

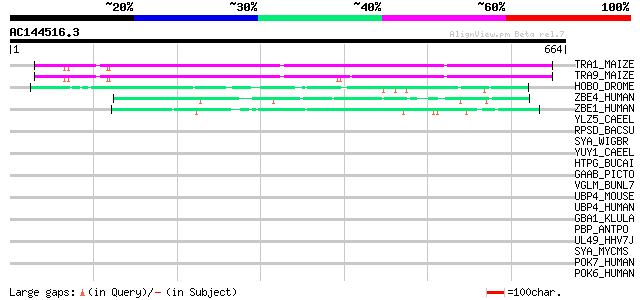
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144516.3 + phase: 0
(664 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TRA1_MAIZE (P08770) Putative AC transposase (ORFA) 358 2e-98
TRA9_MAIZE (P03010) Putative AC9 transposase 342 2e-93
HOBO_DROME (P12258) Transposable element Hobo transposase 77 2e-13
ZBE4_HUMAN (O75132) Zinc finger BED domain containing protein 4 65 6e-10
ZBE1_HUMAN (O96006) Zinc finger BED domain containing protein 1 ... 65 7e-10
YLZ5_CAEEL (P34418) Hypothetical protein F42H10.5 in chromosome III 40 0.020
RPSD_BACSU (P10726) RNA polymerase sigma-D factor (Sigma-28) 37 0.22
SYA_WIGBR (Q8D2W8) Alanyl-tRNA synthetase (EC 6.1.1.7) (Alanine-... 35 0.82
YUY1_CAEEL (P54073) Hypothetical protein F44G4.1 in chromosome II 34 1.1
HTPG_BUCAI (P57555) Chaperone protein htpG (Heat shock protein h... 34 1.4
GAAB_PICTO (Q6L1Q1) GMP synthase [glutamine-hydrolyzing] subunit... 33 3.1
VGLM_BUNL7 (P09612) M polyprotein precursor [Contains: Glycoprot... 32 4.1
UBP4_MOUSE (P35123) Ubiquitin carboxyl-terminal hydrolase 4 (EC ... 32 4.1
UBP4_HUMAN (Q13107) Ubiquitin carboxyl-terminal hydrolase 4 (EC ... 32 4.1
GBA1_KLULA (Q9Y7B7) Guanine nucleotide-binding protein alpha-1 s... 32 4.1
PBP_ANTPO (P20797) Pheromone-binding protein precursor (PBP) 32 7.0
UL49_HHV7J (P52442) Protein U33 31 9.1
SYA_MYCMS (P61704) Alanyl-tRNA synthetase (EC 6.1.1.7) (Alanine-... 31 9.1
POK7_HUMAN (Q9QC07) HERV-K_1q23.3 provirus ancestral Pol protein... 31 9.1
POK6_HUMAN (P63133) HERV-K_8p23.1 provirus ancestral Pol protein... 31 9.1
>TRA1_MAIZE (P08770) Putative AC transposase (ORFA)
Length = 806
Score = 358 bits (920), Expect = 2e-98
Identities = 213/643 (33%), Positives = 348/643 (53%), Gaps = 32/643 (4%)
Query: 30 DPKDKKRKGKAKAKDKSKGKALTSDVWLYLVKVGI---VDGVE------KCRCKACHKLL 80
+P+ + + + + + K TSDVW + K I VDG + C C
Sbjct: 117 EPQPQPQPEPEEEAPQKRAKKCTSDVWQHFTKKEIEVEVDGKKYVQVWGHCNFPNCKAKY 176
Query: 81 TCESGSGTSHLKRHVRSCSKTIKNHDVGEMMIDVE---GK----LRKKKFDPMANREFLA 133
E GTS + H+R+ +K G++ + E GK + K+D + + + L
Sbjct: 177 RAEGHHGTSGFRNHLRTSHSLVK----GQLCLKSEKDHGKDINLIEPYKYDEVVSLKKLH 232
Query: 134 RIMITHGAPFNMVEWKVFREYQKFLNDDCVFVSRNTIAKEILNVYRDEKQKLKSQLAQIR 193
+I H PFN+VE + F E+ K L SR T K I+++Y +EK+KL +L ++
Sbjct: 233 LAIIMHEYPFNIVEHEYFVEFVKSLRPHFPIKSRVTARKYIMDLYLEEKEKLYGKLKDVQ 292
Query: 194 GRVCLTSDCWTACSNEGYISLTAHYVNVNWKLESKILAFAHMEPPHSGRDLALKVLEMLD 253
R T D WT+C N+ Y+ +T H+++ +W L+ +I+ F H+E H+G+ L+ ++
Sbjct: 293 SRFSTTMDMWTSCQNKSYMCVTIHWIDDDWCLQKRIVGFFHVEGRHTGQRLSQTFTAIMV 352
Query: 254 DWGIEKKIFSITLDNASANNSMANFLKEHLSLSNS-LLLDGEFFHIRCSAHILNLIVQDG 312
W IEKK+F+++LDNASAN + + E L ++S L+ DG FFH+RC+ HILNL+ +DG
