

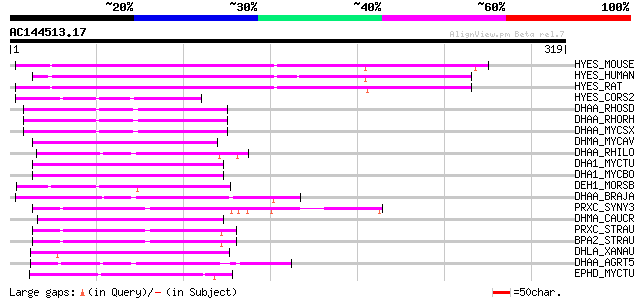
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144513.17 + phase: 0
(319 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
HYES_MOUSE (P34914) Soluble epoxide hydrolase (SEH) (EC 3.3.2.3)... 177 2e-44
HYES_HUMAN (P34913) Soluble epoxide hydrolase (SEH) (EC 3.3.2.3)... 177 4e-44
HYES_RAT (P80299) Soluble epoxide hydrolase (SEH) (EC 3.3.2.3) (... 174 3e-43
HYES_CORS2 (O52866) Soluble epoxide hydrolase (EC 3.3.2.3) (SEH)... 79 1e-14
DHAA_RHOSD (P59336) Haloalkane dehalogenase (EC 3.8.1.5) 76 1e-13
DHAA_RHORH (Q53042) Haloalkane dehalogenase (EC 3.8.1.5) 76 1e-13
DHAA_MYCSX (Q9ZER0) Haloalkane dehalogenase (EC 3.8.1.5) 76 1e-13
DHMA_MYCAV (Q93K00) Haloalkane dehalogenase (EC 3.8.1.5) 69 1e-11
DHAA_RHILO (Q98C03) Haloalkane dehalogenase (EC 3.8.1.5) 69 1e-11
DHA1_MYCTU (P64301) Haloalkane dehalogenase 1 (EC 3.8.1.5) 68 3e-11
DHA1_MYCBO (P64302) Haloalkane dehalogenase 1 (EC 3.8.1.5) 68 3e-11
DEH1_MORSB (Q01398) Haloacetate dehalogenase H-1 (EC 3.8.1.3) 67 6e-11
DHAA_BRAJA (P59337) Haloalkane dehalogenase (EC 3.8.1.5) 64 4e-10
PRXC_SYNY3 (Q55921) Putative non-heme chloroperoxidase (EC 1.11.... 63 9e-10
DHMA_CAUCR (Q9A919) Haloalkane dehalogenase (EC 3.8.1.5) 63 9e-10
PRXC_STRAU (O31168) Non-heme chloroperoxidase (EC 1.11.1.10) (Ch... 62 2e-09
BPA2_STRAU (P29715) Non-haem bromoperoxidase BPO-A2 (EC 1.11.1.-... 62 2e-09
DHLA_XANAU (P22643) Haloalkane dehalogenase (EC 3.8.1.5) 60 6e-09
DHAA_AGRT5 (Q8U671) Haloalkane dehalogenase (EC 3.8.1.5) 59 2e-08
EPHD_MYCTU (P66777) Probable oxidoreductase ephD (EC 1.-.-.-) 59 2e-08
>HYES_MOUSE (P34914) Soluble epoxide hydrolase (SEH) (EC 3.3.2.3)
(Epoxide hydratase) (Cytosolic epoxide hydrolase) (CEH)
Length = 554
Score = 177 bits (450), Expect = 2e-44
Identities = 96/278 (34%), Positives = 155/278 (55%), Gaps = 8/278 (2%)
Query: 4 IEHRTVEVN-GIKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRG 62
+ H V V GI++H E G GP + HGFPE W+SWR+QI AL G+R +A D++G
Sbjct: 235 VSHGYVTVKPGIRLHFVEMGS-GPALCLCHGFPESWFSWRYQIPALAQAGFRVLAIDMKG 293
Query: 63 YGDTDVPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKAY 122
YGD+ P I Y + ++V+ +D LG+ Q +GHD ++ W + +F PER++A
Sbjct: 294 YGDSSSPPEIEEYAMELLCKEMVTFLDKLGIPQAVFIGHDWAGVMVWNMALFYPERVRAV 353
Query: 123 VCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNILTTRK 182
L+ PF+ +P + + +R++ +Y + FQEPG EA++ + + + +
