

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144517.6 + phase: 0
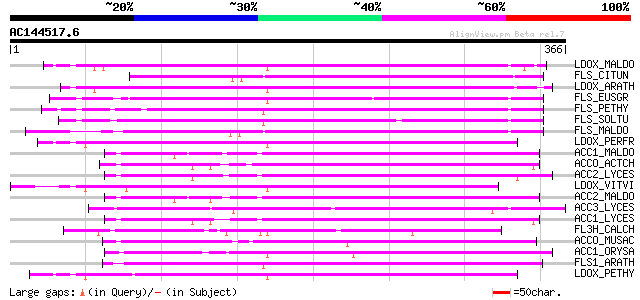
(366 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
LDOX_MALDO (P51091) Leucoanthocyanidin dioxygenase (EC 1.14.11.1... 172 2e-42
FLS_CITUN (Q9ZWQ9) Flavonol synthase (EC 1.14.11.-) (FLS) (CitFLS) 169 8e-42
LDOX_ARATH (Q96323) Leucoanthocyanidin dioxygenase (EC 1.14.11.1... 168 2e-41
FLS_EUSGR (Q9M547) Flavonol synthase (EC 1.14.11.-) (FLS) 167 4e-41
FLS_PETHY (Q07512) Flavonol synthase (EC 1.14.11.-) (FLS) 163 6e-40
FLS_SOLTU (Q41452) Flavonol synthase (EC 1.14.11.-) (FLS) 163 7e-40
FLS_MALDO (Q9XHG2) Flavonol synthase (EC 1.14.11.-) (FLS) 160 5e-39
LDOX_PERFR (O04274) Leucoanthocyanidin dioxygenase (EC 1.14.11.1... 158 2e-38
ACC1_MALDO (Q00985) 1-aminocyclopropane-1-carboxylate oxidase 1 ... 157 4e-38
ACCO_ACTCH (P31237) 1-aminocyclopropane-1-carboxylate oxidase (E... 157 5e-38
ACC2_LYCES (P07920) 1-aminocyclopropane-1-carboxylate oxidase 2 ... 156 9e-38
LDOX_VITVI (P51093) Leucoanthocyanidin dioxygenase (EC 1.14.11.1... 154 3e-37
ACC2_MALDO (O48882) 1-aminocyclopropane-1-carboxylate oxidase 2 ... 154 3e-37
ACC3_LYCES (P10967) 1-aminocyclopropane-1-carboxylate oxidase ho... 154 4e-37
ACC1_LYCES (P05116) 1-aminocyclopropane-1-carboxylate oxidase 1 ... 153 6e-37
FL3H_CALCH (Q05963) Naringenin,2-oxoglutarate 3-dioxygenase (EC ... 152 1e-36
ACCO_MUSAC (Q9FR99) 1-aminocyclopropane-1-carboxylate oxidase (E... 152 1e-36
ACC1_ORYSA (Q40634) 1-aminocyclopropane-1-carboxylate oxidase 1 ... 152 1e-36
FLS1_ARATH (Q96330) Flavonol synthase 1 (EC 1.14.11.-) (FLS 1) 152 2e-36
LDOX_PETHY (P51092) Leucoanthocyanidin dioxygenase (EC 1.14.11.1... 151 2e-36
>LDOX_MALDO (P51091) Leucoanthocyanidin dioxygenase (EC 1.14.11.19)
(LDOX) (Leucocyanidin oxygenase) (Leucoanthocyanidin
hydroxylase) (Anthocyanidin synthase)
Length = 357
Score = 172 bits (435), Expect = 2e-42
Identities = 112/347 (32%), Positives = 181/347 (51%), Gaps = 21/347 (6%)
Query: 23 VKTLSESPDFNSIPSSYTYTTNPHDENEIVAD---QDEVND--PIPVIDYSLLINGNHDQ 77
V+TL+ S ++IP Y P DE + D Q++ N+ +P ID + + N
Sbjct: 11 VETLAGS-GISTIPKEYI---RPKDELVNIGDIFEQEKNNEGPQVPTIDLKEIESDNEKV 66
Query: 78 RTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDS 137
R K + KA +WG L NH +S LM+K+ AFF+L E K+ YA+ + +
Sbjct: 67 RAKCREKLKKAAVDWGVMHLVNHGISDELMDKVRKAGKAFFDLPIEQKEKYANDQASGKI 126
Query: 138 IKYGTSFNVSGDKNLFWRDFIKIIVHPKFHS-----PDKPSGFRETSAEYSRKTWKLGRE 192
YG+ + L W D+ V+P+ P P+ + E +AEY+++ +L +
Sbjct: 127 QGYGSKLANNASGQLEWEDYFFHCVYPEDKRDLSIWPQTPADYIEATAEYAKQLRELATK 186
Query: 193 LLKGISESLGLEVNYIDKTMN-LDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLI 251
+LK +S LGL+ ++K + L+ L + N YP CPQP+LA+G+ H+D L ++
Sbjct: 187 VLKVLSLGLGLDEGRLEKEVGGLEELLLQMKINYYPKCPQPELALGVEAHTDVSALTFIL 246
Query: 252 QNGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLA 311
N V GLQ+ + GKW+ N ++ + D LEI+SNGKYKS++HR V+ R+S A
Sbjct: 247 HNMVPGLQLFYEGKWVTAKCVPNSIVMHIGDTLEILSNGKYKSILHRGMVNKEKVRISWA 306
Query: 312 TVIAPSLDTVV-EPASELLDNESNPAAY---VGMKHIDYMKLQRNNQ 354
P + ++ +P E + +E PA + +HI + KL R +Q
Sbjct: 307 VFCEPPKEKIILKPLPETV-SEDEPAMFPPRTFAEHIQH-KLFRKSQ 351
>FLS_CITUN (Q9ZWQ9) Flavonol synthase (EC 1.14.11.-) (FLS) (CitFLS)
Length = 335
Score = 169 bits (429), Expect = 8e-42
Identities = 99/279 (35%), Positives = 148/279 (52%), Gaps = 8/279 (2%)
Query: 80 KTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIK 139
+ + I +A EWG F +TNH + L+ K+ FF L +E+K+VY+ D
Sbjct: 55 RLVRSIAEASREWGIFQVTNHGIPSDLICKLQAVGKEFFELPQEEKEVYSRPADAKDVQG 114
Query: 140 YGTSFN--VSGDKN----LFWRDFIKIIVHPKFHSPDKPSGFRETSAEYSRKTWKLGREL 193