Sbjct: 353 KWNIEKKLFALSLDNASANEVAVHDIIEDLQDTDSNLVCDGAFFHVRCACHILNLVAKDG 412
Query: 313 LKVVSDALHKIRQSVAYVRVTEGRTLLFSECVRIVG--DIDTTIGLRLDCVTRWNSTYIM 370
L V++ + KI+ A V + L + E ++ D+D + G+ D TRWNSTY+M
Sbjct: 413 LAVIAGTIEKIK---AIVLAVKSSPLQWEELMKCASECDLDKSKGISYDVSTRWNSTYLM 469
Query: 371 LQSALVYRRAFYSLSLRDSN--FKCCPTSEEWRRAEIMCEILKPFFTITNLISGSSYPTS 428
L+ AL Y+ A L D CP +EEW+ A + + LK FF +T L+SG+ Y T+
Sbjct: 470 LRDALYYKPALIRLKTSDPRRYDAICPKAEEWKMALTLFKCLKKFFDLTELLSGTQYSTA 529
Query: 429 NLYFGEIWKIECLIRSYLTSEDLLIQKMAENMKVKFDKYWSDYNVVLAVGAVLDPTKKFN 488
NL++ +I+ LI + E +I++MA M KF+KYW N+ LAV LDP K
Sbjct: 530 NLFYKGFCEIKDLIDQWCVHEKFVIRRMAVAMSEKFEKYWKVSNIALAVACFLDPRYKKI 589
Query: 489 FLKFAYEKLDPLTSEEKLKKVKMTLGKLFSEYIKNGIPSNLSSSQVQPSYGGGTRITSSS 548
++F +K + + + + KL+ Y + + ++ + + +
Sbjct: 590 LIEFYMKKFHGDSYKVHVDDFVRVIRKLYQFY--SSCSPSAPKTKTTTNDSMDDTLMENE 647
Query: 549 YDEFEEY--ESQSSNNTGKSELDTYLDELRMPLSQEFDVLAFWKERSRRSPNLARMACDI 606
DEF+ Y E + + +ELD Y+ E + S +FD+L++W+ R P L ++A D+
Sbjct: 648 DDEFQNYLHELKDYDQVESNELDKYMSEPLLKHSGQFDILSWWRGRVAEYPILTQIARDV 707
Query: 607 LSIPITTVASESAFSIGARVVNRYRSSMKDDSVQALLCARSWL 649
L+I ++TVASESAFS G RVV+ YR+ + + V+AL+C + W+
Sbjct: 708 LAIQVSTVASESAFSAGGRVVDPYRNRLGSEIVEALICTKDWV 750
>TRA9_MAIZE (P03010) Putative AC9 transposase
Length = 839
Score = 342 bits (876), Expect = 2e-93
Identities = 213/669 (31%), Positives = 344/669 (50%), Gaps = 60/669 (8%)
Query: 30 DPKDKKRKGKAKAKDKSKGKALTSDVWLYLVKVGI---VDGVE------KCRCKACHKLL 80
+P+ + + + + + K TSDVW + K I VDG + C C
Sbjct: 63 EPQPQPQPEPEEEAPQKRAKKCTSDVWQHFTKKEIEVEVDGKKYVQVWGHCNFPNCKAKY 122
Query: 81 TCESGSGTSHLKRHVRSCSKTIKNHDVGEMMIDVE---GK----LRKKKFDPMANREFLA 133
E GTS + H+R+ +K G++ + E GK + K+D + + + L
Sbjct: 123 RAEGHHGTSGFRNHLRTSHSLVK----GQLCLKSEKDHGKDINLIEPYKYDEVVSLKKLH 178
Query: 134 RIMITHGAPFNMVEWKVFREYQKFLNDDCVFVSRNTIAKEILNVYRDEKQKLKSQLAQIR 193
+I H PFN+VE + F E+ K L SR T K I+++Y +EK+KL +L ++
Sbjct: 179 LAIIMHEYPFNIVEHEYFVEFVKSLRPHFPIKSRVTARKYIMDLYLEEKEKLYGKLKDVQ 238
Query: 194 GRVCLTSDCWTACSNEGYISLTAHYVNVNWKLESKILAFAHMEPPHSGRDLALKVLEMLD 253
R T D WT+C N+ Y+ +T H+++ +W L+ +I+ F H+E H+G+ L+ ++
Sbjct: 239 SRFSTTMDMWTSCQNKSYMCVTIHWIDDDWCLQKRIVGFFHVEGRHTGQRLSQTFTAIMV 298
Query: 254 DWGIEKKIFSITLDNASANNSMANFLKEHLSLSNS-LLLDGEFFHIRCSAHILNLIVQDG 312
W IEKK+F+++LDNASAN + + E L ++S L+ DG FFH+RC+ HILNL+ +DG
Sbjct: 299 KWNIEKKLFALSLDNASANEVAVHDIIEDLQDTDSNLVCDGAFFHVRCACHILNLVAKDG 358
Query: 313 LKVVSDALHKIRQSVAYVRVTEGRTLLFSECVRIVG--DIDTTIGLRLDCVTRWNSTYIM 370
L V++ + KI+ A V + L + E ++ D+D + G+ D TRWNSTY+M
Sbjct: 359 LAVIAGTIEKIK---AIVLAVKSSPLQWEELMKCASECDLDKSKGISYDVSTRWNSTYLM 415
Query: 371 LQSALVYRRAFYSLSLRDSN----FKCC------------------------PTSEEWRR 402
L+ AL Y+ A L D CC P S W
Sbjct: 416 LRDALYYKPALIRLKTSDPRRYVCLNCCTCHHYKFSINQMSIIVGTMQFVLKPRSGRWHL 475
