

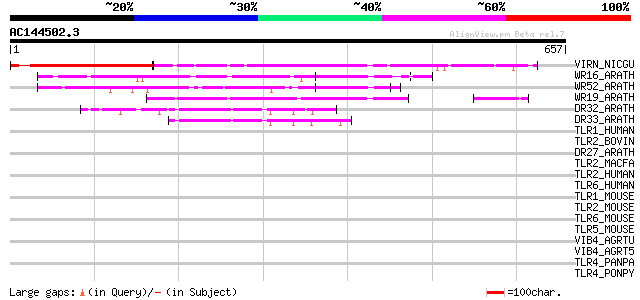
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144502.3 + phase: 0 /pseudo
(657 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
VIRN_NICGU (Q40392) TMV resistance protein N 149 3e-35
WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY ... 142 2e-33
WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY ... 110 1e-23
WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY ... 105 3e-22
DR32_ARATH (Q9FG91) Probable disease resistance protein At5g43730 50 2e-05
DR33_ARATH (Q9FG90) Probable disease resistance protein At5g43740 47 2e-04
TLR1_HUMAN (Q15399) Toll-like receptor 1 precursor (Toll/interle... 44 0.002
TLR2_BOVIN (Q95LA9) Toll-like receptor 2 precursor 43 0.003
DR27_ARATH (Q9T048) Putative disease resistance protein At4g27190 41 0.009
TLR2_MACFA (Q95M53) Toll-like receptor 2 precursor 41 0.011
TLR2_HUMAN (O60603) Toll-like receptor 2 precursor (Toll/interle... 41 0.011
TLR6_HUMAN (Q9Y2C9) Toll-like receptor 6 precursor 39 0.043
TLR1_MOUSE (Q9EPQ1) Toll-like receptor 1 precursor (Toll/interle... 39 0.056
TLR2_MOUSE (Q9QUN7) Toll-like receptor 2 precursor 36 0.28
TLR6_MOUSE (Q9EPW9) Toll-like receptor 6 precursor 36 0.37
TLR5_MOUSE (Q9JLF7) Toll-like receptor 5 precursor 35 0.81
VIB4_AGRTU (P05353) VirB4 protein precursor 34 1.1
VIB4_AGRT5 (P17794) VirB4 protein precursor 34 1.1
TLR4_PANPA (Q9TTN0) Toll-like receptor 4 precursor 34 1.1
TLR4_PONPY (Q8SPE9) Toll-like receptor 4 precursor 34 1.4
>VIRN_NICGU (Q40392) TMV resistance protein N
Length = 1144
Score = 149 bits (375), Expect = 3e-35
Identities = 79/171 (46%), Positives = 105/171 (61%), Gaps = 13/171 (7%)
Query: 1 MASSNNSSSSALVTVTLPRRKNYYDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDDTN 60
MASS++SS + YD F++FRGEDTR FT HL++ N +GI F+DD
Sbjct: 1 MASSSSSSRWS------------YDVFLSFRGEDTRKTFTSHLYEVLNDKGIKTFQDDKR 48
Query: 61 LPKGESIASELLRAIEDSYIFVAVLSRNYASSIWCLQELEKILECVHVSKKHVLPVFYDV 120
L G +I EL +AIE+S + V S NYA+S WCL EL KI+EC K+ V+P+FYDV
Sbjct: 49 LEYGATIPGELCKAIEESQFAIVVFSENYATSRWCLNELVKIMECKTRFKQTVIPIFYDV 108
Query: 121 DPPVVRKQSGIYCEAFVKHEQIFQQDSQMVLRWREALTQVAGLSG-CDLRD 170
DP VR Q + +AF +HE ++ D + + RWR AL + A L G CD RD
Sbjct: 109 DPSHVRNQKESFAKAFEEHETKYKDDVEGIQRWRIALNEAANLKGSCDNRD 159
Score = 139 bits (350), Expect = 2e-32
Identities = 132/478 (27%), Positives = 219/478 (45%), Gaps = 47/478 (9%)
Query: 170 DNLYDRI--SHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNLSTASNL 227
D L R+ S+QF CF+ D+ + R G +Q +L + E+ N N +
Sbjct: 231 DTLLGRMDSSYQFDGACFLKDIKENKR---GMHSLQNALLSELLREKANYN-NEEDGKHQ 286
Query: 228 IRRRLCRQRVLLIFDNVD-KVEQLEKI-GVCVW*MVRWGSKIIIISRDEHILKFFGVDEV 285
+ RL ++VL++ D++D K LE + G W GS+III +RD+H+++ D +
Sbjct: 287 MASRLRSKKVLIVLDDIDNKDHYLEYLAGDLDW--FGNGSRIIITTRDKHLIE--KNDII 342
