

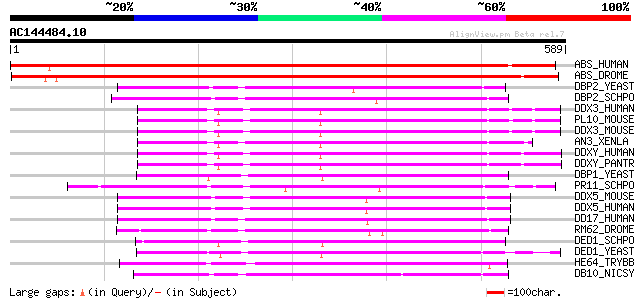
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144484.10 - phase: 0
(589 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ABS_HUMAN (Q9UJV9) DEAD-box protein abstrakt homolog (DEAD-box p... 708 0.0
ABS_DROME (Q9V3C0) DEAD-box protein abstrakt 667 0.0
DBP2_YEAST (P24783) P68-like protein 303 6e-82
DBP2_SCHPO (P24782) P68-like protein 300 7e-81
DDX3_HUMAN (O00571) DEAD-box protein 3 (Helicase-like protein 2)... 296 1e-79
PL10_MOUSE (P16381) Putative ATP-dependent RNA helicase PL10 295 2e-79
DDX3_MOUSE (Q62167) DEAD-box protein 3 (DEAD-box RNA helicase DE... 293 7e-79
AN3_XENLA (P24346) Putative ATP-dependent RNA helicase An3 293 9e-79
DDXY_HUMAN (O15523) DEAD-box protein 3, Y-chromosomal 293 1e-78
DDXY_PANTR (Q6GVM6) DEAD-box protein 3, Y-chromosomal 291 3e-78
DBP1_YEAST (P24784) Probable ATP-dependent RNA helicase DBP1 (He... 291 3e-78
PR11_SCHPO (Q9P7C7) Probable ATP-dependent RNA helicase prp11 291 4e-78
DDX5_MOUSE (Q61656) Probable RNA-dependent helicase p68 (DEAD-bo... 290 9e-78
DDX5_HUMAN (P17844) Probable RNA-dependent helicase p68 (DEAD-bo... 290 9e-78
DD17_HUMAN (Q92841) Probable RNA-dependent helicase p72 (DEAD-bo... 286 1e-76
RM62_DROME (P19109) ATP-dependent RNA helicase P62 286 1e-76
DED1_SCHPO (O13370) ATP-dependent RNA helicase ded1 285 2e-76
DED1_YEAST (P06634) Probable ATP-dependent RNA helicase DED1 281 3e-75
HE64_TRYBB (Q26696) Putative DEAD-box RNA helicase HEL64 280 7e-75
DB10_NICSY (P46942) RNA helicase-like protein DB10 278 4e-74
>ABS_HUMAN (Q9UJV9) DEAD-box protein abstrakt homolog (DEAD-box
protein 41)
Length = 622
Score = 708 bits (1828), Expect = 0.0
Identities = 358/590 (60%), Positives = 459/590 (77%), Gaps = 15/590 (2%)
Query: 2 EEEDDYEEYIPVAKRRAMEAQKILQRKGKATAAIQEDDS--------EKLKV-VETKPSL 52
E+++DY Y+P+ +RR + QK+LQR+ K A ++ DS + + + ++ SL
Sbjct: 28 EDDEDYVPYVPLRQRRQLLLQKLLQRRRKGAAEEEQQDSGSEPRGDEDDIPLGPQSNVSL 87
Query: 53 LVKASQLKK--DQPEISVTEQIVQQEKEMIENLSDKKTLMSVRELAKGITYTEPLPTGWK 110
L + LK+ + + S E+ +++E++++E++++ + LMSV+E+AKGITY +P+ T W
Sbjct: 88 LDQHQHLKEKAEARKESAKEKQLKEEEKILESVAEGRALMSVKEMAKGITYDDPIKTSWT 147
Query: 111 PPLHIRRMSKKDCDLIQKQWHIIVNGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPI 170
PP ++ MS++ + ++K++HI+V G+ IPPPIK+FK+M+FP IL+ LK KGI PTPI
Sbjct: 148 PPRYVLSMSEERHERVRKKYHILVEGDGIPPPIKSFKEMKFPAAILRGLKKKGIHHPTPI 207
Query: 171 QVQGLPVILSGRDMIGIAFTGSGKTLVFVLPMIMMAMQEEIMMPIVPGEGPFGLIICPSR 230
Q+QG+P ILSGRDMIGIAFTGSGKTLVF LP+IM +++E +P EGP+GLIICPSR
Sbjct: 208 QIQGIPTILSGRDMIGIAFTGSGKTLVFTLPVIMFCLEQEKRLPFSKREGPYGLIICPSR 267
Query: 231 ELARQTYEVIEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDM 290
ELARQT+ ++E + L+E P LR LCIGG+ ++ Q+E ++ GVH++VATPGRL D+
Sbjct: 268 ELARQTHGILEYYCRLLQEDSSPLLRCALCIGGMSVKEQMETIRHGVHMMVATPGRLMDL 327
Query: 291 LAKKKMNLDNCRYLTLDEADRLVDLGFEDDIREVFDHFKAQRQTLLFSATMPTKIQNFAR 350
L KK ++LD CRYL LDEADR++D+GFE DIR +F +FK QRQTLLFSATMP KIQNFA+
Sbjct: 328 LQKKMVSLDICRYLALDEADRMIDMGFEGDIRTIFSYFKGQRQTLLFSATMPKKIQNFAK 387
Query: 351 SALVKPIIVNVGRAGAANLDVIQEVEYVKQEAKIVYLLECLQKTPPPVLIFCENKADVDD 410
SALVKP+ +NVGRAGAA+LDVIQEVEYVK+EAK+VYLLECLQKTPPPVLIF E KADVD
Sbjct: 388 SALVKPVTINVGRAGAASLDVIQEVEYVKEEAKMVYLLECLQKTPPPVLIFAEKKADVDA 447
Query: 411 IHEYLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINY 470
IHEYLLLKGVEAVAIHGGKDQEER AI +F+ GKKDVLVATDVASKGLDFP IQHVINY
Sbjct: 448 IHEYLLLKGVEAVAIHGGKDQEERTKAIEAFREGKKDVLVATDVASKGLDFPAIQHVINY 507
Query: 471 DMPAEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAELV 530
DMP EIENYVHRIGRTGR G TGIATTFINK E+ L+DLK LL EAKQ++PPVL L
Sbjct: 508 DMPEEIENYVHRIGRTGRSGNTGIATTFINKACDESVLMDLKALLLEAKQKVPPVLQVL- 566
Query: 531 DPMEDNEEITGISGVKGCAYCGGLGHRIRDCPKLEHQKSVAIAN-NRKDY 579
+E + I G +GCA+CGGLGHRI DCPKLE ++ ++N RKDY
Sbjct: 567 --HCGDESMLDIGGERGCAFCGGLGHRITDCPKLEAMQTKQVSNIGRKDY 614
>ABS_DROME (Q9V3C0) DEAD-box protein abstrakt
Length = 619
Score = 667 bits (1720), Expect = 0.0
Identities = 338/597 (56%), Positives = 437/597 (72%), Gaps = 19/597 (3%)
Query: 3 EEDDYEEYIPVAKRRAMEAQKILQRKGKATAAIQ---------EDDSEKLKVVET----- 48
+ +DY Y+PV +R+ K+ + + Q EDDS+ VET
Sbjct: 20 DNEDYVPYVPVKERKKQHMIKLGRIVQLVSETAQPKSSSENENEDDSQGAHDVETWGRKY 79
Query: 49 KPSLLVKASQLKK--DQPEISVTEQIVQQEKEMIENLSDKKTLMSVRELAKGITYTEPLP 106
SLL + ++LKK + ++S E+ +++E++++E+++ +K LM V ELAKGI Y +P+
Sbjct: 80 NISLLDQHTELKKIAEAKKLSAVEKQLREEEKIMESIAQQKALMGVAELAKGIQYEQPIK 139
Query: 107 TGWKPPLHIRRMSKKDCDLIQKQWHIIVNGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQ 166
T WKPP +IR MS+++ + ++ + I+V GE PPI++F++M+FP IL L KGI
Sbjct: 140 TAWKPPRYIREMSEEEREAVRHELRILVEGETPSPPIRSFREMKFPKGILNGLAAKGIKN 199
Query: 167 PTPIQVQGLPVILSGRDMIGIAFTGSGKTLVFVLPMIMMAMQEEIMMPIVPGEGPFGLII 226
