

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144478.5 + phase: 0
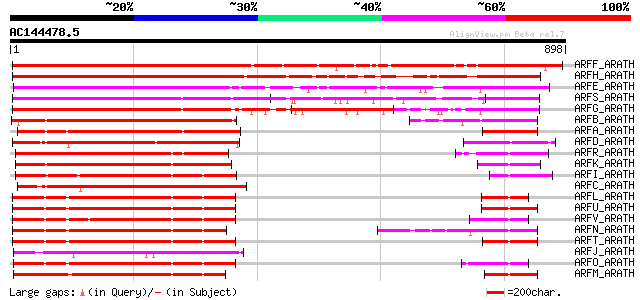
(898 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ARFF_ARATH (Q9ZTX8) Auxin response factor 6 1241 0.0
ARFH_ARATH (Q9FGV1) Auxin response factor 8 867 0.0
ARFE_ARATH (P93024) Auxin response factor 5 (Transcription facto... 615 e-175
ARFS_ARATH (Q8RYC8) Auxin response factor 19 (Auxin-responsive p... 568 e-161
ARFG_ARATH (P93022) Auxin response factor 7 (Non-phototropic hyp... 560 e-159
ARFB_ARATH (Q94JM3) Auxin response factor 2 (ARF1-binding protei... 396 e-109
ARFA_ARATH (Q8L7G0) Auxin response factor 1 388 e-107
ARFD_ARATH (Q9ZTX9) Auxin response factor 4 376 e-103
ARFR_ARATH (Q9C5W9) Auxin response factor 18 359 2e-98
ARFK_ARATH (Q9ZPY6) Auxin response factor 11 357 6e-98
ARFI_ARATH (Q9XED8) Auxin response factor 9 345 3e-94
ARFC_ARATH (O23661) Auxin response factor 3 (ETTIN protein) 337 1e-91
ARFL_ARATH (Q9XID4) Putative auxin response factor 12 289 2e-77
ARFU_ARATH (Q9C8N9) Putative auxin response factor 21 288 6e-77
ARFV_ARATH (Q9C8N7) Putative auxin response factor 22 287 8e-77
ARFN_ARATH (Q9LQE8) Putative auxin response factor 14 287 1e-76
ARFT_ARATH (Q9C7I9) Putative auxin response factor 20 286 1e-76
ARFJ_ARATH (Q9SKN5) Auxin response factor 10 280 9e-75
ARFO_ARATH (Q9LQE3) Putative auxin response factor 15 279 2e-74
ARFM_ARATH (Q9FX25) Putative auxin response factor 13 266 2e-70
>ARFF_ARATH (Q9ZTX8) Auxin response factor 6
Length = 933
Score = 1241 bits (3210), Expect = 0.0
Identities = 668/933 (71%), Positives = 741/933 (78%), Gaps = 62/933 (6%)
Query: 5 GEKRVLDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQ 64
GEKRVL+SELWHACAGPLVSLP VGSRVVYFPQGHSEQVA STNKEVDAHIPNYPSL PQ
Sbjct: 15 GEKRVLNSELWHACAGPLVSLPPVGSRVVYFPQGHSEQVAASTNKEVDAHIPNYPSLHPQ 74
Query: 65 LICQLHNLTMHADVETDEVYAQMTLQPLNAQEQKEAYLPAELGTPSKQPTNYFCKTLTAS 124
LICQLHN+TMHADVETDEVYAQMTLQPLNAQEQK+ YLPAELG PS+QPTNYFCKTLTAS
Sbjct: 75 LICQLHNVTMHADVETDEVYAQMTLQPLNAQEQKDPYLPAELGVPSRQPTNYFCKTLTAS 134
Query: 125 DTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFRGQPKRHLLTT 184
DTSTHGGFSVPRRAAEKVFPPLD+SQQPPAQEL+ARDLH NEWKFRHIFRGQPKRHLLTT