Query: 286 YKVPLLDRTNSLQLLCRKAFKLDHILSSMKGWSMAYYIMLRAHR*QLK-YRAHFCGRDIF 344
Y+V L S+QL + AF + + + S+ + LK + + +
Sbjct: 343 YEVTALPDHESIQLFKQHAFGKEVPNENFEKLSLEVVNYAKGLPLALKVWGSLLHNLRLT 402
Query: 345 EWKSALARLRDSPDKDVMDVLRLSFDGLEESEKEIFLHIACFFNPSMEKYVKNVLNCCGF 404
EWKSA+ ++++ ++D L++S+DGLE ++E+FL IACF + Y+ +L C
Sbjct: 403 EWKSAIEHMKNNSYSGIIDKLKISYDGLEPKQQEMFLDIACFLRGEEKDYILQILESCHI 462
Query: 405 HADIGLRVLIDKSLISIDESFSSLKEESISMHGLLEELGRKIVQENSSKEPRKWSRLWLE 464
A+ GLR+LIDKSL+ I E + MH L++++G+ IV N K+P + SRLWL
Sbjct: 463 GAEYGLRILIDKSLVFISE------YNQVQMHDLIQDMGKYIV--NFQKDPGERSRLWLA 514
Query: 465 TQVDNVMLEKMFFILCNALGKACRSHTFKKKNSQQR**KE------GYDSRTLSF----- 513
+V+ VM + A+ + S T + N + K G S +
Sbjct: 515 KEVEEVMSNNTGTMAMEAIWVSSYSSTLRFSNQAVKNMKRLRVFNMGRSSTHYAIDYLPN 574
Query: 514 -*RIKIC*ME*ISFQVFAVKFSAKSTC*VDLEE*QHRTIMGRQEEFPNLERLDLEGCIKL 572
R +C ++ F F K + L R + + P+L R+DL +L
Sbjct: 575 NLRCFVC--TNYPWESFPSTFELKMLVHLQLRHNSLRHLWTETKHLPSLRRIDLSWSKRL 632
Query: 573 VQIDPSIGLLTKLVYLNLKDCK------HIISLLSNIFGLSCLDDLNIYVLQSKEFEC 624
+ P + L Y+NL C H + S + GL D ++ K F C
Sbjct: 633 TR-TPDFTGMPNLEYVNLYQCSNLEEVHHSLGCCSKVIGLYLNDCKSL-----KRFPC 684
>WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY
DNA-binding protein 16)
Length = 1372
Score = 142 bits (359), Expect = 2e-33
Identities = 129/488 (26%), Positives = 223/488 (45%), Gaps = 74/488 (15%)
Query: 33 EDTRNNFTYHLFDAFNREGILAFRDDTNLPKGESIASELLRAIEDSYIFVAVLSRNYASS 92
E+ R +F HL A R+G+ +D + +S+++E +E + + V +L N S
Sbjct: 14 EEVRYSFVSHLSKALQRKGV----NDVFIDSDDSLSNESQSMVERARVSVMILPGNRTVS 69
Query: 93 IWCLQELEKILECVHVSKKHVLPVFYDVDPPVVRKQSGIYCEAF--VKHEQIFQQDSQMV 150
L +L K+L+C + V+PV Y V S + + F V H + DSQ+V
Sbjct: 70 ---LDKLVKVLDCQKNKDQVVVPVLYGVRSSETEWLSALDSKGFSSVHHSRKECSDSQLV 126
Query: 151 ----------LRWRE-------------------------ALTQVAGLSGCDLRDNLYDR 175
L + E + + G+ L ++D+
Sbjct: 127 KETVRDVYEKLFYMERIGIYSKLLEIEKMINKQPLDIRCVGIWGMPGIGKTTLAKAVFDQ 186
Query: 176 ISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNLSTASNLIRRRLCRQ 235
+S +F A CFI+D +K + +++Q L + G T +L+R RL +
Sbjct: 187 MSGEFDAHCFIEDYTKAIQEKGVYCLLEEQFLKENAGAS-----GTVTKLSLLRDRLNNK 241
Query: 236 RVLLIFDNVDKVEQLEK-IGVCVW*MVRWGSK--IIIISRDEHILKFFGVDEVYKVPLLD 292
RVL++ D+V +E +G W +G K III S+D+ + + V+++Y+V L+
Sbjct: 242 RVLVVLDDVRSPLVVESFLGGFDW----FGPKSLIIITSKDKSVFRLCRVNQIYEVQGLN 297
Query: 293 RTNSLQLLCRKAFKLDHILSSMKGWSMAYYIMLRAHR*QLKYRAHFCGRDIF------EW 346
+LQL A D ++ SM H L + GR++ E
Sbjct: 298 EKEALQLFSLCASIDDMAEQNLHEVSMKVIKYANGHPLAL----NLYGRELMGKKRPPEM 353
Query: 347 KSALARLRDSPDKDVMDVLRLSFDGLEESEKEIFLHIACFFNPSMEKYVKNVLNCCGFHA 406
+ A +L++ P +D ++ S+D L + EK IFL IACFF YV +L CGF
Sbjct: 354 EIAFLKLKECPPAIFVDAIKSSYDTLNDREKNIFLDIACFFQGENVDYVMQLLEGCGFFP 413
Query: 407 DIGLRVLIDKSLISIDESFSSLKEESISMHGLLEELGRKIVQENSSKEPRKWSRLWLETQ 466