Query: 403 AEIMCEILKPFFTITNLISGSSYPTSNLYFGEIWKIECLIRSYLTSEDLLIQKMAENMKV 462
C LK FF +T L+SG+ Y T+NL++ +I+ LI + E +I++MA M
Sbjct: 476 TLFKC--LKKFFDLTELLSGTQYSTANLFYKGFCEIKDLIDQWCCHEKFVIRRMAVAMSE 533
Query: 463 KFDKYWSDYNVVLAVGAVLDPTKKFNFLKFAYEKLDPLTSEEKLKKVKMTLGKLFSEYIK 522
KF+KYW N+ LAV LDP K ++F +K + + + + KL+ Y
Sbjct: 534 KFEKYWKVSNIALAVACFLDPRYKKILIEFYMKKFHGDSYKVHVDDFVRVIRKLYQFY-- 591
Query: 523 NGIPSNLSSSQVQPSYGGGTRITSSSYDEFEEY--ESQSSNNTGKSELDTYLDELRMPLS 580
+ + ++ + + + DEF+ Y E + + +ELD Y+ E + S
Sbjct: 592 SSCSPSAPKTKTTTNDSMDDTLMENEDDEFQNYLHELKDYDQVESNELDKYMSEPLLKHS 651
Query: 581 QEFDVLAFWKERSRRSPNLARMACDILSIPITTVASESAFSIGARVVNRYRSSMKDDSVQ 640
+FD+L++W+ R P L ++A D+L+I ++TVASESAFS G RVV+ YR+ + + V+
Sbjct: 652 GQFDILSWWRGRVAEYPILTQIARDVLAIQVSTVASESAFSAGGRVVDPYRNRLGSEIVE 711
Query: 641 ALLCARSWL 649
AL+C + W+
Sbjct: 712 ALICTKDWV 720
>HOBO_DROME (P12258) Transposable element Hobo transposase
Length = 644
Score = 76.6 bits (187), Expect = 2e-13
Identities = 131/631 (20%), Positives = 257/631 (39%), Gaps = 75/631 (11%)
Query: 26 EVSSDPKDKKRKGKAKAKDKSKGKALTSDVWLYLVKVG--IVDGVEKCRCKACHKLLTCE 83
E + K+K G +K KGK++ + ++K ++DG CR C K+L
Sbjct: 52 EAADFVKNKINNGTYSVANKHKGKSVIWSILCDILKEDETVLDGWLFCR--QCQKVLKFL 109
Query: 84 SGSGTSHLKRHVRSCSKTIKNHDVGEMMIDVEGKLRKKKFDPMANREFLARIMITHGAPF 143
TS+L RH C T++ +++ + + K+ +K ++ +T
Sbjct: 110 H-KNTSNLSRH--KCCLTLRRPTELKIVSENDKKVAIEKCTQWVVQDCRPFSAVTGAGFK 166
Query: 144 NMVEW--KVFREYQKFLNDDCVFVSRNTIAKEILNVYRDEKQKLKSQLAQI--RGRVCLT 199
N+V++ ++ Y + ++ D + T++++ + +++ + S++ + GR T
Sbjct: 167 NLVKFFLQIGAIYGEQVDVDDLLPDPTTLSRKAKSDAEEKRSLISSEIKKAVDSGRASAT 226
Query: 200 SDCWT-ACSNEGYISLTAHYVNVNWKLESKILAFAHMEPPHS-GRDLALKVLEMLDDWGI 257
D WT ++ +T HY +KL IL M S ++ +K+ + ++ +
Sbjct: 227 VDMWTDQYVQRNFLGITFHYEK-EFKLCDMILGLKSMNFQKSTAENILMKIKGLFSEFNV 285
Query: 258 EKKIFSITLDNASANNSMANFLKEHLSLSNSLLLDGEFFHIRCSAHILNLIVQDGLKVVS 317
E +DN +K+ L + L CS+H+L+ +++ +
Sbjct: 286 EN------IDNVKFVTDRGANIKKALEGNTRL---------NCSSHLLSNVLEKSFNEAN 330
Query: 318 DALHKIRQSVAYVRVTEGRTLLFSECVRIVGDIDTTIGLRLDCVTRWNSTYIMLQSAL-V 376
+ ++ V+ + L + ++TT L+ C TRWNS Y M++S L
Sbjct: 331 ELKKIVKSCKKIVKYCKKSNLQHT--------LETT--LKSACPTRWNSNYKMMKSILDN 380
Query: 377 YRRAFYSLSLRDSNFKCCPTSEEWRRAEIMCEILKPFFTITNLISGSSYPTSNLYFGEIW 436
+R L D + +S +++ +IL F I + SS P+ I
Sbjct: 381 WRSVDKILGEADIHVDFNKSS-----LKVVVDILGDFERIFKKLQTSSSPSICFVLPSIS 435
Query: 437 KIECLIRSY---LTSEDLLIQKMAENM------------KVKFDKYWSDYNV-------- 473
KI L L++ LL +++ EN+ K F Y ++
Sbjct: 436 KILELCEPNILDLSAAALLKERILENIRKIWMANLSIWHKAAFFLYPPAAHLQEEDILEI 495
Query: 474 -VLAVGAVLDPTKKFNFLKFAYEKLDPLTSEEKLKKVKMTLGKLFSEYIKNGIPSNLSSS 532
V + + P L+ P T E + P +L S
Sbjct: 496 KVFCISQIQVPISYTLSLESTETPRTPETPETPETPETPETPETPETPETPETPESLESP 555