PTPIQVQGLP +L+GRD+IGIAFTGSGKTLVFVLP+IM A+++E +P EGP+GLII
Sbjct: 200 PTPIQVQGLPTVLAGRDLIGIAFTGSGKTLVFVLPVIMFALEQEYSLPFERNEGPYGLII 259
Query: 227 CPSRELARQTYEVIEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGR 286
CPSRELA+QT+E+I+ + L+ G PE+R L +GG+ + L+++ +GVHIVVATPGR
Sbjct: 260 CPSRELAKQTHEIIQHYSKHLQACGMPEIRSCLAMGGLPVSEALDVISRGVHIVVATPGR 319
Query: 287 LKDMLAKKKMNLDNCRYLTLDEADRLVDLGFEDDIREVFDHFKAQRQTLLFSATMPTKIQ 346
L DML KK + LD CRYL +DEADR++D+GFE+D+R +F FK QRQTLLFSATMP KIQ
Sbjct: 320 LMDMLDKKILTLDMCRYLCMDEADRMIDMGFEEDVRTIFSFFKGQRQTLLFSATMPKKIQ 379
Query: 347 NFARSALVKPIIVNVGRAGAANLDVIQEVEYVKQEAKIVYLLECLQKTPPPVLIFCENKA 406
NFARSALVKP+ +NVGRAGAA+++V Q+VEYVKQEAK+VYLL+CLQKT PPVLIF E K
Sbjct: 380 NFARSALVKPVTINVGRAGAASMNVTQQVEYVKQEAKVVYLLDCLQKTAPPVLIFAEKKQ 439
Query: 407 DVDDIHEYLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQH 466
DVD IHEYLLLKGVEAVAIHGGKDQEER A+ +++ GKKDVLVATDVASKGLDFP++QH
Sbjct: 440 DVDCIHEYLLLKGVEAVAIHGGKDQEERSRAVDAYRVGKKDVLVATDVASKGLDFPNVQH 499
Query: 467 VINYDMPAEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVL 526
VINYDMP +IENYVHRIGRTGR G+ATT INK ++ LLDLKHLL E KQ +P L
Sbjct: 500 VINYDMPDDIENYVHRIGRTGRSNTKGLATTLINKTTEQSVLLDLKHLLIEGKQEVPDFL 559
Query: 527 AELVDPMEDNEEITGISGVKGCAYCGGLGHRIRDCPKLEHQKSVAIAN-NRKDYFGS 582
EL E G S GC YCGGLGHRI +CPKLE ++ +N R+DY +
Sbjct: 560 DELAPETEHQHLDLGDS--HGCTYCGGLGHRITECPKLEAVQNKQASNIGRRDYLSN 614
>DBP2_YEAST (P24783) P68-like protein
Length = 546
Score = 303 bits (777), Expect = 6e-82
Identities = 174/417 (41%), Positives = 244/417 (57%), Gaps = 15/417 (3%)
Query: 115 IRRMSKKDCDLIQKQWHIIVNGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQG 174
+R S + +K+ + ++G +IP PI F + FPD +L +K +G +PT IQ QG
Sbjct: 84 VRDRSDSEIAQFRKENEMTISGHDIPKPITTFDEAGFPDYVLNEVKAEGFDKPTGIQCQG 143
Query: 175 LPVILSGRDMIGIAFTGSGKTLVFVLPMIMMAMQEEIMMPIVPGEGPFGLIICPSRELAR 234
P+ LSGRDM+GIA TGSGKTL + LP I+ + ++ PG+GP L++ P+RELA
Sbjct: 144 WPMALSGRDMVGIAATGSGKTLSYCLPGIVHINAQPLL---APGDGPIVLVLAPTRELAV 200
Query: 235 QTYEVIEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKK 294
Q +F +R GG+ Q+ + +G IV+ATPGRL DML
Sbjct: 201 QIQTECSKF------GHSSRIRNTCVYGGVPKSQQIRDLSRGSEIVIATPGRLIDMLEIG 254
Query: 295 KMNLDNCRYLTLDEADRLVDLGFEDDIREVFDHFKAQRQTLLFSATMPTKIQNFARSALV 354
K NL YL LDEADR++D+GFE IR++ D + RQTL++SAT P +++ A L
Sbjct: 255 KTNLKRVTYLVLDEADRMLDMGFEPQIRKIVDQIRPDRQTLMWSATWPKEVKQLAADYLN 314
Query: 355 KPIIVNVGR---AGAANLDVIQEV--EYVKQEAKIVYLLECLQKTPPPVLIFCENKADVD 409
PI V VG + + N+ I EV ++ K++ YL Q LIF K D
Sbjct: 315 DPIQVQVGSLELSASHNITQIVEVVSDFEKRDRLNKYLETASQDNEYKTLIFASTKRMCD 374
Query: 410 DIHEYLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVIN 469
DI +YL G A+AIHG KDQ ER++ + F+ G+ ++VATDVA++G+D I +VIN
Sbjct: 375 DITKYLREDGWPALAIHGDKDQRERDWVLQEFRNGRSPIMVATDVAARGIDVKGINYVIN 434
Query: 470 YDMPAEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVL 526
YDMP IE+YVHRIGRTGR G TG A +F + Q++ L +++EA Q IPP L
Sbjct: 435 YDMPGNIEDYVHRIGRTGRAGATGTAISFFTE-QNKGLGAKLISIMREANQNIPPEL 490
>DBP2_SCHPO (P24782) P68-like protein
Length = 550
Score = 300 bits (768), Expect = 7e-81
Identities = 175/426 (41%), Positives = 245/426 (57%), Gaps = 15/426 (3%)
Query: 109 WKPPLHIRRMSKKDCDLIQKQWHIIVNGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPT 168
+K ++R S + +K+ I+V+G +P P+ F++ FP+ +LK +K G PT
Sbjct: 87 YKEHENVRNRSDAEVTEYRKEKEIVVHGLNVPKPVTTFEEAGFPNYVLKEVKQLGFEAPT 146
Query: 169 PIQVQGLPVILSGRDMIGIAFTGSGKTLVFVLPMIMMAMQEEIMMPIVPGEGPFGLIICP 228
PIQ Q P+ +SGRDM+GI+ TGSGKTL + LP I+ + ++ P G+GP L++ P
Sbjct: 147 PIQQQAWPMAMSGRDMVGISATGSGKTLSYCLPAIVHINAQPLLSP---GDGPIVLVLAP 203
Query: 229 SRELARQTYEVIEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLK 288
+RELA Q + +F +R GG+ Q+ + +GV I +ATPGRL
Sbjct: 204 TRELAVQIQQECTKF------GKSSRIRNTCVYGGVPRGPQIRDLIRGVEICIATPGRLL 257
Query: 289 DMLAKKKMNLDNCRYLTLDEADRLVDLGFEDDIREVFDHFKAQRQTLLFSATMPTKIQNF 348
DML K NL YL LDEADR++D+GFE IR++ D + RQT++FSAT P ++Q
Sbjct: 258 DMLDSNKTNLRRVTYLVLDEADRMLDMGFEPQIRKIVDQIRPDRQTVMFSATWPKEVQRL 317
Query: 349 ARSALVKPIIVNVGRAG-AANLDVIQEVEYVKQEAKIVYL----LECLQKTPPPVLIFCE 403
AR L I V VG AA+ ++ Q VE V K L E L+ VLIF
Sbjct: 318 ARDYLNDYIQVTVGSLDLAASHNIKQIVEVVDNADKRARLGKDIEEVLKDRDNKVLIFTG 377
Query: 404 NKADVDDIHEYLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPD 463
K DDI +L G A+AIHG K Q+ER++ ++ F+ GK ++VATDVAS+G+D
Sbjct: 378 TKRVADDITRFLRQDGWPALAIHGDKAQDERDWVLNEFRTGKSPIMVATDVASRGIDVKG 437
Query: 464 IQHVINYDMPAEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIP 523
I HV NYD P E+YVHRIGRTGR G G A T+ + ++ +L +L EAKQ I
Sbjct: 438 ITHVFNYDFPGNTEDYVHRIGRTGRAGAKGTAYTYFTSDNAKQA-RELVSILSEAKQDID 496
Query: 524 PVLAEL 529
P L E+
Sbjct: 497 PKLEEM 502
>DDX3_HUMAN (O00571) DEAD-box protein 3 (Helicase-like protein 2)