YGT V G K+ LF R + ++ +F P P +R + EY++ ++ +L
Sbjct: 115 YGTKLQKEVEGKKSWVDHLFHRVWPPSSINYRFW-PKNPPSYRAVNEEYAKYMREVVDKL 173
Query: 194 LKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQN 253
+S LG+E + + D ML N YPPCP+PDLA+G+ H+D L +L+ N
Sbjct: 174 FTYLSLGLGVEGGVLKEAAGGDDIEYMLKINYYPPCPRPDLALGVVAHTDLSALTVLVPN 233
Query: 254 GVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATV 313
V GLQV + +WI+ N ++ + D +EI+SNGKYK+V+HR V+ TRMS
Sbjct: 234 EVPGLQVFKDDRWIDAKYIPNALVIHIGDQIEILSNGKYKAVLHRTTVNKDKTRMSWPVF 293
Query: 314 IAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRN 352
+ P DTVV P +L+D+E NP Y K DY + N
Sbjct: 294 LEPPADTVVGPLPQLVDDE-NPPKYKAKKFKDYSYCKLN 331
>LDOX_ARATH (Q96323) Leucoanthocyanidin dioxygenase (EC 1.14.11.19)
(LDOX) (Leucocyanidin oxygenase) (Leucoanthocyanidin
hydroxylase) (Anthocyanidin synthase) (ANS)
Length = 356
Score = 168 bits (425), Expect = 2e-41
Identities = 104/337 (30%), Positives = 171/337 (49%), Gaps = 20/337 (5%)
Query: 34 SIPSSYTYTTNPHDENEIVAD-----QDEVNDPIPVIDYSLLINGNHDQRTKTIHDIGKA 88
SIP Y P +E E + D + E +P ID + + + R I ++ KA
Sbjct: 17 SIPKEYI---RPKEELESINDVFLEEKKEDGPQVPTIDLKNIESDDEKIRENCIEELKKA 73
Query: 89 CEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYGTSFNVSG 148
+WG L NH + LME++ FF+L E+K+ YA+ + T YG+ +
Sbjct: 74 SLDWGVMHLINHGIPADLMERVKKAGEEFFSLSVEEKEKYANDQATGKIQGYGSKLANNA 133
Query: 149 DKNLFWRDFIKIIVHPKFHS-----PDKPSGFRETSAEYSRKTWKLGRELLKGISESLGL 203
L W D+ + +P+ P PS + E ++EY++ L ++ K +S LGL
Sbjct: 134 SGQLEWEDYFFHLAYPEEKRDLSIWPKTPSDYIEATSEYAKCLRLLATKVFKALSVGLGL 193
Query: 204 EVNYIDKTMN-LDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGVSGLQVLH 262
E + ++K + L+ L + N YP CPQP+LA+G+ H+D L ++ N V GLQ+ +
Sbjct: 194 EPDRLEKEVGGLEELLLQMKINYYPKCPQPELALGVEAHTDVSALTFILHNMVPGLQLFY 253
Query: 263 NGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVV 322
GKW+ + ++ + D LEI+SNGKYKS++HR V+ R+S A P D +V
Sbjct: 254 EGKWVTAKCVPDSIVMHIGDTLEILSNGKYKSILHRGLVNKEKVRISWAVFCEPPKDKIV 313
Query: 323 -EPASELLDNESNPAAYVGMKHIDYMKLQRNNQLYGK 358
+P E++ ES PA + +++ ++L+GK
Sbjct: 314 LKPLPEMVSVES-PAKFPPRTFAQHIE----HKLFGK 345
>FLS_EUSGR (Q9M547) Flavonol synthase (EC 1.14.11.-) (FLS)
Length = 334
Score = 167 bits (423), Expect = 4e-41
Identities = 108/331 (32%), Positives = 165/331 (49%), Gaps = 16/331 (4%)
Query: 27 SESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTKTIHDIG 86
S S ++IP+ Y + N E +++ V +PVID S + D++ K + +
Sbjct: 11 SLSKVIDTIPAEYIRSEN---EQPVISTVHGVVLEVPVIDLS-----DSDEK-KIVGLVS 61
Query: 87 KACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYGTSFNV 146
+A +EWG F + NH + ++ K+ + FF L +E+K++ A E + YGT
Sbjct: 62 EASKEWGIFQVVNHGIPNEVIRKLQEVGKHFFELPQEEKELIAKPEGSQSIEGYGTRLQK 121
Query: 147 SGDKNLFWRDFIKIIVHPKFHS-----PDKPSGFRETSAEYSRKTWKLGRELLKGISESL 201
D W D + + P P P +RE + EY+++ + L K +S L
Sbjct: 122 EVDGKKGWVDHLFHKIWPPSAINYQFWPKNPPAYREANEEYAKRLQLVVDNLFKYLSLGL 181
Query: 202 GLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGVSGLQVL 261
LE N D + ++ N YPPCP+PDLA+G+ H+D + +L+ N V GLQV
Sbjct: 182 DLEPNSFKDGAGGDDLVYLMKINYYPPCPRPDLALGVA-HTDMSAITVLVPNEVPGLQVY 240
Query: 262 HNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTV 321
+G W + N +V + D +EIMSNGKYKSV HR V+ TRMS + P D
Sbjct: 241 KDGHWYDCKYIPNALIVHIGDQVEIMSNGKYKSVYHRTTVNKEKTRMSWPVFLEPPPDHE 300
Query: 322 VEPASELLDNESNPAAYVGMKHIDYMKLQRN 352
V P +L+ NE NPA + K+ DY + N
Sbjct: 301 VGPIPKLV-NEENPAKFKTKKYKDYAYCKLN 330
>FLS_PETHY (Q07512) Flavonol synthase (EC 1.14.11.-) (FLS)
Length = 348
Score = 163 bits (413), Expect = 6e-40
Identities = 108/337 (32%), Positives = 170/337 (50%), Gaps = 18/337 (5%)
Query: 22 SVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTKT 81