Query: 533 QVQPSYGGGTRITSSSYDEFEEYESQSSNNTGKS---ELDTYLDELRMPLSQEFDVLAFW 589
+ P I+S + F + ++S++N +S E++ Y+ + R+PLSQ F+V+ +W
Sbjct: 556 NLFPK--KNKTISSENEFFFPKPVTESNSNFNESPLDEIERYIRQ-RVPLSQNFEVIEWW 612
Query: 590 KERSRRSPNLARMACDILSIPITTVASESAF 620
K + P L+++A +LSIP ++ + F
Sbjct: 613 KNNANLYPQLSKLALKLLSIPASSAELKECF 643
>ZBE4_HUMAN (O75132) Zinc finger BED domain containing protein 4
Length = 1171
Score = 65.1 bits (157), Expect = 6e-10
Identities = 110/525 (20%), Positives = 210/525 (39%), Gaps = 69/525 (13%)
Query: 125 PMANR--EFLARIMITHGAPFNMVEWKVFREYQKFLNDDCVFVSRNTIAKEIL-NVYRDE 181
P+A + +A ++ P++ V+ F ++L + + ++ + +Y +
Sbjct: 653 PVAKKITSLIAEMIALDLQPYSFVDNVGFNRLLEYLKPQYSLPAPSYFSRTAIPGMYDNV 712
Query: 182 KQKLKSQLAQIR-GRVCLTSDCWTACSNEGYISLTAHYVNVNWKLE--------SKILAF 232
KQ + S L + G + TS W + Y++LTAH+V+ S +L
Sbjct: 713 KQIIMSHLKEAESGVIHFTSGIWMSNQTREYLTLTAHWVSFESPARPRCDDHHCSALLDV 772
Query: 233 AHMEPPHSGRDLALKVLEMLDDWGIEKKI-FSITL-DNASANNSMANFLKEHLSLSNSLL 290
+ ++ +SG + ++ + W + IT+ DNAS + L
Sbjct: 773 SQVDCDYSGNSIQKQLECWWEAWVTSTGLQVGITVTDNASIGKT---------------L 817
Query: 291 LDGEFFHIRCSAHILNLIVQDGLK---VVSDALHKIRQSVAYVRVTEGRTLLFSECVRIV 347
+GE ++C +H +NLIV + +K +V + L R+ V + +E R
Sbjct: 818 NEGEHSSVQCFSHTVNLIVSEAIKSQRMVQNLLSLARKICERVHRSPKAKEKLAELQREY 877
Query: 348 GDIDTTIGLRLDCVTRWNSTYIMLQSALVYRRAFYSLSLRDSNFKCCPTSEEWRRAEIMC 407
L D ++W++++ ML+ + +RA +S+ + NF+ + ++W + +C
Sbjct: 878 A--LPQHHLIQDVPSKWSTSFHMLERLIEQKRAINEMSV-ECNFRELISCDQWEVMQSVC 934
Query: 408 EILKPFFTITNLISGSSYPTSNLY-FGEIW--KIECLIRSYLTSEDLLIQKMAENMKVKF 464
LKPF + +S S + I K+E L + D +++ + E M +
Sbjct: 935 RALKPFEAASREMSTQMSTLSQVIPMVHILNRKVEMLFEETM-GIDTMLRSLKEAMVSRL 993
Query: 465 DKYWSDYNVVLAVGAVLDPTKKFNFLKFAYEKLDPLTSEEKLKKVKMTLGKLFSEYIKNG 524
D V A +LDP K + L +EE+ ++ K L I+
Sbjct: 994 SATLHDPRYVFA--TLLDPRYKAS-----------LFTEEEAEQYKQDL-------IREL 1033
Query: 525 IPSNLSSSQVQPSY---GGGTRITSSSYDEFEEYESQSSNNTGKSELD-----TYLDELR 576
N +S V S+ G S++ + ++ + +L YL+E
Sbjct: 1034 ELMNSTSEDVAASHRCDAGSPSKDSAAEENLWSLVAKVKKKDPREKLPEAMVLAYLEE-- 1091
Query: 577 MPLSQEFDVLAFWKERSRRSPNLARMACDILSIPITTVASESAFS 621
L D L +W + P L+ +A L P + V SE F+
Sbjct: 1092 EVLEHSCDPLTYWNLKKASWPGLSALAVRFLGCPPSIVPSEKLFN 1136
>ZBE1_HUMAN (O96006) Zinc finger BED domain containing protein 1
(dREF homolog) (Putative Ac-like transposable element)
Length = 694
Score = 64.7 bits (156), Expect = 7e-10
Identities = 114/543 (20%), Positives = 220/543 (39%), Gaps = 55/543 (10%)
Query: 123 FDPMANREFLARIM--ITHGA-PFNMVEWKVFREYQKFLNDDCVFVSRNTIA-KEILNVY 178
+D +E A ++ I G P ++V+ F+ K + SR I+ K I Y
Sbjct: 118 YDSKKQQELTAAVLGLICEGLYPASIVDEPTFKVLLKTADPRYELPSRKYISTKAIPEKY 177
Query: 179 RDEKQKLKSQLAQIRGRVC-LTSDCWTACS-NEGYISLTAHYVNVN----WKLESKILAF 232