+G+ VL++KSL++I E+ + MH L++++GR+I+ + + R+ SRLW
Sbjct: 414 HVGIDVLVEKSLVTISEN-------RVRMHNLIQDVGRQIINRETRQTKRR-SRLWEPCS 465
Query: 467 VDNVMLEK 474
+ ++ +K
Sbjct: 466 IKYLLEDK 473
Score = 46.6 bits (109), Expect = 2e-04
Identities = 39/139 (28%), Positives = 65/139 (46%), Gaps = 20/139 (14%)
Query: 363 DVLRLSFDGLEESEKEIFLHIACFFNPSMEKYVKNVL-NCCGFHADIGLRVLIDKSLISI 421
+VLR+ + GL+E K +FL+IA FN V ++ N GL+VL +SLI +
Sbjct: 1049 EVLRVRYAGLQEIYKALFLYIAGLFNDEDVGLVAPLIANIIDMDVSYGLKVLAYRSLIRV 1108
Query: 422 DESFSSLKEESISMHGLLEELGRKIVQENSSKEPRKWSRLWLETQVDNVMLEKMFFILCN 481
+ I MH LL ++G++I+ S K + VDN+ ++
Sbjct: 1109 SSN------GEIVMHYLLRQMGKEILHTESKK---------TDKLVDNIQSS----MIAT 1149
Query: 482 ALGKACRSHTFKKKNSQQR 500
+ RS + +K N ++R
Sbjct: 1150 KEIEITRSKSRRKNNKEKR 1168
Score = 31.2 bits (69), Expect = 9.0
Identities = 18/39 (46%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query: 560 NLERLDLEGCIKLVQIDPSIGLLTKLVYLNLKDCKHIIS 598
NLE +DL+GC +L Q P+ G L L +NL C I S
Sbjct: 616 NLEVVDLQGCTRL-QSFPATGQLLHLRVVNLSGCTEIKS 653
>WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY
DNA-binding protein 52) (Disease resistance protein
RRS1) (Resistance to Ralstonia solanacearum 1 protein)
Length = 1378
Score = 110 bits (275), Expect = 1e-23
Identities = 122/480 (25%), Positives = 212/480 (43%), Gaps = 78/480 (16%)
Query: 33 EDTRNNFTYHLFDAFNREGILAFRDDTNLPKGESIASELLRAIEDSYIFVAVLSRNYASS 92
E+ R +F HL +A R+GI D ++ + + E IE + + V VL N S
Sbjct: 17 EEVRYSFVSHLSEALRRKGINNVVVDVDID--DLLFKESQAKIEKAGVSVMVLPGNCDPS 74
Query: 93 IWCLQELEKILECVHVSKKH-VLPVFY-----------DVDPPVVRK--QSGIYCEAFVK 138
L + K+LEC +K V+ V Y ++D + + QS C +
Sbjct: 75 EVWLDKFAKVLECQRNNKDQAVVSVLYGDSLLRDQWLSELDFRGLSRIHQSRKECSDSIL 134
Query: 139 HEQIFQ-------------------QDSQMVLRWREALTQVA-----GLSGCDLRDNLYD 174
E+I + + MV + + V G+ L ++D
Sbjct: 135 VEEIVRDVYETHFYVGRIGIYSKLLEIENMVNKQPIGIRCVGIWGMPGIGKTTLAKAVFD 194
Query: 175 RISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNLSTASNLIRRRLCR 234
++S F A CFI+D K +++Q+L I LS+ +R RL
Sbjct: 195 QMSSAFDASCFIEDYDKSIHEKGLYCLLEEQLLPGNDAT----IMKLSS----LRDRLNS 246
Query: 235 QRVLLIFDNVDKVEQLEKIGVCVW*MVRWGSKIIIISRDEHILKFFGVDEVYKVPLLDRT 294
+RVL++ D+V E + + + GS III SRD+ + + G++++Y+V L+
Sbjct: 247 KRVLVVLDDVCNALVAESF-LEGFDWLGPGSLIIITSRDKQVFRLCGINQIYEVQGLNEK 305
Query: 295 NSLQLLCRKAFKLD------------HILSSMKGWSMAYYIMLRAHR*QLKYRAHFCGRD 342
+ QL A ++ ++S G +A + R +LK + +
Sbjct: 306 EARQLFLLSASIMEDMGEQNLHELSVRVISYANGNPLAISVYGR----ELKGK-----KK 356
Query: 343 IFEWKSALARLRDSPDKDVMDVLRLSFDGLEESEKEIFLHIACFFNPSMEKYVKNVLNCC 402
+ E ++A +L+ P ++D + S+D L ++EK IFL IACFF YV +L C
Sbjct: 357 LSEMETAFLKLKRRPPFKIVDAFKSSYDTLSDNEKNIFLDIACFFQGENVNYVIQLLEGC 416
Query: 403 GFHADIGLRVLIDKSLISIDESFSSLKEESISMHGLLEELGRKIVQENSSKEPRKWSRLW 462
