

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144513.3 - phase: 0 /pseudo
(1115 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
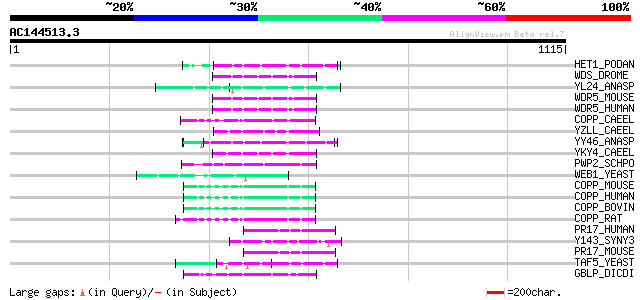
HET1_PODAN (Q00808) Vegetatible incompatibility protein HET-E-1 78 2e-13
WDS_DROME (Q9V3J8) Will die slowly protein 63 4e-09
YL24_ANASP (Q8YV57) Hypothetical WD-repeat protein all2124 62 9e-09
WDR5_MOUSE (P61965) WD-repeat protein 5 (WD-repeat protein BIG-3) 61 1e-08
WDR5_HUMAN (P61964) WD-repeat protein 5 61 1e-08
COPP_CAEEL (Q20168) Probable coatomer beta' subunit (Beta'-coat ... 61 2e-08
YZLL_CAEEL (Q93847) Hypothetical WD-repeat protein K04G11.4 IN c... 60 3e-08
YY46_ANASP (Q8YRI1) Hypothetical WD-repeat protein alr3466 60 3e-08
YKY4_CAEEL (Q17963) Hypothetical WD-repeat protein C14B1.4 in ch... 59 6e-08
PWP2_SCHPO (Q9C1X1) Periodic tryptophan protein 2 homolog 59 7e-08
WEB1_YEAST (P38968) WEB1 protein (Protein transport protein SEC31) 58 1e-07
COPP_MOUSE (O55029) Coatomer beta' subunit (Beta'-coat protein) ... 57 2e-07
COPP_HUMAN (P35606) Coatomer beta' subunit (Beta'-coat protein) ... 57 2e-07
COPP_BOVIN (P35605) Coatomer beta' subunit (Beta'-coat protein) ... 57 2e-07
COPP_RAT (O35142) Coatomer beta' subunit (Beta'-coat protein) (B... 57 4e-07
PR17_HUMAN (O60508) Pre-mRNA splicing factor PRP17 (hPRP17) (Cel... 54 2e-06
Y143_SYNY3 (P74442) Hypothetical WD-repeat protein slr0143 53 4e-06
PR17_MOUSE (Q9DC48) Pre-mRNA splicing factor PRP17 (Cell divisio... 53 4e-06
TAF5_YEAST (P38129) Transcription initiation factor TFIID subuni... 53 5e-06
GBLP_DICDI (P46800) Guanine nucleotide-binding protein beta subu... 53 5e-06
>HET1_PODAN (Q00808) Vegetatible incompatibility protein HET-E-1
Length = 1356
Score = 77.8 bits (190), Expect = 2e-13
Identities = 87/323 (26%), Positives = 131/323 (39%), Gaps = 44/323 (13%)
Query: 347 GSSVTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISA--CSLPFQAAV 404
GSSV S+ F P Q V S +G+ K KIWD ++ C+ +
Sbjct: 967 GSSVLSVAFSPDGQR---VASGSGD--------------KTIKIWDTASGTCTQTLEG-- 1007
Query: 405 VKDTPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHP 464
SV V +S DG V I I+ S Q +E H G V + F+
Sbjct: 1008 --HGGSVWSVAFSPDGQRVASGSDDKTIKIWD-TASGTCTQTLE--GHGGWVQSVVFSPD 1062
Query: 465 NKQLCVVTCGDDKLIKVWD-LTGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIK 523
++ V + DD IK+WD ++G EGH V+S+ + Q + S ++DG IK
Sbjct: 1063 GQR--VASGSDDHTIKIWDAVSGTCTQTLEGHGDSVWSVA--FSPDGQRVASGSIDGTIK 1118
Query: 524 AWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYN 583
W + + G W ++ +S DG R+ S D + W+ + G +T
Sbjct: 1119 IWDAASGTCTQTLEGHGGWVHSVAFSPDGQRVASGSI----DGTIKIWDAASGTCTQTLE 1174
Query: 584 GFRKKSAGVVQ---FDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFN 640
G G VQ F R + D IK WD + + E GG + + F+
Sbjct: 1175 GH----GGWVQSVAFSPDGQRVASGSSDKTIKIWDTASGTCTQTLEGHGG--WVQSVAFS 1228
Query: 641 KEGNLLAVTTADNGFKILANAGG 663
+G +A ++DN KI A G
Sbjct: 1229 PDGQRVASGSSDNTIKIWDTASG 1251
Score = 62.4 bits (150), Expect = 7e-09
Identities = 67/249 (26%), Positives = 102/249 (40%), Gaps = 15/249 (6%)
Query: 410 SVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLC 469
SV V +S DG V I I+ S Q +E H G V +AF+ ++
Sbjct: 843 SVLSVAFSADGQRVASGSDDKTIKIWD-TASGTGTQTLE--GHGGSVWSVAFSPDRER-- 897
Query: 470 VVTCGDDKLIKVWDL-TGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLYD 528
V + DDK IK+WD +G EGH V S+ + Q + S + D IK W
Sbjct: 898 VASGSDDKTIKIWDAASGTCTQTLEGHGGRVQSVA--FSPDGQRVASGSDDHTIKIWDAA 955
Query: 529 NMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKK 588
+ + G ++ +S DG R+ S GD + W+ + G +T G
Sbjct: 956 SGTCTQTLEGHGSSVLSVAFSPDGQRV----ASGSGDKTIKIWDTASGTCTQTLEG-HGG 1010
Query: 589 SAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAV 648
S V F R + +D IK WD + + E GG + + F+ +G +A
Sbjct: 1011 SVWSVAFSPDGQRVASGSDDKTIKIWDTASGTCTQTLEGHGG--WVQSVVFSPDGQRVAS 1068
Query: 649 TTADNGFKI 657
+ D+ KI
Sbjct: 1069 GSDDHTIKI 1077
Score = 39.3 bits (90), Expect = 0.060
Identities = 31/119 (26%), Positives = 51/119 (42%), Gaps = 7/119 (5%)
Query: 545 TMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLA 604
++ +SADG R+ S D + W+ + G +T G S V F + R +
Sbjct: 846 SVAFSADGQRV----ASGSDDKTIKIWDTASGTGTQTLEG-HGGSVWSVAFSPDRERVAS 900
Query: 605 AGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAVTTADNGFKILANAGG 663
+D IK WD + + E GG + + F+ +G +A + D+ KI A G
Sbjct: 901 GSDDKTIKIWDAASGTCTQTLEGHGG--RVQSVAFSPDGQRVASGSDDHTIKIWDAASG 957
>WDS_DROME (Q9V3J8) Will die slowly protein
Length = 361
Score = 63.2 bits (152), Expect = 4e-09
Identities = 59/212 (27%), Positives = 98/212 (45%), Gaps = 18/212 (8%)
Query: 408 TPSVSRVTWSLDGSFVGVAFTKHLIHIY-AYNGSNELAQRVEIDAHIGGVNDLAFAHPNK 466
T +VS V +S +G ++ + LI I+ AY+G E I H G++D+A++ ++
Sbjct: 72 TKAVSAVKFSPNGEWLASSSADKLIKIWGAYDGKFEKT----ISGHKLGISDVAWSSDSR 127
Query: 467 QLCVVTCGDDKLIKVWDL-TGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAW 525
L V+ DDK +KVW+L TG+ L +GH Y C + I S + D ++ W
Sbjct: 128 LL--VSGSDDKTLKVWELSTGKSLKTLKGHSN--YVFCCNFNPQSNLIVSGSFDESVRIW 183
Query: 526 LYDNMGSRVDYDAPGHW--CTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYN 583
D + P H + + ++ DG+ + S +S DG + W+ + G +T
Sbjct: 184 --DVRTGKCLKTLPAHSDPVSAVHFNRDGSLIVS--SSYDGLCRI--WDTASGQCLKTLI 237