Sbjct: 135 DTSTHGGFSVPRRAAEKVFPPLDYSQQPPAQELMARDLHDNEWKFRHIFRGQPKRHLLTT 194
Query: 185 GWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHLGLLAAAA 244
GWSVFVSAKRLVAGDSVLFIWN+KNQLLLGIRRA+RPQTVMPSSVLSSDSMHLGLLAAAA
Sbjct: 195 GWSVFVSAKRLVAGDSVLFIWNDKNQLLLGIRRANRPQTVMPSSVLSSDSMHLGLLAAAA 254
Query: 245 HAAATNSRFTIFYNPRACPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESSVRRYMG 304
HAAATNSRFTIFYNPRA PSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESSVRRYMG
Sbjct: 255 HAAATNSRFTIFYNPRASPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESSVRRYMG 314
Query: 305 TITGICDLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEPLTTFPMYPSPFPLRLK 364
TITGICDLD RW NSHWRSVKVGWDESTAGERQPRVSLWEIEPLTTFPMYPSPFPLRLK
Sbjct: 315 TITGICDLDPTRWANSHWRSVKVGWDESTAGERQPRVSLWEIEPLTTFPMYPSPFPLRLK 374
Query: 365 RPWPPGLPSFHGMKDDDFG--MSSPLMWLRDTDRGLQSLNYQGIGVNPWMQPRFDPA-ML 421
RPWPPGLPSFHG+K+DD G MSSPLMW DRGLQSLN+QG+GVNPWMQPR D + +L
Sbjct: 375 RPWPPGLPSFHGLKEDDMGMSMSSPLMW----DRGLQSLNFQGMGVNPWMQPRLDTSGLL 430
Query: 422 NMQTDMYQAVAAAALQDMRTVVDPSKQLPGSLLQFQQPPNFPNRTAALMQAQMLQQ--SQ 479
MQ D+YQA+AAAALQDMR +DP+K SLLQFQ P F ++ +L+Q QMLQQ SQ
Sbjct: 431 GMQNDVYQAMAAAALQDMRG-IDPAK-AAASLLQFQNSPGFSMQSPSLVQPQMLQQQLSQ 488
Query: 480 PQQAFQNNNQENQNLSQSQPQAQTNPQQHPQHQHSFNNQLHHHSQQQ------------- 526
QQ Q+ Q LSQ Q Q Q + QQ Q QL QQQ
Sbjct: 489 QQQQLSQQQQQQQQLSQQQ-QQQLSQQQQQQLSQQQQQQLSQQQQQQAYLGVPETHQPQS 547
Query: 527 ----------QQTQQQVVDNNQQISGS---VSTMSQFVSATQPQSPPPMQALSSLCHQQS 573
Q QQQVVDN+ + S VS MSQF SA+QP + P+Q+++SLCHQQS
Sbjct: 548 QAQSQSNNHLSQQQQQVVDNHNPSASSAAVVSAMSQFGSASQPNT-SPLQSMTSLCHQQS 606
Query: 574 FSDSNVNSSTTIVSPLHSIMGSSFPHDESSLLMSLPRTSSWVPVQNSTGWPSKRIAVDPL 633
FSD+N ++ +SPLH+++ S+F DESS L+ L RT+S S+GWPSKR AVD
Sbjct: 607 FSDTNGGNNP--ISPLHTLL-SNFSQDESSQLLHLTRTNS---AMTSSGWPSKRPAVDSS 660
Query: 634 LS-SGASQCILPQV-EQLGQARNS-MSQNAITLPPFP-GRECSIDQEGS-NDPQSNLLFG 688
SGA V EQLGQ+ S + NA++LPPFP GRECSI+QEGS +DP S+LLFG
Sbjct: 661 FQHSGAGNNNTQSVLEQLGQSHTSNVPPNAVSLPPFPGGRECSIEQEGSASDPHSHLLFG 720
Query: 689 VNIDPSSLLLHNGMSNFKGISGNNNDSSTMSYHQSSSYMNTAGADSSLNHGVTPS--IGE 746
VNID SSLL+ NGMSN + I DS+T+ + SS++ N + ++ TPS I E
Sbjct: 721 VNIDSSSLLMPNGMSNLRSIGIEGGDSTTLPF-TSSNFNNDFSGNLAM---TTPSSCIDE 776