++ +LS+ D +IPS Y + N E V +PVID L + + ++ K
Sbjct: 20 AIASLSKCMD--TIPSEYIRSEN---EQPAATTLHGVVLQVPVID---LRDPDENKMVKL 71
Query: 82 IHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFF-NLKEEDKQVYADKEVTDDSIKY 140
I D K EWG F L NH + + + FF ++ +E+K++ A ++D Y
Sbjct: 72 IADASK---EWGIFQLINHGIPDEAIADLQKVGKEFFEHVPQEEKELIAKTPGSNDIEGY 128
Query: 141 GTSFNVSGDKNLFWRDFIKIIVHPKF-----HSPDKPSGFRETSAEYSRKTWKLGRELLK 195
GTS + W D + + P + P P +RE + EY ++ ++ + K
Sbjct: 129 GTSLQKEVEGKKGWVDHLFHKIWPPSAVNYRYWPKNPPSYREANEEYGKRMREVVDRIFK 188
Query: 196 GISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGV 255
+S LGLE + + + D + +L N YPPCP+PDLA+G+ H+D + +L+ N V
Sbjct: 189 SLSLGLGLEGHEMIEAAGGDEIVYLLKINYYPPCPRPDLALGVVAHTDMSYITILVPNEV 248
Query: 256 SGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIA 315
GLQV +G W +V N +V + D +EI+SNGKYKSV HR V+ TRMS +
Sbjct: 249 QGLQVFKDGHWYDVKYIPNALIVHIGDQVEILSNGKYKSVYHRTTVNKDKTRMSWPVFLE 308
Query: 316 PSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRN 352
P + V P +LL +E+NP + K+ DY+ + N
Sbjct: 309 PPSEHEVGPIPKLL-SEANPPKFKTKKYKDYVYCKLN 344
>FLS_SOLTU (Q41452) Flavonol synthase (EC 1.14.11.-) (FLS)
Length = 349
Score = 163 bits (412), Expect = 7e-40
Identities = 105/326 (32%), Positives = 163/326 (49%), Gaps = 17/326 (5%)
Query: 33 NSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLLINGNHDQRTKTIHDIGKACEEW 92
++IPS Y + N E V +PVID I+ D K + +I +A +EW
Sbjct: 31 DTIPSEYIRSEN---EQPAATTLQGVVLEVPVID----ISNVDDDEEKLVKEIVEASKEW 83
Query: 93 GFFILTNHSVSKSLMEKMVDQVFAFFN-LKEEDKQVYADKEVTDDSIKYGTSFNVSGDKN 151
G F + NH + ++E + FF + +E+K++ A K YGTS +
Sbjct: 84 GIFQVINHGIPDEVIENLQKVGKEFFEEVPQEEKELIAKKPGAQSLEGYGTSLQKEIEGK 143
Query: 152 LFWRDFIKIIVHPKF-----HSPDKPSGFRETSAEYSRKTWKLGRELLKGISESLGLEVN 206
W D + + P + P P +RE + EY++ K+ + + +S LGLE +
Sbjct: 144 KGWVDHLFHKIWPPSAINYRYWPKNPPSYREANEEYAKWLRKVADGIFRSLSLGLGLEGH 203
Query: 207 YIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGVSGLQVLHNGKW 266
+ + + + ML N YPPCP+PDLA+G+ H+D + LL+ N V QV +G W
Sbjct: 204 EMMEAAGSEDIVYMLKINYYPPCPRPDLALGVVAHTDMSYITLLVPNEV---QVFKDGHW 260
Query: 267 INVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVVEPAS 326
+V+ N +V + D +EI+SNGKYKSV HR V+ TRMS + PS + V P
Sbjct: 261 YDVNYIPNAIIVHIGDQVEILSNGKYKSVYHRTTVNKYKTRMSWPVFLEPSSEHEVGPIP 320
Query: 327 ELLDNESNPAAYVGMKHIDYMKLQRN 352
L+ NE+NP + K+ DY+ + N
Sbjct: 321 NLI-NEANPPKFKTKKYKDYVYCKLN 345
>FLS_MALDO (Q9XHG2) Flavonol synthase (EC 1.14.11.-) (FLS)
Length = 337
Score = 160 bits (405), Expect = 5e-39
Identities = 105/352 (29%), Positives = 167/352 (46%), Gaps = 38/352 (10%)
Query: 11 QKSNNGITPFTSVKTLSESPDFNSIPSSYTYTTNPHDENEIVADQDEVNDPIPVIDYSLL 70
++SN G P +++ +E P ++ +P+ID+S
Sbjct: 10 RESNEGTIPAEFIRSENEQPGITTVHGKVL--------------------EVPIIDFS-- 47
Query: 71 INGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYAD 130
K I I +A WG + + NH + ++ K+ FF L +E+K+ YA
Sbjct: 48 ----DPDEEKLIVQITEASSNWGMYQIVNHDIPSEVISKLQAVGKEFFELPQEEKEAYAK 103
Query: 131 KEVTDDSIKYGTSF--NVS-GDK-------NLFWRDFIKIIVHPKFHSPDKPSGFRETSA 180
+ YGT +S GD NLF + + +V+ +F P P +RE +
Sbjct: 104 PPDSASIEGYGTKLFKEISEGDTTKKGWVDNLFNKIWPPSVVNYQFW-PKNPPSYREANE 162
Query: 181 EYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLAMGMPP 240
EY++ + +L + +S LGLE + K D+ +L N YPPCP+PDLA+G+
Sbjct: 163 EYAKHLHNVVEKLFRLLSLGLGLEGQELKKAAGGDNLEYLLKINYYPPCPRPDLALGVVA 222
Query: 241 HSDHGLLNLLIQNGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAA 300
H+D + +L+ N V GLQ +G+W +V N ++ + D +EIMSNGKY SV+HR
Sbjct: 223 HTDMSTVTILVPNDVQGLQACKDGRWYDVKYIPNALVIHIGDQMEIMSNGKYTSVLHRTT 282
Query: 301 VSNGATRMSLATVIAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRN 352