Query: 584 GFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWD 615
V+F LAA D+ +K WD
Sbjct: 238 DDDNPPVSFVKFSPNGKYILAATLDNTLKLWD 269
Score = 39.3 bits (90), Expect = 0.060
Identities = 42/177 (23%), Positives = 72/177 (39%), Gaps = 15/177 (8%)
Query: 490 FNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAW-LYDNMGSRVDYDAPGH--WCTTM 546
F GH V ++ N +++ S++ D IK W YD + + GH + +
Sbjct: 66 FTLAGHTKAVSAV--KFSPNGEWLASSSADKLIKIWGAYDG---KFEKTISGHKLGISDV 120
Query: 547 LYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAG 606
+S+D +RL G+ D L W S G +T G F+ N ++
Sbjct: 121 AWSSD-SRLLVSGSD---DKTLKVWELSTGKSLKTLKG-HSNYVFCCNFNPQSNLIVSGS 175
Query: 607 EDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAVTTADNGFKILANAGG 663
D ++ WD+ L + A H FN++G+L+ ++ D +I A G
Sbjct: 176 FDESVRIWDVRTGKCLKTLPAHSDPVSAVH--FNRDGSLIVSSSYDGLCRIWDTASG 230
Score = 37.7 bits (86), Expect = 0.17
Identities = 33/145 (22%), Positives = 60/145 (40%), Gaps = 12/145 (8%)
Query: 389 KIWDISACSLPFQAAVVKDTPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVE 448
+IWD +A + + D P VS V +S +G ++ A + + ++ Y+ L
Sbjct: 223 RIWD-TASGQCLKTLIDDDNPPVSFVKFSPNGKYILAATLDNTLKLWDYSKGKCLK---- 277
Query: 449 IDAHIGGVNDLAFAHPNKQLC----VVTCGDDKLIKVWDLTGRRLFN-FEGHEAPVYSIC 503
+ G N+ N + +V+ +D ++ +W+L + + +GH V
Sbjct: 278 --TYTGHKNEKYCIFANFSVTGGKWIVSGSEDNMVYIWNLQSKEVVQKLQGHTDTVLCTA 335
Query: 504 PHHKENIQFIFSTAVDGKIKAWLYD 528
H ENI + D IK W D
Sbjct: 336 CHPTENIIASAALENDKTIKLWKSD 360
>YL24_ANASP (Q8YV57) Hypothetical WD-repeat protein all2124
Length = 1683
Score = 62.0 bits (149), Expect = 9e-09
Identities = 58/224 (25%), Positives = 87/224 (37%), Gaps = 15/224 (6%)
Query: 442 ELAQRVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLTGRRLFNFEGHEAPVYS 501
E+ +R ++ H GV ++ + + + + DK IK+W GR GHE VYS
Sbjct: 1061 EMQERNRLEGHKDGVISISISRDGQTIASGSL--DKTIKLWSRDGRLFRTLNGHEDAVYS 1118
Query: 502 ICPHHKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTS 561
+ + Q I S D IK W + + +S DG L S
Sbjct: 1119 V--SFSPDGQTIASGGSDKTIKLWQTSDGTLLKTITGHEQTVNNVYFSPDGKNL----AS 1172
Query: 562 KDGDSFLVEWNESEGAIKRTYNGFRKKSAGV--VQFDTTQNRFLAAGEDSQIKFWDMDNV 619
D + W+ + G + T G SAGV V+F A ED +K W +
Sbjct: 1173 ASSDHSIKLWDTTSGQLLMTLTG---HSAGVITVRFSPDGQTIAAGSEDKTVKLWHRQDG 1229
Query: 620 NPLTSTEAEGGLQGLPHLRFNKEGNLLAVTTADNGFKILANAGG 663
L + G + L F+ +G LA +AD K+ A G
Sbjct: 1230 KLLKT--LNGHQDWVNSLSFSPDGKTLASASADKTIKLWRIADG 1271
Score = 60.5 bits (145), Expect = 3e-08
Identities = 92/396 (23%), Positives = 155/396 (38%), Gaps = 42/396 (10%)
Query: 293 TTPGMVEYQSADHEQLMKRLRPAPSVEEVSYPSARQAS--WSLDDLPRTVAMSLHQGSSV 350
TT G + H + +R +P + ++ S + W D ++ HQ V
Sbjct: 1184 TTSGQLLMTLTGHSAGVITVRFSPDGQTIAAGSEDKTVKLWHRQDGKLLKTLNGHQ-DWV 1242
Query: 351 TSMDFHPSHQTLLLVGSNNGEISLWELG---MRERLVSKPFKIWDISACSLPFQAAVVKD 407
S+ F P +TL ++ I LW + + + L +WD++ S A+
Sbjct: 1243 NSLSFSPDGKTLASASADK-TIKLWRIADGKLVKTLKGHNDSVWDVNFSS--DGKAIASA 1299
Query: 408 TPSVSRVTWSLDGSFVGVAFTKHLIHIYAYN---GSNELAQR----------------VE 448
+ + W+ G + FT H +YA N SN +A +E
Sbjct: 1300 SRDNTIKLWNRHGIELET-FTGHSGGVYAVNFLPDSNIIASASLDNTIRLWQRPLISPLE 1358
Query: 449 IDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVW-DLTGRRLFNFEGHEAPVYSICPHHK 507
+ A GV ++F H + T G D I++W G L G++A +Y I +
Sbjct: 1359 VLAGNSGVYAVSFLHDGS--IIATAGADGNIQLWHSQDGSLLKTLPGNKA-IYGISFTPQ 1415
Query: 508 ENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSF 567
++ I S D +K W + + + + +S DG L S D+
Sbjct: 1416 GDL--IASANADKTVKIWRVRDGKALKTLIGHDNEVNKVNFSPDGKTLASASR----DNT 1469
Query: 568 LVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEA 627
+ WN S+G K+T G + V F +A D I+ WD + N + S A
Sbjct: 1470 VKLWNVSDGKFKKTLKGHTDEVFWV-SFSPDGKIIASASADKTIRLWDSFSGNLIKSLPA 1528
Query: 628 EGGLQGLPHLRFNKEGNLLAVTTADNGFKILANAGG 663
L + + FN +G++LA T+AD K+ + G
Sbjct: 1529 HNDL--VYSVNFNPDGSMLASTSADKTVKLWRSHDG 1562
Score = 45.1 bits (105), Expect = 0.001
Identities = 55/250 (22%), Positives = 98/250 (39%), Gaps = 26/250 (10%)
Query: 414 VTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLCVVTC 473
++++ G + A + I+ L + H VN + F+ K L +
Sbjct: 1410 ISFTPQGDLIASANADKTVKIWRVRDGKALKTLI---GHDNEVNKVNFSPDGKTLA--SA 1464
Query: 474 GDDKLIKVWDLT-GRRLFNFEGHEAPVY--SICPHHKENIQFIFSTAVDGKIKAWLYDNM 530
D +K+W+++ G+ +GH V+ S P K I S + D I+ W D+
Sbjct: 1465 SRDNTVKLWNVSDGKFKKTLKGHTDEVFWVSFSPDGK----IIASASADKTIRLW--DSF 1518
Query: 531 GSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSA 590
+ P H ++YS + S S D + W +G + T++G +
Sbjct: 1519 SGNLIKSLPAH--NDLVYSVNFNPDGSMLASTSADKTVKLWRSHDGHLLHTFSGH----S 1572
Query: 591 GVVQFDT--TQNRFLA-AGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLA 647
VV + R++A A ED +K W +D T + + G+ F+ +G L
Sbjct: 1573 NVVYSSSFSPDGRYIASASEDKTVKIWQIDGHLLTTLPQHQAGVMS---AIFSPDGKTLI 1629
Query: 648 VTTADNGFKI 657
+ D KI
Sbjct: 1630 SGSLDTTTKI 1639
Score = 36.2 bits (82), Expect = 0.51
Identities = 29/152 (19%), Positives = 63/152 (41%), Gaps = 17/152 (11%)
Query: 386 KPFKIWDISACSLPFQAAVVKDTPSVSRVTWSL----DGSFVGVAFTKHLIHIYAYNGSN 441
K ++WD F ++K P+ + + +S+ DGS + + ++ + +
Sbjct: 1510 KTIRLWD------SFSGNLIKSLPAHNDLVYSVNFNPDGSMLASTSADKTVKLWRSHDGH 1563
Query: 442 ELAQRVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLTGRRLFNFEGHEAPVYS 501
