

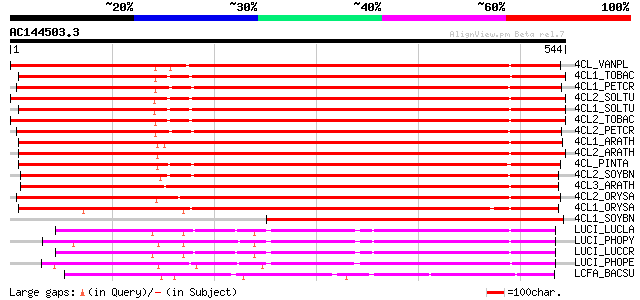
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144503.3 + phase: 0
(544 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
4CL_VANPL (O24540) 4-coumarate--CoA ligase (EC 6.2.1.12) (4CL) (... 832 0.0
4CL1_TOBAC (O24145) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL... 829 0.0
4CL1_PETCR (P14912) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL... 828 0.0
4CL2_SOLTU (P31685) 4-coumarate--CoA ligase 2 (EC 6.2.1.12) (4CL... 828 0.0
4CL1_SOLTU (P31684) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL... 828 0.0
4CL2_TOBAC (O24146) 4-coumarate--CoA ligase 2 (EC 6.2.1.12) (4CL... 827 0.0
4CL2_PETCR (P14913) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL... 826 0.0
4CL1_ARATH (Q42524) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL... 785 0.0
4CL2_ARATH (Q9S725) 4-coumarate--CoA ligase 2 (EC 6.2.1.12) (4CL... 770 0.0
4CL_PINTA (P41636) 4-coumarate--CoA ligase (EC 6.2.1.12) (4CL) (... 711 0.0
4CL2_SOYBN (P31687) 4-coumarate--CoA ligase 2 (EC 6.2.1.12) (4CL... 694 0.0
4CL3_ARATH (Q9S777) 4-coumarate--CoA ligase 3 (EC 6.2.1.12) (4CL... 683 0.0
4CL2_ORYSA (Q42982) 4-coumarate--CoA ligase 2 (EC 6.2.1.12) (4CL... 630 e-180
4CL1_ORYSA (P17814) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL... 617 e-176
4CL1_SOYBN (P31686) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL... 502 e-141
LUCI_LUCLA (Q01158) Luciferin 4-monooxygenase (EC 1.13.12.7) (Lu... 284 5e-76
LUCI_PHOPY (P08659) Luciferin 4-monooxygenase (EC 1.13.12.7) (Lu... 283 1e-75
LUCI_LUCCR (P13129) Luciferin 4-monooxygenase (EC 1.13.12.7) (Lu... 282 1e-75
LUCI_PHOPE (Q27757) Luciferin 4-monooxygenase (EC 1.13.12.7) (Lu... 274 4e-73
LCFA_BACSU (P94547) Long-chain-fatty-acid--CoA ligase (EC 6.2.1.... 244 3e-64
>4CL_VANPL (O24540) 4-coumarate--CoA ligase (EC 6.2.1.12) (4CL)
(4-coumaroyl-CoA synthase)
Length = 553
Score = 832 bits (2149), Expect = 0.0
Identities = 414/546 (75%), Positives = 472/546 (85%), Gaps = 9/546 (1%)
Query: 1 MTIQTMEQELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQL 60
+ I+ ++++IF+SKLPDIYIPK+LPLHSYCFEN+SK+ SRPCLIN T EI+TY DV+L
Sbjct: 5 VAIEEQKKDIIFRSKLPDIYIPKNLPLHSYCFENISKFSSRPCLINGATDEIFTYADVEL 64
Query: 61 TAQKVASGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQA 120
+++V SGL+KLGI+QGD IM+LLPN PEFVFAFLGASF G+I T ANPFFTS E+ KQA
Sbjct: 65 ISRRVGSGLSKLGIKQGDTIMILLPNSPEFVFAFLGASFIGSISTMANPFFTSTEVIKQA 124
Query: 121 KASNTKLLVTQACYYDKVKDL---ENVKLVFVDSSPEED---NHMHFSELIQADQNEMEE 174
KASN KL++TQ CY DKVKD VK++ +D++ D N +HFSEL AD+NEM
Sbjct: 125 KASNAKLIITQGCYVDKVKDYACENGVKIISIDTTTTADDAANILHFSELTGADENEMP- 183
Query: 175 VKVNIKPDDVVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVL 234
KV I PD VVALPYSSGTTGLPKGVMLTHKGLVTS+AQQVDGENPNLY HS+DV+LCVL
Sbjct: 184 -KVEISPDGVVALPYSSGTTGLPKGVMLTHKGLVTSVAQQVDGENPNLYMHSDDVLLCVL 242
Query: 235 PMFHIYSLNSVLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPE 294
P+FHIYSLNSVLLCGLRA + IL+M KF+I F L+ KYKVTI P VPPIVLAIAKS
Sbjct: 243 PLFHIYSLNSVLLCGLRAGSGILIMQKFEIVPFLELIQKYKVTIGPFVPPIVLAIAKSTV 302
Query: 295 LDKYDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPI 354
+D YDLSS+R + SG APLGKELED VRAKFP AKLGQGYGMTEAGPVL MCL+FAKEP
Sbjct: 303 VDNYDLSSVRTVMSGAAPLGKELEDAVRAKFPNAKLGQGYGMTEAGPVLAMCLAFAKEPF 362
Query: 355 DVKSGACGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKE 414
D+KSGACGTVVRNAEMKIVDP+ SSLPRN PGEICIRGDQIMKGYLN+PEAT TIDKE
Sbjct: 363 DIKSGACGTVVRNAEMKIVDPETGSSLPRNHPGEICIRGDQIMKGYLNDPEATARTIDKE 422
Query: 415 GWLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDE 474
GWLHTGDIG+IDDDDELFIVDRLKELIKYKGFQVAPAELEA++L+HP ISD AVVPM DE
Sbjct: 423 GWLHTGDIGYIDDDDELFIVDRLKELIKYKGFQVAPAELEALLLTHPCISDAAVVPMKDE 482
Query: 475 AAGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDL 534
AAGEVPVAFVV+SNG + TED+IK+F+SKQV+FYKRINRVFF++AIPK+PSGKILRKDL
Sbjct: 483 AAGEVPVAFVVKSNGH-NITEDEIKQFISKQVIFYKRINRVFFVEAIPKAPSGKILRKDL 541
Query: 535 RAKLAA 540
RA+LAA
Sbjct: 542 RARLAA 547
>4CL1_TOBAC (O24145) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL 1)
(4-coumaroyl-CoA synthase 1)
Length = 547
Score = 829 bits (2142), Expect = 0.0
Identities = 408/539 (75%), Positives = 467/539 (85%), Gaps = 8/539 (1%)
Query: 9 ELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVASG 68
+LIF+SKLPDIYIPKHLPLHSYCFEN+S++ SRPCLIN +IYTY +V+LT +KVA G
Sbjct: 14 DLIFRSKLPDIYIPKHLPLHSYCFENISEFSSRPCLINGANDQIYTYAEVELTCRKVAVG 73
Query: 69 LNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLL 128
LNKLGIQQ D IM+LLPN PEFVFAF+GAS+ GAI T ANP FT AE+ KQAKAS+ K++
Sbjct: 74 LNKLGIQQKDTIMILLPNSPEFVFAFMGASYLGAISTMANPLFTPAEVVKQAKASSAKII 133
Query: 129 VTQACYYDKVKDL---ENVKLVFVDSSPEEDNHMHFSELIQADQNEMEEVKVNIKPDDVV 185
+TQ+C+ KVKD +VK++ +DS+PE +HFSEL Q+D++E+ EVK I+PDDVV
Sbjct: 134 ITQSCFVGKVKDYASENDVKVICIDSAPE--GCLHFSELTQSDEHEIPEVK--IQPDDVV 189
Query: 186 ALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLNSV 245
ALPYSSGTTGLPKGVMLTHKGLVTS+AQQVDGEN NLY HSEDV++CVLP+FHIYSLNS+
Sbjct: 190 ALPYSSGTTGLPKGVMLTHKGLVTSVAQQVDGENANLYMHSEDVLMCVLPLFHIYSLNSI 249
Query: 246 LLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIRV 305
LLCGLR A+IL+M KFDI F L+ KYKV+I P VPPIVLAIAKSP +D YDLSS+R
Sbjct: 250 LLCGLRVGAAILIMQKFDIAPFLELIQKYKVSIGPFVPPIVLAIAKSPIVDSYDLSSVRT 309
Query: 306 LKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTVV 365
+ SG APLGKELED VR KFP AKLGQGYGMTEAGPVL MCL+FAKEP D+KSGACGTVV
Sbjct: 310 VMSGAAPLGKELEDAVRTKFPNAKLGQGYGMTEAGPVLAMCLAFAKEPFDIKSGACGTVV 369