GF + + VL+DK L++I E+ + +H L +++GR+I+ + + R+ RLW
Sbjct: 417 GFFPHVEIDVLVDKCLVTISEN-------RVWLHKLTQDIGREIINGETVQIERR-RRLW 468
Score = 53.5 bits (127), Expect = 2e-06
Identities = 31/89 (34%), Positives = 49/89 (54%), Gaps = 6/89 (6%)
Query: 363 DVLRLSFDGLEESEKEIFLHIACFFNPSMEKYVKNVLNCCGFHADIGLRVLIDKSLISID 422
+VLR+S+D L+E +K +FL+IA FN +V ++ GL+VL D SLIS+
Sbjct: 1087 EVLRVSYDDLQEMDKVLFLYIASLFNDEDVDFVAPLIAGIDLDVSSGLKVLADVSLISVS 1146
Query: 423 ESFSSLKEESISMHGLLEELGRKIVQENS 451
+ I MH L ++G++I+ S
Sbjct: 1147 SN------GEIVMHSLQRQMGKEILHGQS 1169
Score = 33.1 bits (74), Expect = 2.4
Identities = 19/41 (46%), Positives = 26/41 (63%), Gaps = 1/41 (2%)
Query: 560 NLERLDLEGCIKLVQIDPSIGLLTKLVYLNLKDCKHIISLL 600
NLE +DL+GC +L Q P+ G L +L +NL C I S+L
Sbjct: 623 NLEVIDLQGCTRL-QNFPAAGRLLRLRVVNLSGCIKIKSVL 662
>WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY
DNA-binding protein 19)
Length = 1895
Score = 105 bits (263), Expect = 3e-22
Identities = 83/312 (26%), Positives = 149/312 (47%), Gaps = 14/312 (4%)
Query: 162 GLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNL 221
G+ + + ++ +IS Q+ C + D+ K + G V++ L + E + I
Sbjct: 847 GIGKTTIAEEIFRKISVQYETCVVLKDLHKEVEVK-GHDAVRENFLSEVLEVEPHVIRIS 905
Query: 222 STASNLIRRRLCRQRVLLIFDNVDKVEQLEK-IGVCVW*MVRWGSKIIIISRDEHILKFF 280
++ +R RL R+R+L+I D+V+ ++ +G + GS+II+ SR+ +
Sbjct: 906 DIKTSFLRSRLQRKRILVILDDVNDYRDVDTFLGTLNY--FGPGSRIIMTSRNRRVFVLC 963
Query: 281 GVDEVYKVPLLDRTNSLQLLCRKAFKLDHILSSMKGWSMAYYIMLRAHR*QLKYRAHFCG 340
+D VY+V LD SL LL R ++ K S+ + L++ +
Sbjct: 964 KIDHVYEVKPLDIPKSLLLLDRGTCQIVLSPEVYKTLSLELVKFSNGNPQVLQFLSSIDR 1023
Query: 341 RDIFEWKSALARLRDSPDKDVMDVLRLSFDGLEESEKEIFLHIACFFNPSMEKYVKNVLN 400
EW ++ + + + S GL+++E+ IFL IACFFN + V +L+
Sbjct: 1024 ----EWNKLSQEVKTTSPIYIPGIFEKSCCGLDDNERGIFLDIACFFNRIDKDNVAMLLD 1079
Query: 401 CCGFHADIGLRVLIDKSLISIDESFSSLKEESISMHGLLEELGRKIVQENSSKEPRKWSR 460
CGF A +G R L+DKSL++I + + M ++ GR+IV++ S+ P SR
Sbjct: 1080 GCGFSAHVGFRGLVDKSLLTISQ------HNLVDMLSFIQATGREIVRQESADRPGDRSR 1133
Query: 461 LWLETQVDNVML 472
LW + +V +
Sbjct: 1134 LWNADYIRHVFI 1145
Score = 47.8 bits (112), Expect = 9e-05
Identities = 27/65 (41%), Positives = 38/65 (57%), Gaps = 1/65 (1%)
Query: 550 TIMGRQEEFPNLERLDLEGCIKLVQIDPSIGLLTKLVYLNLKDCKHIISLLSNIFGLSCL 609
T + R NLE +DLEGC L+ + SI L KLV+LNLK C + ++ S + L L
Sbjct: 1272 TKIPRLSSATNLEHIDLEGCNSLLSLSQSISYLKKLVFLNLKGCSKLENIPSMV-DLESL 1330
Query: 610 DDLNI 614
+ LN+
Sbjct: 1331 EVLNL 1335
Score = 35.4 bits (80), Expect = 0.48
Identities = 35/148 (23%), Positives = 57/148 (37%), Gaps = 26/148 (17%)
Query: 24 YDAFVTFRGEDTRN-NFTYHLFDAFNREGILAFRDDTNLPKGESIASELLRAIEDSYIFV 82
YD + + D N +F HL + R GI + + A+ + +
Sbjct: 668 YDVVIRYGRADISNEDFISHLRASLCRRGISVYEKFNEVD-----------ALPKCRVLI 716
Query: 83 AVLSRNYASSIWCLQELEKILECVHVSKKHVLPVFYDVDPPVVRKQSGIYCEAFVKHEQI 142