++ + +LA+ C +++D W + + N Y++L AH++ + + S+ L
Sbjct: 178 GAVREVILKELAEATW--CGISTDMWRSENQNRAYVTLAAHFLGLGAPNCLSMGSRCLKT 235
Query: 233 AHMEPPHSGRDLALKVLEMLDDWGIEKKIFSITLDNASANNSMANFLKEHLSLSNSLLLD 292
+ ++ + + E+ +WGI K+F T N+ K+ + + LLD
Sbjct: 236 FEVPEENTAETITRVLYEVFIEWGISAKVFGAT----------TNYGKDIVKACS--LLD 283
Query: 293 GEFFHIRCSAHILNLIVQDGLKV--VSDALHKIRQSVAYVRVTEGRTLLFSECVRIVGDI 350
H+ C H N +Q ++ + L + R+ V Y + + + E +
Sbjct: 284 VAV-HMPCLGHTFNAGIQQAFQLPKLGALLSRCRKLVEYFQQSAVAMYMLYEKQKQQNVA 342
Query: 351 DTTIGLRLDCVTRWNSTYIMLQSALVYRRAFYSLSLRDSN-FKCCPTSEEWRRAEIMCEI 409
L + V+ W ST MLQ + + + DSN + EW E + E+
Sbjct: 343 HCM--LVSNRVSWWGSTLAMLQRLKEQQFVIAGVLVEDSNNHHLMLEASEWATIEGLVEL 400
Query: 410 LKPFFTITNLISGSSYPTSNLYFGEIWKIECLIRSYLTSEDLLIQKMAENMKVKFDKYWS 469
L+PF + ++S S YPT ++ + + + ++ + E + + K +
Sbjct: 401 LQPFKQVAEMLSASRYPTISMVKPLLHMLLNTTLNIKETDSKELSMAKEVIAKELSKTYQ 460
Query: 470 ---DYNVVLAVGAVLDPT-KKFNFLK-FAYEKLDPLTSEEK---LKKVK-----MTLGKL 516
+ ++ L V LDP K+ FL F ++++ EE L KVK K+
Sbjct: 461 ETPEIDMFLNVATFLDPRYKRLPFLSAFERQQVENRVVEEAKGLLDKVKDGGYRPAEDKI 520
Query: 517 FSEYIKNGIPSNLSSSQVQPSYGGGTRIT-----SSSYDEFEEYESQSSNNTGKSELDTY 571
F + + + +S P+ + + ++ EE+ +Q EL +
Sbjct: 521 FPVPEEPPVKKLMRTSTPPPASVINNMLAEIFCQTGGVEDQEEWHAQV-----VEELSNF 575
Query: 572 LDELRMPLSQEFDVLAFWKERSRRSPNLARMACDILSIPITTVASESAFSIGARVVNRYR 631
+ + L++ D L +W +R P L ++ + T VA E F A VV+ R
Sbjct: 576 KSQKVLGLNE--DPLKWWSDRLALFPLLPKVLQKYWCVTATRVAPERLFGSAANVVSAKR 633
Query: 632 SSM 634
+ +
Sbjct: 634 NRL 636
>YLZ5_CAEEL (P34418) Hypothetical protein F42H10.5 in chromosome III
Length = 810
Score = 40.0 bits (92), Expect = 0.020
Identities = 60/270 (22%), Positives = 109/270 (40%), Gaps = 33/270 (12%)
Query: 375 LVYRRAFYSLSLRDSNFKCCPTSEEWRRA---EIMCEILKPFFTITNLIS--GSSYPT-- 427
LV R Y +++ + EW + + I KPF T + ++ + PT
Sbjct: 510 LVAHRDIYQMNMEGITLI---SEREWNKVTGIHHLMNIFKPFMTYSTDMTTVDTVIPTIV 566
Query: 428 --SNLYFGEIWKIECLIRSYLTSEDLLIQKMAENMKVKFDKYWSDYNVVLAVGAVLDPTK 485
N+ +I+ + + LTS L Q +A M + + + S Y A+ L T
Sbjct: 567 QIQNVLEKDIYHLGDIGSDLLTS---LKQTVAPIMNPEHENFDSTYIQATALNPQLAVTL 623
Query: 486 KFNFLKFAYEKLDPLTSEEKLKKVKMTLGKLFSEYIKNGIPSNLSSSQVQPSYG--GGTR 543
+ + A ++ S K K K + G+ S L++ + G GG
Sbjct: 624 TSDQMTTAKSLIETEISRRTKKMRKAQSDKKLAM----GVDSLLANVMRKNDGGSDGGCE 679
Query: 544 ITSSSYDEFEEYESQSSNNTGKSELDTYLDELRMPLSQEFDV------------LAFWKE 591
+ Y + + + +S+ + ++ ++ Y DE+ S E L++WK
Sbjct: 680 TALAIYGDLFQSITGNSSESKENIVNQYFDEISSTTSVESMFMLRTFGNPMQAPLSYWKS 739
Query: 592 RSRRSPNLARMACDILSIPITTVASESAFS 621
S R L+ +A ++LSIPI T+ +E S
Sbjct: 740 CSSRCSELSDLATELLSIPIFTLTAERVLS 769
>RPSD_BACSU (P10726) RNA polymerase sigma-D factor (Sigma-28)
Length = 254
Score = 36.6 bits (83), Expect = 0.22