Sbjct: 354 ASLNTPFMPPDPDVSPMKVIRSIPVFNYQL-YFQEPGVAEAELEKNMSRTFKSFFRASDE 412
Query: 183 TGPPIFPKGEYGTGFNPDTPD--NLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNFNLNW 240
TG K G +TP+ NL TE+++ +++ +F+KTGF G LN+YRN NW
Sbjct: 413 TGFIAVHKATEIGGILVNTPEDPNLSKITTEEEIEFYIQQFKKTGFRGPLNWYRNTERNW 472
Query: 241 ELTAPWTGVKIKVPVKFITGELDMVY---TSFNLKEYI 275
+ + G KI VP +T E D+V S N++++I
Sbjct: 473 KWSCKGLGRKILVPALMVTAEKDIVLRPEMSKNMEKWI 510
>HYES_HUMAN (P34913) Soluble epoxide hydrolase (SEH) (EC 3.3.2.3)
(Epoxide hydratase) (Cytosolic epoxide hydrolase) (CEH)
Length = 554
Score = 177 bits (448), Expect = 4e-44
Identities = 90/254 (35%), Positives = 142/254 (55%), Gaps = 6/254 (2%)
Query: 14 IKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYGDTDVPSSIS 73
+++H E G P V HGFPE WYSWR+QI AL GYR +A D++GYG++ P I
Sbjct: 248 VRLHFVELG--WPAVCLCHGFPESWYSWRYQIPALAQAGYRVLAMDMKGYGESSAPPEIE 305
Query: 74 SYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKAYVCLSVPFLHRN 133
Y + ++V+ +D LG+ Q +GHD G ++ WY+ +F PER++A L+ PF+ N
Sbjct: 306 EYCMEVLCKEMVTFLDKLGLSQAVFIGHDWGGMLVWYMALFYPERVRAVASLNTPFIPAN 365
Query: 134 PKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNILTTRKTGPPIFPKGEY 193
P + ++ ++A DY + FQEPG EA++ E + K++ K
Sbjct: 366 PNMSPLESIKANPVFDYQL-YFQEPGVAEAEL-EQNLSRTFKSLFRASDESVLSMHKVCE 423
Query: 194 GTGFNPDTPD--NLPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNFNLNWELTAPWTGVKI 251
G ++P+ +L +TE+++ ++V +F+K+GF G LN+YRN NW+ G KI
Sbjct: 424 AGGLFVNSPEEPSLSRMVTEEEIQFYVQQFKKSGFRGPLNWYRNMERNWKWACKSLGRKI 483
Query: 252 KVPVKFITGELDMV 265
+P +T E D V
Sbjct: 484 LIPALMVTAEKDFV 497
>HYES_RAT (P80299) Soluble epoxide hydrolase (SEH) (EC 3.3.2.3)
(Epoxide hydratase) (Cytosolic epoxide hydrolase) (CEH)
Length = 554
Score = 174 bits (441), Expect = 3e-43
Identities = 90/265 (33%), Positives = 147/265 (54%), Gaps = 5/265 (1%)
Query: 4 IEHRTVEVN-GIKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRG 62
+ H V V GI++H E G GP + HGFPE W+SWR+QI AL G+R +A D++G
Sbjct: 235 VSHGYVTVKPGIRLHFVEMGS-GPAICLCHGFPESWFSWRYQIPALAQAGFRVLAIDMKG 293
Query: 63 YGDTDVPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKAY 122
YGD+ P I Y + ++V+ ++ LG+ Q +GHD ++ W + +F PER++A
Sbjct: 294 YGDSSSPPEIEEYAMELLCEEMVTFLNKLGIPQAVFIGHDWAGVLVWNMALFHPERVRAV 353
Query: 123 VCLSVPFLHRNPKIRTVDGMRAVYGDDYYICRFQEPGEMEAQMAEVGTTYVMKNILTTRK 182
L+ P + NP++ ++ +R++ +Y + FQEPG EA++ + + T+
