

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144503.2 + phase: 0
(322 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
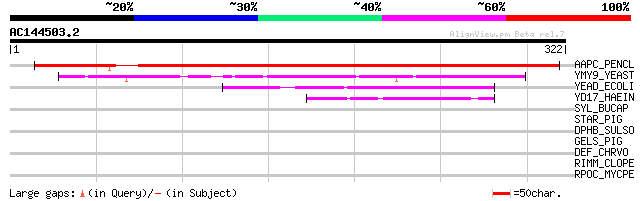
Score E
Sequences producing significant alignments: (bits) Value
AAPC_PENCL (Q40784) Possible apospory-associated protein C 403 e-112
YMY9_YEAST (Q03161) Hypothetical UPF0010 protein YMR099c 94 5e-19
YEAD_ECOLI (P39173) UPF0010 protein yeaD (Unknown protein from 2... 66 1e-10
YD17_HAEIN (P44160) Hypothetical UPF0010 protein HI1317 54 5e-07
SYL_BUCAP (Q8K9B9) Leucyl-tRNA synthetase (EC 6.1.1.4) (Leucine-... 35 0.19
STAR_PIG (Q28996) Steroidogenic acute regulatory protein, mitoch... 33 0.96
DPHB_SULSO (Q97TX8) Probable diphthine synthase (EC 2.1.1.98) (D... 32 2.1
GELS_PIG (P20305) Gelsolin precursor, plasma (Actin-depolymerizi... 32 2.8
DEF_CHRVO (Q7NQ75) Peptide deformylase (EC 3.5.1.88) (PDF) (Poly... 32 2.8
RIMM_CLOPE (Q8XJP6) Probable 16S rRNA processing protein rimM 30 6.2
RPOC_MYCPE (Q8EWX0) DNA-directed RNA polymerase beta' chain (EC ... 30 8.1
>AAPC_PENCL (Q40784) Possible apospory-associated protein C
Length = 329
Score = 403 bits (1035), Expect = e-112
Identities = 194/309 (62%), Positives = 238/309 (76%), Gaps = 16/309 (5%)
Query: 15 NGLEKIILRENRGFSAEVYLYGGQVTSWKNERGEELLFVSSK----PEKEVKKTNQYNCF 70
+GLEK++LR R AE+YLYGGQVTSWKN+ GEELLF+SSK P K ++
Sbjct: 31 SGLEKVVLRGARNCCAEIYLYGGQVTSWKNDNGEELLFLSSKAIFKPPKAIR-------- 82
Query: 71 DMNNNKCRLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSEDD 130
+ L + Q+ F +N+ W +D +PPP P N +AFVDL+L+ +E+D
Sbjct: 83 ----GGIPICLPQFGTHGNLEQHGFARNRFWSIDNDPPPLPVNPAIKAFVDLILRPAEED 138
Query: 131 TKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIEG 190
K WPH +EFRLR+ALGP+GDL LTSRIRNTN DG+ F++TFAYHTY +V+DISEVR+EG
Sbjct: 139 LKIWPHSFEFRLRVALGPSGDLSLTSRIRNTNTDGRPFSYTFAYHTYFFVSDISEVRVEG 198
Query: 191 LETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLPD 250
LET+DYLD+LK +ERFTEQGDAI FE EVDKVYL+ P+KIAIIDHE+K+TF + K+GLPD
Sbjct: 199 LETMDYLDNLKAKERFTEQGDAIVFESEVDKVYLAAPSKIAIIDHEKKKTFVVTKEGLPD 258
Query: 251 AVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWKGRQEISAVPSSYCS 310
AVVWNPWDKK+K + D GD EYK+MLCV+PA VEK ITLKPGEEW+GR +SAVPSSYCS
Sbjct: 259 AVVWNPWDKKAKAMQDFGDAEYKNMLCVEPAAVEKPITLKPGEEWRGRIALSAVPSSYCS 318
Query: 311 GQLDPRKVL 319
GQLDP KVL
Sbjct: 319 GQLDPLKVL 327
>YMY9_YEAST (Q03161) Hypothetical UPF0010 protein YMR099c
Length = 297
Score = 94.0 bits (232), Expect = 5e-19
Identities = 78/280 (27%), Positives = 122/280 (42%), Gaps = 25/280 (8%)
Query: 29 SAEVYLYGGQVTSWKNERGEELLFVSSKPEKEVKKTNQ------YNCFDMNNNKCRLILL 82
S + YG V SWK + EE L++S+ + + K + + F N+ L L
Sbjct: 21 SVHILKYGATVYSWKL-KSEEQLWLSTAAKLDGSKPVRGGIPLVFPVFGKNSTDEHLSKL 79
Query: 83 LQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSEDDTKNWPHRYEFRL 142
Q A + FL NPP + N E TK WP Y L
Sbjct: 80 PQHGLARNSTWEFLGQT----KENPPTVQFGLKPE------IANPEL-TKLWPMDYLLIL 128
Query: 143 RIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDISEVRIEGLETLDYLDSLKN 202
+ LG D + T+ K F + +HTY + DI + L + D L
Sbjct: 129 TVELG--SDYLKTAIEVENTSSSKELKFNWLFHTYFRIEDIEGTMVSNLAGMKLYDQLL- 185