Identities = 24/78 (30%), Positives = 40/78 (50%), Gaps = 12/78 (15%)
Query: 453 IQKMAENMKVKFDKYWSDYNVVLAVGAVLDPTKKFNFLKFAYEKLDPLTSEEKLKKVKMT 512
++K + +KFD Y S GA++D +K ++L P TS EK KKV+
Sbjct: 67 LEKFDPSRDLKFDTYAS----FRIRGAIIDGLRKEDWL--------PRTSREKTKKVEAA 114
Query: 513 LGKLFSEYIKNGIPSNLS 530
+ KL Y++N P+ ++
Sbjct: 115 IEKLEQRYLRNVSPAEIA 132
>SYA_WIGBR (Q8D2W8) Alanyl-tRNA synthetase (EC 6.1.1.7)
(Alanine--tRNA ligase) (AlaRS)
Length = 873
Score = 34.7 bits (78), Expect = 0.82
Identities = 42/175 (24%), Positives = 74/175 (42%), Gaps = 22/175 (12%)
Query: 377 YRRAFYSLSLRDSNFKCCPTSEEWRRAEIMCEILKPFFTI---TNLISGSSYPTSNLY-- 431
Y+ L++ D + + C + R +I C ++ F I T I + T+ +
Sbjct: 644 YKDLVRVLNIGDFSIELCGGTHANRTGDIGCFVITNFSKISSDTYRIKAITGETAIAFIQ 703
Query: 432 --FGEIWKIECLIRSYLTSEDLLIQKMAENMKVKFDKYWSDYNVVLA--VGAVLDPTKKF 487
F +I I LI+S I+K+ EN+K+ K + N ++ + + + KK
Sbjct: 704 KKFNDINTISSLIKSNSNEIVNKIEKITENLKILEKKNKNLKNKIIKNYIKLIFNKIKKI 763
Query: 488 N----FLKFAYEKLDPLTSEEKLKKVKMTLG---------KLFSEYIKNGIPSNL 529
N + + E++D + E L KVK L K + YI G+ +NL
Sbjct: 764 NNFEIIMDYFNEEIDKIILREILNKVKTNLKNGIVILASIKKDNLYIVTGVTNNL 818
>YUY1_CAEEL (P54073) Hypothetical protein F44G4.1 in chromosome II
Length = 384
Score = 34.3 bits (77), Expect = 1.1
Identities = 24/99 (24%), Positives = 46/99 (46%), Gaps = 3/99 (3%)
Query: 87 GTSHLKRHVRSCSKTIKNHD---VGEMMIDVEGKLRKKKFDPMANREFLARIMITHGAPF 143
G S ++ V ++++ +D V E +VE +F P NRE ++MIT
Sbjct: 128 GESAPQKEVPKTIESMREYDATMVNEEDDEVEHDEANDEFAPYFNRETSPKVMITMTPKA 187
Query: 144 NMVEWKVFREYQKFLNDDCVFVSRNTIAKEILNVYRDEK 182
+ +K E QK + + +F +N + K I+ ++ +
Sbjct: 188 KITTFKFCFELQKCIPNSEIFTRKNVLLKTIIEQAKERE 226
>HTPG_BUCAI (P57555) Chaperone protein htpG (Heat shock protein
htpG) (High temperature protein G)
Length = 624
Score = 33.9 bits (76), Expect = 1.4
Identities = 39/152 (25%), Positives = 62/152 (40%), Gaps = 27/152 (17%)
Query: 439 ECLIRSYLTSEDLLIQKMAENMKVKFDKYWSDYNVVLAVGAVLDP---TKKFNFLKFAYE 495
E L +S + +++K+A+ K+ +W+ + +VL G D K N L+F
Sbjct: 346 EKLRKSLIKKSLSMLEKLAQKNNEKYQIFWNQFGLVLKEGPAEDHENLNKIANLLRFTSM 405
Query: 496 KLDPLTSEEKLKKVKMTLGKLFSEYIKNGIPSNLSSSQVQPSYGGGTRITSSSY---DEF 552
K + SE+K+ EYI SN++ Q + Y IT+ SY +
Sbjct: 406 KSN--NSEQKMS---------LKEYI-----SNMNEQQEKIYY-----ITADSYSSANNS 444
Query: 553 EEYESQSSNNTGKSELDTYLDELRMPLSQEFD 584
E NN L +DE M EF+
Sbjct: 445 PHLELFKKNNIDVLLLSDRIDEWMMNYLSEFE 476
>GAAB_PICTO (Q6L1Q1) GMP synthase [glutamine-hydrolyzing] subunit B
(EC 6.3.5.2) (GMP synthetase)
Length = 305
Score = 32.7 bits (73), Expect = 3.1
Identities = 41/159 (25%), Positives = 64/159 (39%), Gaps = 22/159 (13%)
Query: 463 KFDKYWSDYNVVLAVGAVLDPTKKFNFLKFAYEKLDPLTSEEKLKKVKMTLGKLFSEYIK 522
+ + +S YNV G +LD + F + DP EEK K + T ++F E K
Sbjct: 63 RIKEIFSSYNVN---GRILDRSSLF--ISRLRNVTDP---EEKRKIIGRTFIEVFEEEAK 114