Query: 366 RNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFI 425
RNAEMKIVDP SLPRNQPGEICIRGDQIMKGYLN+PEAT TIDKEGWLHTGDIGFI
Sbjct: 370 RNAEMKIVDPDTGCSLPRNQPGEICIRGDQIMKGYLNDPEATTRTIDKEGWLHTGDIGFI 429
Query: 426 DDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVV 485
D+DDELFIVDRLKELIKYKGFQVAPAE+EA++L+HP ISD AVVPM DE AGEVPVAFVV
Sbjct: 430 DEDDELFIVDRLKELIKYKGFQVAPAEIEALLLNHPNISDAAVVPMKDEQAGEVPVAFVV 489
Query: 486 RSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKLAAGVPN 544
RSNGS TED++K F+SKQV+FYKR+ RVFF++ +PKSPSGKILRKDLRA+LAAGVPN
Sbjct: 490 RSNGSA-ITEDEVKDFISKQVIFYKRVKRVFFVETVPKSPSGKILRKDLRARLAAGVPN 547
>4CL1_PETCR (P14912) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL 1)
(4-coumaroyl-CoA synthase 1)
Length = 544
Score = 828 bits (2139), Expect = 0.0
Identities = 406/538 (75%), Positives = 474/538 (87%), Gaps = 8/538 (1%)
Query: 7 EQELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVA 66
+++LIF+SKLPDIYIPKHLPLH+YCFEN+SK G + CLIN T E +TY V+L ++KVA
Sbjct: 8 KEDLIFRSKLPDIYIPKHLPLHTYCFENISKVGDKSCLINGATGETFTYSQVELLSRKVA 67
Query: 67 SGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTK 126
SGLNKLGIQQGD IM+LLPN PE+ FAFLGAS+RGAI T ANPFFTSAE+ KQ KAS K
Sbjct: 68 SGLNKLGIQQGDTIMLLLPNSPEYFFAFLGASYRGAISTMANPFFTSAEVIKQLKASQAK 127
Query: 127 LLVTQACYYDKVKDL---ENVKLVFVDSSPEEDNHMHFSELIQADQNEMEEVKVNIKPDD 183
L++TQACY DKVKD +N++++ +D +P++ +HFS+L++AD++EM EV +N DD
Sbjct: 128 LIITQACYVDKVKDYAAEKNIQIICIDDAPQDC--LHFSKLMEADESEMPEVVIN--SDD 183
Query: 184 VVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLN 243
VVALPYSSGTTGLPKGVMLTHKGLVTS+AQQVDG+NPNLY HSEDV++C+LP+FHIYSLN
Sbjct: 184 VVALPYSSGTTGLPKGVMLTHKGLVTSVAQQVDGDNPNLYMHSEDVMICILPLFHIYSLN 243
Query: 244 SVLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSI 303
+VL CGLRA +IL+M KFDI F L+ KYKVTI P VPPIVLAIAKSP +DKYDLSS+
Sbjct: 244 AVLCCGLRAGVTILIMQKFDIVPFLELIQKYKVTIGPFVPPIVLAIAKSPVVDKYDLSSV 303
Query: 304 RVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGT 363
R + SG APLGKELED VRAKFP AKLGQGYGMTEAGPVL MCL+FAKEP ++KSGACGT
Sbjct: 304 RTVMSGAAPLGKELEDAVRAKFPNAKLGQGYGMTEAGPVLAMCLAFAKEPYEIKSGACGT 363
Query: 364 VVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIG 423
VVRNAEMKIVDP+ ++SLPRNQ GEICIRGDQIMKGYLN+PE+TR TID+EGWLHTGDIG
Sbjct: 364 VVRNAEMKIVDPETNASLPRNQRGEICIRGDQIMKGYLNDPESTRTTIDEEGWLHTGDIG 423
Query: 424 FIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAF 483
FIDDDDELFIVDRLKE+IKYKGFQVAPAELEA++L+HP ISD AVVPM+DE AGEVPVAF
Sbjct: 424 FIDDDDELFIVDRLKEIIKYKGFQVAPAELEALLLTHPTISDAAVVPMIDEKAGEVPVAF 483
Query: 484 VVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKLAAG 541
VVR+NG TTE++IK+FVSKQVVFYKRI RVFF+DAIPKSPSGKILRKDLRA++A+G
Sbjct: 484 VVRTNG-FTTTEEEIKQFVSKQVVFYKRIFRVFFVDAIPKSPSGKILRKDLRARIASG 540
>4CL2_SOLTU (P31685) 4-coumarate--CoA ligase 2 (EC 6.2.1.12) (4CL 2)
(4-coumaroyl-CoA synthase 2)
Length = 545
Score = 828 bits (2138), Expect = 0.0
Identities = 413/548 (75%), Positives = 475/548 (86%), Gaps = 9/548 (1%)
Query: 1 MTIQTMEQ-ELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQ 59
M I+T + +LIF+SKLPDIYIPKHLPLHSYCFENLS++ SRPCLI+ IYTY +V+
Sbjct: 3 MDIETKQSGDLIFRSKLPDIYIPKHLPLHSYCFENLSEFNSRPCLIDGANDRIYTYAEVE 62
Query: 60 LTAQKVASGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQ 119
LT++KVA GLNKLGIQQ D IM+LLPNCPEFVFAF+GAS+ GAI T ANP FT AE+ KQ
Sbjct: 63 LTSRKVAVGLNKLGIQQKDTIMILLPNCPEFVFAFIGASYLGAISTMANPLFTPAEVVKQ 122
Query: 120 AKASNTKLLVTQACYYDKVKD--LEN-VKLVFVDSSPEEDNHMHFSELIQADQNEMEEVK 176
AKAS+ K+++TQAC+ KVKD +EN +K++ VDS+PE +HFSELIQ+D++E+ +VK
Sbjct: 123 AKASSAKIVITQACFAGKVKDYAIENDLKVICVDSAPE--GCVHFSELIQSDEHEIPDVK 180
Query: 177 VNIKPDDVVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPM 236
I+PDDVVALPYSSGTTGLPKGVMLTHKGLVTS+AQQVDGEN NLY HS+DV++CVLP+
Sbjct: 181 --IQPDDVVALPYSSGTTGLPKGVMLTHKGLVTSVAQQVDGENANLYMHSDDVLMCVLPL 238
Query: 237 FHIYSLNSVLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELD 296
FHIYSLNSVLLC LR A+IL+M KFDI F L+ K+KVTI P VPPIVLAIAKSP +
Sbjct: 239 FHIYSLNSVLLCALRVGAAILIMQKFDIAQFLELIPKHKVTIGPFVPPIVLAIAKSPLVH 298
Query: 297 KYDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDV 356
YDLSS+R + SG APLGKELED VRAKFP AKLGQGYGMTEAGPVL MCL+FAKEP D+
Sbjct: 299 NYDLSSVRTVMSGAAPLGKELEDAVRAKFPNAKLGQGYGMTEAGPVLAMCLAFAKEPFDI 358
Query: 357 KSGACGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGW 416
KSGACGTVVRNAEMKIVDP SLPRNQPGEICIRGDQIMKGYLN+PEAT TI+KEGW
Sbjct: 359 KSGACGTVVRNAEMKIVDPDTGCSLPRNQPGEICIRGDQIMKGYLNDPEATARTIEKEGW 418
Query: 417 LHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAA 476
LHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEA++++HP ISD AVVPM+DE A
Sbjct: 419 LHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEALLINHPDISDAAVVPMIDEQA 478
Query: 477 GEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRA 536
GEVPVAFVVRSNGS TED++K F+SKQV+FYKRI RVFF++ +PKSPSGKILRKDLRA
Sbjct: 479 GEVPVAFVVRSNGS-TITEDEVKDFISKQVIFYKRIKRVFFVETVPKSPSGKILRKDLRA 537
Query: 537 KLAAGVPN 544
+LAAG+ N
Sbjct: 538 RLAAGISN 545
>4CL1_SOLTU (P31684) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL 1)
(4-coumaroyl-CoA synthase 1)
Length = 545
Score = 828 bits (2138), Expect = 0.0
Identities = 411/539 (76%), Positives = 470/539 (86%), Gaps = 8/539 (1%)
Query: 9 ELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVASG 68
+LIF+SKLPDIYIPKHLPLHSYCFENLS++ SRPCLI+ IYTY +V+LT++KVA G
Sbjct: 12 DLIFRSKLPDIYIPKHLPLHSYCFENLSEFNSRPCLIDGANDRIYTYAEVELTSRKVAVG 71