Sbjct: 354 ASLNTPLMPPNPEVSPMEVIRSIPVFNYQL-YFQEPGVAEAELEKNMSRTFKSFFRTSDD 412
Query: 183 TGPPIFPKGEYGTGFNPDTPDN--LPSWLTEDDLAYFVSKFEKTGFTGGLNYYRNFNLNW 240
G K G TP++ + TE+++ Y++ +F+K+GF G LN+YRN NW
Sbjct: 413 MGLLTVNKATEMGGILVGTPEDPKVSKITTEEEIEYYIQQFKKSGFRGPLNWYRNTERNW 472
Query: 241 ELTAPWTGVKIKVPVKFITGELDMV 265
+ + G KI VP +T E D+V
Sbjct: 473 KWSCKALGRKILVPALMVTAEKDIV 497
>HYES_CORS2 (O52866) Soluble epoxide hydrolase (EC 3.3.2.3) (SEH)
(Epoxide hydratase) (Cytosolic epoxide hydrolase) (cEH)
Length = 285
Score = 79.3 bits (194), Expect = 1e-14
Identities = 40/107 (37%), Positives = 62/107 (57%), Gaps = 4/107 (3%)
Query: 4 IEHRTVEVNGIKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGY 63
I H +NG +MH G P+VL LHG+P+ WY WR+ I AL + +APDLRG
Sbjct: 4 ITHHQAMINGYRMHYVTAGSGYPLVL-LHGWPQSWYEWRNVIPALAE-QFTVIAPDLRGL 61
Query: 64 GDTDVPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWY 110
GD++ P ++ + + D+ L+ LG ++V ++GHD G + +Y
Sbjct: 62 GDSEKP--MTGFDKRTMATDVRELVSHLGYDKVGVIGHDWGGSVAFY 106
>DHAA_RHOSD (P59336) Haloalkane dehalogenase (EC 3.8.1.5)
Length = 294
Score = 75.9 bits (185), Expect = 1e-13
Identities = 46/118 (38%), Positives = 66/118 (54%), Gaps = 4/118 (3%)
Query: 9 VEVNGIKMHIAEKG-KEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYGDTD 67
VEV G +MH + G ++G VLFLHG P Y WR+ I + +R +APDL G G +D
Sbjct: 15 VEVLGERMHYVDVGPRDGTPVLFLHGNPTSSYLWRNIIPHVAP-SHRCIAPDLIGMGKSD 73
Query: 68 VPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKAYVCL 125
P Y V + + I+ LG+E+V LV HD G+ +G++ PER+K C+
Sbjct: 74 KPDL--DYFFDDHVRYLDAFIEALGLEEVVLVIHDWGSALGFHWAKRNPERVKGIACM 129
>DHAA_RHORH (Q53042) Haloalkane dehalogenase (EC 3.8.1.5)
Length = 293
Score = 75.9 bits (185), Expect = 1e-13
Identities = 46/118 (38%), Positives = 66/118 (54%), Gaps = 4/118 (3%)
Query: 9 VEVNGIKMHIAEKG-KEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYGDTD 67
VEV G +MH + G ++G VLFLHG P Y WR+ I + +R +APDL G G +D
Sbjct: 15 VEVLGERMHYVDVGPRDGTPVLFLHGNPTSSYLWRNIIPHVAP-SHRCIAPDLIGMGKSD 73
Query: 68 VPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKAYVCL 125
P Y V + + I+ LG+E+V LV HD G+ +G++ PER+K C+
Sbjct: 74 KPDL--DYFFDDHVRYLDAFIEALGLEEVVLVIHDWGSALGFHWAKRNPERVKGIACM 129
>DHAA_MYCSX (Q9ZER0) Haloalkane dehalogenase (EC 3.8.1.5)
Length = 307
Score = 75.9 bits (185), Expect = 1e-13
Identities = 46/118 (38%), Positives = 66/118 (54%), Gaps = 4/118 (3%)