L H V +F+ + + + +DK +K+W + G L H+A V S
Sbjct: 1564 LLHT---FSGHSNVVYSSSFSPDGRY--IASASEDKTVKIWQIDGHLLTTLPQHQAGVMS 1618
Query: 502 ICPHHKENIQFIFSTAVDGKIKAWLYDNMGSR 533
+ + + S ++D K W +D+ ++
Sbjct: 1619 AI--FSPDGKTLISGSLDTTTKIWRFDSQQAK 1648
>WDR5_MOUSE (P61965) WD-repeat protein 5 (WD-repeat protein BIG-3)
Length = 334
Score = 61.2 bits (147), Expect = 1e-08
Identities = 57/212 (26%), Positives = 97/212 (44%), Gaps = 18/212 (8%)
Query: 408 TPSVSRVTWSLDGSFVGVAFTKHLIHIY-AYNGSNELAQRVEIDAHIGGVNDLAFAHPNK 466
T +VS V +S +G ++ + LI I+ AY+G E I H G++D+A++ +
Sbjct: 45 TKAVSSVKFSPNGEWLASSSADKLIKIWGAYDGKFEKT----ISGHKLGISDVAWSSDSN 100
Query: 467 QLCVVTCGDDKLIKVWDLT-GRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAW 525
L V+ DDK +K+WD++ G+ L +GH Y C + I S + D ++ W
Sbjct: 101 LL--VSASDDKTLKIWDVSSGKCLKTLKGHSN--YVFCCNFNPQSNLIVSGSFDESVRIW 156
Query: 526 LYDNMGSRVDYDAPGHW--CTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYN 583
D + P H + + ++ DG+ + S +S DG + W+ + G +T
Sbjct: 157 --DVKTGKCLKTLPAHSDPVSAVHFNRDGSLIVS--SSYDGLCRI--WDTASGQCLKTLI 210
Query: 584 GFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWD 615
V+F LAA D+ +K WD
Sbjct: 211 DDDNPPVSFVKFSPNGKYILAATLDNTLKLWD 242
Score = 42.4 bits (98), Expect = 0.007
Identities = 42/177 (23%), Positives = 70/177 (38%), Gaps = 15/177 (8%)
Query: 490 FNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAW-LYDNMGSRVDYDAPGH--WCTTM 546
F GH V S+ N +++ S++ D IK W YD + + GH + +
Sbjct: 39 FTLAGHTKAVSSV--KFSPNGEWLASSSADKLIKIWGAYDG---KFEKTISGHKLGISDV 93
Query: 547 LYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAG 606
+S+D L S K L W+ S G +T G F+ N ++
Sbjct: 94 AWSSDSNLLVSASDDKT----LKIWDVSSGKCLKTLKG-HSNYVFCCNFNPQSNLIVSGS 148
Query: 607 EDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAVTTADNGFKILANAGG 663
D ++ WD+ L + A H FN++G+L+ ++ D +I A G
Sbjct: 149 FDESVRIWDVKTGKCLKTLPAHSDPVSAVH--FNRDGSLIVSSSYDGLCRIWDTASG 203
Score = 39.7 bits (91), Expect = 0.046
Identities = 35/145 (24%), Positives = 61/145 (41%), Gaps = 12/145 (8%)
Query: 389 KIWDISACSLPFQAAVVKDTPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVE 448
+IWD +A + + D P VS V +S +G ++ A + + ++ Y+ L
Sbjct: 196 RIWD-TASGQCLKTLIDDDNPPVSFVKFSPNGKYILAATLDNTLKLWDYSKGKCLK---- 250
Query: 449 IDAHIGGVNDLAFAHPNKQLC----VVTCGDDKLIKVWDLTGRRLFN-FEGHEAPVYSIC 503
+ G N+ N + +V+ +D L+ +W+L + + +GH V S
Sbjct: 251 --TYTGHKNEKYCIFANFSVTGGKWIVSGSEDNLVYIWNLQTKEIVQKLQGHTDVVISTA 308
Query: 504 PHHKENIQFIFSTAVDGKIKAWLYD 528
H ENI + D IK W D
Sbjct: 309 CHPTENIIASAALENDKTIKLWKSD 333
>WDR5_HUMAN (P61964) WD-repeat protein 5
Length = 334
Score = 61.2 bits (147), Expect = 1e-08
Identities = 57/212 (26%), Positives = 97/212 (44%), Gaps = 18/212 (8%)
Query: 408 TPSVSRVTWSLDGSFVGVAFTKHLIHIY-AYNGSNELAQRVEIDAHIGGVNDLAFAHPNK 466
T +VS V +S +G ++ + LI I+ AY+G E I H G++D+A++ +
Sbjct: 45 TKAVSSVKFSPNGEWLASSSADKLIKIWGAYDGKFEKT----ISGHKLGISDVAWSSDSN 100
Query: 467 QLCVVTCGDDKLIKVWDLT-GRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAW 525
L V+ DDK +K+WD++ G+ L +GH Y C + I S + D ++ W
Sbjct: 101 LL--VSASDDKTLKIWDVSSGKCLKTLKGHSN--YVFCCNFNPQSNLIVSGSFDESVRIW 156
Query: 526 LYDNMGSRVDYDAPGHW--CTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYN 583
D + P H + + ++ DG+ + S +S DG + W+ + G +T
Sbjct: 157 --DVKTGKCLKTLPAHSDPVSAVHFNRDGSLIVS--SSYDGLCRI--WDTASGQCLKTLI 210
Query: 584 GFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWD 615
V+F LAA D+ +K WD
Sbjct: 211 DDDNPPVSFVKFSPNGKYILAATLDNTLKLWD 242
Score = 42.4 bits (98), Expect = 0.007
Identities = 42/177 (23%), Positives = 70/177 (38%), Gaps = 15/177 (8%)
Query: 490 FNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAW-LYDNMGSRVDYDAPGH--WCTTM 546
F GH V S+ N +++ S++ D IK W YD + + GH + +
Sbjct: 39 FTLAGHTKAVSSV--KFSPNGEWLASSSADKLIKIWGAYDG---KFEKTISGHKLGISDV 93
Query: 547 LYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAG 606
+S+D L S K L W+ S G +T G F+ N ++
Sbjct: 94 AWSSDSNLLVSASDDKT----LKIWDVSSGKCLKTLKG-HSNYVFCCNFNPQSNLIVSGS 148
Query: 607 EDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAVTTADNGFKILANAGG 663
D ++ WD+ L + A H FN++G+L+ ++ D +I A G
Sbjct: 149 FDESVRIWDVKTGKCLKTLPAHSDPVSAVH--FNRDGSLIVSSSYDGLCRIWDTASG 203
Score = 39.7 bits (91), Expect = 0.046
Identities = 35/145 (24%), Positives = 61/145 (41%), Gaps = 12/145 (8%)
Query: 389 KIWDISACSLPFQAAVVKDTPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVE 448
+IWD +A + + D P VS V +S +G ++ A + + ++ Y+ L
Sbjct: 196 RIWD-TASGQCLKTLIDDDNPPVSFVKFSPNGKYILAATLDNTLKLWDYSKGKCLK---- 250
Query: 449 IDAHIGGVNDLAFAHPNKQLC----VVTCGDDKLIKVWDLTGRRLFN-FEGHEAPVYSIC 503
+ G N+ N + +V+ +D L+ +W+L + + +GH V S
Sbjct: 251 --TYTGHKNEKYCIFANFSVTGGKWIVSGSEDNLVYIWNLQTKEIVQKLQGHTDVVISTA 308
Query: 504 PHHKENIQFIFSTAVDGKIKAWLYD 528
H ENI + D IK W D
Sbjct: 309 CHPTENIIASAALENDKTIKLWKSD 333
>COPP_CAEEL (Q20168) Probable coatomer beta' subunit (Beta'-coat
protein) (Beta'-COP)
Length = 1000
Score = 60.8 bits (146), Expect = 2e-08
Identities = 70/279 (25%), Positives = 116/279 (41%), Gaps = 42/279 (15%)
Query: 344 LHQGSSVTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAA 403
L + V +D HP +T LL NG + +W + + K F++ C +P +AA
Sbjct: 12 LARSDRVKCVDLHPV-ETWLLAALYNGNVHIWNY--ETQTLVKSFEV-----CDVPVRAA 63
