++++S N+TLVL G+ +E+ ++GF P++ V +G
Sbjct: 563 VWTVFDDATKKSSRNDQHDFMLLSQRNSTLVLQTGQEINEIENTGFTVNQPTIFVGNLGQ 622
Query: 523 DS-LMQVHPNGIRHIR 537
++QV RH+R
Sbjct: 623 QRFIVQV---TTRHVR 635
>PUR5_METJA (Q57656) Phosphoribosylformylglycinamidine cyclo-ligase
(EC 6.3.3.1) (AIRS) (Phosphoribosyl-aminoimidazole
synthetase) (AIR synthase)
Length = 350
Score = 35.4 bits (80), Expect = 0.96
Identities = 21/58 (36%), Positives = 32/58 (54%), Gaps = 3/58 (5%)
Query: 286 IVSAAMHKTKNLKPEEFKLFFLLQTEYGDIFKVTLTDGGGDR--VSELNIKYFDTIAV 341
+VS K ++KP E L + E+GD + V TDG G + V+E+ K FDT+ +
Sbjct: 21 LVSQITFKRSDIKPAELGLHYAGAVEFGDYYLVLSTDGVGSKMIVAEMANK-FDTVGI 77
>NDF1_RAT (Q64289) Neurogenic differentiation factor 1 (NeuroD1)
(Basic helix-loop-helix factor 1) (BHF-1)
Length = 357
Score = 35.0 bits (79), Expect = 1.2
Identities = 21/55 (38%), Positives = 29/55 (52%), Gaps = 7/55 (12%)
Query: 816 EEREAARKEC-FEAAHAGENKTGSEDQMENGGEDEDNDDSLSDEHYGYPKSESDK 869
EE EA +KE EA +A ED + NGGE+ED D+ L +E + + K
Sbjct: 32 EEHEADKKEDELEAMNA------EEDSLRNGGEEEDEDEDLEEEEEEEEEEDDQK 80
>NDF1_HUMAN (Q13562) Neurogenic differentiation factor 1 (NeuroD1)
(NeuroD)
Length = 356
Score = 35.0 bits (79), Expect = 1.2
Identities = 21/55 (38%), Positives = 29/55 (52%), Gaps = 7/55 (12%)
Query: 816 EEREAARKEC-FEAAHAGENKTGSEDQMENGGEDEDNDDSLSDEHYGYPKSESDK 869
EE EA +KE EA +A ED + NGGE+ED D+ L +E + + K
Sbjct: 32 EEHEADKKEDDLEAMNA------EEDSLRNGGEEEDEDEDLEEEEEEEEEDDDQK 80
>LDH_THESA (Q7X5C9) L-lactate dehydrogenase (EC 1.1.1.27) (L-LDH)
Length = 311
Score = 35.0 bits (79), Expect = 1.2
Identities = 19/69 (27%), Positives = 33/69 (47%)
Query: 92 LDYNKQKNVFDKIHQETFGKSGCRRIVPGQYLAIDPKGRAVMIAACEKKKLVYVLNRDSL 151
+ +N+ ++ ++ F K +V Y ID KG A +++V + RD
Sbjct: 189 ISFNEYCSICGRVCNTNFRKEVEEEVVNAAYKIIDKKGATYYAVAVAVRRIVECILRDEN 248
Query: 152 ARLTISSPL 160
+ LT+SSPL
Sbjct: 249 SILTVSSPL 257
>EFTS_MYCGA (Q7NC21) Elongation factor Ts (EF-Ts)
Length = 292
Score = 35.0 bits (79), Expect = 1.2
Identities = 41/162 (25%), Positives = 60/162 (36%), Gaps = 16/162 (9%)
Query: 1064 FARLPQDVSDEIEEDPTGGKIKWEQGKLNGAPNKVEEIVQFHVGDVI---TSLQKASLVP 1120
F ++ D + + T K EQ KL E Q H+ +I SL++ + V
Sbjct: 88 FVAFSNELVDLVHKHETTDVAKIEQLKLASGSTVAE--TQIHLTAIIGEKISLRRVAFVK 145
Query: 1121 --GGGECIVYGTVMGSVGALHAFTSRDDVDFFSHLEMHMRQDNPPLCGRDHMAYRSAYFP 1178
Y +G + + DD +F HL MH+ NP + + SA F
Sbjct: 146 EEANSSLATYLHSNSRIGVIVKTSKTDDKEFLKHLAMHIAASNPKFVSQKDV---SADFI 202
Query: 1179 VKDVIDGDLCEQFPTLPMDLQRKIADELDRTPGEILKKLEEV 1220
K+ Q P + +I D G I K LEEV
Sbjct: 203 AKEREIAAAQAQSENKPKEFIDRIVD------GRINKVLEEV 238
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.138 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 146,674,367
Number of Sequences: 164201
Number of extensions: 6523255
Number of successful extensions: 16703
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 16533
Number of HSP's gapped (non-prelim): 103
length of query: 1225
length of database: 59,974,054
effective HSP length: 122
effective length of query: 1103
effective length of database: 39,941,532
effective search space: 44055509796
effective search space used: 44055509796
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 72 (32.3 bits)
Medicago: description of AC144540.4