VL+ Y S L ILE H + V P+FY + P C + +E+
Sbjct: 717 IVLTSTYVPS-----NLLNILEHQHTEDRVVYPIFYRLSP------YDFVCNS-KNYERF 764
Query: 143 FQQDSQMVLRWREALTQVAGLSGCDLRD 170
+ QD +W+ AL ++ + G L D
Sbjct: 765 YLQDEPK--KWQAALKEITQMPGYTLTD 790
>DR32_ARATH (Q9FG91) Probable disease resistance protein At5g43730
Length = 848
Score = 50.1 bits (118), Expect = 2e-05
Identities = 77/326 (23%), Positives = 146/326 (44%), Gaps = 44/326 (13%)
Query: 84 VLSRNYASSIWCLQELEKILECVHVSKKHVLPVFYDVDPPVVRKQS----GIYCEAFVKH 139
+ S NY + ++ LE++ E + SKK+ V + P +K G+ +
Sbjct: 109 ISSYNYGGKV--MKNLEEVKELL--SKKNFEVVAQKIIPKAEKKHIQTTVGLDTMVGIAW 164
Query: 140 EQIFQQDSQMVLRWREALTQVAGLSGCDLRDNLYDR---ISHQFGACCFIDDVSKMFRLH 196
E + + + + L + G+ L ++L ++ + +F ++ VSK F+L
Sbjct: 165 ESLIDDEIRTL-----GLYGMGGIGKTTLLESLNNKFVELESEFDVVIWVV-VSKDFQLE 218
Query: 197 DGPLGVQKQILYQTHGEEHNQICNLSTASNLIRRRLCRQRVLLIFDNVDKVEQLEKIGVC 256
G+Q QIL + ++ + S ++LI L R++ +L+ D++ L KIGV
Sbjct: 219 ----GIQDQILGRLRPDKEWERETESKKASLINNNLKRKKFVLLLDDLWSEVDLIKIGVP 274
Query: 257 VW*MVRWGSKIIIISRDEHILKFFGVDEVYKVPLLDRTNSLQLLCRKAFKL---DHILSS 313
GSKI+ +R + + K D+ KV L + +L F+L D IL S
Sbjct: 275 PPSREN-GSKIVFTTRSKEVCKHMKADKQIKVDCLSPDEAWEL-----FRLTVGDIILRS 328
Query: 314 MKGW-SMAYYIMLRAHR*QLKY----RAHFCGRDIFEWKSALARLRDSP-------DKDV 361
+ ++A + + H L +A C + EW+ A+ + +SP ++ +
Sbjct: 329 HQDIPALARIVAAKCHGLPLALNVIGKAMVCKETVQEWRHAI-NVLNSPGHKFPGMEERI 387
Query: 362 MDVLRLSFDGLEESE-KEIFLHIACF 386
+ +L+ S+D L+ E K FL+ + F
Sbjct: 388 LPILKFSYDSLKNGEIKLCFLYCSLF 413
>DR33_ARATH (Q9FG90) Probable disease resistance protein At5g43740
Length = 862
Score = 46.6 bits (109), Expect = 2e-04
Identities = 62/234 (26%), Positives = 106/234 (44%), Gaps = 29/234 (12%)
Query: 189 VSKMFRLHDGPLGVQKQILYQTHGEEHNQICNLSTASNLIRRRLCRQRVLLIFDNVDKVE 248
VSK F+ G+Q QIL + ++ + S ++LI L R++ +L+ D++
Sbjct: 210 VSKDFQFE----GIQDQILGRLRSDKEWERETESKKASLIYNNLERKKFVLLLDDLWSEV 265
Query: 249 QLEKIGVCVW*MVRWGSKIIIISRDEHILKFFGVDEVYKVPLLDRTNSLQLLCRKAFKL- 307
+ KIGV GSKI+ +R + K D+ KV L + +L F+L
Sbjct: 266 DMTKIGVPPPTREN-GSKIVFTTRSTEVCKHMKADKQIKVACLSPDEAWEL-----FRLT 319
Query: 308 --DHILSSMKGW-SMAYYIMLRAHR*QLKY----RAHFCGRDIFEWKSALARLRDS---- 356
D IL S + ++A + + H L +A C I EW A+ L +
Sbjct: 320 VGDIILRSHQDIPALARIVAAKCHGLPLALNVIGKAMSCKETIQEWSHAINVLNSAGHEF 379
Query: 357 --PDKDVMDVLRLSFDGLEESE-KEIFLHIACFFNPS---MEKYVKNVLNCCGF 404
++ ++ +L+ S+D L+ E K FL+ + F S EK+++ + C GF
Sbjct: 380 PGMEERILPILKFSYDSLKNGEIKLCFLYCSLFPEDSEIPKEKWIEYWI-CEGF 432
>TLR1_HUMAN (Q15399) Toll-like receptor 1 precursor
(Toll/interleukin-1 receptor-like) (TIL)
Length = 786
Score = 43.5 bits (101), Expect = 0.002
Identities = 26/84 (30%), Positives = 42/84 (49%), Gaps = 7/84 (8%)
Query: 19 RRKNYYDAFVTFRGEDT---RNNFTYHLFDAFNREGILAFRDDTNLPKGESIASELLRAI 75
+R + AF+++ G D+ +N +L +EG+ + N G+SI ++ I