(HLP2) (DEAD-box, X isoform)
Length = 661
Score = 296 bits (758), Expect = 1e-79
Identities = 173/471 (36%), Positives = 263/471 (55%), Gaps = 39/471 (8%)
Query: 136 GEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGIAFTGSGKT 195
G PP I++F D+ + I+ ++ +PTP+Q +P+I RD++ A TGSGKT
Sbjct: 171 GNNCPPHIESFSDVEMGEIIMGNIELTRYTRPTPVQKHAIPIIKEKRDLMACAQTGSGKT 230
Query: 196 LVFVLPMIMMAMQEEIMMPIVPGEG----------------PFGLIICPSRELARQTYEV 239
F+LP++ + PGE P L++ P+RELA Q YE
Sbjct: 231 AAFLLPILSQIYSDG------PGEALRAMKENGRYGRRKQYPISLVLAPTRELAVQIYEE 284
Query: 240 IEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMNLD 299
+F + +RP + GG D+ Q+ +++G H++VATPGRL DM+ + K+ LD
Sbjct: 285 ARKFSYRSR------VRPCVVYGGADIGQQIRDLERGCHLLVATPGRLVDMMERGKIGLD 338
Query: 300 NCRYLTLDEADRLVDLGFEDDIREVFDHF----KAQRQTLLFSATMPTKIQNFARSALVK 355
C+YL LDEADR++D+GFE IR + + K R T++FSAT P +IQ AR L +
Sbjct: 339 FCKYLVLDEADRMLDMGFEPQIRRIVEQDTMPPKGVRHTMMFSATFPKEIQMLARDFLDE 398
Query: 356 PIIVNVGRAGAANLDVIQEVEYVKQEAKIVYLLECLQKTPPP--VLIFCENKADVDDIHE 413
I + VGR G+ + ++ Q+V +V++ K +LL+ L T L+F E K D + +
Sbjct: 399 YIFLAVGRVGSTSENITQKVVWVEESDKRSFLLDLLNATGKDSLTLVFVETKKGADSLED 458
Query: 414 YLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMP 473
+L +G +IHG + Q +RE A+ F++GK +LVAT VA++GLD +++HVIN+D+P
Sbjct: 459 FLYHEGYACTSIHGDRSQRDREEALHQFRSGKSPILVATAVAARGLDISNVKHVINFDLP 518
Query: 474 AEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAELVDPM 533
++IE YVHRIGRTGR G G+AT+F N+ T DL LL EAKQ +P L +
Sbjct: 519 SDIEEYVHRIGRTGRVGNLGLATSFFNERNINIT-KDLLDLLVEAKQEVPSWLENMA--Y 575
Query: 534 EDNEEITGISGVKGCAYCGGLGHRIRDCPKLEHQKSVAIANNRKDYFGSGG 584
E + + + K + GG G RD + S + +++R SGG
Sbjct: 576 EHHYKGSSRGRSKSSRFSGGFG--ARDYRQSSGASSSSFSSSRASSSRSGG 624
>PL10_MOUSE (P16381) Putative ATP-dependent RNA helicase PL10
Length = 660
Score = 295 bits (756), Expect = 2e-79
Identities = 173/471 (36%), Positives = 262/471 (54%), Gaps = 40/471 (8%)
Query: 136 GEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGIAFTGSGKT 195
G PP I++F D+ + I+ ++ +PTP+Q +P+I RD++ A TGSGKT
Sbjct: 171 GNNCPPHIESFSDVEMGEIIMGNIELTRYTRPTPVQKHAIPIIKEKRDLMACAQTGSGKT 230
Query: 196 LVFVLPMIMMAMQEEIMMPIVPGEG----------------PFGLIICPSRELARQTYEV 239
F+LP++ + PGE P L++ P+RELA Q YE
Sbjct: 231 AAFLLPILSQIYTDG------PGEALRAMKENGKYGRRKQYPISLVLAPTRELAVQIYEE 284
Query: 240 IEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMNLD 299
+F + +RP + GG D+ Q+ +++G H++VATPGRL DM+ + K+ LD
Sbjct: 285 ARKFSYRSR------VRPCVVYGGADIGQQIRDLERGCHLLVATPGRLVDMMERGKIGLD 338
Query: 300 NCRYLTLDEADRLVDLGFEDDIREVFDHF----KAQRQTLLFSATMPTKIQNFARSALVK 355
C+YL LDEADR++D+GFE IR + + K R T++FSAT P +IQ AR L +
Sbjct: 339 FCKYLVLDEADRMLDMGFEPQIRRIVEQDTMPPKGVRHTMMFSATFPKEIQMLARDFLDE 398
Query: 356 PIIVNVGRAGAANLDVIQEVEYVKQEAKIVYLLECLQKTPPP--VLIFCENKADVDDIHE 413
I + VGR G+ + ++ Q+V +V++ K +LL+ L T +L+F E K D + +
Sbjct: 399 YIFLAVGRVGSTSENITQKVVWVEEADKRSFLLDLLNATGKDSLILVFVETKKGADSLED 458
Query: 414 YLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMP 473
+L +G +IHG + Q +RE A+ F++GK +LVAT VA++GLD +++HVIN+D+P
Sbjct: 459 FLYHEGYACTSIHGDRSQRDREEALHQFRSGKSPILVATAVAARGLDISNVKHVINFDLP 518
Query: 474 AEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAELVDPM 533
++IE YVHRIGRTGR G G+AT+F N+ T DL LL EAKQ +P L +
Sbjct: 519 SDIEEYVHRIGRTGRVGNLGLATSFFNERNINIT-KDLLDLLVEAKQEVPSWLENMA--- 574
Query: 534 EDNEEITGISGVKGCAYCGGLGHRIRDCPKLEHQKSVAIANNRKDYFGSGG 584
++ G G + GG G RD + S + ++ R SGG
Sbjct: 575 FEHHYKGGSRGRSKSRFSGGFG--ARDYRQSSGASSSSFSSGRASNSRSGG 623
>DDX3_MOUSE (Q62167) DEAD-box protein 3 (DEAD-box RNA helicase
DEAD3) (mDEAD3) (Embryonic RNA helicase) (D1PAS1 related
sequence 2)
Length = 661
Score = 293 bits (751), Expect = 7e-79
Identities = 172/471 (36%), Positives = 263/471 (55%), Gaps = 39/471 (8%)
Query: 136 GEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGIAFTGSGKT 195
G PP I++F D+ + I+ ++ +PTP+Q +P+I RD++ A TGSGKT
Sbjct: 171 GNNCPPHIESFSDVEMGEIIMGNIELTRYTRPTPVQKHAIPIIKEKRDLMACAQTGSGKT 230
Query: 196 LVFVLPMIMMAMQEEIMMPIVPGEG----------------PFGLIICPSRELARQTYEV 239
F+LP++ + PGE P L++ P+RELA Q YE
Sbjct: 231 AAFLLPILSQIYADG------PGEALRAMKENGRYGRRKQYPISLVLAPTRELAVQIYEE 284
Query: 240 IEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMNLD 299
+F + +RP + GG ++ Q+ +++G H++VATPGRL DM+ + K+ LD
Sbjct: 285 ARKFSYRSR------VRPCVVYGGAEIGQQIRDLERGCHLLVATPGRLVDMMERGKIGLD 338
Query: 300 NCRYLTLDEADRLVDLGFEDDIREVFDHF----KAQRQTLLFSATMPTKIQNFARSALVK 355
C+YL LDEADR++D+GFE IR + + K R T++FSAT P +IQ AR L +
Sbjct: 339 FCKYLVLDEADRMLDMGFEPQIRRIVEQDTMPPKGVRHTMMFSATFPKEIQMLARDFLDE 398
Query: 356 PIIVNVGRAGAANLDVIQEVEYVKQEAKIVYLLECLQKTPPP--VLIFCENKADVDDIHE 413
I + VGR G+ + ++ Q+V +V++ K +LL+ L T L+F E K D + +
Sbjct: 399 YIFLAVGRVGSTSENITQKVVWVEEIDKRSFLLDLLNATGKDSLTLVFVETKKGADSLED 458