Query: 404 VVKDTPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAH 463
V R +W + GS +HI +N N L + + +AH + L H
Sbjct: 64 KF-----VPRKSWVVTGS--------DDMHIRVFN-YNTLERVHQFEAHSDYLRSLV-VH 108
Query: 464 PNKQLCVVTCGDDKLIKVWDLTGRRLF--NFEGHEAPVYSICPHHKENIQFIFSTAVDGK 521
P V++ DD L+K+WD + +FEGH V I + K+N F + ++D
Sbjct: 109 PTLPY-VISSSDDMLVKMWDWDNKWAMKQSFEGHTHYVMQIAINPKDNNTFA-TASLDKT 166
Query: 522 IKAWLYDNMGSRV-DYDAPGHW----CTTMLYSADGTRLFSCGTSKDGDSFLVE-WNESE 575
+K W + GS V ++ GH C + + + S D LV+ W+
Sbjct: 167 VKVWQF---GSNVPNFTLEGHEKGVNCVDYYHGGEKPYIIS-----GADDHLVKIWDYQN 218
Query: 576 GAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFW 614
+T +G + + V F + EDS ++ W
Sbjct: 219 KTCVQTLDGHAQNVSSVC-FHPELPLIITGSEDSTVRLW 256
Score = 42.7 bits (99), Expect = 0.005
Identities = 23/98 (23%), Positives = 46/98 (46%), Gaps = 5/98 (5%)
Query: 449 IDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLTGRR-LFNFEGHEAPVYSICPHHK 507
++ H GVN + + H ++ +++ DD L+K+WD + + +GH V S+C H
Sbjct: 181 LEGHEKGVNCVDYYHGGEKPYIISGADDHLVKIWDYQNKTCVQTLDGHAQNVSSVCFH-- 238
Query: 508 ENIQFIFSTAVDGKIKAWLYDNMGSR--VDYDAPGHWC 543
+ I + + D ++ W + ++Y WC
Sbjct: 239 PELPLIITGSEDSTVRLWHANTYRQEHTLNYGLERVWC 276
>YZLL_CAEEL (Q93847) Hypothetical WD-repeat protein K04G11.4 IN
chromosome X
Length = 395
Score = 60.5 bits (145), Expect = 3e-08
Identities = 53/216 (24%), Positives = 98/216 (44%), Gaps = 17/216 (7%)
Query: 410 SVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLC 469
S+S + +S DG ++G I I+ + + H G+N+ +++ +K
Sbjct: 109 SISGIKFSPDGRYMGSGSADCSIKIWRM----DFVYEKTLMGHRLGINEFSWSSDSK--L 162
Query: 470 VVTCGDDKLIKVWDL-TGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLYD 528
+V+C DDKL+KV+D+ +GR + +GH V+ C + + I S + D I+ W
Sbjct: 163 IVSCSDDKLVKVFDVSSGRCVKTLKGHTNYVFCCCFNPSGTL--IASGSFDETIRIWCAR 220
Query: 529 NMGSRVDYDAPGHW--CTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFR 586
N G+ + + PGH +++ ++ DG L S D + W+ + G +T
Sbjct: 221 N-GNTI-FSIPGHEDPVSSVCFNRDGAYL----ASGSYDGIVRIWDSTTGTCVKTLIDEE 274
Query: 587 KKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPL 622
V+F LA+ ++ +K WD + L
Sbjct: 275 HPPITHVKFSPNGKYILASNLNNTLKLWDYQKLRVL 310
Score = 33.1 bits (74), Expect = 4.3
Identities = 30/150 (20%), Positives = 60/150 (40%), Gaps = 28/150 (18%)
Query: 389 KIWDISACSLPFQAAVVKDTPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVE 448
+IWD S + + ++ P ++ V +S +G ++ + + + ++ Y Q++
Sbjct: 257 RIWD-STTGTCVKTLIDEEHPPITHVKFSPNGKYILASNLNNTLKLWDY-------QKLR 308
Query: 449 IDAHIGGVNDLAFAHPNKQLCV------------VTCGDDKLIKVWDLTGRRLFN-FEGH 495
+ G H N + CV V+ +D + +W+L R + +GH
Sbjct: 309 VLKEYTG-------HENSKYCVAANFSVTGGKWIVSGSEDHKVYIWNLQTREILQTLDGH 361
Query: 496 EAPVYSICPHHKENIQFIFSTAVDGKIKAW 525
V H +NI + D +IK W
Sbjct: 362 NTAVMCTDCHPGQNIIASAALEPDMRIKIW 391
>YY46_ANASP (Q8YRI1) Hypothetical WD-repeat protein alr3466
Length = 1526
Score = 60.5 bits (145), Expect = 3e-08
Identities = 71/341 (20%), Positives = 136/341 (39%), Gaps = 52/341 (15%)
Query: 348 SSVTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLV----------------------- 384
S V S+ F P+ +L GS++ + LW++ E L
Sbjct: 949 SRVRSVVFSPN-SLMLASGSSDQTVRLWDISSGECLYIFQGHTGWVYSVAFNLDGSMLAT 1007
Query: 385 ---SKPFKIWDISA--CSLPFQAAVVKDTPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNG 439
+ ++WDIS+ C FQ T V V +S DG+ + + ++ +
Sbjct: 1008 GSGDQTVRLWDISSSQCFYIFQG----HTSCVRSVVFSSDGAMLASGSDDQTVRLWDISS 1063
Query: 440 SNELAQRVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDL-TGRRLFNFEGHEAP 498
N L + H V + F+ L + GDD+++++WD+ +G L+ +G+ +
Sbjct: 1064 GNCL---YTLQGHTSCVRSVVFSPDGAMLA--SGGDDQIVRLWDISSGNCLYTLQGYTSW 1118
Query: 499 VYSICPHHKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGH--WCTTMLYSADGTRLF 556
V + N + + + D ++ W D + Y GH W + +S DG L
Sbjct: 1119 VRFLV--FSPNGVTLANGSSDQIVRLW--DISSKKCLYTLQGHTNWVNAVAFSPDGATL- 1173
Query: 557 SCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDM 616
S GD + W+ S G VV F+ + + D ++ W++
Sbjct: 1174 ---ASGSGDQTVRLWDISSSKCLYILQGHTSWVNSVV-FNPDGSTLASGSSDQTVRLWEI 1229
Query: 617 DNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAVTTADNGFKI 657
++ L + +G + + FN +G++LA ++D ++
Sbjct: 1230 NSSKCLCT--FQGHTSWVNSVVFNPDGSMLASGSSDKTVRL 1268
Score = 58.5 bits (140), Expect = 1e-07
Identities = 58/271 (21%), Positives = 110/271 (40%), Gaps = 19/271 (7%)
Query: 389 KIWDISACSLPFQAAVVKDTPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVE 448
++WDIS+ + + T V+ V ++ DGS + + + ++ N S L
Sbjct: 1183 RLWDISSSKCLY--ILQGHTSWVNSVVFNPDGSTLASGSSDQTVRLWEINSSKCLCT--- 1237
Query: 449 IDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLTGRR-LFNFEGHEAPVYSICPHHK 507
H VN + F L + DK +++WD++ + L F+GH V S+
Sbjct: 1238 FQGHTSWVNSVVFNPDGSMLA--SGSSDKTVRLWDISSSKCLHTFQGHTNWVNSVA--FN 1293
Query: 508 ENIQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSF 567
+ + S + D ++ W + + W +++ +S DGT L S D
Sbjct: 1294 PDGSMLASGSGDQTVRLWEISSSKCLHTFQGHTSWVSSVTFSPDGTML----ASGSDDQT 1349
Query: 568 LVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGE-DSQIKFWDMDNVNPLTSTE 626
+ W+ S G T+ G V+ + LA+G D ++ W + + L + +
Sbjct: 1350 VRLWSISSGECLYTFLGHTNWVGSVI--FSPDGAILASGSGDQTVRLWSISSGKCLYTLQ 1407
Query: 627 AEGGLQGLPHLRFNKEGNLLAVTTADNGFKI 657
G + F+ +G LLA + D ++
Sbjct: 1408 GHNNWVG--SIVFSPDGTLLASGSDDQTVRL 1436