Query: 9 VEVNGIKMHIAEKG-KEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYGDTD 67
VEV G +MH + G ++G VLFLHG P Y WR+ I + +R +APDL G G +D
Sbjct: 15 VEVLGERMHYVDVGPRDGTPVLFLHGNPTSSYLWRNIIPHVAP-SHRCIAPDLIGMGKSD 73
Query: 68 VPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKAYVCL 125
P Y V + + I+ LG+E+V LV HD G+ +G++ PER+K C+
Sbjct: 74 KPDL--DYFFDDHVRYLDAFIEALGLEEVVLVIHDWGSALGFHWAKRNPERVKGIACM 129
>DHMA_MYCAV (Q93K00) Haloalkane dehalogenase (EC 3.8.1.5)
Length = 301
Score = 69.3 bits (168), Expect = 1e-11
Identities = 38/107 (35%), Positives = 59/107 (54%), Gaps = 1/107 (0%)
Query: 14 IKMHIAEKGK-EGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYGDTDVPSSI 72
++MH ++G +GP ++ LHG P Y +R I L G R +APDL G+G +D PS I
Sbjct: 34 LRMHYLDEGPIDGPPIVLLHGEPTWSYLYRTMITPLTDAGNRVLAPDLIGFGRSDKPSRI 93
Query: 73 SSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERI 119
Y+ V +VS + L + V L D G++IG + +P+R+
Sbjct: 94 EDYSYQRHVDWVVSWFEHLNLSDVTLFVQDWGSLIGLRIAAEQPDRV 140
>DHAA_RHILO (Q98C03) Haloalkane dehalogenase (EC 3.8.1.5)
Length = 309
Score = 69.3 bits (168), Expect = 1e-11
Identities = 45/129 (34%), Positives = 63/129 (47%), Gaps = 10/129 (7%)
Query: 16 MHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYGDTDVPSSISSY 75
M E G GP VLFLHG P + WR+ I + G R +APDL GYG + P Y
Sbjct: 27 MSYVEAGASGPTVLFLHGNPTSSHIWRNIIPHVAPFG-RCIAPDLIGYGQSGKPD--IDY 83
Query: 76 TCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERI--KAYVCLSVPF---- 129
F V + + +D L + V LV D G + ++L RP+R+ A++ PF
Sbjct: 84 RFFDHVRYLDAFLDALDIRDVLLVAQDWGTALAFHLAARRPQRVLGLAFMEFIRPFERWE 143
Query: 130 -LHRNPKIR 137
H+ P+ R
Sbjct: 144 DFHQRPQAR 152
>DHA1_MYCTU (P64301) Haloalkane dehalogenase 1 (EC 3.8.1.5)
Length = 300
Score = 68.2 bits (165), Expect = 3e-11
Identities = 38/111 (34%), Positives = 59/111 (52%), Gaps = 1/111 (0%)
Query: 14 IKMHIAEKGK-EGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYGDTDVPSSI 72
++MH ++G +GP ++ LHG P Y +R I L + G+R +APDL G+G +D P+ I
Sbjct: 34 LRMHYVDEGPGDGPPIVLLHGEPTWSYLYRTMIPPLSAAGHRVLAPDLIGFGRSDKPTRI 93
Query: 73 SSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKAYV 123
YT V + S + L + V L D G++IG + +RI V
Sbjct: 94 EDYTYLRHVEWVTSWFENLDLHDVTLFVQDWGSLIGLRIAAEHGDRIARLV 144
>DHA1_MYCBO (P64302) Haloalkane dehalogenase 1 (EC 3.8.1.5)
Length = 300
Score = 68.2 bits (165), Expect = 3e-11
Identities = 38/111 (34%), Positives = 59/111 (52%), Gaps = 1/111 (0%)