Query: 69 LNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLL 128
LNKLGIQQ D IM+LLPNCPEFVFAF+GAS+ GAI T ANP FT AE+ KQAKAS+ K++
Sbjct: 72 LNKLGIQQKDTIMILLPNCPEFVFAFIGASYLGAISTMANPLFTPAEVVKQAKASSAKIV 131
Query: 129 VTQACYYDKVKD--LEN-VKLVFVDSSPEEDNHMHFSELIQADQNEMEEVKVNIKPDDVV 185
+TQAC+ KVKD +EN +K++ VDS PE +HFSELIQ+D++E+ +VK I+PDDVV
Sbjct: 132 ITQACFAGKVKDYAIENDLKVICVDSVPE--GCVHFSELIQSDEHEIPDVK--IQPDDVV 187
Query: 186 ALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLNSV 245
ALPYSSGTTGLPKGVMLTHKGLVTS+AQQVDGEN NLY HS+DV++CVLP+FHIYSLNSV
Sbjct: 188 ALPYSSGTTGLPKGVMLTHKGLVTSVAQQVDGENANLYMHSDDVLMCVLPLFHIYSLNSV 247
Query: 246 LLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIRV 305
LLC LR A+IL+M KFDI F L+ K+KVTI P VPPIVLAIAKSP +D YDLSS+R
Sbjct: 248 LLCALRVGAAILIMQKFDIAQFLELIPKHKVTIGPFVPPIVLAIAKSPLVDNYDLSSVRT 307
Query: 306 LKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTVV 365
+ SG APLGKELED VRAKFP AKLGQGYGMTEAGPVL MCL+FAKEP D+KSGACGTVV
Sbjct: 308 VMSGAAPLGKELEDAVRAKFPNAKLGQGYGMTEAGPVLAMCLAFAKEPFDIKSGACGTVV 367
Query: 366 RNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFI 425
RNAEMKIVDP SLPRNQPGEICIRGDQIMKGYLN+PEAT TI+KEGWLHTGDIGFI
Sbjct: 368 RNAEMKIVDPDTGCSLPRNQPGEICIRGDQIMKGYLNDPEATARTIEKEGWLHTGDIGFI 427
Query: 426 DDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVV 485
DDDDELFIVDRLKELIKYKGFQVAPAELEA++++HP ISD AVVPM+DE AGEVPVAFVV
Sbjct: 428 DDDDELFIVDRLKELIKYKGFQVAPAELEALLINHPDISDAAVVPMIDEQAGEVPVAFVV 487
Query: 486 RSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKLAAGVPN 544
RSNGS TED++K F+SKQV+FYKRI RVFF++ +PKSPSGKILRKDLRA+LAAG+ N
Sbjct: 488 RSNGS-TITEDEVKDFISKQVIFYKRIKRVFFVETVPKSPSGKILRKDLRARLAAGISN 545
>4CL2_TOBAC (O24146) 4-coumarate--CoA ligase 2 (EC 6.2.1.12) (4CL 2)
(4-coumaroyl-CoA synthase 2)
Length = 542
Score = 827 bits (2136), Expect = 0.0
Identities = 411/547 (75%), Positives = 474/547 (86%), Gaps = 8/547 (1%)
Query: 1 MTIQTMEQELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQL 60
M T + ++IF+SKLPDIYIP HLPLHSYCFEN+S++ SRPCLIN +IYTY DV+L
Sbjct: 1 MEKDTKQVDIIFRSKLPDIYIPNHLPLHSYCFENISEFSSRPCLINGANKQIYTYADVEL 60
Query: 61 TAQKVASGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQA 120
++KVA+GL+K GIQ D IM+LLPN PEFVFAF+GAS+ GAI T ANP FT AE+ KQA
Sbjct: 61 NSRKVAAGLHKQGIQPKDTIMILLPNSPEFVFAFIGASYLGAISTMANPLFTPAEVVKQA 120
Query: 121 KASNTKLLVTQACYYDKVKDL---ENVKLVFVDSSPEEDNHMHFSELIQADQNEMEEVKV 177
KAS+ K++VTQAC+ +KVKD +VK++ +DS+PE +HFS L QA+++++ EV+
Sbjct: 121 KASSAKIIVTQACHVNKVKDYAFENDVKIICIDSAPE--GCLHFSVLTQANEHDIPEVE- 177
Query: 178 NIKPDDVVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMF 237
I+PDDVVALPYSSGTTGLPKGVMLTHKGLVTS+AQQVDGENPNLY HSEDV+LCVLP+F
Sbjct: 178 -IQPDDVVALPYSSGTTGLPKGVMLTHKGLVTSVAQQVDGENPNLYIHSEDVMLCVLPLF 236
Query: 238 HIYSLNSVLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDK 297
HIYSLNSVLLCGLR A+IL+M KFDI +F L+ +YKVTI P VPPIVLAIAKSP +D
Sbjct: 237 HIYSLNSVLLCGLRVGAAILIMQKFDIVSFLELIQRYKVTIGPFVPPIVLAIAKSPMVDD 296
Query: 298 YDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVK 357
YDLSS+R + SG APLGKELEDTVRAKFP AKLGQGYGMTEAGPVL MCL+FAKEP ++K
Sbjct: 297 YDLSSVRTVMSGAAPLGKELEDTVRAKFPNAKLGQGYGMTEAGPVLAMCLAFAKEPFEIK 356
Query: 358 SGACGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWL 417
SGACGTVVRNAEMKIVDP+ +SLPRNQ GEICIRGDQIMKGYLN+PEAT TIDKEGWL
Sbjct: 357 SGACGTVVRNAEMKIVDPKTGNSLPRNQSGEICIRGDQIMKGYLNDPEATARTIDKEGWL 416
Query: 418 HTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAG 477
+TGDIG+IDDDDELFIVDRLKELIKYKGFQVAPAELEA++L+HP ISD AVVPM DE AG
Sbjct: 417 YTGDIGYIDDDDELFIVDRLKELIKYKGFQVAPAELEALLLNHPNISDAAVVPMKDEQAG 476
Query: 478 EVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAK 537
EVPVAFVVRSNGS TED++K F+SKQV+FYKRI RVFF+DAIPKSPSGKILRKDLRAK
Sbjct: 477 EVPVAFVVRSNGS-TITEDEVKDFISKQVIFYKRIKRVFFVDAIPKSPSGKILRKDLRAK 535
Query: 538 LAAGVPN 544
LAAG+PN
Sbjct: 536 LAAGLPN 542
>4CL2_PETCR (P14913) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL 1)
(4-coumaroyl-CoA synthase 1)
Length = 544
Score = 826 bits (2134), Expect = 0.0
Identities = 406/538 (75%), Positives = 474/538 (87%), Gaps = 8/538 (1%)
Query: 7 EQELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVA 66
+++LIF+SKLPDIYIPKHLPLH+YCFEN+SK G + CLIN T E +TY V+L ++KVA
Sbjct: 8 KEDLIFRSKLPDIYIPKHLPLHTYCFENISKVGDKSCLINGATGETFTYSQVELLSRKVA 67
Query: 67 SGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTK 126
SGLNKLGIQQGD IM+LLPN PE+ FAFLGAS+RGAI T ANPFFTSAE+ KQ KAS K
Sbjct: 68 SGLNKLGIQQGDTIMLLLPNSPEYFFAFLGASYRGAISTMANPFFTSAEVIKQLKASLAK 127
Query: 127 LLVTQACYYDKVKDL---ENVKLVFVDSSPEEDNHMHFSELIQADQNEMEEVKVNIKPDD 183
L++TQACY DKVKD +N++++ +D +P++ +HFS+L++AD++EM EV ++ DD
Sbjct: 128 LIITQACYVDKVKDYAAEKNIQIICIDDAPQDC--LHFSKLMEADESEMPEVVID--SDD 183
Query: 184 VVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLN 243
VVALPYSSGTTGLPKGVMLTHKGLVTS+AQQVDG+NPNLY HSEDV++C+LP+FHIYSLN
Sbjct: 184 VVALPYSSGTTGLPKGVMLTHKGLVTSVAQQVDGDNPNLYMHSEDVMICILPLFHIYSLN 243
Query: 244 SVLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSI 303
+VL CGLRA +IL+M KFDI F L+ KYKVTI P VPPIVLAIAKSP +DKYDLSS+
Sbjct: 244 AVLCCGLRAGVTILIMQKFDIVPFLELIQKYKVTIGPFVPPIVLAIAKSPVVDKYDLSSV 303
Query: 304 RVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGT 363
R + SG APLGKELED VRAKFP AKLGQGYGMTEAGPVL MCL+FAKEP ++KSGACGT
Sbjct: 304 RTVMSGAAPLGKELEDAVRAKFPNAKLGQGYGMTEAGPVLAMCLAFAKEPYEIKSGACGT 363
Query: 364 VVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIG 423
VVRNAEMKIVDP+ ++SLPRNQ GEICIRGDQIMKGYLN+PE+TR TID+EGWLHTGDIG