Score = 56.2 bits (134), Expect = 5e-07
Identities = 71/308 (23%), Positives = 119/308 (38%), Gaps = 40/308 (12%)
Query: 349 SVTSMDFHPSHQTLLLVGSNNGEISLWELGM-RERLVSKPFKIWDISACSLPFQAAVVKD 407
SV ++ F P + L G + G + WE +E L K W
Sbjct: 866 SVLTVAFSPDGK-LFATGDSGGIVRFWEAATGKELLTCKGHNSW---------------- 908
Query: 408 TPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQ 467
V+ V +S DG + + ++ + L H V + F+ PN
Sbjct: 909 ---VNSVGFSQDGKMLASGSDDQTVRLWDISSGQCLKT---FKGHTSRVRSVVFS-PNS- 960
Query: 468 LCVVTCGDDKLIKVWDLT-GRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWL 526
L + + D+ +++WD++ G L+ F+GH VYS+ N+ G L
Sbjct: 961 LMLASGSSDQTVRLWDISSGECLYIFQGHTGWVYSVA----FNLDGSMLATGSGDQTVRL 1016
Query: 527 YDNMGSRVDYDAPGH--WCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNG 584
+D S+ Y GH ++++S+DG L S D + W+ S G T G
Sbjct: 1017 WDISSSQCFYIFQGHTSCVRSVVFSSDGAML----ASGSDDQTVRLWDISSGNCLYTLQG 1072
Query: 585 FRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGN 644
VV F + G+D ++ WD+ + N L + +G + L F+ G
Sbjct: 1073 HTSCVRSVV-FSPDGAMLASGGDDQIVRLWDISSGNCLYT--LQGYTSWVRFLVFSPNGV 1129
Query: 645 LLAVTTAD 652
LA ++D
Sbjct: 1130 TLANGSSD 1137
>YKY4_CAEEL (Q17963) Hypothetical WD-repeat protein C14B1.4 in
chromosome III
Length = 376
Score = 59.3 bits (142), Expect = 6e-08
Identities = 50/211 (23%), Positives = 91/211 (42%), Gaps = 16/211 (7%)
Query: 408 TPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQ 467
T S+S +S G ++G + + I+ N + + +R + H GVND+A++ ++
Sbjct: 87 TKSISSAKFSPCGKYLGTSSADKTVKIW--NMDHMICERT-LTGHKLGVNDIAWSSDSR- 142
Query: 468 LCVVTCGDDKLIKVWDL-TGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWL 526
CVV+ DDK +K++++ T R +GH Y C + + S + D ++ W
Sbjct: 143 -CVVSASDDKTLKIFEIVTSRMTKTLKGHNN--YVFCCNFNPQSSLVVSGSFDESVRIW- 198
Query: 527 YDNMGSRVDYDAPGHW--CTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNG 584
D P H + + ++ DG+ + S D + W+ + G +T
Sbjct: 199 -DVKTGMCIKTLPAHSDPVSAVSFNRDGSLI----ASGSYDGLVRIWDTANGQCIKTLVD 253
Query: 585 FRKKSAGVVQFDTTQNRFLAAGEDSQIKFWD 615
V+F LA+ DS +K WD
Sbjct: 254 DENPPVAFVKFSPNGKYILASNLDSTLKLWD 284
Score = 38.1 bits (87), Expect = 0.13
Identities = 42/185 (22%), Positives = 72/185 (38%), Gaps = 32/185 (17%)
Query: 357 PSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVVKDTPSVSRVTW 416
P+H + S N + SL G + LV +IWD +A + V + P V+ V +
Sbjct: 210 PAHSDPVSAVSFNRDGSLIASGSYDGLV----RIWD-TANGQCIKTLVDDENPPVAFVKF 264
Query: 417 SLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLCV------ 470
S +G ++ + + ++ ++ L Q H N + C+
Sbjct: 265 SPNGKYILASNLDSTLKLWDFSKGKTLKQYT--------------GHENSKYCIFANFSV 310
Query: 471 ------VTCGDDKLIKVWDLTGRRLFN-FEGHEAPVYSICPHHKENIQFIFSTAVDGKIK 523
++ +D I +W+L R + EGH PV + H +NI + D KI
Sbjct: 311 TGGKWIISGSEDCKIYIWNLQTREIVQCLEGHTQPVLASDCHPVQNIIASGALEPDNKIH 370
Query: 524 AWLYD 528
W D
Sbjct: 371 IWRSD 375
>PWP2_SCHPO (Q9C1X1) Periodic tryptophan protein 2 homolog
Length = 854
Score = 58.9 bits (141), Expect = 7e-08
Identities = 54/273 (19%), Positives = 126/273 (45%), Gaps = 29/273 (10%)
Query: 345 HQGSSVTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAV 404
+Q S + FHP+ LL+VG ++G F I+++ + ++ +Q ++
Sbjct: 252 NQNSKLRCAAFHPT-SNLLVVGFSSGL----------------FGIYELPSFTMLYQLSI 294
Query: 405 VKDTPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHP 464
+ ++ +T + G ++ + +K L + + +E + ++ +H ++ L ++
Sbjct: 295 TQS--NIDTLTVNSTGDWIAIGSSK-LGQLLVWEWQSE-SYVLKQQSHYDALSTLQYSSD 350
Query: 465 NKQLCVVTCGDDKLIKVWDL-TGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIK 523
++ ++T DD IKVWD+ +G + F H + V +C + N+ +FS+++DG ++
Sbjct: 351 GQR--IITGADDGKIKVWDMNSGFCIVTFTQHTSAVSGLCFSKRGNV--LFSSSLDGSVR 406
Query: 524 AWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYN 583
AW + + AP + + + D + C S+D + W+ G + T
Sbjct: 407 AWDLIRYRNFRTFTAPSRVQFSCI-AVDPSGEIVCAGSQDSFEIFM-WSVQTGQLLETLA 464
Query: 584 GFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDM 616
G + + F+++ + + D ++ WD+
Sbjct: 465 G-HEGPVSSLSFNSSGSLLASGSWDKTVRIWDI 496
Score = 38.1 bits (87), Expect = 0.13
Identities = 25/99 (25%), Positives = 47/99 (47%), Gaps = 7/99 (7%)
Query: 559 GTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDN 618
G+SK G + EW +K+ + + +Q+ + R + +D +IK WDM++
Sbjct: 314 GSSKLGQLLVWEWQSESYVLKQQSH---YDALSTLQYSSDGQRIITGADDGKIKVWDMNS 370
Query: 619 -VNPLTSTEAEGGLQGLPHLRFNKEGNLLAVTTADNGFK 656
+T T+ + G L F+K GN+L ++ D +
Sbjct: 371 GFCIVTFTQHTSAVSG---LCFSKRGNVLFSSSLDGSVR 406
>WEB1_YEAST (P38968) WEB1 protein (Protein transport protein SEC31)
Length = 1273
Score = 58.2 bits (139), Expect = 1e-07
Identities = 71/327 (21%), Positives = 133/327 (39%), Gaps = 51/327 (15%)
Query: 256 MANASVSSSVQAAVVTASTIPVPHNQVSILKRPITPSTTPGMVEYQSADHEQLMKRLRPA 315
+ + +VS +V A T S++ + + ++PI ++ D K + A
Sbjct: 23 LVSGTVSGTVDANFSTDSSLELWSLLAADSEKPIASLQVDS--KFNDLDWSHNNKIIAGA 80
Query: 316 PSVEEVSYPSARQASWSLDDLPRTVAMSLHQGSSVTSMDFHPSHQTLLLVGSNNGEISLW 375
+ S +A+ +++ + R S H SSV ++ F+ +L G NNGEI
Sbjct: 81 LDNGSLELYSTNEANNAINSMAR---FSNHS-SSVKTVKFNAKQDNVLASGGNNGEIF-- 134
Query: 376 ELGMRERLVSKPFKIWDISACSLPFQAAVVKDTPSVSRVTWSLDGSFVGVAFTKHLIHIY 435
IWD++ C+ + TP + S + +A+ + L H++
Sbjct: 135 --------------IWDMNKCT----ESPSNYTPLTPGQSMSSVDEVISLAWNQSLAHVF 176