Query: 14 IKMHIAEKGK-EGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYGDTDVPSSI 72
++MH ++G +GP ++ LHG P Y +R I L + G+R +APDL G+G +D P+ I
Sbjct: 34 LRMHYVDEGPGDGPPIVLLHGEPTWSYLYRTMIPPLSAAGHRVLAPDLIGFGRSDKPTRI 93
Query: 73 SSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKAYV 123
YT V + S + L + V L D G++IG + +RI V
Sbjct: 94 EDYTYLRHVEWVTSWFENLDLHDVTLFVQDWGSLIGLRIAAEHGDRIARLV 144
>DEH1_MORSB (Q01398) Haloacetate dehalogenase H-1 (EC 3.8.1.3)
Length = 294
Score = 67.0 bits (162), Expect = 6e-11
Identities = 42/126 (33%), Positives = 63/126 (49%), Gaps = 5/126 (3%)
Query: 5 EHRTVEVNGIKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYG 64
++ TV V+G+ + G EGP VL LHGFP+ W L + V DLRGYG
Sbjct: 7 KNSTVTVDGVDIAYTVSG-EGPPVLMLHGFPQNRAMWARVAPQLAE-HHTVVCADLRGYG 64
Query: 65 DTDVPSSI---SSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKA 121
D+D P + S+Y+ D + ++ LG E+ LVGHD G G + + PE + +
Sbjct: 65 DSDKPKCLPDRSNYSFRTFAHDQLCVMRHLGFERFHLVGHDRGGRTGHRMALDHPEAVLS 124
Query: 122 YVCLSV 127
+ +
Sbjct: 125 LTVMDI 130
>DHAA_BRAJA (P59337) Haloalkane dehalogenase (EC 3.8.1.5)
Length = 310
Score = 64.3 bits (155), Expect = 4e-10
Identities = 49/171 (28%), Positives = 78/171 (44%), Gaps = 12/171 (7%)
Query: 4 IEHRTVEVNGIKMHIAEKG-KEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRG 62
IE R V G M E G ++ PVVLFLHG P + WR+ + + + + +APDL G
Sbjct: 7 IEIRRAPVLGSSMAYRETGAQDAPVVLFLHGNPTSSHIWRNILPLVSPVAH-CIAPDLIG 65
Query: 63 YGDTDVPSSISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKAY 122
+G + P +Y F V + + I+ GV +LV D G + ++L RP+ ++
Sbjct: 66 FGQSGKPD--IAYRFFDHVRYLDAFIEQRGVTSAYLVAQDWGTALAFHLAARRPDFVRGL 123
Query: 123 VCLSVPFLHRNPKIRTVDGMRAVYGDDY------YICRFQEPGEMEAQMAE 167
+ F+ P + D+ +F+ PGE EA + E
Sbjct: 124 AFME--FIRPMPTWQDFHHTEVAEEQDHAEAARAVFRKFRTPGEGEAMILE 172
>PRXC_SYNY3 (Q55921) Putative non-heme chloroperoxidase (EC
1.11.1.10) (Chloride peroxidase)
Length = 276
Score = 63.2 bits (152), Expect = 9e-10
Identities = 64/216 (29%), Positives = 97/216 (44%), Gaps = 31/216 (14%)
Query: 14 IKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYGDTDVPSSIS 73
I ++ + G P+VL +HGFP SW Q+ L + GYR + D RG+G + PSS
Sbjct: 14 IDIYYEDLGAGQPIVL-IHGFPLNGDSWEKQVLVLLNAGYRVITYDRRGFGASSQPSSGY 72
Query: 74 SYTCFHVVGDIVSLIDLLGVEQVFLVGHDMG-AIIGWYLCMFRPERIKAYVCLS--VPFL 130
Y F D+ +L+ L ++ LVG MG + YL + ER++ V ++ PFL
Sbjct: 73 DYDTF--AADLHTLMTKLDLQNTVLVGFSMGTGEVTRYLGKYGSERVQKAVLMAPVPPFL 130