Sbjct: 364 VVRNAEMKIVDPETNASLPRNQRGEICIRGDQIMKGYLNDPESTRTTIDEEGWLHTGDIG 423
Query: 424 FIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAF 483
FIDDDDELFIVDRLKE+IKYKGFQVAPAELEA++L+HP ISD AVVPM+DE AGEVPVAF
Sbjct: 424 FIDDDDELFIVDRLKEIIKYKGFQVAPAELEALLLTHPTISDAAVVPMIDEKAGEVPVAF 483
Query: 484 VVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKLAAG 541
VVR+NG TTE++IK+FVSKQVVFYKRI RVFF+DAIPKSPSGKILRKDLRAK+A+G
Sbjct: 484 VVRTNG-FTTTEEEIKQFVSKQVVFYKRIFRVFFVDAIPKSPSGKILRKDLRAKIASG 540
>4CL1_ARATH (Q42524) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL 1)
(4-coumaroyl-CoA synthase 1)
Length = 561
Score = 785 bits (2028), Expect = 0.0
Identities = 391/541 (72%), Positives = 455/541 (83%), Gaps = 8/541 (1%)
Query: 9 ELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVASG 68
++IF+SKLPDIYIP HL LH Y F+N+S++ ++PCLIN PT +YTY DV + ++++A+
Sbjct: 22 DVIFRSKLPDIYIPNHLSLHDYIFQNISEFATKPCLINGPTGHVYTYSDVHVISRQIAAN 81
Query: 69 LNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLL 128
+KLG+ Q DV+M+LLPNCPEFV +FL ASFRGA TAANPFFT AEIAKQAKASNTKL+
Sbjct: 82 FHKLGVNQNDVVMLLLPNCPEFVLSFLAASFRGATATAANPFFTPAEIAKQAKASNTKLI 141
Query: 129 VTQACYYDKVKDLEN---VKLVFVD---SSPEEDNHMHFSELIQADQNEMEEV-KVNIKP 181
+T+A Y DK+K L+N V +V +D S P + + F+EL Q+ E + V I P
Sbjct: 142 ITEARYVDKIKPLQNDDGVVIVCIDDNESVPIPEGCLRFTELTQSTTEASEVIDSVEISP 201
Query: 182 DDVVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYS 241
DDVVALPYSSGTTGLPKGVMLTHKGLVTS+AQQVDGENPNLY+HS+DVILCVLPMFHIY+
Sbjct: 202 DDVVALPYSSGTTGLPKGVMLTHKGLVTSVAQQVDGENPNLYFHSDDVILCVLPMFHIYA 261
Query: 242 LNSVLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLS 301
LNS++LCGLR A+IL+MPKF+IN L+ + KVT+AP+VPPIVLAIAKS E +KYDLS
Sbjct: 262 LNSIMLCGLRVGAAILIMPKFEINLLLELIQRCKVTVAPMVPPIVLAIAKSSETEKYDLS 321
Query: 302 SIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGAC 361
SIRV+KSG APLGKELED V AKFP AKLGQGYGMTEAGPVL M L FAKEP VKSGAC
Sbjct: 322 SIRVVKSGAAPLGKELEDAVNAKFPNAKLGQGYGMTEAGPVLAMSLGFAKEPFPVKSGAC 381
Query: 362 GTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGD 421
GTVVRNAEMKIVDP SL RNQPGEICIRG QIMKGYLNNP AT ETIDK+GWLHTGD
Sbjct: 382 GTVVRNAEMKIVDPDTGDSLSRNQPGEICIRGHQIMKGYLNNPAATAETIDKDGWLHTGD 441
Query: 422 IGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPV 481
IG IDDDDELFIVDRLKELIKYKGFQVAPAELEA+++ HP I+DVAVV M +EAAGEVPV
Sbjct: 442 IGLIDDDDELFIVDRLKELIKYKGFQVAPAELEALLIGHPDITDVAVVAMKEEAAGEVPV 501
Query: 482 AFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKLAAG 541
AFVV+S S + +EDD+K+FVSKQVVFYKRIN+VFF ++IPK+PSGKILRKDLRAKLA G
Sbjct: 502 AFVVKSKDS-ELSEDDVKQFVSKQVVFYKRINKVFFTESIPKAPSGKILRKDLRAKLANG 560
Query: 542 V 542
+
Sbjct: 561 L 561
>4CL2_ARATH (Q9S725) 4-coumarate--CoA ligase 2 (EC 6.2.1.12) (4CL 2)
(4-coumaroyl-CoA synthase 2)
Length = 556
Score = 770 bits (1988), Expect = 0.0
Identities = 377/538 (70%), Positives = 455/538 (84%), Gaps = 3/538 (0%)
Query: 9 ELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVASG 68
++IF+S+LPDIYIP HLPLH Y FEN+S++ ++PCLIN PT E+YTY DV +T++K+A+G
Sbjct: 20 DVIFRSRLPDIYIPNHLPLHDYIFENISEFAAKPCLINGPTGEVYTYADVHVTSRKLAAG 79
Query: 69 LNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLL 128
L+ LG++Q DV+M+LLPN PE V FL ASF GAI T+ANPFFT AEI+KQAKAS KL+
Sbjct: 80 LHNLGVKQHDVVMILLPNSPEVVLTFLAASFIGAITTSANPFFTPAEISKQAKASAAKLI 139
Query: 129 VTQACYYDKVKDLEN--VKLVFVDSSPEEDNHMHFSELIQADQNEMEEVKVNIKPDDVVA 186
VTQ+ Y DK+K+L+N V +V DS +N + FSEL Q+++ ++ + I P+DVVA
Sbjct: 140 VTQSRYVDKIKNLQNDGVLIVTTDSDAIPENCLRFSELTQSEEPRVDSIPEKISPEDVVA 199
Query: 187 LPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLNSVL 246
LP+SSGTTGLPKGVMLTHKGLVTS+AQQVDGENPNLY++ +DVILCV PMFHIY+LNS++
Sbjct: 200 LPFSSGTTGLPKGVMLTHKGLVTSVAQQVDGENPNLYFNRDDVILCVWPMFHIYALNSIM 259
Query: 247 LCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIRVL 306
LC LR A+IL+MPKF+I + + KVT+A VVPPIVLAIAKSPE +KYDLSS+R++
Sbjct: 260 LCSLRIGATILIMPKFEITLLLEQIQRCKVTVAMVVPPIVLAIAKSPETEKYDLSSVRMV 319
Query: 307 KSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTVVR 366
KSG APLGKELED + AKFP AKLGQGYGMTEAGPVL M L FAKEP VKSGACGTVVR
Sbjct: 320 KSGAAPLGKELEDAISAKFPNAKLGQGYGMTEAGPVLAMSLGFAKEPFPVKSGACGTVVR 379
Query: 367 NAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFID 426
NAEMKI+DP SLPRN+PGEICIRG+QIMKGYLN+P AT TIDK+GWLHTGD+GFID
Sbjct: 380 NAEMKILDPDTGDSLPRNKPGEICIRGNQIMKGYLNDPLATASTIDKDGWLHTGDVGFID 439
Query: 427 DDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVR 486
DDDELFIVDRLKELIKYKGFQVAPAELE++++ HP+I+DVAVV M +E AGEVPVAFVVR
Sbjct: 440 DDDELFIVDRLKELIKYKGFQVAPAELESLLIGHPEINDVAVVAMKEEDAGEVPVAFVVR 499
Query: 487 SNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKLAAGVPN 544
S S + +ED+IK+FVSKQVVFYKRIN+VFF D+IPK+PSGKILRKDLRA+LA G+ N
Sbjct: 500 SKDS-NISEDEIKQFVSKQVVFYKRINKVFFTDSIPKAPSGKILRKDLRARLANGLMN 556
>4CL_PINTA (P41636) 4-coumarate--CoA ligase (EC 6.2.1.12) (4CL)
(4-coumaroyl-CoA synthase)
Length = 537
Score = 711 bits (1834), Expect = 0.0
Identities = 345/534 (64%), Positives = 439/534 (81%), Gaps = 8/534 (1%)
Query: 9 ELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVASG 68
E +++SKLPDI I HLPLHSYCFE ++++ RPCLI+ T Y + +V+L ++KVA+G
Sbjct: 9 EHLYRSKLPDIEISDHLPLHSYCFERVAEFADRPCLIDGATDRTYCFSEVELISRKVAAG 68
Query: 69 LNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLL 128
L KLG+QQG V+M+LLPNC EF F F+GAS RGAI+T ANPF+ EIAKQAKA+ +++
Sbjct: 69 LAKLGLQQGQVVMLLLPNCIEFAFVFMGASVRGAIVTTANPFYKPGEIAKQAKAAGARII 128