Query: 436 AYNGSNELAQRVEIDAHIGGVNDLAFAHPN----KQLCVVT--------------CGDDK 477
A GS+ A ++ A V L++ PN +QL VV +D
Sbjct: 177 ASAGSSNFASIWDLKAK-KEVIHLSYTSPNSGIKQQLSVVEWHPKNSTRVATATGSDNDP 235
Query: 478 LIKVWDL----TGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLYDNMGSR 533
I +WDL T + N +GH+ + S+ H++ + S+ D + W ++
Sbjct: 236 SILIWDLRNANTPLQTLN-QGHQKGILSLDWCHQDE-HLLLSSGRDNTVLLWNPESAEQL 293
Query: 534 VDYDAPGHWCTTMLYSADGTRLFSCGT 560
+ A G+WC ++ + LF+C +
Sbjct: 294 SQFPARGNWCFKTKFAPEAPDLFACAS 320
>COPP_MOUSE (O55029) Coatomer beta' subunit (Beta'-coat protein)
(Beta'-COP) (p102)
Length = 905
Score = 57.4 bits (137), Expect = 2e-07
Identities = 64/268 (23%), Positives = 109/268 (39%), Gaps = 32/268 (11%)
Query: 350 VTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVVKDTP 409
V S+D HP+ + +L NG + +W + + K F++ C LP +AA
Sbjct: 18 VKSVDLHPT-EPWMLASLYNGSVCVWN--HETQTLVKTFEV-----CDLPVRAAKF---- 65
Query: 410 SVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEI-DAHIGGVNDLAFAHPNKQL 468
V+R W + G+ I ++ YN +RV + +AH + +A HP +
Sbjct: 66 -VARKNWVVTGA------DDMQIRVFNYN----TLERVHMFEAHSDYIRCIA-VHPTQPF 113
Query: 469 CVVTCGDDKLIKVWDLTGRRLFN--FEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWL 526
++T DD LIK+WD + + FEGH V I + K+N QF S ++D IK W
Sbjct: 114 -ILTSSDDMLIKLWDWDKKWSCSQVFEGHTHYVMQIVINPKDNNQFA-SASLDRTIKVWQ 171
Query: 527 YDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFR 586
+ + + Y + G + + S D + W+ +T G
Sbjct: 172 LGSSSPNFTLEGHEKGVNCIDYYSGGDKPYL--ISGADDRLVKIWDYQNKTCVQTLEG-H 228
Query: 587 KKSAGVVQFDTTQNRFLAAGEDSQIKFW 614
++ F + ED ++ W
Sbjct: 229 AQNVSCASFHPELPIIITGSEDGTVRIW 256
Score = 45.1 bits (105), Expect = 0.001
Identities = 32/139 (23%), Positives = 58/139 (41%), Gaps = 10/139 (7%)
Query: 449 IDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLTGRR-LFNFEGHEAPVYSICPHHK 507
++ H GVN + + + +++ DD+L+K+WD + + EGH V C
Sbjct: 181 LEGHEKGVNCIDYYSGGDKPYLISGADDRLVKIWDYQNKTCVQTLEGHAQNV--SCASFH 238
Query: 508 ENIQFIFSTAVDGKIKAWLYD--NMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGD 565
+ I + + DG ++ W + S ++Y WC L ++ L D
Sbjct: 239 PELPIIITGSEDGTVRIWHSSTYRLESTLNYGMERVWCVASLRGSNNVAL-----GYDEG 293
Query: 566 SFLVEWNESEGAIKRTYNG 584
S +V+ E A+ NG
Sbjct: 294 SIIVKLGREEPAMSMDANG 312
>COPP_HUMAN (P35606) Coatomer beta' subunit (Beta'-coat protein)
(Beta'-COP) (p102)
Length = 905
Score = 57.4 bits (137), Expect = 2e-07
Identities = 64/268 (23%), Positives = 109/268 (39%), Gaps = 32/268 (11%)
Query: 350 VTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVVKDTP 409
V S+D HP+ + +L NG + +W + + K F++ C LP +AA
Sbjct: 17 VKSVDLHPT-EPWMLASLYNGSVCVWN--HETQTLVKTFEV-----CDLPVRAAKF---- 64
Query: 410 SVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEI-DAHIGGVNDLAFAHPNKQL 468
V+R W + G+ I ++ YN +RV + +AH + +A HP +
Sbjct: 65 -VARKNWVVTGA------DDMQIRVFNYN----TLERVHMFEAHSDYIRCIA-VHPTQPF 112
Query: 469 CVVTCGDDKLIKVWDLTGRRLFN--FEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWL 526
++T DD LIK+WD + + FEGH V I + K+N QF S ++D IK W
Sbjct: 113 -ILTSSDDMLIKLWDWDKKWSCSQVFEGHTHYVMQIVINPKDNNQFA-SASLDRTIKVWQ 170
Query: 527 YDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFR 586
+ + + Y + G + + S D + W+ +T G
Sbjct: 171 LGSSSPNFTLEGHEKGVNCIDYYSGGDKPYL--ISGADDRLVKIWDYQNKTCVQTLEG-H 227
Query: 587 KKSAGVVQFDTTQNRFLAAGEDSQIKFW 614
++ F + ED ++ W
Sbjct: 228 AQNVSCASFHPELPIIITGSEDGTVRIW 255
Score = 45.1 bits (105), Expect = 0.001
Identities = 32/139 (23%), Positives = 58/139 (41%), Gaps = 10/139 (7%)
Query: 449 IDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLTGRR-LFNFEGHEAPVYSICPHHK 507
++ H GVN + + + +++ DD+L+K+WD + + EGH V C
Sbjct: 180 LEGHEKGVNCIDYYSGGDKPYLISGADDRLVKIWDYQNKTCVQTLEGHAQNV--SCASFH 237
Query: 508 ENIQFIFSTAVDGKIKAWLYD--NMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGD 565
+ I + + DG ++ W + S ++Y WC L ++ L D
Sbjct: 238 PELPIIITGSEDGTVRIWHSSTYRLESTLNYGMERVWCVASLRGSNNVAL-----GYDEG 292
Query: 566 SFLVEWNESEGAIKRTYNG 584
S +V+ E A+ NG
Sbjct: 293 SIIVKLGREEPAMSMDANG 311
>COPP_BOVIN (P35605) Coatomer beta' subunit (Beta'-coat protein)
(Beta'-COP) (p102)
Length = 905
Score = 57.4 bits (137), Expect = 2e-07
Identities = 64/268 (23%), Positives = 109/268 (39%), Gaps = 32/268 (11%)
Query: 350 VTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVVKDTP 409
V S+D HP+ + +L NG + +W + + K F++ C LP +AA
Sbjct: 17 VKSVDLHPT-EPWMLASLYNGSVCVWN--HETQTLVKTFEV-----CDLPVRAAKF---- 64
Query: 410 SVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEI-DAHIGGVNDLAFAHPNKQL 468
V+R W + G+ I ++ YN +RV + +AH + +A HP +
Sbjct: 65 -VARKNWVVTGA------DDMQIRVFNYN----TLERVHMFEAHSDYIRCIA-VHPTQPF 112
Query: 469 CVVTCGDDKLIKVWDLTGRRLFN--FEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWL 526
++T DD LIK+WD + + FEGH V I + K+N QF S ++D IK W
Sbjct: 113 -ILTSSDDMLIKLWDWDKKWSCSQVFEGHTHYVMQIVINPKDNNQFA-SASLDRTIKVWQ 170
Query: 527 YDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFR 586
+ + + Y + G + + S D + W+ +T G
Sbjct: 171 LGSSSPNFTLEGHEKGVNCIDYYSGGDKPYL--ISGADDRLVKIWDYQNKTCVQTLEG-H 227
Query: 587 KKSAGVVQFDTTQNRFLAAGEDSQIKFW 614
++ F + ED ++ W
Sbjct: 228 AQNVSCASFHPELPIIITGSEDGTVRIW 255
Score = 45.1 bits (105), Expect = 0.001
Identities = 32/139 (23%), Positives = 58/139 (41%), Gaps = 10/139 (7%)