Query: 131 ---HRNPK---IRTVDGMRAVYGDD---YYICRFQEPGEMEAQMAEVGTTYVMKNILTTR 181
+ NP+ DG+ DD Y+ F+E ++ + E R
Sbjct: 131 LKTNDNPEGVDQSVFDGIMKAIVDDRPAYFSAFFKEFFNVDVLLGE-------------R 177
Query: 182 KTGPPIFPKGEYGTGFN-PDTPDNLPSWLTE--DDL 214
+ I G + T D +PSWLT+ DDL
Sbjct: 178 ISNEAIQASWNVAAGASAKGTLDCVPSWLTDFRDDL 213
>DHMA_CAUCR (Q9A919) Haloalkane dehalogenase (EC 3.8.1.5)
Length = 302
Score = 63.2 bits (152), Expect = 9e-10
Identities = 37/107 (34%), Positives = 56/107 (51%)
Query: 17 HIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYGDTDVPSSISSYT 76
H+ E K+ +L +HG P Y +R IA L + G+R VAPDL G+G +D P+ + YT
Sbjct: 38 HVDEGPKDQRPILLMHGEPSWAYLYRKVIAELVAKGHRVVAPDLVGFGRSDKPAKRTDYT 97
Query: 77 CFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKAYV 123
V + + ++ ++ + L D G +IG L PER A V
Sbjct: 98 YERHVAWMSAWLEQNDLKDIVLFCQDWGGLIGLRLVAAFPERFSAVV 144
>PRXC_STRAU (O31168) Non-heme chloroperoxidase (EC 1.11.1.10)
(Chloride peroxidase) (CPO-T) (Chloroperoxidase T)
Length = 278
Score = 61.6 bits (148), Expect = 2e-09
Identities = 40/120 (33%), Positives = 62/120 (51%), Gaps = 6/120 (5%)
Query: 14 IKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYGDTDVPSSIS 73
I ++ + G PVVL +HGFP +SW Q AAL GYR + D RG+G + P++
Sbjct: 14 IDLYYEDHGAGQPVVL-IHGFPLSGHSWERQSAALLDAGYRVITYDRRGFGQSSQPTTGY 72
Query: 74 SYTCFHVVGDIVSLIDLLGVEQVFLVGHDMG-AIIGWYLCMFRPERIK--AYVCLSVPFL 130
Y F D+ ++++ L ++ LVG MG + Y+ + RI A++ PFL
Sbjct: 73 DYDTF--AADLNTVLETLDLQDAVLVGFSMGTGEVARYVSSYGTARIAKVAFLASLEPFL 130
>BPA2_STRAU (P29715) Non-haem bromoperoxidase BPO-A2 (EC 1.11.1.-)
(Bromide peroxidase) (BPO2)
Length = 277
Score = 61.6 bits (148), Expect = 2e-09
Identities = 40/120 (33%), Positives = 62/120 (51%), Gaps = 6/120 (5%)
Query: 14 IKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYGDTDVPSSIS 73
I ++ + G PVVL +HGFP +SW Q AAL GYR + D RG+G + P++
Sbjct: 13 IDLYYEDHGTGQPVVL-IHGFPLSGHSWERQSAALLDAGYRVITYDRRGFGQSSQPTTGY 71
Query: 74 SYTCFHVVGDIVSLIDLLGVEQVFLVGHDMG-AIIGWYLCMFRPERIK--AYVCLSVPFL 130
Y F D+ ++++ L ++ LVG MG + Y+ + RI A++ PFL
Sbjct: 72 DYDTF--AADLNTVLETLDLQDAVLVGFSMGTGEVARYVSSYGTARIAKVAFLASLEPFL 129
>DHLA_XANAU (P22643) Haloalkane dehalogenase (EC 3.8.1.5)
Length = 310
Score = 60.5 bits (145), Expect = 6e-09
Identities = 36/116 (31%), Positives = 54/116 (46%), Gaps = 2/116 (1%)
Query: 13 GIKMHIAEKGKEGP--VVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYGDTDVPS 70