Query: 414 YLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMP 473
+L +G +IHG + Q +RE A+ F++GK +LVAT VA++GLD +++HVIN+D+P
Sbjct: 459 FLYHEGYACTSIHGDRSQRDREEALHQFRSGKSPILVATAVAARGLDISNVKHVINFDLP 518
Query: 474 AEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAELVDPM 533
++IE YVHRIGRTGR G G+AT+F N+ T DL LL EAKQ +P L +
Sbjct: 519 SDIEEYVHRIGRTGRVGNLGLATSFFNERNINIT-KDLLDLLVEAKQEVPSWLENMA--F 575
Query: 534 EDNEEITGISGVKGCAYCGGLGHRIRDCPKLEHQKSVAIANNRKDYFGSGG 584
E + + + K + GG G RD + S + +++R SGG
Sbjct: 576 EHHYKGSSRGRSKSSRFSGGFG--ARDYRQSSGASSSSFSSSRASSSRSGG 624
>AN3_XENLA (P24346) Putative ATP-dependent RNA helicase An3
Length = 697
Score = 293 bits (750), Expect = 9e-79
Identities = 167/442 (37%), Positives = 251/442 (56%), Gaps = 38/442 (8%)
Query: 136 GEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGIAFTGSGKT 195
G PP I++F D+ + I+ ++ +PTP+Q +P+I+ RD++ A TGSGKT
Sbjct: 213 GSNCPPHIESFHDVTMGEIIMGNIQLTRYTRPTPVQKHAIPIIIEKRDLMACAQTGSGKT 272
Query: 196 LVFVLPMIMMAMQEEIMMPIVPGEG----------------PFGLIICPSRELARQTYEV 239
F+LP++ + PG+ P L++ P+RELA Q YE
Sbjct: 273 AAFLLPILSQIYADG------PGDAMKHLQENGRYGRRKQFPLSLVLAPTRELAVQIYEE 326
Query: 240 IEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMNLD 299
+F A +RP + GG D+ Q+ +++G H++VATPGRL DM+ + K+ LD
Sbjct: 327 ARKF------AYRSRVRPCVVYGGADIGQQIRDLERGCHLLVATPGRLVDMMERGKIGLD 380
Query: 300 NCRYLTLDEADRLVDLGFEDDIREVFDHF----KAQRQTLLFSATMPTKIQNFARSALVK 355
C+YL LDEADR++D+GFE IR + + K RQT++FSAT P +IQ AR L +
Sbjct: 381 FCKYLVLDEADRMLDMGFEPQIRRIVEQDTMPPKGVRQTMMFSATFPKEIQILARDFLDE 440
Query: 356 PIIVNVGRAGAANLDVIQEVEYVKQEAKIVYLLECLQKTPPP--VLIFCENKADVDDIHE 413
I + VGR G+ + ++ Q+V +V++ K +LL+ L T L+F E K D + +
Sbjct: 441 YIFLAVGRVGSTSENITQKVVWVEEMDKRSFLLDLLNATGKDSLTLVFVETKKGADALED 500
Query: 414 YLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMP 473
+L +G +IHG + Q +RE A+ F++GK +LVAT VA++GLD +++HVIN+D+P
Sbjct: 501 FLYHEGYACTSIHGDRSQRDREEALHQFRSGKSPILVATAVAARGLDISNVKHVINFDLP 560
Query: 474 AEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAELVDPM 533
++IE YVHRIGRTGR G G+AT+F N+ T DL LL EAKQ +P L +
Sbjct: 561 SDIEEYVHRIGRTGRVGNLGLATSFFNEKNINIT-KDLLDLLVEAKQEVPSWLENMAYEQ 619
Query: 534 EDNEEITGISGVKGCAYCGGLG 555
G S + + GG G
Sbjct: 620 HHKSSSRGRSKSR---FSGGFG 638
>DDXY_HUMAN (O15523) DEAD-box protein 3, Y-chromosomal
Length = 660
Score = 293 bits (749), Expect = 1e-78
Identities = 173/472 (36%), Positives = 254/472 (53%), Gaps = 37/472 (7%)
Query: 136 GEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGIAFTGSGKT 195
G PP I+NF D+ + I+ ++ +PTP+Q +P+I RD++ A TGSGKT
Sbjct: 170 GSNCPPHIENFSDIDMGEIIMGNIELTRYTRPTPVQKHAIPIIKGKRDLVACAQTGSGKT 229
Query: 196 LVFVLPMIMMAMQEEIMMPIVPGEG----------------PFGLIICPSRELARQTYEV 239
F+LP++ + PGE P L++ P+RELA Q YE
Sbjct: 230 AAFLLPILSQIYTDG------PGEALKAVKENGRYGRRKQYPISLVLAPTRELAVQIYEE 283
Query: 240 IEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMNLD 299
+F + +RP + GG D+ Q+ +++G H++VATPGRL DM+ + K+ LD
Sbjct: 284 ARKFSYRSR------VRPCVVYGGADIGQQIRDLERGCHLLVATPGRLVDMMERGKIGLD 337
Query: 300 NCRYLTLDEADRLVDLGFEDDIREVFDHF----KAQRQTLLFSATMPTKIQNFARSALVK 355
C+YL LDEADR++D+GFE IR + + K R T++FSAT P +IQ AR L +
Sbjct: 338 FCKYLVLDEADRMLDMGFEPQIRRIVEQDTMPPKGVRHTMMFSATFPKEIQMLARDFLDE 397
Query: 356 PIIVNVGRAGAANLDVIQEVEYVKQEAKIVYLLECLQKTPPP--VLIFCENKADVDDIHE 413
I + VGR G+ + ++ Q+V +V+ K +LL+ L T L+F E K D + +
Sbjct: 398 YIFLAVGRVGSTSENITQKVVWVEDLDKRSFLLDILGATGSDSLTLVFVETKKGADSLED 457
Query: 414 YLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMP 473
+L +G +IHG + Q +RE A+ F++GK +LVAT VA++GLD +++HVIN+D+P
Sbjct: 458 FLYHEGYACTSIHGDRSQRDREEALHQFRSGKSPILVATAVAARGLDISNVRHVINFDLP 517
Query: 474 AEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAELVDPM 533
++IE YVHRIGRTGR G G+AT+F N+ T DL LL EAKQ +P L +
Sbjct: 518 SDIEEYVHRIGRTGRVGNLGLATSFFNEKNMNIT-KDLLDLLVEAKQEVPSWLENMAYEH 576
Query: 534 EDNEEITGISGVKGCAYCGGLGHRIRDCPKLEHQKSVAIANNRKDYFGSGGY 585
G S K + GG G R + G GGY
Sbjct: 577 HYKGGSRGRS--KSNRFSGGFGARDYRQSSGSSSSGFGASRGSSSRSGGGGY 626
>DDXY_PANTR (Q6GVM6) DEAD-box protein 3, Y-chromosomal
Length = 660
Score = 291 bits (745), Expect = 3e-78
Identities = 173/472 (36%), Positives = 255/472 (53%), Gaps = 37/472 (7%)
Query: 136 GEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGIAFTGSGKT 195
G PP I+NF D+ + I+ ++ +PTP+Q +P+I RD++ A TGSGKT
Sbjct: 170 GSNCPPHIENFGDIDMGEIIMGNIQLTRYTRPTPVQKHAIPIIKGKRDLMACAQTGSGKT 229
Query: 196 LVFVLPMIMMAMQEEIMMPIVPGEG----------------PFGLIICPSRELARQTYEV 239
F+LP++ + PGE P L++ P+RELA Q YE
Sbjct: 230 AAFLLPILSQIYTDG------PGEALKAVKENGRYGRRKQYPISLVLAPTRELAVQIYEE 283
Query: 240 IEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMNLD 299
+F + +RP + GG D+ Q+ +++G H++VATPGRL DM+ + K+ LD