Query: 449 IDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLTGRR-LFNFEGHEAPVYSICPHHK 507
++ H GVN + + + +++ DD+L+K+WD + + EGH V C
Sbjct: 180 LEGHEKGVNCIDYYSGGDKPYLISGADDRLVKIWDYQNKTCVQTLEGHAQNV--SCASFH 237
Query: 508 ENIQFIFSTAVDGKIKAWLYD--NMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGD 565
+ I + + DG ++ W + S ++Y WC L ++ L D
Sbjct: 238 PELPIIITGSEDGTVRIWHSSTYRLESTLNYGMERVWCVASLRGSNNVAL-----GYDEG 292
Query: 566 SFLVEWNESEGAIKRTYNG 584
S +V+ E A+ NG
Sbjct: 293 SIIVKLGREEPAMSMDANG 311
>COPP_RAT (O35142) Coatomer beta' subunit (Beta'-coat protein)
(Beta'-COP) (p102)
Length = 904
Score = 56.6 bits (135), Expect = 4e-07
Identities = 69/285 (24%), Positives = 115/285 (40%), Gaps = 36/285 (12%)
Query: 333 LDDLPRTVAMSLHQGSSVTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWD 392
LD + AMS V S+D HP+ + +L NG + +W + + K F++
Sbjct: 4 LDIKRKLTAMS----DRVKSVDLHPT-EPWMLASLYNGSVCVWN--HETQTLVKTFEV-- 54
Query: 393 ISACSLPFQAAVVKDTPSVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEI-DA 451
C LP +AA V+R W + G+ I ++ YN +RV + +A
Sbjct: 55 ---CDLPVRAAKF-----VARKNWVVTGA------DDMQIRVFNYN----TLERVHMFEA 96
Query: 452 HIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLTGRRLFN--FEGHEAPVYSICPHHKEN 509
H + +A HP + ++T DD LIK+WD + + FEGH V I + K+N
Sbjct: 97 HSDYIRCIA-VHPTQPF-ILTSSDDMLIKLWDWDKKWSCSQVFEGHTHYVMQIVINPKDN 154
Query: 510 IQFIFSTAVDGKIKAWLYDNMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLV 569
QF S ++D IK W + + + Y + G + + S D +
Sbjct: 155 NQFA-SASLDRTIKVWQLGSSSPNFTLEGHEKGVNCIDYYSGGDKPYL--ISGADDRLVK 211
Query: 570 EWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFW 614
W+ +T G ++ F + ED ++ W
Sbjct: 212 IWDYQNKTCVQTPEG-HAQNVSCATFHPELPIIITGSEDGTVRIW 255
Score = 44.3 bits (103), Expect = 0.002
Identities = 34/139 (24%), Positives = 59/139 (41%), Gaps = 10/139 (7%)
Query: 449 IDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLTGRRLFNF-EGHEAPVYSICPHHK 507
++ H GVN + + + +++ DD+L+K+WD + EGH V S H
Sbjct: 180 LEGHEKGVNCIDYYSGGDKPYLISGADDRLVKIWDYQNKTCVQTPEGHAQNV-SCATFHP 238
Query: 508 ENIQFIFSTAVDGKIKAWLYD--NMGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGD 565
E + I + + DG ++ W + S ++Y WC L ++ L D
Sbjct: 239 E-LPIIITGSEDGTVRIWHSSTYRLESTLNYGMERVWCVASLRGSNNVAL-----GYDEG 292
Query: 566 SFLVEWNESEGAIKRTYNG 584
S +V+ E A+ NG
Sbjct: 293 SIIVKLGREEPAMSMDANG 311
>PR17_HUMAN (O60508) Pre-mRNA splicing factor PRP17 (hPRP17) (Cell
division cycle 40 homolog) (EH-binding protein 3) (Ehb3)
Length = 579
Score = 53.9 bits (128), Expect = 2e-06
Identities = 48/189 (25%), Positives = 80/189 (41%), Gaps = 14/189 (7%)
Query: 470 VVTCGDDKLIKVWDLTGRR--LFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLY 527
+++C D IK+W++ G R L F GH V IC + QF+ S A D +K W
Sbjct: 304 LLSCSMDCKIKLWEVYGERRCLRTFIGHSKAVRDIC-FNTAGTQFL-SAAYDRYLKLWDT 361
Query: 528 DN---MGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNG 584
+ + + P +C D LF G S D +V+W+ G I + Y+
Sbjct: 362 ETGQCISRFTNRKVP--YCVKFNPDEDKQNLFVAGMS---DKKIVQWDIRSGEIVQEYDR 416
Query: 585 FRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGN 644
+ + F RF++ +D ++ W+ D + AE + +P + + G
Sbjct: 417 -HLGAVNTIVFVDENRRFVSTSDDKSLRVWEWD-IPVDFKYIAEPSMHSMPAVTLSPNGK 474
Query: 645 LLAVTTADN 653
LA + DN
Sbjct: 475 WLACQSMDN 483
Score = 44.7 bits (104), Expect = 0.001
Identities = 42/183 (22%), Positives = 74/183 (39%), Gaps = 17/183 (9%)
Query: 440 SNELAQRVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWD----LTGRRLFNFEGH 495
S E+ Q E D H+G VN + F N++ V+ DDK ++VW+ + + + H
Sbjct: 407 SGEIVQ--EYDRHLGAVNTIVFVDENRRF--VSTSDDKSLRVWEWDIPVDFKYIAEPSMH 462
Query: 496 EAPVYSICPHHKENIQFIFSTAVDGKIKAWLYDN---MGSRVDYDAPGHWCTTMLYSADG 552
P ++ P+ K ++ ++D +I + N + + + GH D
Sbjct: 463 SMPAVTLSPNGK----WLACQSMDNQILIFGAQNRFRLNKKKIF--KGHMVAGYACQVDF 516
Query: 553 TRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIK 612
+ S S DG+ L W+ + + K G V ++ + G D IK
Sbjct: 517 SPDMSYVISGDGNGKLNIWDWKTTKLYSRFKAHDKVCIGAVWHPHETSKVITCGWDGLIK 576
Query: 613 FWD 615
WD
Sbjct: 577 LWD 579
>Y143_SYNY3 (P74442) Hypothetical WD-repeat protein slr0143
Length = 1191
Score = 53.1 bits (126), Expect = 4e-06
Identities = 60/237 (25%), Positives = 98/237 (41%), Gaps = 35/237 (14%)
Query: 441 NELAQRVEIDAHIGGVNDLAFA-HPNKQLCVVTCGDDKLIKVWDLTGRRLFNFEGHEAPV 499
+++A++ + H GV +A + H N + + D + +W G L F GH +
Sbjct: 548 SQIAEKNVLTGHRDGVTSVAISSHKN---LIASASRDGTVHLWTPQGEFLREFTGHTGSI 604
Query: 500 YSI--CPHHKENIQFIFSTA-VDGKIKAWLYDNMGSRVDYDAPGHWCT--TMLYSADGTR 554
Y + P+ K IF+TA D +K W ++ + GH + ++ +S DG
Sbjct: 605 YRVDFSPNGK-----IFATAGQDQTVKIW---DLDGNLLQTLKGHQDSVYSVSFSPDGEI 656
Query: 555 LFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKFW 614
L S TS+D L W + G KS QF ++ D QI+ W
Sbjct: 657 LAS--TSRDRTVRLWHWRSGKTL---AVLGGHTKSVDDAQFSPDGQTLVSVCRDGQIRLW 711
Query: 615 DMDNVNPLTSTEAEGGLQGLPHLRF-----NKEGNLLAVTTADNGFKILANAGGLRS 666
D+D N + GLP + F + GNLLAV D ++ G +++
Sbjct: 712 DLDG-NLIRQF-------GLPEVAFFGVNWHPNGNLLAVAADDGTVRLWTPQGEIKA 760
>PR17_MOUSE (Q9DC48) Pre-mRNA splicing factor PRP17 (Cell division
cycle 40 homolog)
Length = 579
Score = 53.1 bits (126), Expect = 4e-06
Identities = 48/189 (25%), Positives = 80/189 (41%), Gaps = 14/189 (7%)