V+ TR+S + P D VV P +L+ N N Y K+ DY+ + N
Sbjct: 283 VNKDKTRISWPVFLEPPADHVVGPHPQLV-NAVNQPKYKTKKYGDYVYCKIN 333
>LDOX_PERFR (O04274) Leucoanthocyanidin dioxygenase (EC 1.14.11.19)
(LDOX) (Leucocyanidin oxygenase) (Leucoanthocyanidin
hydroxylase)
Length = 362
Score = 158 bits (399), Expect = 2e-38
Identities = 95/330 (28%), Positives = 163/330 (48%), Gaps = 17/330 (5%)
Query: 19 PFTSVKTLSESPDFNSIPSSYTYTTNPHDE-----NEIVADQDEVNDP-IPVIDYSLLIN 72
P V+ L+ S ++IP Y P +E I+A++ P +P ID + +
Sbjct: 8 PSPRVEELARS-GLDTIPKDYV---RPEEELKSIIGNILAEEKSSEGPQLPTIDLEEMDS 63
Query: 73 GNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKE 132
+ + R K ++ KA +WG L NH + + L++++ FF L E+K+ YA+ +
Sbjct: 64 RDEEGRKKCHEELKKAATDWGVMHLINHGIPEELIDRVKAAGKEFFELPVEEKEAYANDQ 123
Query: 133 VTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHS-----PDKPSGFRETSAEYSRKTW 187
+ YG+ + L W D+ V+P+ + P KP + ++EY+++
Sbjct: 124 AAGNVQGYGSKLANNASGQLEWEDYFFHCVYPEHKTDLSIWPTKPPDYIPATSEYAKQLR 183
Query: 188 KLGRELLKGISESLGLEVNYIDKTMN-LDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGL 246
L ++L +S LGLE ++K + + + + N YP CPQP+LA+G H+D
Sbjct: 184 ALATKILSVLSIGLGLEKGRLEKEVGGAEDLIVQMKINFYPKCPQPELALGWEAHTDVSA 243
Query: 247 LNLLIQNGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGAT 306
L ++ N V GLQ+ + KW+ N ++ + D LEI+SNGKYKS++HR V+
Sbjct: 244 LTFILHNMVPGLQLFYEDKWVTAKCVPNSIIMHIGDTLEILSNGKYKSILHRGLVNKEKV 303
Query: 307 RMSLATVIAPSLDTVV-EPASELLDNESNP 335
R+S A P + +V +P E + P
Sbjct: 304 RISWAVFCEPPKEKIVLQPLPETVSEVEPP 333
>ACC1_MALDO (Q00985) 1-aminocyclopropane-1-carboxylate oxidase 1 (EC
1.14.17.4) (ACC oxidase 1) (Ethylene-forming enzyme)
(EFE) (Protein AP4) (PAE12)
Length = 314
Score = 157 bits (397), Expect = 4e-38
Identities = 100/293 (34%), Positives = 156/293 (53%), Gaps = 16/293 (5%)
Query: 63 PVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLM---EKMVDQVFAFFN 119
PV+D SL+ N ++R T+ I ACE WGFF L NH +S L+ EKM +
Sbjct: 5 PVVDLSLV---NGEERAATLEKINDACENWGFFELVNHGMSTELLDTVEKMTKDHYKK-T 60
Query: 120 LKEEDKQVYADKEVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHSPDKPSGFRETS 179
+++ K++ A K + D + ++ + F R + PD +R+T
Sbjct: 61 MEQRFKEMVAAKGLDDVQSEI---HDLDWESTFFLRHLPSSNIS---EIPDLEEEYRKTM 114
Query: 180 AEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQM-LAANLYPPCPQPDLAMGM 238
E++ + KL +LL + E+LGLE Y+ K G + YPPCP+PDL G+
Sbjct: 115 KEFAVELEKLAEKLLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPCPKPDLIKGL 174
Query: 239 PPHSDHGLLNLLIQNG-VSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVH 297
HSD G + LL Q+ VSGLQ+L +G+W++V + ++ + D +E+++NGKYKSV+H
Sbjct: 175 RAHSDAGGIILLFQDDKVSGLQLLKDGEWVDVPPMHHSIVINLGDQIEVITNGKYKSVMH 234
Query: 298 RAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPA-AYVGMKHIDYMKL 349
R + TRMS+A+ P D+ + PA +L+ ++ A Y DYMKL
Sbjct: 235 RVIAQSDGTRMSIASFYNPGNDSFISPAPAVLEKKTEDAPTYPKFVFDDYMKL 287
>ACCO_ACTCH (P31237) 1-aminocyclopropane-1-carboxylate oxidase (EC
1.14.17.4) (ACC oxidase) (Ethylene-forming enzyme) (EFE)
Length = 319
Score = 157 bits (396), Expect = 5e-38
Identities = 103/298 (34%), Positives = 152/298 (50%), Gaps = 19/298 (6%)
Query: 60 DPIPVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFN 119
+ PVID L N ++R T+ I ACE WGFF L NH +S LM+ + +N
Sbjct: 2 EAFPVIDMEKL---NGEERAPTMEKIKDACENWGFFELVNHGISHELMDTVERLTKEHYN 58
Query: 120 --LKEEDKQVYADK--EVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHSPDKPSGF 175
+++ K++ A K E I ++ + F R + V PD
Sbjct: 59 KCMEQRFKEMVATKGLEAVQSEIN-----DLDWESTFFLRH---LPVSNISEIPDLEQDH 110
Query: 176 RETSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQM-LAANLYPPCPQPDL 234