G++ H ++G V L LHG P Y +R I G R +APD G+G +D P
Sbjct: 33 GLRAHYLDEGNSDAEDVFLCLHGEPTWSYLYRKMIPVFAESGARVIAPDFFGFGKSDKPV 92
Query: 71 SISSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKAYVCLS 126
YT +++LI+ L + + LV D G +G L M P R K + ++
Sbjct: 93 DEEDYTFEFHRNFLLALIERLDLRNITLVVQDWGGFLGLTLPMADPSRFKRLIIMN 148
>DHAA_AGRT5 (Q8U671) Haloalkane dehalogenase (EC 3.8.1.5)
Length = 304
Score = 58.9 bits (141), Expect = 2e-08
Identities = 48/150 (32%), Positives = 67/150 (44%), Gaps = 11/150 (7%)
Query: 13 GIKMHIAEKGKEGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYGDTDVPSSI 72
G+++H E G P+V FLHG P Y WRH L G R +A DL GYG + P
Sbjct: 29 GLQIHTVEHGSGAPIV-FLHGNPTSSYLWRHIFRRLHGHG-RLLAVDLIGYGQSSKPD-- 84
Query: 73 SSYTCFHVVGDIVSLIDLLGVEQVFLVGHDMGAIIGWYLCMFRPERIKAYVCLSVPFLHR 132
YT + + + D L + V LV D GA G P+R++A V F
Sbjct: 85 IEYTLENQQRYVDAWFDALDLRNVTLVLQDYGAAFGLNWASRNPDRVRA-----VAFF-- 137
Query: 133 NPKIRTVDGMRAVYGDDYYICRFQEPGEME 162
P +R +D + + ++PGE E
Sbjct: 138 EPVLRNIDSVDLSPEFVTRRAKLRQPGEGE 167
>EPHD_MYCTU (P66777) Probable oxidoreductase ephD (EC 1.-.-.-)
Length = 592
Score = 58.5 bits (140), Expect = 2e-08
Identities = 38/122 (31%), Positives = 65/122 (53%), Gaps = 7/122 (5%)
Query: 12 NGIKMHIAEKGK-EGPVVLFLHGFPELWYSWRHQIAALGSLGYRAVAPDLRGYGDTDVPS 70
+G+++ + +G +GP V+ +HGFP+ W + L +R V D RG G + VP
Sbjct: 15 DGVRIAVYHEGNPDGPTVVLVHGFPDSHVLWDGVVPLLAER-FRIVRYDNRGVGRSSVPK 73
Query: 71 SISSYTCFHVVGDIVSLI-DLLGVEQVFLVGHDMGAIIGWYLCMFRP---ERIKAYVCLS 126
IS+YT H D ++I +L E V ++ HD G++ W + RP +R+ ++ +S
Sbjct: 74 PISAYTMAHFADDFDAVIGELSPGEPVHVLAHDWGSVGVWEY-LRRPGASDRVASFTSVS 132
Query: 127 VP 128
P
Sbjct: 133 GP 134
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.142 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,148,798
Number of Sequences: 164201
Number of extensions: 2027172
Number of successful extensions: 3754
Number of sequences better than 10.0: 100
Number of HSP's better than 10.0 without gapping: 45
Number of HSP's successfully gapped in prelim test: 55
Number of HSP's that attempted gapping in prelim test: 3660
Number of HSP's gapped (non-prelim): 101
length of query: 319
length of database: 59,974,054
effective HSP length: 110
effective length of query: 209
effective length of database: 41,911,944
effective search space: 8759596296
effective search space used: 8759596296
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC144513.17