Query: 523 NGIPSNLSSSQVQPSY---GGGTRITSSSYDEFEEYESQSSNNTGKSELDTYLDELR--- 576
N +L + P + GGG+R T S+ + + + D Y DE+R
Sbjct: 115 NYGAESLLQGTIAPDWIESGGGSRDTIKSHHNVGGLPEKMNLRLVEPLRDLYKDEVREVS 174
Query: 577 ----MPLSQEFD----VLAFWKERSRRSPNLARMACDIL 607
+P Q F + E + N+ R A DI+
Sbjct: 175 RYLGLPEVQPFPGPGLAIRIIGEVTEEKLNILRRATDIV 213
>VGLM_BUNL7 (P09612) M polyprotein precursor [Contains: Glycoprotein
G2; Nonstructural protein NS-M; Glycoprotein G1]
Length = 1441
Score = 32.3 bits (72), Expect = 4.1
Identities = 19/51 (37%), Positives = 28/51 (54%), Gaps = 6/51 (11%)
Query: 158 LNDDCVFVSRNTIAKEILNVYRDEKQKLKSQLAQIRGRVCLTSDCWTACSN 208
++D CVF S I KE L+VYR E +++ + +CLT T C+N
Sbjct: 1076 VSDGCVFGSCQDIIKEELSVYRKETEEVTNV------ELCLTFSDKTYCTN 1120
>UBP4_MOUSE (P35123) Ubiquitin carboxyl-terminal hydrolase 4 (EC
3.1.2.15) (Ubiquitin thiolesterase 4)
(Ubiquitin-specific processing protease 4)
(Deubiquitinating enzyme 4) (Ubiquitous nuclear protein)
Length = 962
Score = 32.3 bits (72), Expect = 4.1
Identities = 23/69 (33%), Positives = 33/69 (47%), Gaps = 8/69 (11%)
Query: 508 KVKMTLGKLF-------SEYIKNGIPSNLSSSQVQPSYGGGTRITSSSYDEFEEYESQSS 560
K K+TL L+ S YIK +P SS ++P G+R +S DE EE + Q
Sbjct: 611 KHKLTLESLYQAVCDRISRYIKQPLPDEFLSSPLEPGACNGSR-SSYEGDEEEEMDHQEE 669
Query: 561 NNTGKSELD 569
SE++
Sbjct: 670 GKEQLSEVE 678
>UBP4_HUMAN (Q13107) Ubiquitin carboxyl-terminal hydrolase 4 (EC
3.1.2.15) (Ubiquitin thiolesterase 4)
(Ubiquitin-specific processing protease 4)
(Deubiquitinating enzyme 4) (Ubiquitous nuclear protein
homolog)
Length = 963
Score = 32.3 bits (72), Expect = 4.1
Identities = 22/69 (31%), Positives = 31/69 (44%), Gaps = 8/69 (11%)
Query: 508 KVKMTLGKLF-------SEYIKNGIPSNLSSSQVQPSYGGGTRITSSSYDEFEEYESQSS 560
K K+TL L+ S Y+K +P SS ++P G+R + DE EE E Q
Sbjct: 611 KHKLTLESLYQAVCDRISRYVKQPLPDEFGSSPLEPGACNGSRNSCEGEDE-EEMEHQEE 669
Query: 561 NNTGKSELD 569
SE +
Sbjct: 670 GKEQLSETE 678
>GBA1_KLULA (Q9Y7B7) Guanine nucleotide-binding protein alpha-1
subunit (GP1-alpha)
Length = 446
Score = 32.3 bits (72), Expect = 4.1
Identities = 16/53 (30%), Positives = 27/53 (50%), Gaps = 6/53 (11%)
Query: 612 TTVASESAFSIGARVVNRYRSSMKDDSVQALLCARSWLHGFEGISSIIFTIVL 664
TT +E++F IG Y + + R W+H FEGI++++F I +
Sbjct: 274 TTGITENSFKIGPSTFKVYDAGGQRSE------RRKWIHCFEGITAVVFVIAI 320
>PBP_ANTPO (P20797) Pheromone-binding protein precursor (PBP)
Length = 163
Score = 31.6 bits (70), Expect = 7.0
Identities = 20/55 (36%), Positives = 29/55 (52%), Gaps = 1/55 (1%)
Query: 560 SNNTGKSELDTYLDELRMPLSQEFDVLAFWKERSRRSPNLARMACDILSIPITTV 614
SNN GK+ +D DEL +P S D+ FWK+ + LA A + L+ + V
Sbjct: 30 SNNFGKA-MDQCKDELSLPDSVVADLYNFWKDDYVMTDRLAGCAINCLATKLDVV 83
>UL49_HHV7J (P52442) Protein U33
Length = 477