R+ E++ K KL +LL + E++GLE Y+ K G + YPPCP+P+L
Sbjct: 111 RKAMKEFAEKLEKLAEQLLDLLCENVGLEKGYLKKAFYGSKGPTFGTKVSNYPPCPRPEL 170
Query: 235 AMGMPPHSDHGLLNLLIQ-NGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYK 293
G+ H+D G + LL Q N VSGLQ+L +G+WI+V + ++ + D LE+++NGKYK
Sbjct: 171 IKGLRAHTDAGGIILLFQDNKVSGLQLLKDGEWIDVPPMKHSIVINIGDQLEVITNGKYK 230
Query: 294 SVVHRAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPAAYVGMKHI--DYMKL 349
SV+HR RMS+A+ P D V+ PA L+D E + V K + DYMKL
Sbjct: 231 SVMHRVIAQPDGNRMSIASFYNPGSDAVMYPAPALVDKEEDQQKQVYPKFVFEDYMKL 288
>ACC2_LYCES (P07920) 1-aminocyclopropane-1-carboxylate oxidase 2 (EC
1.14.17.4) (ACC oxidase 2) (Ethylene-forming enzyme)
(EFE) (Protein GTOMA)
Length = 316
Score = 156 bits (394), Expect = 9e-38
Identities = 100/303 (33%), Positives = 153/303 (50%), Gaps = 16/303 (5%)
Query: 63 PVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFN--L 120
P+I+ L N +R T+ I ACE WGFF L NH + +M+ + + +
Sbjct: 5 PIINLEKL---NGAERVATMEKINDACENWGFFELVNHGIPHEVMDTVEKLTKGHYKKCM 61
Query: 121 KEEDKQVYADKEVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHSPDKPSGFRETSA 180
++ K++ A K + ++ ++ + F R + PD +RE
Sbjct: 62 EQRFKELVAKKGLEGVEVEVT---DMDWESTFFLRHLPSSNIS---QLPDLDDVYREVMR 115
Query: 181 EYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQM-LAANLYPPCPQPDLAMGMP 239
++ ++ KL ELL + E+LGLE +Y+ T G + YPPCP+PDL G+
Sbjct: 116 DFRKRLEKLAEELLDLLCENLGLEKSYLKNTFYGSKGPNFGTKVSNYPPCPKPDLIKGLR 175
Query: 240 PHSDHGLLNLLIQNG-VSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHR 298
H+D G + LL Q+ VSGLQ+L +G+WI+V + +V + D LE+++NGKYKSV+HR
Sbjct: 176 AHTDAGGIILLFQDDKVSGLQLLKDGRWIDVPPMRHSIVVNLGDQLEVITNGKYKSVMHR 235
Query: 299 AAVSNGATRMSLATVIAPSLDTVVEPASELLDNES---NPAAYVGMKHIDYMKLQRNNQL 355
TRMSLA+ P D ++ PA L+D E+ N Y DYMKL N +
Sbjct: 236 VIAQKDGTRMSLASFYNPGNDALIYPAPALVDKEAEEHNKQVYPKFMFDDYMKLYANLKF 295
Query: 356 YGK 358
K
Sbjct: 296 QAK 298
>LDOX_VITVI (P51093) Leucoanthocyanidin dioxygenase (EC 1.14.11.19)
(LDOX) (Leucocyanidin oxygenase) (Leucoanthocyanidin
hydroxylase)
Length = 362
Score = 154 bits (390), Expect = 3e-37
Identities = 96/337 (28%), Positives = 160/337 (46%), Gaps = 33/337 (9%)
Query: 1 MASTLPPQVNQKSNNGITPFTSVKTLSESPDFNSIPSSYTYTTNPHDE-----NEIVADQ 55
M +++ P+V S++GI SIP Y P +E N ++
Sbjct: 1 MVTSVAPRVESLSSSGI---------------QSIPKEYI---RPQEELTSIGNVFEEEK 42
Query: 56 DEVNDPIPVIDYSLLINGNH----DQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMV 111
+ +P ID + + + + R + ++ KA EWG L NH +S L+ ++
Sbjct: 43 KDEGPQVPTIDLKDIESEDEVVRREIRERCREELKKAAMEWGVMHLVNHGISDDLINRVK 102
Query: 112 DQVFAFFNLKEEDKQVYADKEVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHS--- 168
FFNL E+K+ YA+ + + YG+ + L W D+ ++ P+
Sbjct: 103 VAGETFFNLPMEEKEKYANDQASGKIAGYGSKLANNASGQLEWEDYFFHLIFPEDKRDMT 162
Query: 169 --PDKPSGFRETSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMN-LDSGLQMLAANL 225
P PS + + EYS K L ++L +S LGLE ++K + ++ L N
Sbjct: 163 IWPKTPSDYVPATCEYSVKLRSLATKILSVLSLGLGLEEGRLEKEVGGMEELLLQKKINY 222
Query: 226 YPPCPQPDLAMGMPPHSDHGLLNLLIQNGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLE 285
YP CPQP+LA+G+ H+D L ++ N V GLQ+ + GKW+ N ++ + D +E
Sbjct: 223 YPKCPQPELALGVEAHTDVSALTFILHNMVPGLQLFYEGKWVTAKCVPNSIIMHIGDTIE 282
Query: 286 IMSNGKYKSVVHRAAVSNGATRMSLATVIAPSLDTVV 322
I+SNGKYKS++HR V+ R+S A P + ++
Sbjct: 283 ILSNGKYKSILHRGLVNKEKVRISWAVFCEPPKEKII 319
>ACC2_MALDO (O48882) 1-aminocyclopropane-1-carboxylate oxidase 2 (EC
1.14.17.4) (ACC oxidase 2) (Ethylene-forming enzyme)
(EFE)
Length = 330
Score = 154 bits (390), Expect = 3e-37
Identities = 100/295 (33%), Positives = 153/295 (50%), Gaps = 20/295 (6%)
Query: 63 PVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLM---EKMVDQVFAFFN 119