Query: 747 SGFLHTQENGEQGNNPLNKTFVKVYKSGSFGRSLDITKFSSYNELRSELARMFGLEGELE 806
SGFL + EN NP + TFVKVYKSGSFGRSLDI+KFSSY+ELRSELARMFGLEG+LE
Sbjct: 777 SGFLQSSEN-LGSENPQSNTFVKVYKSGSFGRSLDISKFSSYHELRSELARMFGLEGQLE 835
Query: 807 DPVRSGWQLVFVDRENDVLLLGDGPWPEFVNSVWCIKILSPEEVQQMGNTGLGLLNSVP- 865
DPVRSGWQLVFVDRENDVLLLGD PWPEFV+SVWCIKILSP+EVQQMG GL LLNS P
Sbjct: 836 DPVRSGWQLVFVDRENDVLLLGDDPWPEFVSSVWCIKILSPQEVQQMGKRGLELLNSAPS 895
Query: 866 ---IQRL-SNSICDDYVSRQDSRNLSSGITTVG 894
+ +L SN CDD+ +R D RNL +GI +VG
Sbjct: 896 SNNVDKLPSNGNCDDFGNRSDPRNLGNGIASVG 928
>ARFH_ARATH (Q9FGV1) Auxin response factor 8
Length = 811
Score = 867 bits (2240), Expect = 0.0
Identities = 493/864 (57%), Positives = 586/864 (67%), Gaps = 88/864 (10%)
Query: 5 GEKRVLDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQ 64
GEK L+SELWHACAGPLVSLP+ GSRVVYFPQGHSEQVA +TNKEVD HIPNYPSLPPQ
Sbjct: 15 GEK-CLNSELWHACAGPLVSLPSSGSRVVYFPQGHSEQVAATTNKEVDGHIPNYPSLPPQ 73
Query: 65 LICQLHNLTMHADVETDEVYAQMTLQPLNAQEQKEAYLPAELGTPSKQPTNYFCKTLTAS 124
LICQLHN+TMHADVETDEVYAQMTLQPL +EQKE ++P ELG PSKQP+NYFCKTLTAS
Sbjct: 74 LICQLHNVTMHADVETDEVYAQMTLQPLTPEEQKETFVPIELGIPSKQPSNYFCKTLTAS 133
Query: 125 DTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFRGQPKRHLLTT 184
DTSTHGGFSVPRRAAEKVFPPLD++ QPPAQELIARDLH EWKFRHIFRGQPKRHLLTT
Sbjct: 134 DTSTHGGFSVPRRAAEKVFPPLDYTLQPPAQELIARDLHDVEWKFRHIFRGQPKRHLLTT 193
Query: 185 GWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHLGLLAAAA 244
GWSVFVSAKRLVAGDSV+FI NEKNQL LGIR A+RPQT++PSSVLSSDSMH+GLLAAAA
Sbjct: 194 GWSVFVSAKRLVAGDSVIFIRNEKNQLFLGIRHATRPQTIVPSSVLSSDSMHIGLLAAAA 253
Query: 245 HAAATNSRFTIFYNPRACPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESSVRRYMG 304
HA+ATNS FT+F++PRA SEFVI L+KY+KAV+HTR+SVGMRFRMLFETEESSVRRYMG
Sbjct: 254 HASATNSCFTVFFHPRASQSEFVIQLSKYIKAVFHTRISVGMRFRMLFETEESSVRRYMG 313
Query: 305 TITGICDLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEPLTTFPMYPSPFPLRLK 364
TITGI DLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEPLTTFPMYPS FPLRLK
Sbjct: 314 TITGISDLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEPLTTFPMYPSLFPLRLK 373
Query: 365 RPWPPGLPSFHGMKDDDFGMSSPLMWLRD---TDRGLQSLNYQGIGVNPWMQPRFDPAML 421
RPW G S + D + S L WLR +GL LNY +G+ PWMQ R D + +
Sbjct: 374 RPWHAGTSSLPDGRGD---LGSGLTWLRGGGGEQQGLLPLNYPSVGLFPWMQQRLDLSQM 430