Score = 31.2 bits (69), Expect = 9.1
Identities = 26/97 (26%), Positives = 45/97 (45%), Gaps = 14/97 (14%)
Query: 405 IMCEILKPFFTITNLISGSSYPTSNLYF-GEIWKIE-CLIRSYLTSEDLLIQKMAENMKV 462
++C ++ P L SY L+ G++ K E C++ ++T++ L + + +N
Sbjct: 7 LLCRLISP------LCKHGSYSHLQLFLIGDVSKSENCVLSMFVTNKKFLSKDLTDNFYR 60
Query: 463 KFDKYW-----SDYNVVLAVGAVLDPTKKFNFLKFAY 494
KF K W N + A+ + TK F FL AY
Sbjct: 61 KFLKVWLKCDLDSRNQIKAIFNKMMMTKNF-FLLLAY 96
>SYA_MYCMS (P61704) Alanyl-tRNA synthetase (EC 6.1.1.7)
(Alanine--tRNA ligase) (AlaRS)
Length = 896
Score = 31.2 bits (69), Expect = 9.1
Identities = 25/93 (26%), Positives = 46/93 (48%), Gaps = 7/93 (7%)
Query: 423 SSYPTSNLYFGEIWKI-----ECLIRSYLTSEDLLIQKMAENMKVKFDKYWSDYNVVLAV 477
+S+ N Y E +K+ E +I Y ++DLL EN+ ++ + + +L +
Sbjct: 710 TSFRAVNEYLNEQFKVYKDQAESIIDKYNQNKDLLKNDQLENIYLQIKNTTINKDNLLVI 769
Query: 478 GAVLDPTKKFNFLKFAYEKLDPLTSEEKLKKVK 510
+LD +K+ N K +K++ L + KL K K
Sbjct: 770 KNLLDQSKEVN--KDYDKKVNDLITANKLLKYK 800
>POK7_HUMAN (Q9QC07) HERV-K_1q23.3 provirus ancestral Pol protein
(HERV-K18 Pol protein) (HERV-K110 Pol protein)
(HERV-K(C1a) Pol protein) [Includes: Reverse
transcriptase (EC 2.7.7.49); Ribonuclease H (EC
3.1.26.4) (RNase H)]
Length = 812
Score = 31.2 bits (69), Expect = 9.1
Identities = 21/66 (31%), Positives = 35/66 (52%), Gaps = 9/66 (13%)
Query: 81 TCESGSGTSHLKRHVRSCSKTIKNHDVGEMMIDVEGKLRKKKFDPMANREFLARIMITH- 139
TC++G TSH+K+H+ SC + V E + G + A ++FL++ I+H
Sbjct: 680 TCQTGESTSHVKKHLLSCFAVM---GVPEKIKTDNG----PGYCSKAFQKFLSQWKISHT 732
Query: 140 -GAPFN 144
G P+N
Sbjct: 733 TGIPYN 738
>POK6_HUMAN (P63133) HERV-K_8p23.1 provirus ancestral Pol protein
(HERV-K115 Pol protein) [Includes: Reverse transcriptase
(RT) (EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4) (RNase
H); Integrase (IN)]
Length = 956
Score = 31.2 bits (69), Expect = 9.1
Identities = 21/66 (31%), Positives = 35/66 (52%), Gaps = 9/66 (13%)
Query: 81 TCESGSGTSHLKRHVRSCSKTIKNHDVGEMMIDVEGKLRKKKFDPMANREFLARIMITH- 139
TC++G TSH+K+H+ SC + V E + G + A ++FL++ I+H
Sbjct: 680 TCQTGESTSHVKKHLLSCFAVM---GVPEKIKTDNG----PGYCSKAFQKFLSQWKISHT 732
Query: 140 -GAPFN 144
G P+N
Sbjct: 733 TGIPYN 738
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 75,355,196
Number of Sequences: 164201
Number of extensions: 3107695
Number of successful extensions: 9689
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 9662
Number of HSP's gapped (non-prelim): 30
length of query: 664
length of database: 59,974,054
effective HSP length: 117
effective length of query: 547
effective length of database: 40,762,537
effective search space: 22297107739
effective search space used: 22297107739
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 69 (31.2 bits)
Medicago: description of AC144516.3