Query: 203 RERFTEQGDAITFEGEVDKVY--LSTPTKIAIIDHERKRTFEIRKDGLPDAVVWNPWDKK 260
+E + ++ +TF E D +Y +S I I+D + + +++ LPD VVWNPW +K
Sbjct: 186 KESYVDKHPVVTFNQETDVIYQNVSAERAIQIVD-KGVQIHTLKRYNLPDTVVWNPWIEK 244
Query: 261 SKIISDLGDDE-YKHMLCVQPACVEKAITLKPGEEWKGRQ 299
S+ ++D Y+ M+C++P V I+L PG++W Q
Sbjct: 245 SQGMADFEPKTGYQQMICIEPGHVHDFISLAPGKKWNAYQ 284
>YEAD_ECOLI (P39173) UPF0010 protein yeaD (Unknown protein from
2D-page spots T26/PR37)
Length = 294
Score = 66.2 bits (160), Expect = 1e-10
Identities = 46/158 (29%), Positives = 67/158 (42%), Gaps = 9/158 (5%)
Query: 124 LKNSEDDTKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYLYVTDI 183
L SE+ K WPH + +G T ++ L S F T A HTY V DI
Sbjct: 121 LTQSEETKKFWPHDFTLLAHFRVGKTCEIDLESH--------GEFETTSALHTYFNVGDI 172
Query: 184 SEVRIEGLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERKRTFEI 243
++V + GL ++D + + + TF D+VYL+ I D R +
Sbjct: 173 AKVSVSGLGDR-FIDKVNDAKENVLTDGIQTFPDRTDRVYLNPQDCSVINDEALNRIIAV 231
Query: 244 RKDGLPDAVVWNPWDKKSKIISDLGDDEYKHMLCVQPA 281
+ V WNP S + D+ DD YK +CV+ A
Sbjct: 232 GHQHHLNVVGWNPGPALSISMGDMPDDGYKTFVCVETA 269
>YD17_HAEIN (P44160) Hypothetical UPF0010 protein HI1317
Length = 271
Score = 53.9 bits (128), Expect = 5e-07
Identities = 33/109 (30%), Positives = 55/109 (50%), Gaps = 6/109 (5%)
Query: 173 AYHTYLYVTDISEVRIEGLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAI 232
A HTY + DI++V ++GL + +SL ++ I+ VD +Y + + I
Sbjct: 149 ALHTYFNIGDINQVEVQGLPETCF-NSLNQQQENVPSPRHIS--ENVDCIYSAENMQNQI 205
Query: 233 IDHERKRTFEIRKDGLPDAVVWNPWDKKSKIISDLGDDEYKHMLCVQPA 281
+D RT + V+WNPW KK+ +S+ G Y+ MLC++ A
Sbjct: 206 LDKSFNRTIALHHHNASQFVLWNPWHKKTSGMSETG---YQKMLCLETA 251
>SYL_BUCAP (Q8K9B9) Leucyl-tRNA synthetase (EC 6.1.1.4)
(Leucine--tRNA ligase) (LeuRS)
Length = 861
Score = 35.4 bits (80), Expect = 0.19
Identities = 27/111 (24%), Positives = 47/111 (42%), Gaps = 7/111 (6%)
Query: 124 LKNSEDDTKNWPHRYEFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAY---HTYLYV 180
L N +D +NWP + + R +G + + + N+NK K+FT TY+ +
Sbjct: 202 LHNDLNDLENWPEKVKNMQRNWIGRSKGFEIVFNVLNSNKKIKAFTNRLDLIMGATYISI 261
Query: 181 TDISEVRI----EGLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTP 227
+ E + E E ++D N E + I FEG +++ P
Sbjct: 262 SPCHEFSVHVSKEKKEIQKFIDQKINNSASQEDIEKIKFEGINSNMFVIHP 312
>STAR_PIG (Q28996) Steroidogenic acute regulatory protein,
mitochondrial precursor (StAR) (StARD1)
Length = 285
Score = 33.1 bits (74), Expect = 0.96
Identities = 34/135 (25%), Positives = 52/135 (38%), Gaps = 8/135 (5%)
Query: 139 EFRLRIALGPTGDLMLTSRIRNTNKDGKSFTFTFAYHTYL-YVTDISEVRIEGLETLDYL 197
E R GPT + R ++ G TF L Y+ E L+ L
Sbjct: 34 ELNRRALGGPTSGSWINQVRRRSSLLGSQLEDTFYSDQDLAYIQQGEEAMQRALDILSNQ 93
Query: 198 DSLKNRERFTEQGDAITFE--GEVDKVYLSTPTKIAIIDHERKRTFEIRKDGLPDAVVWN 255
+ K R E GD + + +V KV+ ++D +R +E + + WN
Sbjct: 94 EGWKKESR-QENGDEVLSKVIPDVGKVFRLE----VVVDQPMERLYEELVERMEAMGEWN 148
Query: 256 PWDKKSKIISDLGDD 270
P KK KI+ +G D
Sbjct: 149 PSVKKIKILQKIGKD 163
>DPHB_SULSO (Q97TX8) Probable diphthine synthase (EC 2.1.1.98)
(Diphthamide biosynthesis methyltransferase)