Sbjct: 284 ARKFSYRSR------VRPCVVYGGADIGQQIRDLERGCHLLVATPGRLVDMMERGKIGLD 337
Query: 300 NCRYLTLDEADRLVDLGFEDDIREVFDHF----KAQRQTLLFSATMPTKIQNFARSALVK 355
C+YL LDEADR++D+GFE IR + + K R T++FSAT P +IQ AR L +
Sbjct: 338 FCKYLVLDEADRMLDMGFEPQIRRIVEQDTMPPKGVRHTMMFSATFPKEIQMLARDFLDE 397
Query: 356 PIIVNVGRAGAANLDVIQEVEYVKQEAKIVYLLECLQK--TPPPVLIFCENKADVDDIHE 413
I + VGR G+ + ++ Q+V +V+ K +LL+ L T L+F E K D + +
Sbjct: 398 YIFLAVGRVGSTSENITQKVVWVEDLDKRSFLLDILGAAGTDSLTLVFVETKKGADSLED 457
Query: 414 YLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMP 473
+L +G +IHG + Q +RE A+ F++GK +LVAT VA++GLD +++HVIN+D+P
Sbjct: 458 FLYHEGYACTSIHGDRSQRDREEALHQFRSGKSPILVATAVAARGLDISNVRHVINFDLP 517
Query: 474 AEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAELVDPM 533
++IE YVHRIGRTGR G G+AT+F N+ T DL LL EAKQ +P L +
Sbjct: 518 SDIEEYVHRIGRTGRVGNLGLATSFFNEKNINIT-KDLLDLLVEAKQEVPSWLENMAYEH 576
Query: 534 EDNEEITGISGVKGCAYCGGLGHRIRDCPKLEHQKSVAIANNRKDYFGSGGY 585
+ G S K + GG G R + G GGY
Sbjct: 577 QYKGGSRGRS--KSNRFSGGFGARDYRQSSGSSSSGFGASRGSSSRSGGGGY 626
>DBP1_YEAST (P24784) Probable ATP-dependent RNA helicase DBP1
(Helicase CA1)
Length = 617
Score = 291 bits (745), Expect = 3e-78
Identities = 160/407 (39%), Positives = 244/407 (59%), Gaps = 19/407 (4%)
Query: 135 NGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGIAFTGSGK 194
+G+++P PI +F + +++ +K +PTP+Q +P++ GRD++ A TGSGK
Sbjct: 145 SGKDVPEPILDFSSPPLDELLMENIKLASFTKPTPVQKYSIPIVTKGRDLMACAQTGSGK 204
Query: 195 TLVFVLPMIMMAMQE------EIMMPIVPGEG-PFGLIICPSRELARQTYEVIEEFLLPL 247
T F+ P+ + E +G P L++ P+RELA Q +E +F
Sbjct: 205 TGGFLFPLFTELFRSGPSPVPEKAQSFYSRKGYPSALVLAPTRELATQIFEEARKFTYR- 263
Query: 248 KEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMNLDNCRYLTLD 307
+RP + GG + +Q+ V +G ++VATPGRL D+L + K++L N +YL LD
Sbjct: 264 -----SWVRPCVVYGGAPIGNQMREVDRGCDLLVATPGRLNDLLERGKVSLANIKYLVLD 318
Query: 308 EADRLVDLGFEDDIREVFDHFKA----QRQTLLFSATMPTKIQNFARSALVKPIIVNVGR 363
EADR++D+GFE IR + + RQTL+FSAT P IQ+ AR L I ++VGR
Sbjct: 319 EADRMLDMGFEPQIRHIVEECDMPSVENRQTLMFSATFPVDIQHLARDFLDNYIFLSVGR 378
Query: 364 AGAANLDVIQEVEYVKQEAKIVYLLECLQKTPPPV-LIFCENKADVDDIHEYLLLKGVEA 422
G+ + ++ Q + YV K LL+ L + LIF E K D + ++L+++ +A
Sbjct: 379 VGSTSENITQRILYVDDMDKKSALLDLLSAEHKGLTLIFVETKRMADQLTDFLIMQNFKA 438
Query: 423 VAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMPAEIENYVHR 482
AIHG + Q ERE A+S+FKA D+LVAT VA++GLD P++ HVINYD+P++I++YVHR
Sbjct: 439 TAIHGDRTQAERERALSAFKANVADILVATAVAARGLDIPNVTHVINYDLPSDIDDYVHR 498
Query: 483 IGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAEL 529
IGRTGR G TG+AT+F N N ++ + L +L EA Q +P L++L
Sbjct: 499 IGRTGRAGNTGVATSFFNSN-NQNIVKGLMEILNEANQEVPTFLSDL 544
>PR11_SCHPO (Q9P7C7) Probable ATP-dependent RNA helicase prp11
Length = 1014
Score = 291 bits (744), Expect = 4e-78
Identities = 182/529 (34%), Positives = 280/529 (52%), Gaps = 30/529 (5%)
Query: 62 DQPEISVTEQIVQQEK--EMIENLSDKKTLMSVRELAKGITYTEPLPTGWKPPLHIRRMS 119
D + ++E + ++E + S KK +++V I Y + + P ++ +S
Sbjct: 335 DDERMVISETLEEEENLLALAAKRSKKKDVITVDH--SKINYEDFKKDFYVEPEELKNLS 392
Query: 120 KKDCDLIQKQWH-IIVNGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVI 178
+ D ++ I + G + P P+ ++ + ++ + G +PT IQ Q +P I
Sbjct: 393 PAEVDELRASLDGIKIRGIDCPKPVTSWSQCGLSAQTISVINSLGYEKPTSIQAQAIPAI 452
Query: 179 LSGRDMIGIAFTGSGKTLVFVLPMIMMAMQEEIMMPIVPGEGPFGLIICPSRELARQTYE 238
SGRD+IG+A TGSGKT+ F+LPM + P+ GEGP +I+ P+RELA Q +
Sbjct: 453 TSGRDVIGVAKTGSGKTIAFLLPMFRHIKDQR---PLKTGEGPIAIIMTPTRELAVQIFR 509
Query: 239 VIEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDML---AKKK 295
+ FL L +R GG ++ Q+ +K+G IVV TPGR+ D+L A +
Sbjct: 510 ECKPFLKLLN------IRACCAYGGAPIKDQIADLKRGAEIVVCTPGRMIDVLSANAGRV 563
Query: 296 MNLDNCRYLTLDEADRLVDLGFEDDIREVFDHFKAQRQTLLFSATMPTKIQNFARSALVK 355
NL C YL LDEADR+ DLGFE + + ++ + RQT+LFSAT P ++ AR L K
Sbjct: 564 TNLHRCTYLVLDEADRMFDLGFEPQVMRIINNIRPDRQTVLFSATFPRAMEALARKVLKK 623
Query: 356 PIIVNVGRAGAANLDVIQEVEYVKQEAKIVYLLECL-----QKTPPPVLIFCENKADVDD 410
P+ + VG +V Q VE +E+K LLE L + L+F + + D
Sbjct: 624 PVEITVGGRSVVASEVEQIVEVRPEESKFSRLLELLGELYNNQLDVRTLVFVDRQESADA 683
Query: 411 IHEYLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINY 470
+ L+ +G + +IHGGKDQ +R+ IS +KAG DVL+AT V ++GLD +Q V+NY
Sbjct: 684 LLSDLMKRGYTSNSIHGGKDQHDRDSTISDYKAGVFDVLIATSVVARGLDVKSLQLVVNY 743
Query: 471 DMPAEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAELV 530
D P +E+YVHR+GRTGR G TG+A TFI Q E +D+ L+ +KQ +P L L
Sbjct: 744 DCPNHMEDYVHRVGRTGRAGHTGVAVTFITPEQ-EKYAVDIAKALKMSKQPVPKELQTLA 802
Query: 531 DPMEDNEEITGISGVKGCAYCGGLGHRIRDCPKLEHQKSVAIANNRKDY 579
++ + + K A GG G + +L+ ++ RK Y
Sbjct: 803 -----SQFLEKVKAGKEKAAGGGFGG--KGLSRLDETRNAERKMQRKAY 844
>DDX5_MOUSE (Q61656) Probable RNA-dependent helicase p68 (DEAD-box