Sbjct: 632 QRNLQFHAFISYSGHDSFWVKNELLPNL----EKEGMQICLHERNFVPGKSIVENIITCI 687
Query: 76 EDSYIFVAVLSRNYASSIWCLQEL 99
E SY + VLS N+ S WC EL
Sbjct: 688 EKSYKSIFVLSPNFVQSEWCHYEL 711
>TLR2_BOVIN (Q95LA9) Toll-like receptor 2 precursor
Length = 784
Score = 42.7 bits (99), Expect = 0.003
Identities = 28/85 (32%), Positives = 41/85 (47%), Gaps = 3/85 (3%)
Query: 18 PRRKNYYDAFVTFRGEDTR--NNFTYHLFDAFNREGILAFRDDTNLPKGESIASELLRAI 75
PRR YDAFV++ D+ N + FN L +P G+ I ++ +I
Sbjct: 635 PRRDICYDAFVSYSERDSYWVENLMVQELEHFNPPFKLCLHKRDFIP-GKWIIDNIIDSI 693
Query: 76 EDSYIFVAVLSRNYASSIWCLQELE 100
E S+ + VLS N+ S WC EL+
Sbjct: 694 EKSHKTIFVLSENFVKSEWCKYELD 718
>DR27_ARATH (Q9T048) Putative disease resistance protein At4g27190
Length = 985
Score = 41.2 bits (95), Expect = 0.009
Identities = 68/282 (24%), Positives = 117/282 (41%), Gaps = 37/282 (13%)
Query: 180 FGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNLSTASNLIRRRLCRQRVLL 239
FG F+ VSK F P VQKQI + + + A + + ++ LL
Sbjct: 197 FGLVIFVI-VSKEF----DPREVQKQIAERLDIDTQMEESEEKLARRIYVGLMKERKFLL 251
Query: 240 IFDNVDKVEQLEKIGVCVW*MVRWGSKIIIISRDEHILKFFGVDEVYKVPLLDRTNSLQL 299
I D+V K L+ +G+ + GSK+I+ SR + + D +V L ++ +L
Sbjct: 252 ILDDVWKPIDLDLLGIPRTEENK-GSKVILTSRFLEVCRSMKTDLDVRVDCLLEEDAWEL 310
Query: 300 LCRKA---FKLDHILSSMK-------GWSMAYYIMLRAHR*QLKYRAHFCGRDIFEWKSA 349
C+ A + DH+ K G +A + A R + +++ W
Sbjct: 311 FCKNAGDVVRSDHVRKIAKAVSQECGGLPLAIITVGTAMRGK---------KNVKLWNHV 361
Query: 350 LARLRDSP------DKDVMDVLRLSFDGLEESEKEIFLHIACFFNPSMEKYVKNVLNCCG 403
L++L S ++ + L+LS+D LE+ K FL A F E Y V
Sbjct: 362 LSKLSKSVPWIKSIEEKIFQPLKLSYDFLEDKAKFCFLLCALF----PEDYSIEVTEVVR 417
Query: 404 FHADIGLRVLIDKSLISIDESFSSLKEESISMHGLLEELGRK 445
+ G + S++E +++ ES+ + LLE+ R+
Sbjct: 418 YWMAEGFMEELGSQEDSMNEGITTV--ESLKDYCLLEDGDRR 457
>TLR2_MACFA (Q95M53) Toll-like receptor 2 precursor
Length = 784
Score = 40.8 bits (94), Expect = 0.011
Identities = 28/85 (32%), Positives = 39/85 (44%), Gaps = 3/85 (3%)
Query: 18 PRRKNYYDAFVTFRGEDTR--NNFTYHLFDAFNREGILAFRDDTNLPKGESIASELLRAI 75
P R YDAFV++ D N + FN L +P G+ I ++ +I
Sbjct: 635 PNRDICYDAFVSYSERDAYWVENLMVQELENFNPPFKLCLHKRDFIP-GKWIIDNIIDSI 693
Query: 76 EDSYIFVAVLSRNYASSIWCLQELE 100
E S+ V VLS N+ S WC EL+
Sbjct: 694 EKSHKTVFVLSENFVKSEWCKYELD 718
>TLR2_HUMAN (O60603) Toll-like receptor 2 precursor
(Toll/interleukin 1 receptor-like protein 4)
Length = 784
Score = 40.8 bits (94), Expect = 0.011
Identities = 28/85 (32%), Positives = 39/85 (44%), Gaps = 3/85 (3%)
Query: 18 PRRKNYYDAFVTFRGEDTR--NNFTYHLFDAFNREGILAFRDDTNLPKGESIASELLRAI 75
P R YDAFV++ D N + FN L +P G+ I ++ +I
Sbjct: 635 PSRNICYDAFVSYSERDAYWVENLMVQELENFNPPFKLCLHKRDFIP-GKWIIDNIIDSI 693
Query: 76 EDSYIFVAVLSRNYASSIWCLQELE 100
E S+ V VLS N+ S WC EL+
Sbjct: 694 EKSHKTVFVLSENFVKSEWCKYELD 718
>TLR6_HUMAN (Q9Y2C9) Toll-like receptor 6 precursor