Query: 129 VTQACYYDKVKDLEN--VKLVFVDSSPEEDNHMHFSELIQADQNEMEEVKVNIKPDDVVA 186
VT A Y +K+ DL++ V ++ +D +P+E H S L +AD+ + VK++ PDDVVA
Sbjct: 129 VTLAAYVEKLADLQSHDVLVITIDDAPKE-GCQHISVLTEADETQCPAVKIH--PDDVVA 185
Query: 187 LPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLNSVL 246
LPYSSGTTGLPKGVMLTHKGLV+S+AQQVDGENPNLY+HS+DVILCVLP+FHIYSLNSVL
Sbjct: 186 LPYSSGTTGLPKGVMLTHKGLVSSVAQQVDGENPNLYFHSDDVILCVLPLFHIYSLNSVL 245
Query: 247 LCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIRVL 306
LC LRA A+ L+M KF++ L+ KYKVT+AP+VPPIVL I KSP + +YD+SS+R++
Sbjct: 246 LCALRAGAATLIMQKFNLTTCLELIQKYKVTVAPIVPPIVLDITKSPIVSQYDVSSVRII 305
Query: 307 KSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTVVR 366
SG APLGKELED +R +FPKA GQGYGMTEAGPVL M L+FAK P VKSG+CGTVVR
Sbjct: 306 MSGAAPLGKELEDALRERFPKAIFGQGYGMTEAGPVLAMNLAFAKNPFPVKSGSCGTVVR 365
Query: 367 NAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFID 426
NA++KI+D + SLP NQ GEICIRG +IMKGY+N+PE+T TID+EGWLHTGD+ +ID
Sbjct: 366 NAQIKILDTETGESLPHNQAGEICIRGPEIMKGYINDPESTAATIDEEGWLHTGDVEYID 425
Query: 427 DDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVR 486
DD+E+FIVDR+KE+IKYKGFQVAPAELEA++++HP I+D AVVP E AGEVPVAFVV+
Sbjct: 426 DDEEIFIVDRVKEIIKYKGFQVAPAELEALLVAHPSIADAAVVPQKHEEAGEVPVAFVVK 485
Query: 487 SNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKLAA 540
S+ + +E +IK+FV+KQV+FYK+I+RV+F+DAIPKSPSGKILRKDLR++LAA
Sbjct: 486 SS---EISEQEIKEFVAKQVIFYKKIHRVYFVDAIPKSPSGKILRKDLRSRLAA 536
>4CL2_SOYBN (P31687) 4-coumarate--CoA ligase 2 (EC 6.2.1.12) (4CL 2)
(4-coumaroyl-CoA synthase 2) (Clone 4CL16)
Length = 562
Score = 694 bits (1790), Expect = 0.0
Identities = 339/534 (63%), Positives = 429/534 (79%), Gaps = 11/534 (2%)
Query: 11 IFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVASGLN 70
+FKSKLPDI I HLPLHSYCF+NLS++ RPCLI P ++ +TY D L + K+A+GL+
Sbjct: 26 VFKSKLPDIPISNHLPLHSYCFQNLSQFAHRPCLIVGPASKTFTYADTHLISSKIAAGLS 85
Query: 71 KLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLLVT 130
LGI +GDV+M+LL N +FVF+FL S GA+ T ANPF+T+ EI KQ S KL++T
Sbjct: 86 NLGILKGDVVMILLQNSADFVFSFLAISMIGAVATTANPFYTAPEIFKQFTVSKAKLIIT 145
Query: 131 QACYYDKVKDLENVKL------VFVDSSPEEDNHMHFSELIQADQNEMEEVKVNIKPDDV 184
QA Y DK+++ + KL V VD PE N +HFS L +A+++++ EV+++ PDD
Sbjct: 146 QAMYVDKLRNHDGAKLGEDFKVVTVDDPPE--NCLHFSVLSEANESDVPEVEIH--PDDA 201
Query: 185 VALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLNS 244
VA+P+SSGTTGLPKGV+LTHK L TS+AQQVDGENPNLY +EDV+LCVLP+FHI+SLNS
Sbjct: 202 VAMPFSSGTTGLPKGVILTHKSLTTSVAQQVDGENPNLYLTTEDVLLCVLPLFHIFSLNS 261
Query: 245 VLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIR 304
VLLC LRA +++LLM KF+I L+ +++V++A VVPP+VLA+AK+P + +DLSSIR
Sbjct: 262 VLLCALRAGSAVLLMQKFEIGTLLELIQRHRVSVAMVVPPLVLALAKNPMVADFDLSSIR 321
Query: 305 VLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTV 364
++ SG APLGKELE+ +R + P+A LGQGYGMTEAGPVL+MCL FAK+P KSG+CGTV
Sbjct: 322 LVLSGAAPLGKELEEALRNRMPQAVLGQGYGMTEAGPVLSMCLGFAKQPFQTKSGSCGTV 381
Query: 365 VRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGF 424
VRNAE+K+VDP+ SL NQPGEICIRG QIMKGYLN+ AT TID EGWLHTGD+G+
Sbjct: 382 VRNAELKVVDPETGRSLGYNQPGEICIRGQQIMKGYLNDEAATASTIDSEGWLHTGDVGY 441
Query: 425 IDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFV 484
+DDDDE+FIVDR+KELIKYKGFQV PAELE +++SHP I+D AVVP D AAGEVPVAFV
Sbjct: 442 VDDDDEIFIVDRVKELIKYKGFQVPPAELEGLLVSHPSIADAAVVPQKDVAAGEVPVAFV 501
Query: 485 VRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKL 538
VRSNG D TE+ +K+F++KQVVFYKR+++V+F+ AIPKSPSGKILRKDLRAKL
Sbjct: 502 VRSNG-FDLTEEAVKEFIAKQVVFYKRLHKVYFVHAIPKSPSGKILRKDLRAKL 554
>4CL3_ARATH (Q9S777) 4-coumarate--CoA ligase 3 (EC 6.2.1.12) (4CL 3)
(4-coumaroyl-CoA synthase 3)
Length = 561
Score = 683 bits (1762), Expect = 0.0
Identities = 339/529 (64%), Positives = 426/529 (80%), Gaps = 3/529 (0%)
Query: 11 IFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVASGLN 70
IF+SKLPDI IP HLPLH+YCFE LS +PCLI T + YTY + L ++VASGL
Sbjct: 34 IFRSKLPDIDIPNHLPLHTYCFEKLSSVSDKPCLIVGSTGKSYTYGETHLICRRVASGLY 93
Query: 71 KLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLLVT 130
KLGI++GDVIM+LL N EFVF+F+GAS GA+ T ANPF+TS E+ KQ K+S KL++T
Sbjct: 94 KLGIRKGDVIMILLQNSAEFVFSFMGASMIGAVSTTANPFYTSQELYKQLKSSGAKLIIT 153
Query: 131 QACYYDKVKDL-ENVKLVFVDSSPEEDNHMHFSELIQADQNEMEEVKVNIKPDDVVALPY 189
+ Y DK+K+L EN+ L+ D P +N + FS LI D+ + V+I DD ALP+
Sbjct: 154 HSQYVDKLKNLGENLTLITTDE-PTPENCLPFSTLITDDETNPFQETVDIGGDDAAALPF 212
Query: 190 SSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLNSVLLCG 249
SSGTTGLPKGV+LTHK L+TS+AQQVDG+NPNLY S DVILCVLP+FHIYSLNSVLL
Sbjct: 213 SSGTTGLPKGVVLTHKSLITSVAQQVDGDNPNLYLKSNDVILCVLPLFHIYSLNSVLLNS 272
Query: 250 LRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIRVLKSG 309
LR+ A++LLM KF+I A L+ +++VTIA +VPP+V+A+AK+P ++ YDLSS+R + SG
Sbjct: 273 LRSGATVLLMHKFEIGALLDLIQRHRVTIAALVPPLVIALAKNPTVNSYDLSSVRFVLSG 332
Query: 310 GAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTVVRNAE 369
APLGKEL+D++R + P+A LGQGYGMTEAGPVL+M L FAKEPI KSG+CGTVVRNAE
Sbjct: 333 AAPLGKELQDSLRRRLPQAILGQGYGMTEAGPVLSMSLGFAKEPIPTKSGSCGTVVRNAE 392
Query: 370 MKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDDD 429
+K+V + SL NQPGEICIRG QIMK YLN+PEAT TID+EGWLHTGDIG++D+DD
Sbjct: 393 LKVVHLETRLSLGYNQPGEICIRGQQIMKEYLNDPEATSATIDEEGWLHTGDIGYVDEDD 452
Query: 430 ELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNG 489
E+FIVDRLKE+IK+KGFQV PAELE+++++H I+D AVVP DE AGEVPVAFVVRSNG
Sbjct: 453 EIFIVDRLKEVIKFKGFQVPPAELESLLINHHSIADAAVVPQNDEVAGEVPVAFVVRSNG 512