PV+D L+ N ++R T+ I ACE WGFF L NH +S L+ EKM +
Sbjct: 5 PVVDMDLI---NGEERAATLEKINDACENWGFFELVNHGISTELLDTVEKMNKDHYKK-T 60
Query: 120 LKEEDKQVYADK--EVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHSPDKPSGFRE 177
+++ K++ A K E I Y + + F R + PD +R+
Sbjct: 61 MEQRFKEMVAAKGLEAVQSEIHY-----LDWESTFFLRHLPSSNIS---EIPDLEEDYRK 112
Query: 178 TSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQM-LAANLYPPCPQPDLAM 236
T E++ + KL +LL + E+LGLE Y+ K G + YPPCP+PDL
Sbjct: 113 TMKEFAVELEKLAEKLLDLLCENLGLEKGYLKKAFYGSKGPNFGTKVSNYPPCPKPDLIK 172
Query: 237 GMPPHSDHGLLNLLIQNG-VSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSV 295
G+ H+D G + LL Q+ VSGLQ+L +G+W++V + ++ + D +E+++NGKYKS+
Sbjct: 173 GLRAHTDAGGIILLFQDDKVSGLQLLKDGEWMDVPPVHHSIVINLGDQIEVITNGKYKSI 232
Query: 296 VHRAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPA-AYVGMKHIDYMKL 349
+HR + TRMS+A+ P D + PA LL+ +S + Y DYMKL
Sbjct: 233 MHRVIAQSDGTRMSIASFYNPGDDAFISPAPALLEEKSEVSPTYPKFLFDDYMKL 287
>ACC3_LYCES (P10967) 1-aminocyclopropane-1-carboxylate oxidase
homolog (Protein E8)
Length = 363
Score = 154 bits (388), Expect = 4e-37
Identities = 104/320 (32%), Positives = 164/320 (50%), Gaps = 11/320 (3%)
Query: 53 ADQDEVNDPIPVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVD 112
A + E + PVID I+ + + + + + A E+WGFF + NH + S++++ +
Sbjct: 49 AKKCETHFVFPVIDLQG-IDEDPIKHKEIVDKVRDASEKWGFFQVVNHGIPTSVLDRTLQ 107
Query: 113 QVFAFFNLKEEDKQVYADKEVTDDSIKYGTSFNV--SGDKNLFWRDFIKIIVHPKFHSPD 170
FF E K+ Y ++ T + Y ++ ++ S WRD I + P S
Sbjct: 108 GTRQFFEQDNEVKKQYYTRD-TAKKVVYTSNLDLYKSSVPAASWRDTIFCYMAPNPPSLQ 166
Query: 171 K-PSGFRETSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPC 229
+ P+ E+ ++S+ KLG LL+ +SE LGL+ +Y+ M D + N YPPC
Sbjct: 167 EFPTPCGESLIDFSKDVKKLGFTLLELLSEGLGLDRSYLKDYM--DCFHLFCSCNYYPPC 224
Query: 230 PQPDLAMGMPPHSDHGLLNLLIQNGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSN 289
PQP+L MG H+D G + +L+Q+ + GLQVLH W++V T +V + D L+++SN
Sbjct: 225 PQPELTMGTIQHTDIGFVTILLQDDMGGLQVLHQNHWVDVPPTPGSLVVNIGDFLQLLSN 284
Query: 290 GKYKSVVHRAAVSNGATRMSLATVIAPS---LDTVVEPASELLDNESNPAAYVGMKHIDY 346
KY SV HRA +N +RMS+ S + P +ELL +E NP Y D+
Sbjct: 285 DKYLSVEHRAISNNVGSRMSITCFFGESPYQSSKLYGPITELL-SEDNPPKYRATTVKDH 343
Query: 347 MKLQRNNQLYGKSVLNKVKI 366
N L G S L++ KI
Sbjct: 344 TSYLHNRGLDGTSALSRYKI 363
>ACC1_LYCES (P05116) 1-aminocyclopropane-1-carboxylate oxidase 1 (EC
1.14.17.4) (ACC oxidase 1) (Ethylene-forming enzyme)
(EFE) (Protein pTOM 13)
Length = 315
Score = 153 bits (387), Expect = 6e-37
Identities = 101/300 (33%), Positives = 151/300 (49%), Gaps = 29/300 (9%)
Query: 63 PVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFN--L 120
P+I+ L N D+R T+ I ACE WGFF L NH + +M+ + + +
Sbjct: 5 PIINLEKL---NGDERANTMEMIKDACENWGFFELVNHGIPHEVMDTVEKMTKGHYKKCM 61
Query: 121 KEEDKQVYADK-------EVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHSPDKPS 173
++ K++ A K EVTD + + F R + PD
Sbjct: 62 EQRFKELVASKGLEAVQAEVTD----------LDWESTFFLRHLPTSNIS---QVPDLDE 108
Query: 174 GFRETSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQM-LAANLYPPCPQP 232
+RE +++++ KL ELL + E+LGLE Y+ G + YPPCP+P
Sbjct: 109 EYREVMRDFAKRLEKLAEELLDLLCENLGLEKGYLKNAFYGSKGPNFGTKVSNYPPCPKP 168
Query: 233 DLAMGMPPHSDHGLLNLLIQNG-VSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGK 291
DL G+ H+D G + LL Q+ VSGLQ+L + +WI+V + +V + D LE+++NGK
Sbjct: 169 DLIKGLRAHTDAGGIILLFQDDKVSGLQLLKDEQWIDVPPMRHSIVVNLGDQLEVITNGK 228
Query: 292 YKSVVHRAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPAAYVGMKHI--DYMKL 349
YKSV+HR TRMSLA+ P D V+ PA L++ E+ + V K + DYMKL
Sbjct: 229 YKSVLHRVIAQTDGTRMSLASFYNPGSDAVIYPAKTLVEKEAEESTQVYPKFVFDDYMKL 288