Length = 796
Score = 38.9 bits (89), Expect = 0.043
Identities = 24/81 (29%), Positives = 37/81 (45%), Gaps = 1/81 (1%)
Query: 19 RRKNYYDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDDTNLPKGESIASELLRAIEDS 78
+R + AF+++ D+ L +E I + N G+SI ++ IE S
Sbjct: 637 QRNLQFHAFISYSEHDSAW-VKSELVPYLEKEDIQICLHERNFVPGKSIVENIINCIEKS 695
Query: 79 YIFVAVLSRNYASSIWCLQEL 99
Y + VLS N+ S WC EL
Sbjct: 696 YKSIFVLSPNFVQSEWCHYEL 716
>TLR1_MOUSE (Q9EPQ1) Toll-like receptor 1 precursor
(Toll/interleukin-1 receptor-like) (TIL)
Length = 795
Score = 38.5 bits (88), Expect = 0.056
Identities = 24/81 (29%), Positives = 38/81 (46%), Gaps = 1/81 (1%)
Query: 19 RRKNYYDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDDTNLPKGESIASELLRAIEDS 78
+R + AFV++ G D+ L ++ I + N G+SI ++ IE S
Sbjct: 635 QRNLQFHAFVSYSGHDSAW-VKNELLPNLEKDDIQICLHERNFVPGKSIVENIINFIEKS 693
Query: 79 YIFVAVLSRNYASSIWCLQEL 99
Y + VLS ++ S WC EL
Sbjct: 694 YKSIFVLSPHFIQSEWCHYEL 714
>TLR2_MOUSE (Q9QUN7) Toll-like receptor 2 precursor
Length = 784
Score = 36.2 bits (82), Expect = 0.28
Identities = 26/85 (30%), Positives = 40/85 (46%), Gaps = 3/85 (3%)
Query: 18 PRRKNYYDAFVTFRGEDTR--NNFTYHLFDAFNREGILAFRDDTNLPKGESIASELLRAI 75
P R YDAFV++ +D+ N + + L +P G+ I ++ +I
Sbjct: 635 PCRDVCYDAFVSYSEQDSHWVENLMVQQLENSDPPFKLCLHKRDFVP-GKWIIDNIIDSI 693
Query: 76 EDSYIFVAVLSRNYASSIWCLQELE 100
E S+ V VLS N+ S WC EL+
Sbjct: 694 EKSHKTVFVLSENFVRSEWCKYELD 718
>TLR6_MOUSE (Q9EPW9) Toll-like receptor 6 precursor
Length = 795
Score = 35.8 bits (81), Expect = 0.37
Identities = 23/81 (28%), Positives = 37/81 (45%), Gaps = 1/81 (1%)
Query: 19 RRKNYYDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDDTNLPKGESIASELLRAIEDS 78
+R + AFV++ D+ L ++ I + N G+SI ++ IE S
Sbjct: 637 QRNLQFHAFVSYSEHDSAW-VKNELLPNLEKDDIRVCLHERNFVPGKSIVENIINFIEKS 695
Query: 79 YIFVAVLSRNYASSIWCLQEL 99
Y + VLS ++ S WC EL
Sbjct: 696 YKAIFVLSPHFIQSEWCHYEL 716
>TLR5_MOUSE (Q9JLF7) Toll-like receptor 5 precursor
Length = 859
Score = 34.7 bits (78), Expect = 0.81
Identities = 44/159 (27%), Positives = 66/159 (40%), Gaps = 24/159 (15%)
Query: 24 YDAFVTFRGED---TRNNFTYHL---FDAFNREGILAFRDDTNLPKGESIASELLRAIED 77
YDA+ F +D +N HL + + NR L F + +P GE+ S + A+
Sbjct: 694 YDAYFCFSSKDFEWAQNALLKHLDAHYSSRNRLR-LCFEERDFIP-GENHISNIQAAVWG 751
Query: 78 SYIFVAVLSRNYASSIWCLQELEKILECVHVSKKHVLPVFYDVDPPVVRKQSGIYCEAFV 137
S V ++SR++ WCL+ K +L +V + +
Sbjct: 752 SRKTVCLVSRHFLKDGWCLEAFRYAQSRSLSDLKSIL---------IVVVVGSLSQYQLM 802
Query: 138 KHEQI--FQQDSQMVLRWREALTQVA----GLSGCDLRD 170
+HE I F Q Q LRW E L V LSGC L++
Sbjct: 803 RHETIRGFLQ-KQQYLRWPEDLQDVGWFLDKLSGCILKE 840
>VIB4_AGRTU (P05353) VirB4 protein precursor
Length = 789
Score = 34.3 bits (77), Expect = 1.1
Identities = 26/107 (24%), Positives = 48/107 (44%), Gaps = 8/107 (7%)