Query: 490 SIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKL 538
+ D TE+D+K++V+KQVVFYKR+++VFF+ +IPKSPSGKILRKDL+AKL
Sbjct: 513 N-DITEEDVKEYVAKQVVFYKRLHKVFFVASIPKSPSGKILRKDLKAKL 560
>4CL2_ORYSA (Q42982) 4-coumarate--CoA ligase 2 (EC 6.2.1.12) (4CL 2)
(4-coumaroyl-CoA synthase 2)
Length = 569
Score = 630 bits (1626), Expect = e-180
Identities = 318/548 (58%), Positives = 408/548 (74%), Gaps = 16/548 (2%)
Query: 7 EQELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVA 66
E +F+SKLPDI IP HLPLH YCF ++ PCLI A T YT+ + +L ++ A
Sbjct: 23 EAVTVFRSKLPDIDIPSHLPLHEYCFARAAELPDAPCLIAAATGRTYTFAETRLLCRRAA 82
Query: 67 SGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTK 126
+ L++LG+ GD +MVLL NC EF AF ASF GA+ TAANPF T EI KQ K S K
Sbjct: 83 AALHRLGVGHGDRVMVLLQNCVEFAVAFFAASFLGAVTTAANPFCTPQEIHKQFKGSGVK 142
Query: 127 LLVTQACYYDKVKDLE-------------NVKLVFVDSSPEEDNH-MHFSELIQADQNEM 172
L++TQ+ Y DK++ E + ++ +D + F +LI AD +E
Sbjct: 143 LILTQSVYVDKLRQHEAFPRIDACTVGDDTLTVITIDDDEATPKACLPFWDLI-ADADEG 201
Query: 173 EEVKVNIKPDDVVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILC 232
+V I PDD VALP+SSGTTGL KGV+LTH+ +V+ +A +VDGENPNL+ + DV LC
Sbjct: 202 SVPEVAISPDDPVALPFSSGTTGLAKGVVLTHRSVVSGVAHEVDGENPNLHMGAGDVALC 261
Query: 233 VLPMFHIYSLNSVLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKS 292
VLP+FHI+SLNSVLL +R ++ LMP+F++ A G + +++VT+ VVPP+V+A+AK+
Sbjct: 262 VLPLFHIFSLNSVLLSRVRPAPAVALMPRFEMGAMLGAIERWRVTVGAVVPPLVVALAKN 321
Query: 293 PELDKYDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKE 352
P ++++DLSSIR++ SG APLGKELED +RA+ P+A GQGYGMTEAGPVL+MC +FAKE
Sbjct: 322 PFVERHDLSSIRIVLSGAAPLGKELEDALRARLPQAIFGQGYGMTEAGPVLSMCPAFAKE 381
Query: 353 PIDVKSGACGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETID 412
P KSG+CGTVVRNAE+K+VDP SL RN PGEICIRG QIMKGYLN+PEAT TID
Sbjct: 382 PTPAKSGSCGTVVRNAELKVVDPDTGFSLGRNLPGEICIRGPQIMKGYLNDPEATAATID 441
Query: 413 KEGWLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPML 472
EGWLHTG+IG++DDDDE+FIVDR+KELIK+KGFQV PAELE+++++HP I D AVVP
Sbjct: 442 VEGWLHTGNIGYVDDDDEVFIVDRVKELIKFKGFQVPPAELESLLIAHPSIRDAAVVPQK 501
Query: 473 DEAAGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRK 532
D+ AGEVPVAFVVR+ S D TE+ IK+F+SKQVVFYKR+++V FI AIPKS SGKILR+
Sbjct: 502 DDVAGEVPVAFVVRAADS-DITEESIKEFISKQVVFYKRLHKVHFIHAIPKSASGKILRR 560
Query: 533 DLRAKLAA 540
+LRAKLAA
Sbjct: 561 ELRAKLAA 568
>4CL1_ORYSA (P17814) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL 1)
(4-coumaroyl-CoA synthase 1)
Length = 563
Score = 617 bits (1590), Expect = e-176
Identities = 313/538 (58%), Positives = 402/538 (74%), Gaps = 13/538 (2%)
Query: 9 ELIFKSKLPDIYIPKHLPLHSYCFENLSKYGSRPCLINAPTAEIYTYYDVQLTAQKVASG 68
E+IF+SKL DI I LPLH YCFE L + +RPCLI+ T + TY DV ++++A+
Sbjct: 21 EIIFRSKLQDIAITNTLPLHRYCFERLPEVAARPCLIDGATGGVLTYADVDRLSRRLAAA 80
Query: 69 LNK--LGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTK 126
L + LG+++G V+M LL N PEFV +F AS GA +T ANP T EI Q A+
Sbjct: 81 LRRAPLGLRRGGVVMSLLRNSPEFVLSFFAASRVGAAVTTANPMSTPHEIESQLAAAGAT 140
Query: 127 LLVTQACYYDKVKDLENVKLVFVDSSPEEDNHMHF-SELIQADQ-----NEMEEVKVNIK 180
+++T++ DK+ + L V D +HF +L+ D+ + ++ KV
Sbjct: 141 VVITESMAADKLPSHSHGALTVVLIDERRDGCLHFWDDLMSEDEASPLAGDEDDEKV-FD 199
Query: 181 PDDVVALPYSSGTTGLPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIY 240
PDDVVALPYSSGTTGLPKGVMLTH+ L TS+AQQVDGENPN+ H+ DVILC LPMFHIY
Sbjct: 200 PDDVVALPYSSGTTGLPKGVMLTHRSLSTSVAQQVDGENPNIGLHAGDVILCALPMFHIY 259
Query: 241 SLNSVLLCGLRAKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDL 300
SLN++++CGLR A+I++M +FD+ A LV +++VTIAP+VPPIV+A+AKS DL
Sbjct: 260 SLNTIMMCGLRVGAAIVVMRRFDLAAMMDLVERHRVTIAPLVPPIVVAVAKSEAAAARDL 319
Query: 301 SSIRVLKSGGAPLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGA 360
SS+R++ SG AP+GK++ED AK P A LGQGYGMTEAGPVL+MCL+FAKEP VKSGA
Sbjct: 320 SSVRMVLSGAAPMGKDIEDAFMAKLPGAVLGQGYGMTEAGPVLSMCLAFAKEPFKVKSGA 379
Query: 361 CGTVVRNAEMKIVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTG 420
CGTVVRNAE+KI+DP SL RN GEICIRG QIMKGYLNNPEAT+ TID EGWLHTG
Sbjct: 380 CGTVVRNAELKIIDPDTGKSLGRNLRGEICIRGQQIMKGYLNNPEATKNTIDAEGWLHTG 439
Query: 421 DIGFIDDDDELFIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVP 480
DIG++DDDDE+FIVDRLKE+IKY+GFQVAPAELEA++ +HP I+D AVV + GE+P
Sbjct: 440 DIGYVDDDDEIFIVDRLKEIIKYRGFQVAPAELEALLNTHPSIADAAVVGL---KFGEIP 496
Query: 481 VAFVVRSNGSIDTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKL 538
VAFV ++ GS + +EDD+K+FV+K+V++YK+I VFF+D IPK+PSGKILRK+LR +L
Sbjct: 497 VAFVAKTEGS-ELSEDDVKQFVAKEVIYYKKIREVFFVDKIPKAPSGKILRKELRKQL 553
>4CL1_SOYBN (P31686) 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL 1)
(4-coumaroyl-CoA synthase 1) (Clone 4CL14) (Fragment)
Length = 293
Score = 502 bits (1292), Expect = e-141
Identities = 251/292 (85%), Positives = 271/292 (91%)
Query: 252 AKASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIRVLKSGGA 311
AKA+ILLMPKFDIN+ L+ K+KVTIAPVVPPIVLAI+KSP+L KYDLSSIRVLKSGGA
Sbjct: 1 AKATILLMPKFDINSLLALIHKHKVTIAPVVPPIVLAISKSPDLHKYDLSSIRVLKSGGA 60
Query: 312 PLGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTVVRNAEMK 371
PLGKELEDT+RAKFP AKLGQGYGMTEAGPVLTM L+FAKEPIDVK GACGTVVRNAEMK
Sbjct: 61 PLGKELEDTLRAKFPNAKLGQGYGMTEAGPVLTMSLAFAKEPIDVKPGACGTVVRNAEMK 120
Query: 372 IVDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDDDEL 431
IVDP+ SLPRNQ GEICIRGDQIMKGYLN+ EAT TIDK+GWLHTGDIG+IDDDDEL
Sbjct: 121 IVDPETGHSLPRNQSGEICIRGDQIMKGYLNDGEATERTIDKDGWLHTGDIGYIDDDDEL 180
Query: 432 FIVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNGSI 491