protein p68) (DEAD-box protein 5) (DEAD-box RNA helicase
DEAD1) (mDEAD1)
Length = 614
Score = 290 bits (741), Expect = 9e-78
Identities = 163/422 (38%), Positives = 246/422 (57%), Gaps = 15/422 (3%)
Query: 115 IRRMSKKDCDLIQKQWHIIVNGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQG 174
+ R + ++ D ++ I V G P P+ NF + FP ++ ++ +PT IQ QG
Sbjct: 65 LARRTAQEVDTYRRSKEITVRGHNCPKPVLNFYEANFPANVMDVIARHNFTEPTAIQAQG 124
Query: 175 LPVILSGRDMIGIAFTGSGKTLVFVLPMIMMAMQEEIMMPIVPGEGPFGLIICPSRELAR 234
PV LSG DM+G+A TGSGKTL ++LP I+ + G+GP L++ P+RELA+
Sbjct: 125 WPVALSGLDMVGVAQTGSGKTLSYLLPAIVHINHHPFLER---GDGPICLVLAPTRELAQ 181
Query: 235 QTYEVIEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKK 294
Q +V E+ + L+ GG Q+ +++GV I +ATPGRL D L
Sbjct: 182 QVQQVAAEYCRACR------LKSTCIYGGAPKGPQIRDLERGVEICIATPGRLIDFLECG 235
Query: 295 KMNLDNCRYLTLDEADRLVDLGFEDDIREVFDHFKAQRQTLLFSATMPTKIQNFARSALV 354
K NL YL LDEADR++D+GFE IR++ D + RQTL++SAT P +++ A L
Sbjct: 236 KTNLRRTTYLVLDEADRMLDMGFEPQIRKIVDQIRPDRQTLMWSATWPKEVRQLAEDFLK 295
Query: 355 KPIIVNVGRAG-AANLDVIQEVEY---VKQEAKIVYLLE-CLQKTPPPVLIFCENKADVD 409
I +N+G +AN +++Q V+ V+++ K++ L+E + + ++F E K D
Sbjct: 296 DYIHINIGALELSANHNILQIVDVCHDVEKDEKLIRLMEEIMSEKENKTIVFVETKRRCD 355
Query: 410 DIHEYLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVIN 469
++ + G A+ IHG K Q+ER++ ++ FK GK +L+ATDVAS+GLD D++ VIN
Sbjct: 356 ELTRKMRRDGWPAMGIHGDKSQQERDWVLNEFKHGKAPILIATDVASRGLDVEDVKFVIN 415
Query: 470 YDMPAEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAEL 529
YD P E+Y+HRIGRT R KTG A TF N + + DL +L+EA Q I P L +L
Sbjct: 416 YDYPNSSEDYIHRIGRTARSTKTGTAYTFFTPNNIK-QVSDLISVLREANQAINPKLLQL 474
Query: 530 VD 531
V+
Sbjct: 475 VE 476
>DDX5_HUMAN (P17844) Probable RNA-dependent helicase p68 (DEAD-box
protein p68) (DEAD-box protein 5)
Length = 614
Score = 290 bits (741), Expect = 9e-78
Identities = 162/422 (38%), Positives = 248/422 (58%), Gaps = 15/422 (3%)
Query: 115 IRRMSKKDCDLIQKQWHIIVNGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQG 174
+ R + ++ + ++ I V G P P+ NF + FP ++ ++ + +PT IQ QG
Sbjct: 65 LARRTAQEVETYRRSKEITVRGHNCPKPVLNFYEANFPANVMDVIARQNFTEPTAIQAQG 124
Query: 175 LPVILSGRDMIGIAFTGSGKTLVFVLPMIMMAMQEEIMMPIVPGEGPFGLIICPSRELAR 234
PV LSG DM+G+A TGSGKTL ++LP I+ + + G+GP L++ P+RELA+
Sbjct: 125 WPVALSGLDMVGVAQTGSGKTLSYLLPAIVHINHQPFLER---GDGPICLVLAPTRELAQ 181
Query: 235 QTYEVIEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKK 294
Q +V E+ + L+ GG Q+ +++GV I +ATPGRL D L
Sbjct: 182 QVQQVAAEYCRACR------LKSTCIYGGAPKGPQIRDLERGVEICIATPGRLIDFLECG 235
Query: 295 KMNLDNCRYLTLDEADRLVDLGFEDDIREVFDHFKAQRQTLLFSATMPTKIQNFARSALV 354
K NL YL LDEADR++D+GFE IR++ D + RQTL++SAT P +++ A L
Sbjct: 236 KTNLRRTTYLVLDEADRMLDMGFEPQIRKIVDQIRPDRQTLMWSATWPKEVRQLAEDFLK 295
Query: 355 KPIIVNVGRAG-AANLDVIQEVEY---VKQEAKIVYLLE-CLQKTPPPVLIFCENKADVD 409
I +N+G +AN +++Q V+ V+++ K++ L+E + + ++F E K D
Sbjct: 296 DYIHINIGALELSANHNILQIVDVCHDVEKDEKLIRLMEEIMSEKENKTIVFVETKRRCD 355
Query: 410 DIHEYLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVIN 469
++ + G A+ IHG K Q+ER++ ++ FK GK +L+ATDVAS+GLD D++ VIN
Sbjct: 356 ELTRKMRRDGWPAMGIHGDKSQQERDWVLNEFKHGKAPILIATDVASRGLDVEDVKFVIN 415
Query: 470 YDMPAEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAEL 529
YD P E+Y+HRIGRT R KTG A TF N + + DL +L+EA Q I P L +L
Sbjct: 416 YDYPNSSEDYIHRIGRTARSTKTGTAYTFFTPNNIK-QVSDLISVLREANQAINPKLLQL 474
Query: 530 VD 531
V+
Sbjct: 475 VE 476
>DD17_HUMAN (Q92841) Probable RNA-dependent helicase p72 (DEAD-box
protein p72) (DEAD-box protein 17)
Length = 650
Score = 286 bits (732), Expect = 1e-76
Identities = 163/423 (38%), Positives = 247/423 (57%), Gaps = 16/423 (3%)
Query: 115 IRRMSKKDCDLIQKQWHIIVNGEEI-PPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQ 173
+ R++ + D ++++ I V G ++ P P+ F FP ++ +L + +PTPIQ Q
Sbjct: 62 VARLTPYEVDELRRKKEITVRGGDVCPKPVFAFHHANFPQYVMDVLMDQHFTEPTPIQCQ 121
Query: 174 GLPVILSGRDMIGIAFTGSGKTLVFVLPMIMMAMQEEIMMPIVPGEGPFGLIICPSRELA 233
G P+ LSGRDM+GIA TGSGKTL ++LP I+ + + G+GP L++ P+RELA
Sbjct: 122 GFPLALSGRDMVGIAQTGSGKTLAYLLPAIVHINHQPYLER---GDGPICLVLAPTRELA 178
Query: 234 RQTYEVIEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAK 293
+Q +V +++ L+ GG Q+ +++GV I +ATPGRL D L
Sbjct: 179 QQVQQVADDY------GKCSRLKSTCIYGGAPKGPQIRDLERGVEICIATPGRLIDFLES 232
Query: 294 KKMNLDNCRYLTLDEADRLVDLGFEDDIREVFDHFKAQRQTLLFSATMPTKIQNFARSAL 353
K NL C YL LDEADR++D+GFE IR++ D + RQTL++SAT P +++ A L
Sbjct: 233 GKTNLRRCTYLVLDEADRMLDMGFEPQIRKIVDQIRPDRQTLMWSATWPKEVRQLAEDFL 292
Query: 354 VKPIIVNVGRAG-AANLDVIQEVEYV---KQEAKIVYLLE-CLQKTPPPVLIFCENKADV 408
+NVG +AN +++Q V+ +++ K++ L+E + + +IF E K
Sbjct: 293 RDYTQINVGNLELSANHNILQIVDVCMESEKDHKLIQLMEEIMAEKENKTIIFVETKRRC 352
Query: 409 DDIHEYLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVI 468