Query: 88 NYASSIWCLQELEKILECVHVSKKHVLPVFYDVDPPVVRKQSGIYCEAFVKHEQIFQQDS 147
+Y SI+ +E ++ +L +D P+V QS + H ++ + S
Sbjct: 233 SYVGSIYSFREYPA------KTRPGMLNALLSLDFPLVLTQSFSFLTRPQAHAKLSLKSS 286
Query: 148 QMVLRWREALTQVAGLSGCD--LRDNLYDRISHQFGACCFIDDVSKM 192
QM+ +A+TQ+ LS + L N + SH C + DD++ +
Sbjct: 287 QMLSSGDKAVTQIGKLSEAEDALASNEFVMGSHHLSLCVYADDLNSL 333
>VIB4_AGRT5 (P17794) VirB4 protein precursor
Length = 789
Score = 34.3 bits (77), Expect = 1.1
Identities = 25/107 (23%), Positives = 50/107 (46%), Gaps = 8/107 (7%)
Query: 88 NYASSIWCLQELEKILECVHVSKKHVLPVFYDVDPPVVRKQSGIYCEAFVKHEQIFQQDS 147
+Y SI+ +E ++ +L V +D P+V QS + H ++ + S
Sbjct: 233 SYVGSIYSFREYPA------TTRPGMLNVLLSLDFPLVLTQSFSFLTRSQAHSKLSLKSS 286
Query: 148 QMVLRWREALTQVAGLSGCD--LRDNLYDRISHQFGACCFIDDVSKM 192
QM+ +A+TQ++ LS + L N + +H C + +D++ +
Sbjct: 287 QMLSSGDKAVTQISKLSEAEDALASNEFVLGAHHVSLCIYANDLNNL 333
>TLR4_PANPA (Q9TTN0) Toll-like receptor 4 precursor
Length = 839
Score = 34.3 bits (77), Expect = 1.1
Identities = 31/123 (25%), Positives = 56/123 (45%), Gaps = 14/123 (11%)
Query: 19 RRKNYYDAFVTFRGED---TRNNFTYHLFDAFNREGILAFR---DDTNLPKGESIASELL 72
R +N YDAFV + +D RN +L EG+ F+ + G +IA+ ++
Sbjct: 669 RGENIYDAFVIYSSQDEDWVRNELVKNL-----EEGVPPFQLCLHYRDFIPGVAIAANII 723
Query: 73 -RAIEDSYIFVAVLSRNYASSIWCLQELEKILECVHVSKKH--VLPVFYDVDPPVVRKQS 129
S + V+S+++ S WC+ E E +S + + V V+ ++R+Q
Sbjct: 724 HEGFHKSRKVIVVVSQHFIQSRWCIFEYEIAQTWQFLSSRAGIIFIVLQKVEKTLLRRQV 783
Query: 130 GIY 132
+Y
Sbjct: 784 ELY 786
>TLR4_PONPY (Q8SPE9) Toll-like receptor 4 precursor
Length = 828
Score = 33.9 bits (76), Expect = 1.4
Identities = 31/123 (25%), Positives = 56/123 (45%), Gaps = 14/123 (11%)
Query: 19 RRKNYYDAFVTFRGED---TRNNFTYHLFDAFNREGILAFR---DDTNLPKGESIASELL 72
R +N YDAFV + +D RN +L EG+ F+ + G +IA+ ++
Sbjct: 667 RGENTYDAFVIYSSQDEDWVRNELVKNL-----EEGVPTFQLCLHYRDFIPGVAIAANII 721
Query: 73 -RAIEDSYIFVAVLSRNYASSIWCLQELEKILECVHVSKKH--VLPVFYDVDPPVVRKQS 129
S + V+S+++ S WC+ E E +S + + V V+ ++R+Q
Sbjct: 722 HEGFHKSRKVIVVVSQHFIQSRWCIFEYEIAQTWQFLSSRAGIIFIVLQKVEKTLLRQQV 781
Query: 130 GIY 132
+Y
Sbjct: 782 ELY 784
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.333 0.145 0.451
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 72,040,108
Number of Sequences: 164201
Number of extensions: 2935648
Number of successful extensions: 7835
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 27
Number of HSP's that attempted gapping in prelim test: 7750
Number of HSP's gapped (non-prelim): 97
length of query: 657
length of database: 59,974,054
effective HSP length: 117
effective length of query: 540
effective length of database: 40,762,537
effective search space: 22011769980
effective search space used: 22011769980
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 69 (31.2 bits)
Medicago: description of AC144502.3