FIVDRLKELIKYKGFQVAPAELEA++L+HP+ISD AVVPM DEAAGEVPVAFVV SNG
Sbjct: 181 FIVDRLKELIKYKGFQVAPAELEALLLTHPKISDAAVVPMKDEAAGEVPVAFVVISNGYT 240
Query: 492 DTTEDDIKKFVSKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKLAAGVP 543
DTTED+IK+F+SKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAK+AA VP
Sbjct: 241 DTTEDEIKQFISKQVVFYKRINRVFFIDAIPKSPSGKILRKDLRAKIAASVP 292
>LUCI_LUCLA (Q01158) Luciferin 4-monooxygenase (EC 1.13.12.7)
(Luciferase)
Length = 548
Score = 284 bits (726), Expect = 5e-76
Identities = 186/505 (36%), Positives = 265/505 (51%), Gaps = 27/505 (5%)
Query: 46 NAPTAEIYTYYDVQLTAQKVASGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMT 105
NA T YTY + + + L G+ I + NC EF L F G +
Sbjct: 46 NALTGVDYTYAEYLEKSCCLGEALKNYGLVVDGRIALCSENCEEFFIPVLAGLFIGVGVA 105
Query: 106 AANPFFTSAEIAKQAKASNTKLLVTQACYYDKV----KDLENVK-LVFVDSSPEEDNHMH 160
N +T E+ S ++ + DKV K + +K +V +DS + +
Sbjct: 106 PTNEIYTLRELVHSLGISKPTIVFSSKKGLDKVITVQKTVTAIKTIVILDSKVDYRGYQS 165
Query: 161 FSELIQADQ------NEMEEVKVNIKPDDVVALPYSSGTTGLPKGVMLTHKGLVTSIAQQ 214
I+ + + + V+VN K + V + SSG+TGLPKGV LTH+ VT +
Sbjct: 166 MDNFIKKNTPQGFKGSSFKTVEVNRK-EQVALIMNSSGSTGLPKGVQLTHENAVTRFSHA 224
Query: 215 VDGENPNLYYHSEDVILCVLPMFH---IYSLNSVLLCGLRAKASILLMPKFDINAFFGLV 271
D N IL V+P H +++ L CG R I+++ KFD F +
Sbjct: 225 RDPIYGN-QVSPGTAILTVVPFHHGFGMFTTLGYLTCGFR----IVMLTKFDEETFLKTL 279
Query: 272 TKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLG 331
YK + +VP + + +S LDKYDLS++ + SGGAPL KE+ + V +F +
Sbjct: 280 QDYKCSSVILVPTLFAILNRSELLDKYDLSNLVEIASGGAPLSKEIGEAVARRFNLPGVR 339
Query: 332 QGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTVVRNAEMKIVDPQNDSSLPRNQPGEICI 391
QGYG+TE T + E D K GA G VV + K++D +L N+ GE+C+
Sbjct: 340 QGYGLTET----TSAIIITPEGDD-KPGASGKVVPLFKAKVIDLDTKKTLGPNRRGEVCV 394
Query: 392 RGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPA 451
+G +MKGY++NPEATRE ID+EGWLHTGDIG+ D++ FIVDRLK LIKYKG+QV PA
Sbjct: 395 KGPMLMKGYVDNPEATREIIDEEGWLHTGDIGYYDEEKHFFIVDRLKSLIKYKGYQVPPA 454
Query: 452 ELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKR 511
ELE+++L HP I D V + D AGE+P A VV G TE ++ +V+ QV KR
Sbjct: 455 ELESVLLQHPNIFDAGVAGVPDPIAGELPGAVVVLEKGK-SMTEKEVMDYVASQVSNAKR 513
Query: 512 I-NRVFFIDAIPKSPSGKILRKDLR 535
+ V F+D +PK +GKI K +R
Sbjct: 514 LRGGVRFVDEVPKGLTGKIDGKAIR 538
>LUCI_PHOPY (P08659) Luciferin 4-monooxygenase (EC 1.13.12.7)
(Luciferase)
Length = 550
Score = 283 bits (723), Expect = 1e-75
Identities = 185/524 (35%), Positives = 270/524 (51%), Gaps = 32/524 (6%)
Query: 33 ENLSKYGSRPCLINAPTAEIYTYYDVQLT-------AQKVASGLNKLGIQQGDVIMVLLP 85
E L K R L+ A + +V +T + ++A + + G+ I+V
Sbjct: 24 EQLHKAMKRYALVPGTIAFTDAHIEVNITYAEYFEMSVRLAEAMKRYGLNTNHRIVVCSE 83
Query: 86 NCPEFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLLVTQACYYDKVKDLENV- 144
N +F LGA F G + AN + E+ S ++ K+ +++
Sbjct: 84 NSLQFFMPVLGALFIGVAVAPANDIYNERELLNSMNISQPTVVFVSKKGLQKILNVQKKL 143
Query: 145 ----KLVFVDSSPEEDNHMHFSELIQADQ----NEMEEVKVNIKPDDVVALPY-SSGTTG 195
K++ +DS + + + NE + V + D +AL SSG+TG
Sbjct: 144 PIIQKIIIMDSKTDYQGFQSMYTFVTSHLPPGFNEYDFVPESFDRDKTIALIMNSSGSTG 203
Query: 196 LPKGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFH---IYSLNSVLLCGLRA 252
LPKGV L H+ + D N + IL V+P H +++ L+CG R
Sbjct: 204 LPKGVALPHRTACVRFSHARDPIFGNQII-PDTAILSVVPFHHGFGMFTTLGYLICGFR- 261
Query: 253 KASILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIRVLKSGGAP 312
++LM +F+ F + YK+ A +VP + AKS +DKYDLS++ + SGGAP
Sbjct: 262 ---VVLMYRFEEELFLRSLQDYKIQSALLVPTLFSFFAKSTLIDKYDLSNLHEIASGGAP 318
Query: 313 LGKELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTVVRNAEMKI 372
L KE+ + V +F + QGYG+TE T + E D K GA G VV E K+
Sbjct: 319 LSKEVGEAVAKRFHLPGIRQGYGLTET----TSAILITPEGDD-KPGAVGKVVPFFEAKV 373
Query: 373 VDPQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDDDELF 432
VD +L NQ GE+C+RG IM GY+NNPEAT IDK+GWLH+GDI + D+D+ F
Sbjct: 374 VDLDTGKTLGVNQRGELCVRGPMIMSGYVNNPEATNALIDKDGWLHSGDIAYWDEDEHFF 433
Query: 433 IVDRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNGSID 492
IVDRLK LIKYKG+QVAPAELE+I+L HP I D V + D+ AGE+P A VV +G
Sbjct: 434 IVDRLKSLIKYKGYQVAPAELESILLQHPNIFDAGVAGLPDDDAGELPAAVVVLEHGK-T 492
Query: 493 TTEDDIKKFVSKQVVFYKRI-NRVFFIDAIPKSPSGKILRKDLR 535
TE +I +V+ QV K++ V F+D +PK +GK+ + +R
Sbjct: 493 MTEKEIVDYVASQVTTAKKLRGGVVFVDEVPKGLTGKLDARKIR 536
>LUCI_LUCCR (P13129) Luciferin 4-monooxygenase (EC 1.13.12.7)
(Luciferase)
Length = 548
Score = 282 bits (722), Expect = 1e-75
Identities = 182/505 (36%), Positives = 268/505 (53%), Gaps = 27/505 (5%)
Query: 46 NAPTAEIYTYYDVQLTAQKVASGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMT 105
NA T Y+Y + + + L G+ I + NC EF + F G +
Sbjct: 46 NAVTGVDYSYAEYLEKSCCLGKALQNYGLVVDGRIALCSENCEEFFIPVIAGLFIGVGVA 105
Query: 106 AANPFFTSAEIAKQAKASNTKLLVTQACYYDKV----KDLENVK-LVFVDSSPEEDNHMH 160
N +T E+ S ++ + DKV K + +K +V +DS + +
Sbjct: 106 PTNEIYTLRELVHSLGISKPTIVFSSKKGLDKVITVQKTVTTIKTIVILDSKVDYRGYQC 165
Query: 161 FSELIQADQ------NEMEEVKVNIKPDDVVALPYSSGTTGLPKGVMLTHKGLVTSIAQQ 214
I+ + + + V+V+ K + V + SSG+TGLPKGV LTH+ VT +
Sbjct: 166 LDTFIKRNTPPGFQASSFKTVEVDRK-EQVALIMNSSGSTGLPKGVQLTHENTVTRFSHA 224
Query: 215 VDGENPNLYYHSEDVILCVLPMFH---IYSLNSVLLCGLRAKASILLMPKFDINAFFGLV 271
D N +L V+P H +++ L+CG R ++++ KFD F +
Sbjct: 225 RDPIYGN-QVSPGTAVLTVVPFHHGFGMFTTLGYLICGFR----VVMLTKFDEETFLKTL 279
Query: 272 TKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIRVLKSGGAPLGKELEDTVRAKFPKAKLG 331
YK T +VP + + KS L+KYDLS++ + SGGAPL KE+ + V +F +