>FL3H_CALCH (Q05963) Naringenin,2-oxoglutarate 3-dioxygenase (EC
1.14.11.9) (Flavonone-3-hydroxylase) (F3H) (FHT)
Length = 356
Score = 152 bits (384), Expect = 1e-36
Identities = 100/307 (32%), Positives = 164/307 (52%), Gaps = 28/307 (9%)
Query: 36 PSSYTYTTNPHDENEIVADQDE--------VNDPIPVIDYSLLINGNHDQRTKTIHDIGK 87
P S + + EN+ V D+DE ++ IPVI L + +R + +I K
Sbjct: 4 PISLKWEEHSLHENKFVRDEDERPKVPYNTFSNEIPVIS---LAGIDGCRRAEICDEIVK 60
Query: 88 ACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVYADKEVTDDSIKYG--TSFN 145
ACE+WG F + +H V L+ M FF+L ++K + ++T K G S +
Sbjct: 61 ACEDWGIFQVVDHGVDTKLLSDMTGLARDFFHLPTQEKLRF---DMTGGK-KGGFIVSSH 116
Query: 146 VSGDKNLFWRDFIKIIVHP---KFHS--PDKPSGFRETSAEYSRKTWKLGRELLKGISES 200
+ G+ WR+ + +P + +S PDKP+ +R + EYS+ L +LL+ +SE+
Sbjct: 117 LQGEAVQDWREIVTYFSYPIKARDYSRWPDKPNEWRAVTEEYSKVLMGLACKLLEVLSEA 176
Query: 201 LGLEVNYIDKT-MNLDSGLQMLAANLYPPCPQPDLAMGMPPHSDHGLLNLLIQNGVSGLQ 259
+GLE + K +++D Q + N YP CPQPDL +G+ H+D G + LL+Q+ V GLQ
Sbjct: 177 MGLEKEALTKACVDMD---QKVVVNYYPKCPQPDLTLGLKRHTDPGTITLLLQDQVGGLQ 233
Query: 260 VLHNG--KWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVSNGATRMSLATVIAPS 317
+G WI V F+V + DH +SNG++K+ H+A V++ +R+S+AT P+
Sbjct: 234 ATRDGGESWITVKPVEGAFVVNLGDHGHYLSNGRFKNADHQAVVNSSTSRLSIATFQNPA 293
Query: 318 LDTVVEP 324
+ +V P
Sbjct: 294 PEAIVYP 300
>ACCO_MUSAC (Q9FR99) 1-aminocyclopropane-1-carboxylate oxidase (EC
1.14.17.4) (ACC oxidase) (Ethylene-forming enzyme) (EFE)
Length = 306
Score = 152 bits (384), Expect = 1e-36
Identities = 95/291 (32%), Positives = 147/291 (49%), Gaps = 13/291 (4%)
Query: 62 IPVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLK 121
IPVID+S L + +R +T+ I CEEWGFF L NH + L+E++ + L+
Sbjct: 3 IPVIDFSKL---DGKERAETMARIANGCEEWGFFQLVNHGIPVELLERVKKVSSECYKLR 59
Query: 122 EEDKQVYADKEVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHSPDKPSGFRETSAE 181
EE + ++ D +K G + N+ W D ++ P P F ET E
Sbjct: 60 EERFEGSKPVQLLDTLVKEGDGQRLD---NVDWEDVF--VLQDDNEWPSNPPDFEETMKE 114
Query: 182 YSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQML---AANLYPPCPQPDLAMGM 238
Y + KL ++++ + E+LG E I K + D + YPPCP+ DL G+
Sbjct: 115 YREEIRKLAEKMMEVMDENLGFEKGCIKKAFSGDGQHPPFFGTKVSHYPPCPRLDLVKGL 174
Query: 239 PPHSDHGLLNLLIQNG-VSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVH 297
H+D G + LL Q+ V GLQ+L +G+WI+V ++ ++ D +E++SNG+YKS H
Sbjct: 175 RAHTDAGGVILLFQDDQVGGLQMLKDGRWIDVQPLADAIVINTGDQIEVLSNGRYKSAWH 234
Query: 298 RAAVSNGATRMSLATVIAPSLDTVVEPASELLDNE-SNPAAYVGMKHIDYM 347
R ++ R S+A+ PSL + PA+ E + PA Y DYM
Sbjct: 235 RVLATSHGNRRSIASFYNPSLKATIAPAAGAATEEAAPPALYPKFLFGDYM 285
>ACC1_ORYSA (Q40634) 1-aminocyclopropane-1-carboxylate oxidase 1 (EC
1.14.17.4) (ACC oxidase 1) (Ethylene-forming enzyme)
(EFE)
Length = 322
Score = 152 bits (384), Expect = 1e-36
Identities = 94/302 (31%), Positives = 148/302 (48%), Gaps = 14/302 (4%)
Query: 63 PVIDYSLLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKE 122
PVI+ LL ++R + + ACE WGFF + NH +S LM+++ +
Sbjct: 8 PVINMELLAG---EERPAAMEQLDDACENWGFFEILNHGISTELMDEVEKMTKDHYKRVR 64
Query: 123 EDKQVYADKEVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHS---PDKPSGFRETS 179
E + + E ++K G +V+ + L W + P+ + PD +R
Sbjct: 65 EQRFL----EFASKTLKEGCD-DVNKAEKLDWESTFFVRHLPESNIADIPDLDDDYRRLM 119
Query: 180 AEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLAANL--YPPCPQPDLAMG 237
++ + L LL + E+LGLE Y+ K +G + YPPCP+PDL G
Sbjct: 120 KRFAAELETLAERLLDLLCENLGLEKGYLTKAFRGPAGAPTFGTKVSSYPPCPRPDLVKG 179
Query: 238 MPPHSDHGLLNLLIQN-GVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVV 296
+ H+D G + LL Q+ V GLQ+L +G+W++V + +V + D LE+++NG+YKSV+