DD+ + G A+ IHG K Q ER++ ++ F++GK +L+ATDVAS+GLD D++ VI
Sbjct: 353 DDLTRRMRRDGWPAMCIHGDKSQPERDWVLNEFRSGKAPILIATDVASRGLDVEDVKFVI 412
Query: 469 NYDMPAEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAE 528
NYD P E+YVHRIGRT R G A TF + +L +L+EA Q I P L +
Sbjct: 413 NYDYPNSSEDYVHRIGRTARSTNKGTAYTFFTPGNLKQA-RELIKVLEEANQAINPKLMQ 471
Query: 529 LVD 531
LVD
Sbjct: 472 LVD 474
>RM62_DROME (P19109) ATP-dependent RNA helicase P62
Length = 719
Score = 286 bits (731), Expect = 1e-76
Identities = 161/423 (38%), Positives = 246/423 (58%), Gaps = 18/423 (4%)
Query: 114 HIRRMSKKDCDLIQKQWHIIVNGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQ 173
++ S + +++ I V G+ +P PI++F ++ PD ++K ++ +G PT IQ Q
Sbjct: 252 NVANRSPYEVQRYREEQEITVRGQ-VPNPIQDFSEVHLPDYVMKEIRRQGYKAPTAIQAQ 310
Query: 174 GLPVILSGRDMIGIAFTGSGKTLVFVLPMIMMAMQEEIMMPIVPGEGPFGLIICPSRELA 233
G P+ +SG + +GIA TGSGKTL ++LP I+ ++ P+ G+GP L++ P+RELA
Sbjct: 311 GWPIAMSGSNFVGIAKTGSGKTLGYILPAIVHINNQQ---PLQRGDGPIALVLAPTRELA 367
Query: 234 RQTYEVIEEFLLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAK 293
+Q +V EF +R GG Q+ +++G IV+ATPGRL D L+
Sbjct: 368 QQIQQVATEF------GSSSYVRNTCVFGGAPKGGQMRDLQRGCEIVIATPGRLIDFLSA 421
Query: 294 KKMNLDNCRYLTLDEADRLVDLGFEDDIREVFDHFKAQRQTLLFSATMPTKIQNFARSAL 353
NL C YL LDEADR++D+GFE IR++ + RQTL++SAT P +++ A L
Sbjct: 422 GSTNLKRCTYLVLDEADRMLDMGFEPQIRKIVSQIRPDRQTLMWSATWPKEVKQLAEDFL 481
Query: 354 VKPIIVNVGRAG-AANLDVIQEVEYVKQ---EAKIVYLLECLQKT---PPPVLIFCENKA 406
I +N+G +AN ++ Q V+ + E K+ LL + T P ++IF E K
Sbjct: 482 GNYIQINIGSLELSANHNIRQVVDVCDEFSKEEKLKTLLSDIYDTSESPGKIIIFVETKR 541
Query: 407 DVDDIHEYLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQH 466
VD++ ++ GV AIHG K Q ER++ + F++GK ++LVATDVA++GLD I++
Sbjct: 542 RVDNLVRFIRSFGVRCGAIHGDKSQSERDFVLREFRSGKSNILVATDVAARGLDVDGIKY 601
Query: 467 VINYDMPAEIENYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVL 526
VIN+D P E+Y+HRIGRTGR G + F KN ++ L +L+EA Q I P L
Sbjct: 602 VINFDYPQNSEDYIHRIGRTGRSNTKGTSFAFFTKNNAKQAKA-LVDVLREANQEINPAL 660
Query: 527 AEL 529
L
Sbjct: 661 ENL 663
>DED1_SCHPO (O13370) ATP-dependent RNA helicase ded1
Length = 636
Score = 285 bits (730), Expect = 2e-76
Identities = 162/409 (39%), Positives = 240/409 (58%), Gaps = 24/409 (5%)
Query: 134 VNGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGIAFTGSG 193
V+G +I P + F +L+ +K G QPTP+Q +P++ SGRD++ A TGSG
Sbjct: 160 VSGGDIEP-VNEFTSPPLNSHLLQNIKLSGYTQPTPVQKNSIPIVTSGRDLMACAQTGSG 218
Query: 194 KTLVFVLPMIMMAMQE-EIMMPIVPGEG---------PFGLIICPSRELARQTYEVIEEF 243
KT F+ P++ +A + +P+ G P LI+ P+REL Q +E +F
Sbjct: 219 KTAGFLFPILSLAFDKGPAAVPVDQDAGMGYRPRKAYPTTLILAPTRELVCQIHEESRKF 278
Query: 244 LLPLKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMNLDNCRY 303
+RP GG D+R+Q+ + +G ++ ATPGRL D++ + +++L N ++
Sbjct: 279 CYR------SWVRPCAVYGGADIRAQIRQIDQGCDLLSATPGRLVDLIDRGRISLANIKF 332
Query: 304 LTLDEADRLVDLGFEDDIREVFDHFKA----QRQTLLFSATMPTKIQNFARSALVKPIIV 359
L LDEADR++D+GFE IR + + +RQTL+FSAT P IQ AR L + +
Sbjct: 333 LVLDEADRMLDMGFEPQIRHIVEGADMTSVEERQTLMFSATFPRDIQLLARDFLKDYVFL 392
Query: 360 NVGRAGAANLDVIQEVEYVKQEAKIVYLLECLQKTPPP--VLIFCENKADVDDIHEYLLL 417
+VGR G+ + ++ Q+V +V+ K YLL+ L PP LIF E K D + +YLL
Sbjct: 393 SVGRVGSTSENITQKVVHVEDSEKRSYLLDILHTLPPEGLTLIFVETKRMADTLTDYLLN 452
Query: 418 KGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMPAEIE 477
A +IHG + Q ERE A+ F++G+ ++VAT VAS+GLD P++ HVINYD+P +I+
Sbjct: 453 SNFPATSIHGDRTQRERERALELFRSGRTSIMVATAVASRGLDIPNVTHVINYDLPTDID 512
Query: 478 NYVHRIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVL 526
+YVHRIGRTGR G TG A F N+N ++ +L LLQEA Q P L
Sbjct: 513 DYVHRIGRTGRAGNTGQAVAFFNRN-NKGIAKELIELLQEANQECPSFL 560
>DED1_YEAST (P06634) Probable ATP-dependent RNA helicase DED1
Length = 604
Score = 281 bits (719), Expect = 3e-75
Identities = 164/463 (35%), Positives = 261/463 (55%), Gaps = 47/463 (10%)
Query: 135 NGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGIAFTGSGK 194
+G+++P PI F +L+ +K +PTP+Q +P++ +GRD++ A TGSGK
Sbjct: 133 SGKDVPEPITEFTSPPLDGLLLENIKLARFTKPTPVQKYSVPIVANGRDLMACAQTGSGK 192
Query: 195 TLVFVLPMIMMAMQEEIMMPIVPGEGPF--------GLIICPSRELARQTYEVIEEFLLP 246
T F+ P++ + + P +G F +I+ P+RELA Q ++ ++F
Sbjct: 193 TGGFLFPVLSESFKTG-PSPQPESQGSFYQRKAYPTAVIMAPTRELATQIFDEAKKFTYR 251
Query: 247 LKEAGYPELRPLLCIGGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMNLDNCRYLTL 306
++ + GG + +QL +++G ++VATPGRL D+L + K++L N +YL L
Sbjct: 252 ------SWVKACVVYGGSPIGNQLREIERGCDLLVATPGRLNDLLERGKISLANVKYLVL 305
Query: 307 DEADRLVDLGFEDDIREVFDHFK----AQRQTLLFSATMPTKIQNFARSALVKPIIVNVG 362
DEADR++D+GFE IR + + +RQTL+FSAT P IQ+ AR L I ++VG
Sbjct: 306 DEADRMLDMGFEPQIRHIVEDCDMTPVGERQTLMFSATFPADIQHLARDFLSDYIFLSVG 365
Query: 363 RAGAANLDVIQEVEYVKQEAKIVYLLECLQ-KTPPPVLIFCENKADVDDIHEYLLLKGVE 421