Query: 470 VVTCGDDKLIKVWDLTGRR--LFNFEGHEAPVYSICPHHKENIQFIFSTAVDGKIKAWLY 527
+++C D IK+W++ G R L F GH V IC + QF+ S A D +K W
Sbjct: 304 LLSCSMDCKIKLWEVYGDRRCLRTFIGHSKAVRDIC-FNTAGTQFL-SAAYDRYLKLWDT 361
Query: 528 DN---MGSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNG 584
+ + + P +C D LF G S D +V+W+ G I + Y+
Sbjct: 362 ETGQCISRFTNRKVP--YCVKFNPDEDKQNLFVAGMS---DKKIVQWDIRSGEIVQEYDR 416
Query: 585 FRKKSAGVVQFDTTQNRFLAAGEDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGN 644
+ + F RF++ +D ++ W+ D + AE + +P + + G
Sbjct: 417 -HLGAVNTIVFVDENRRFVSTSDDKSLRVWEWD-IPVDFKYIAEPSMHSMPAVTLSPNGK 474
Query: 645 LLAVTTADN 653
LA + DN
Sbjct: 475 WLACQSMDN 483
Score = 44.7 bits (104), Expect = 0.001
Identities = 44/182 (24%), Positives = 73/182 (39%), Gaps = 15/182 (8%)
Query: 440 SNELAQRVEIDAHIGGVNDLAFAHPNKQLCVVTCGDDKLIKVWDLTGRRLFNFEG----H 495
S E+ Q E D H+G VN + F N++ V+ DDK ++VW+ F + H
Sbjct: 407 SGEIVQ--EYDRHLGAVNTIVFVDENRRF--VSTSDDKSLRVWEWDIPVDFKYIAEPSMH 462
Query: 496 EAPVYSICPHHKENIQFIFSTAVDGKIKAWLYDNMGSRVDYDA--PGHWCTTMLYSADGT 553
P ++ P+ K ++ ++D +I + N R++ GH D +
Sbjct: 463 SMPAVTLSPNGK----WLACQSMDNQILIFGAQNR-FRLNKKKIFKGHMVAGYACQVDFS 517
Query: 554 RLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSAGVVQFDTTQNRFLAAGEDSQIKF 613
S S DG+ L W+ + + K G V ++ + G D IK
Sbjct: 518 PDMSYVISGDGNGKLNIWDWKTTKLYSRFKAHDKVCIGAVWHPHETSKVITCGWDGLIKL 577
Query: 614 WD 615
WD
Sbjct: 578 WD 579
>TAF5_YEAST (P38129) Transcription initiation factor TFIID subunit 5
(TBP-associated factor 5) (TBP-associated factor 90 kDa)
(TAFII-90)
Length = 798
Score = 52.8 bits (125), Expect = 5e-06
Identities = 55/216 (25%), Positives = 85/216 (38%), Gaps = 31/216 (14%)
Query: 334 DDLPRTVAMSLHQGSSVTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVS---KPFKI 390
D+ P + H G+ V S F P ++ LL GS + + LW + LVS +
Sbjct: 513 DEDPTCKTLVGHSGT-VYSTSFSPDNK-YLLSGSEDKTVRLWSMDTHTALVSYKGHNHPV 570
Query: 391 WDISACSLPFQAAVVKDTPSVSRVTWSLDGSFVGVAFTKHLIHI----------YAYNGS 440
WD+S L A + WS D + F HL + Y + GS
Sbjct: 571 WDVSFSPLGHYFATASHDQTAR--LWSCDHIYPLRIFAGHLNDVDCVSFHPNGCYVFTGS 628
Query: 441 NELAQRVEIDAHIGGVNDLAFAHPNKQLCVVTCGD---------DKLIKVWDL-TGRRLF 490
++ R+ D G L H + + C D D +I VWD+ TG+RL
Sbjct: 629 SDKTCRM-WDVSTGDSVRLFLGHTAPVISIAVCPDGRWLSTGSEDGIINVWDIGTGKRLK 687
Query: 491 NFEGH-EAPVYSICPHHKENIQFIFSTAVDGKIKAW 525
GH + +YS+ + N+ + S D ++ W
Sbjct: 688 QMRGHGKNAIYSLSYSKEGNV--LISGGADHTVRVW 721
Score = 47.4 bits (111), Expect = 2e-04
Identities = 61/249 (24%), Positives = 107/249 (42%), Gaps = 28/249 (11%)
Query: 416 WSLDGSFVG---VAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLCVVT 472
WSLDGS + +A N +E + H G V +F+ NK L ++
Sbjct: 493 WSLDGSSLNNPNIALNN--------NDKDEDPTCKTLVGHSGTVYSTSFSPDNKYL--LS 542
Query: 473 CGDDKLIKVWDL-TGRRLFNFEGHEAPVYSICPHHKENIQFIFSTAV-DGKIKAWLYDNM 530
+DK +++W + T L +++GH PV+ + + F+TA D + W D++
Sbjct: 543 GSEDKTVRLWSMDTHTALVSYKGHNHPVWDVS---FSPLGHYFATASHDQTARLWSCDHI 599
Query: 531 GSRVDYDAPGHWCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFRKKSA 590
+ + + + +G +F+ G+S D W+ S G R + G +A
Sbjct: 600 YPLRIFAGHLNDVDCVSFHPNGCYVFT-GSS---DKTCRMWDVSTGDSVRLFLGH---TA 652
Query: 591 GVVQFDTTQN-RFLAAG-EDSQIKFWDMDNVNPLTSTEAEGGLQGLPHLRFNKEGNLLAV 648
V+ + R+L+ G ED I WD+ L G + L ++KEGN+L
Sbjct: 653 PVISIAVCPDGRWLSTGSEDGIINVWDIGTGKRLKQMRGHGK-NAIYSLSYSKEGNVLIS 711
Query: 649 TTADNGFKI 657
AD+ ++
Sbjct: 712 GGADHTVRV 720
>GBLP_DICDI (P46800) Guanine nucleotide-binding protein beta
subunit-like protein
Length = 329
Score = 52.8 bits (125), Expect = 5e-06
Identities = 66/270 (24%), Positives = 112/270 (41%), Gaps = 29/270 (10%)
Query: 350 VTSMDFHPSHQTLLLVGSNNGEISLWELGMRERLVSKPFKIWDISACSLPFQAAVVKDTP 409
VTS+ P + ++ S + + +W+L + + P K + SL + V+D
Sbjct: 24 VTSIAVSPENPDTIISSSRDKTVMVWQLTPTD--ATSPGK----AHRSLKGHSHFVQD-- 75
Query: 410 SVSRVTWSLDGSFVGVAFTKHLIHIYAYNGSNELAQRVEIDAHIGGVNDLAFAHPNKQLC 469
V S DG F + + ++ + ++ R+ H V +AF+ N+Q
Sbjct: 76 ----VVISHDGQFALSGSWDNTLRLWDI--TKGVSTRL-FKGHTQDVMSVAFSSDNRQ-- 126
Query: 470 VVTCGDDKLIKVWDLTGRRLFNFEGHEA-PVYSICPHHKENIQFIFSTAVDGKIKAWLYD 528
+++ D IKVW+ G F EG EA + C N I S + D K+K W D
Sbjct: 127 IISGSRDATIKVWNTLGECKFTLEGPEAHQDWVSCIRFSPNTPTIVSGSWDNKVKIW--D 184
Query: 529 NMGSRVDYDAPGH--WCTTMLYSADGTRLFSCGTSKDGDSFLVEWNESEGAIKRTYNGFR 586
+ ++ H + T+ S DG+ S G D+F W S G K Y
Sbjct: 185 IKSFKCNHTLTDHTGYVNTVTISPDGSLCASGGK----DTFACLWELSSG--KPLYKLEA 238
Query: 587 KKSAGVVQFDTTQNRFLAAGEDSQIKFWDM 616
+ + + F + +L+A D +I WD+
Sbjct: 239 RNTINALAF-SPNKYWLSAATDDKIIIWDL 267
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.333 0.143 0.463
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 125,313,551
Number of Sequences: 164201
Number of extensions: 5220330
Number of successful extensions: 22505
Number of sequences better than 10.0: 290
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 273
Number of HSP's that attempted gapping in prelim test: 21529
Number of HSP's gapped (non-prelim): 975
length of query: 1115
length of database: 59,974,054
effective HSP length: 121
effective length of query: 994
effective length of database: 40,105,733
effective search space: 39865098602
effective search space used: 39865098602
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 71 (32.0 bits)
Medicago: description of AC144513.3