Sbjct: 180 LRAHTDRGGIILLFQDDSVGGLQLLKDGEWVDVPPMRHSIVVNLGDQLEVITNGRYKSVM 239
Query: 297 HRAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPAAYVGMKHIDYMKLQRNNQLY 356
HR RMS+A+ P D V+ PA L+ E AY DYMKL ++
Sbjct: 240 HRVVAQTDGNRMSIASFYNPGSDGVISPAPALVKEEEAVVAYPKFVFEDYMKLYVRHKFE 299
Query: 357 GK 358
K
Sbjct: 300 AK 301
>FLS1_ARATH (Q96330) Flavonol synthase 1 (EC 1.14.11.-) (FLS 1)
Length = 336
Score = 152 bits (383), Expect = 2e-36
Identities = 93/297 (31%), Positives = 144/297 (48%), Gaps = 14/297 (4%)
Query: 62 IPVIDYSLLINGNHDQRTKTIHD-IGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNL 120
IPV+D S D +++ + KA EEWG F + NH + L+ ++ D FF L
Sbjct: 43 IPVVDLS-------DPDEESVRRAVVKASEEWGLFQVVNHGIPTELIRRLQDVGRKFFEL 95
Query: 121 KEEDKQVYADKEVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKF-----HSPDKPSGF 175
+K+ A E + D YGT + W D + + P P P +
Sbjct: 96 PSSEKESVAKPEDSKDIEGYGTKLQKDPEGKKAWVDHLFHRIWPPSCVNYRFWPKNPPEY 155
Query: 176 RETSAEYSRKTWKLGRELLKGISESLGLEVNYIDKTMNLDSGLQMLAANLYPPCPQPDLA 235
RE + EY+ KL LL +S+ LGL+ + + + + + M+ N YPPCP+PDLA
Sbjct: 156 REVNEEYAVHVKKLSETLLGILSDGLGLKRDALKEGLGGEMAEYMMKINYYPPCPRPDLA 215
Query: 236 MGMPPHSDHGLLNLLIQNGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSV 295
+G+P H+D + LL+ N V GLQV + W + + +V + D + +SNG+YK+V
Sbjct: 216 LGVPAHTDLSGITLLVPNEVPGLQVFKDDHWFDAEYIPSAVIVHIGDQILRLSNGRYKNV 275
Query: 296 VHRAAVSNGATRMSLATVIAPSLDTVVEPASELLDNESNPA-AYVGMKHIDYMKLQR 351
+HR V TRMS + P + +V P EL +++ P K Y KL +
Sbjct: 276 LHRTTVDKEKTRMSWPVFLEPPREKIVGPLPELTGDDNPPKFKPFAFKDYSYRKLNK 332
>LDOX_PETHY (P51092) Leucoanthocyanidin dioxygenase (EC 1.14.11.19)
(LDOX) (Leucocyanidin oxygenase) (Leucoanthocyanidin
hydroxylase)
Length = 430
Score = 151 bits (382), Expect = 2e-36
Identities = 94/334 (28%), Positives = 163/334 (48%), Gaps = 17/334 (5%)
Query: 14 NNGITPFTSVKTLSESPDFNSIPSSYTYTTNPHDE-----NEIVADQDEVNDPIPVIDYS 68
N +T + V++L++S +IP Y P +E N ++ + +P ID
Sbjct: 3 NAVVTTPSRVESLAKS-GIQAIPKEYV---RPQEELNGIGNIFEEEKKDEGPQVPTIDLK 58
Query: 69 LLINGNHDQRTKTIHDIGKACEEWGFFILTNHSVSKSLMEKMVDQVFAFFNLKEEDKQVY 128
+ + + + R K H + KA EWG L NH +S L+ ++ FF+ E+K+ Y
Sbjct: 59 EIDSEDKEIREKC-HQLKKAAMEWGVMHLVNHGISDELINRVKVAGETFFDQPVEEKEKY 117
Query: 129 ADKEVTDDSIKYGTSFNVSGDKNLFWRDFIKIIVHPKFHS-----PDKPSGFRETSAEYS 183
A+ + + YG+ S L W D+ P+ P P+ + ++EY+
Sbjct: 118 ANDQANGNVQGYGSKLANSACGQLEWEDYFFHCAFPEDKRDLSIWPKNPTDYTPATSEYA 177
Query: 184 RKTWKLGRELLKGISESLGLEVNYIDKTMN-LDSGLQMLAANLYPPCPQPDLAMGMPPHS 242
++ L ++L +S LGLE ++K + ++ L + N YP CPQP+LA+G+ H+
Sbjct: 178 KQIRALATKILTVLSIGLGLEEGRLEKEVGGMEDLLLQMKINYYPKCPQPELALGVEAHT 237
Query: 243 DHGLLNLLIQNGVSGLQVLHNGKWINVSSTSNCFLVLVSDHLEIMSNGKYKSVVHRAAVS 302
D L ++ N V GLQ+ + G+W+ N ++ + D +EI+SNGKYKS++HR V+
Sbjct: 238 DVSALTFILHNMVPGLQLFYEGQWVTAKCVPNSIIMHIGDTIEILSNGKYKSILHRGVVN 297
Query: 303 NGATRMSLATVIAPSLDTVV-EPASELLDNESNP 335
R S A P + ++ +P E + P
Sbjct: 298 KEKVRFSWAIFCEPPKEKIILKPLPETVTEAEPP 331
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.132 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 44,029,279
Number of Sequences: 164201
Number of extensions: 1872710
Number of successful extensions: 4306
Number of sequences better than 10.0: 91
Number of HSP's better than 10.0 without gapping: 66
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 4073
Number of HSP's gapped (non-prelim): 97
length of query: 366
length of database: 59,974,054
effective HSP length: 112
effective length of query: 254
effective length of database: 41,583,542
effective search space: 10562219668
effective search space used: 10562219668
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 66 (30.0 bits)
Medicago: description of AC144517.6