R G+ + ++ Q+V YV+ + K LL+ L T LIF E K D + ++L+++
Sbjct: 366 RVGSTSENITQKVLYVENQDKKSALLDLLSASTDGLTLIFVETKRMADQLTDFLIMQNFR 425
Query: 422 AVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMPAEIENYVH 481
A AIHG + Q ERE A+++F++G +LVAT VA++GLD P++ HVINYD+P+++++YVH
Sbjct: 426 ATAIHGDRTQSERERALAAFRSGAATLLVATAVAARGLDIPNVTHVINYDLPSDVDDYVH 485
Query: 482 RIGRTGRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAELVDPMEDNEEITG 541
RIGRTGR G TG+AT F N +++ + L +L EA Q +P L + +
Sbjct: 486 RIGRTGRAGNTGLATAFFN-SENSNIVKGLHEILTEANQEVPSFL---------KDAMMS 535
Query: 542 ISGVKGCAYCGGLGHRIRDCPKLEHQKSVAIANNRKDYFGSGG 584
G + + GG G NN +DY +GG
Sbjct: 536 APGSRSNSRRGGFGR-----------------NNNRDYRKAGG 561
>HE64_TRYBB (Q26696) Putative DEAD-box RNA helicase HEL64
Length = 568
Score = 280 bits (716), Expect = 7e-75
Identities = 164/421 (38%), Positives = 254/421 (59%), Gaps = 21/421 (4%)
Query: 117 RMSKKDCDLIQKQWHIIVNGEEIPPPIKNFKDMR--FPDPILKMLKTKGIVQPTPIQVQG 174
++S+++ +++ I + G++ PPP+ +F + P +LK L + PTP+Q Q
Sbjct: 73 QLSEEEATKWREEHVITIFGDDCPPPMSSFDHLCGIVPPYLLKKLTAQNFTAPTPVQAQS 132
Query: 175 LPVILSGRDMIGIAFTGSGKTLVFVLPMIM-MAMQEEIMMPIVPGEGPFGLIICPSRELA 233
PV+LSGRD++G+A TGSGKTL F++P + +A+QE P+ G+GP +++ P+RELA
Sbjct: 133 WPVLLSGRDLVGVAKTGSGKTLGFMVPALAHIAVQE----PLRSGDGPMVVVLAPTRELA 188
Query: 234 RQTYEVIEEFLLPLKEAGYPELRPLLCI-GGIDMRSQLEIVKKGVHIVVATPGRLKDMLA 292
+Q E ++ + G C+ GG QL ++++GVHI+VATPGRL D L
Sbjct: 189 QQIEEETKKVIPGDVYCG--------CVYGGAPKGPQLGLLRRGVHILVATPGRLIDFLD 240
Query: 293 KKKMNLDNCRYLTLDEADRLVDLGFEDDIREVFDHFKAQRQTLLFSATMPTKIQNFARSA 352
K++NL YL LDEADR++D+GFE +R++ + RQT++FSAT P +IQ A
Sbjct: 241 IKRINLHRVTYLVLDEADRMLDMGFEPQVRKICGQIRPDRQTVMFSATWPREIQRLAAEF 300
Query: 353 LVKPIIVNVGRAGA-ANLDVIQEVEYVKQEAKIVYLLECLQK-TPPPVLIFCENKADVDD 410
+ I ++VG AN DV Q ++ AK L + +Q+ VL+FC+ K D+
Sbjct: 301 QKQWIRISVGSTELQANKDVTQRFILTQEFAKQDELRKLMQEHREERVLVFCKMKRTADE 360
Query: 411 IHEYLLLKGVEAVAIHGGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINY 470
+ L G +A+AIHG K+Q +RE+ ++ F+ + LVATDVA++GLD ++ VINY
Sbjct: 361 LERQLRRWGYDAMAIHGDKEQRQREFILARFRKDPRLCLVATDVAARGLDIKQLETVINY 420
Query: 471 DMPAEIENYVHRIGRTGRCGKTGIATTFINKNQSETT---LLDLKHLLQEAKQRIPPVLA 527
D P +I++YVHRIGRTGR G G I K +++ T L +L +L+ A+Q IP +
Sbjct: 421 DFPMQIDDYVHRIGRTGRAGGEGRCVYLITKKEAQITPSVLKELIGILERAQQEIPDWMI 480
Query: 528 E 528
E
Sbjct: 481 E 481
>DB10_NICSY (P46942) RNA helicase-like protein DB10
Length = 607
Score = 278 bits (710), Expect = 4e-74
Identities = 164/403 (40%), Positives = 229/403 (56%), Gaps = 18/403 (4%)
Query: 132 IIVNGEEIPPPIKNFKDMRFPDPILKMLKTKGIVQPTPIQVQGLPVILSGRDMIGIAFTG 191
I V G ++P P+ +F+ FP I++ + G PTPIQ Q P+ L GRD++ IA TG
Sbjct: 133 ISVTGGDVPAPLTSFEATGFPSEIVREMHQAGFSAPTPIQAQSWPIALQGRDIVAIAKTG 192
Query: 192 SGKTLVFVLPMIMMAMQEEIMMPIVPGEGPFGLIICPSRELARQTYEVIEEFLLPLKEAG 251
SGKTL +++P + Q P GP L++ P+RELA Q +F G
Sbjct: 193 SGKTLGYLMPAFIHLQQRRKN----PQLGPTILVLSPTRELATQIQAEAVKF-------G 241
Query: 252 YPELRPLLCI-GGIDMRSQLEIVKKGVHIVVATPGRLKDMLAKKKMNLDNCRYLTLDEAD 310
C+ GG QL + +GV IVVATPGRL D+L ++++L YL LDEAD
Sbjct: 242 KSSRISCTCLYGGAPKGPQLRELSRGVDIVVATPGRLNDILEMRRVSLGQVSYLVLDEAD 301
Query: 311 RLVDLGFEDDIREVFDHFKAQRQTLLFSATMPTKIQNFARSALVKPIIVNVGRAG--AAN 368
R++D+GFE IR++ QRQTL+++AT P ++ A LV + VN+G AN
Sbjct: 302 RMLDMGFEPQIRKIVKEVPVQRQTLMYTATWPKGVRKIAADLLVNSVQVNIGNVDELVAN 361
Query: 369 LDVIQEVEYVKQEAKIVYLLECLQKTPP--PVLIFCENKADVDDIHEYLLLKGVEAVAIH 426
+ Q +E V K + + L+ P ++IFC K D + L + A AIH
Sbjct: 362 KSITQHIEVVLPMEKQRRVEQILRSKEPGSKIIIFCSTKKMCDQLSRNLT-RNFGAAAIH 420
Query: 427 GGKDQEEREYAISSFKAGKKDVLVATDVASKGLDFPDIQHVINYDMPAEIENYVHRIGRT 486
G K Q ER+Y +S F+AG+ VLVATDVA++GLD DI+ VINYD P IE+YVHRIGRT
Sbjct: 421 GDKSQGERDYVLSQFRAGRSPVLVATDVAARGLDIKDIRVVINYDFPTGIEDYVHRIGRT 480
Query: 487 GRCGKTGIATTFINKNQSETTLLDLKHLLQEAKQRIPPVLAEL 529
GR G +G+A TF + +Q LDL +L+ A Q +P L ++
Sbjct: 481 GRAGASGLAYTFFS-DQDSKHALDLVKVLEGANQCVPTELRDM 522
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.138 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 70,543,053
Number of Sequences: 164201
Number of extensions: 3153425
Number of successful extensions: 10734
Number of sequences better than 10.0: 432
Number of HSP's better than 10.0 without gapping: 328
Number of HSP's successfully gapped in prelim test: 106
Number of HSP's that attempted gapping in prelim test: 9588
Number of HSP's gapped (non-prelim): 634
length of query: 589
length of database: 59,974,054
effective HSP length: 116
effective length of query: 473
effective length of database: 40,926,738
effective search space: 19358347074
effective search space used: 19358347074
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 69 (31.2 bits)
Medicago: description of AC144484.10