Sbjct: 280 QDYKCTSVILVPTLFAILNKSELLNKYDLSNLVEIASGGAPLSKEVGEAVARRFNLPGVR 339
Query: 332 QGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTVVRNAEMKIVDPQNDSSLPRNQPGEICI 391
QGYG+TE T + E D K GA G VV + K++D SL N+ GE+C+
Sbjct: 340 QGYGLTET----TSAIIITPEGDD-KPGASGKVVPLFKAKVIDLDTKKSLGPNRRGEVCV 394
Query: 392 RGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDDDELFIVDRLKELIKYKGFQVAPA 451
+G +MKGY+NNPEAT+E ID+EGWLHTGDIG+ D++ FIVDRLK LIKYKG+QV PA
Sbjct: 395 KGPMLMKGYVNNPEATKELIDEEGWLHTGDIGYYDEEKHFFIVDRLKSLIKYKGYQVPPA 454
Query: 452 ELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNGSIDTTEDDIKKFVSKQVVFYKR 511
ELE+++L HP I D V + D AGE+P A VV +G + TE ++ +V+ QV KR
Sbjct: 455 ELESVLLQHPSIFDAGVAGVPDPVAGELPGAVVVLESGK-NMTEKEVMDYVASQVSNAKR 513
Query: 512 I-NRVFFIDAIPKSPSGKILRKDLR 535
+ V F+D +PK +GKI + +R
Sbjct: 514 LRGGVRFVDEVPKGLTGKIDGRAIR 538
>LUCI_PHOPE (Q27757) Luciferin 4-monooxygenase (EC 1.13.12.7)
(Luciferase)
Length = 545
Score = 274 bits (701), Expect = 4e-73
Identities = 185/522 (35%), Positives = 268/522 (50%), Gaps = 30/522 (5%)
Query: 32 FENLSKYGSRP---CLINAPTAEIYTYYDVQLTAQKVASGLNKLGIQQGDVIMVLLPNCP 88
F LS+Y L NA T E Y + + ++A K G++Q D I V N
Sbjct: 26 FYALSRYADISGCIALTNAHTKENVLYEEFLKLSCRLAESFKKYGLKQNDTIAVCSENGL 85
Query: 89 EFVFAFLGASFRGAIMTAANPFFTSAEIAKQAKASNTKLLVTQACYYDKVKDLENV---- 144
+F + + + G I + + E+ +++ + KV ++++
Sbjct: 86 QFFLPLIASLYLGIIAAPVSDKYIERELIHSLGIVKPRIIFCSKNTFQKVLNVKSKLKYV 145
Query: 145 -KLVFVDSSPEEDNHMHFSELIQADQNEMEEVKVNIKP-----DDVVALP-YSSGTTGLP 197
++ +D + + + + I + + +VK KP DD VAL +SSGTTG+
Sbjct: 146 ETIIILDLNEDLGGYQCLNNFISQNSDINLDVK-KFKPNSFNRDDQVALVMFSSGTTGVS 204
Query: 198 KGVMLTHKGLVTSIAQQVDGENPNLYYHSEDVILCVLPMFHIYSLNSVL---LCGLRAKA 254
KGVMLTHK +V + D N + IL V+P H + + + L CG R
Sbjct: 205 KGVMLTHKNIVARFSHCKDPTFGNAI-NPTTAILTVIPFHHGFGMTTTLGYFTCGFR--- 260
Query: 255 SILLMPKFDINAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIRVLKSGGAPLG 314
+ LM F+ F + YKV +VP ++ KS ++KYDLS ++ + SGGAPL
Sbjct: 261 -VALMHTFEEKLFLQSLQDYKVESTLLVPTLMAFFPKSALVEKYDLSHLKEIASGGAPLS 319
Query: 315 KELEDTVRAKFPKAKLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTVVRNAEMKIVD 374
KE+ + V+ +F + QGYG+TE T DV+ G+ G +V +K+VD
Sbjct: 320 KEIGEMVKKRFKLNFVRQGYGLTE-----TTSAVLITPDTDVRPGSTGKIVPFHAVKVVD 374
Query: 375 PQNDSSLPRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDDDELFIV 434
P L N+ GE+ +GD IMK Y NN EAT+ I+K+GWL +GDI + D+D +IV
Sbjct: 375 PTTGKILGPNETGELYFKGDMIMKSYYNNEEATKAIINKDGWLRSGDIAYYDNDGHFYIV 434
Query: 435 DRLKELIKYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNGSIDTT 494
DRLK LIKYKG+QVAPAE+E I+L HP I D V + DEAAGE+P A VV G
Sbjct: 435 DRLKSLIKYKGYQVAPAEIEGILLQHPYIVDAGVTGIPDEAAGELPAAGVVVQTGKY-LN 493
Query: 495 EDDIKKFVSKQVVFYKRI-NRVFFIDAIPKSPSGKILRKDLR 535
E ++ FVS QV K + V F+D IPK +GKI RK LR
Sbjct: 494 EQIVQNFVSSQVSTAKWLRGGVKFLDEIPKGSTGKIDRKVLR 535
>LCFA_BACSU (P94547) Long-chain-fatty-acid--CoA ligase (EC 6.2.1.3)
(Long-chain acyl-CoA synthetase)
Length = 560
Score = 244 bits (624), Expect = 3e-64
Identities = 156/513 (30%), Positives = 255/513 (49%), Gaps = 46/513 (8%)
Query: 54 TYYDVQLTAQKVASGLNKLGIQQGDVIMVLLPNCPEFVFAFLGASFRGAIMTAANPFFTS 113
T++D+ A K+A+ L G+Q+GD + V+LPNCP+ V ++ G F G I+ NP +T
Sbjct: 51 TFHDILTDALKLAAFLQCNGLQKGDRVAVMLPNCPQTVISYYGVLFAGGIVVQTNPLYTE 110
Query: 114 AEIAKQAKASNTKLLVTQACYYDKVKDLENVKLV-------FVDSSPEEDNHMH------ 160
E+ Q + + +++T + K ++ + +V D P N ++
Sbjct: 111 HELEYQLRDAQVSVIITLDLLFPKAIKMKTLSIVDQILITSVKDYLPFPKNILYPLTQKQ 170
Query: 161 --------------FSELIQADQNEMEEVKVNIKPDDVVALPYSSGTTGLPKGVMLTHKG 206
F+ ++ ++ E+ + D+ L Y+ GTTG PKGVMLTH+
Sbjct: 171 KVHIDFDKTANIHTFASCMKQEKTELLTIPKIDPEHDIAVLQYTGGTTGAPKGVMLTHQN 230
Query: 207 LVTSIAQQVDGENPNLYYHSE--DVILCVLPMFHIYSLNSVLLCGLRAKASILLMPKFDI 264
++ + +Y E + +L ++P FH+Y L +V+ ++ ++L+PKFD
Sbjct: 231 ILANTEMCAAW----MYDVKEGAEKVLGIVPFFHVYGLTAVMNYSIKLGFEMILLPKFDP 286
Query: 265 NAFFGLVTKYKVTIAPVVPPIVLAIAKSPELDKYDLSSIRVLKSGGAPLGKELEDTVRAK 324
++ K+K T+ P P I + + PEL YDLSSI+ SG A L E V+ K
Sbjct: 287 LETLKIIDKHKPTLFPGAPTIYIGLLHHPELQHYDLSSIKSCLSGSAALPVE----VKQK 342
Query: 325 FPKA---KLGQGYGMTEAGPVLTMCLSFAKEPIDVKSGACGTVVRNAEMKIVDPQNDSSL 381
F K KL +GYG++EA PV + K K G+ G + + I +
Sbjct: 343 FEKVTGGKLVEGYGLSEASPVTHANFIWGKN----KPGSIGCPWPSTDAAIYSEETGELA 398
Query: 382 PRNQPGEICIRGDQIMKGYLNNPEATRETIDKEGWLHTGDIGFIDDDDELFIVDRLKELI 441
+ GEI ++G Q+MKGY N PE T + ++GWL TGD+G++D++ +I DR K++I
Sbjct: 399 APYEHGEIIVKGPQVMKGYWNKPEETAAVL-RDGWLFTGDMGYMDEEGFFYIADRKKDII 457
Query: 442 KYKGFQVAPAELEAIILSHPQISDVAVVPMLDEAAGEVPVAFVVRSNGSIDTTEDDIKKF 501
G+ + P E+E + H I ++ V + D GE AFVV G+ TE ++ F
Sbjct: 458 IAGGYNIYPREVEEALYEHEAIQEIVVAGVPDSYRGETVKAFVVLKKGAKADTE-ELDAF 516
Query: 502 VSKQVVFYKRINRVFFIDAIPKSPSGKILRKDL 534
++ YK F +PK+ GKILR+ L
Sbjct: 517 ARSRLAPYKVPKAYEFRKELPKTAVGKILRRRL 549
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.137 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 63,343,926
Number of Sequences: 164201
Number of extensions: 2737759
Number of successful extensions: 7951
Number of sequences better than 10.0: 243
Number of HSP's better than 10.0 without gapping: 223
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 6810
Number of HSP's gapped (non-prelim): 429
length of query: 544
length of database: 59,974,054
effective HSP length: 115
effective length of query: 429
effective length of database: 41,090,939
effective search space: 17628012831
effective search space used: 17628012831
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC144503.3