Length = 257
Score = 32.0 bits (71), Expect = 2.1
Identities = 20/58 (34%), Positives = 33/58 (56%), Gaps = 4/58 (6%)
Query: 153 MLTSRIRNTNKDGKSFTFTFAYHTYLYVTD---ISEVRIEGLETLDYLDSLKNRERFT 207
+++ + ++ K GKS T TF Y+ ++ T I + + GL T+ YLD LKN + T
Sbjct: 122 LISKSLLSSYKFGKSVTVTFPYNDFIDPTPYNVIKDNKERGLHTILYLD-LKNEKAMT 178
>GELS_PIG (P20305) Gelsolin precursor, plasma (Actin-depolymerizing
factor) (ADF) (Brevin) (Fragment)
Length = 772
Score = 31.6 bits (70), Expect = 2.8
Identities = 26/116 (22%), Positives = 52/116 (44%), Gaps = 16/116 (13%)
Query: 162 NKDGKSFTFTFAYHTYLYVTDISEVRIEGLETLDYLDSLKNRERFTEQGDAITFEGEVDK 221
++ G + FT YL + ++G E+ +L K+ ++ + G A F+ V
Sbjct: 113 DESGAAAIFTVQLDDYLNGRAVQHREVQGFESATFLGYFKSGLKYKKGGVASGFKHVV-- 170
Query: 222 VYLSTPTKIAIIDHERKRTFEIRKDGLPDA----VVWNPWDKKSKIISDLGDDEYK 273
P ++A+ +R F+++ + A V W +++ I DLG+D Y+
Sbjct: 171 -----PNEVAV-----QRLFQVKGRRVVRATEVPVSWESFNRGDCFILDLGNDIYQ 216
>DEF_CHRVO (Q7NQ75) Peptide deformylase (EC 3.5.1.88) (PDF)
(Polypeptide deformylase)
Length = 167
Score = 31.6 bits (70), Expect = 2.8
Identities = 26/75 (34%), Positives = 36/75 (47%), Gaps = 2/75 (2%)
Query: 174 YHTYLYVTDISEVRIEGLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAII 233
YH L V DISE R E ++ K+ E E+G ++ G DKV + K+
Sbjct: 54 YHRRLVVMDISEERDERRVFINPEIVEKDGETVYEEG-CLSVPGIYDKVTRAERVKVKAQ 112
Query: 234 DHERKRTFEIRKDGL 248
D + K FE+ DGL
Sbjct: 113 DRDGK-PFELEADGL 126
>RIMM_CLOPE (Q8XJP6) Probable 16S rRNA processing protein rimM
Length = 165
Score = 30.4 bits (67), Expect = 6.2
Identities = 31/120 (25%), Positives = 55/120 (45%), Gaps = 11/120 (9%)
Query: 179 YVTDISEVRIEGLETLDYLDSLKNRERFTEQGDAITFEGEVDKVYLSTPTKIAIIDHERK 238
Y D +++EG+ +++ + LK E+ DAI E E D+ ++ ++D E
Sbjct: 52 YFKDKVILKLEGINSIEEAEKLKRTYLEIEREDAI--ELEEDEYFIVDLVGCTVVDTEGF 109
Query: 239 RTFEIRKDGL--PDAVVWNPWDKKSKIISDLGDDEYKHMLCVQPACVEKAITLKPGEEWK 296
+I KD + P V+ KK ++ L D + + EK IT++P EW+
Sbjct: 110 EYGKI-KDVIQTPSNDVYWVQGKKEVLVPVLKD------IVLDINMDEKLITIRPSGEWQ 162
>RPOC_MYCPE (Q8EWX0) DNA-directed RNA polymerase beta' chain (EC
2.7.7.6) (RNAP beta' subunit) (Transcriptase beta'
chain) (RNA polymerase beta' subunit)
Length = 1288
Score = 30.0 bits (66), Expect = 8.1
Identities = 17/56 (30%), Positives = 29/56 (51%), Gaps = 1/56 (1%)
Query: 78 RLILLLQDLYAEAFQYVFLKNKLWILDPNPPPFPTNSNSRAFVDLVLKNSEDDTKN 133
++ L+L Y E Q V+ N + +LD FP N N + +DL + S +T++
Sbjct: 116 KISLVLDISYKEIEQVVYFVNYI-VLDEGNGKFPKNFNFKEVIDLSSQKSSKETRS 170
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.137 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 41,220,365
Number of Sequences: 164201
Number of extensions: 1814787
Number of successful extensions: 4188
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 4173
Number of HSP's gapped (non-prelim): 13
length of query: 322
length of database: 59,974,054
effective HSP length: 110
effective length of query: 212
effective length of database: 41,911,944
effective search space: 8885332128
effective search space used: 8885332128
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC144503.2