Query: 422 NMQTDM-YQAVAAAALQDMRTVVDPSKQLPGSLLQFQQPPN-FPNRTAALMQAQMLQQSQ 479
+ YQA+ AA LQ++ DP +Q +Q Q+P + + ++A+ MLQQ Q
Sbjct: 431 GTDNNQQYQAMLAAGLQNIGGG-DPLRQ---QFVQLQEPHHQYLQQSASHNSDLMLQQQQ 486
Query: 480 PQQAFQNNNQENQNLSQSQPQAQTNPQQHPQHQHSFNNQLHHHSQQQQQTQQQVVDNNQQ 539
QQA ++ +Q+Q ++ PQQ+ + + S NQ QQ QQ Q N +
Sbjct: 487 QQQASRHLMH-----AQTQIMSENLPQQNMRQEVS--NQPAGQQQQLQQPDQNAYLNAFK 539
Query: 540 ISGSVSTMSQFVSATQPQSPPPMQALSSLCHQQSFSDSNVNSSTTIVSPLHSIMGSSFPH 599
+ + Q+ ++ SP M++ F+DS+ N T SP
Sbjct: 540 MQNG--HLQQWQQQSEMPSPSFMKS--------DFTDSS-NKFATTASPASG-------- 580
Query: 600 DESSLLMSLPRTSSWVPVQNST-GWPSKRIAVDPLLSSGASQCILPQVEQLGQARNSMSQ 658
+ +LL S +P Q +T GW K A N+ S+
Sbjct: 581 -DGNLLNFSITGQSVLPEQLTTEGWSPK-------------------------ASNTFSE 614
Query: 659 NAITLPPFPGRECSIDQEGSNDPQSNLLFGVNIDPSSLLLHNGMSNFKGISGNNNDSS-- 716
+PG+ +++ +PQ+ LFGV+ D S L L + + F SG+ S
Sbjct: 615 PLSLPQAYPGKSLALEP---GNPQNPSLFGVDPD-SGLFLPSTVPRFASSSGDAEASPMS 670
Query: 717 -TMSYHQSSSYMNTAGADSSLNHGVTPSIGESGFLHTQENGEQGNNPLNKTFVKVYKSGS 775
T S Q+S Y L HG G+ ++ K FVKVYKSGS
Sbjct: 671 LTDSGFQNSLYSCMQDTTHELLHGA---------------GQINSSNQTKNFVKVYKSGS 715
Query: 776 FGRSLDITKFSSYNELRSELARMFGLEGELEDPVRSGWQLVFVDRENDVLLLGDGPWPEF 835
GRSLDI++FSSY+ELR EL +MF +EG LEDP+RSGWQLVFVD+END+LLLGD PW F
Sbjct: 716 VGRSLDISRFSSYHELREELGKMFAIEGLLEDPLRSGWQLVFVDKENDILLLGDDPWESF 775
Query: 836 VNSVWCIKILSPEEVQQMGNTGLG 859
VN+VW IKILSPE+V QMG+ G G
Sbjct: 776 VNNVWYIKILSPEDVHQMGDHGEG 799
>ARFE_ARATH (P93024) Auxin response factor 5 (Transcription factor
MONOPTEROS) (Auxin-responsive protein IAA24)
Length = 902
Score = 615 bits (1586), Expect = e-175
Identities = 381/895 (42%), Positives = 516/895 (57%), Gaps = 69/895 (7%)
Query: 7 KRVLDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQLI 66
K V++SELWHACAGPLV LP VGS V YF QGHSEQVAVST + +PNYP+LP QL+
Sbjct: 48 KPVINSELWHACAGPLVCLPQVGSLVYYFSQGHSEQVAVSTRRSATTQVPNYPNLPSQLM 107
Query: 67 CQLHNLTMHADVETDEVYAQMTLQPLNAQEQKEAYLPAELGTPSKQPTNYFCKTLTASDT 126
CQ+HN+T+HAD ++DE+YAQM+LQP++++ + SK PT +FCKTLTASDT
Sbjct: 108 CQVHNVTLHADKDSDEIYAQMSLQPVHSERDVFPVPDFGMLRGSKHPTEFFCKTLTASDT 167
Query: 127 STHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFRGQPKRHLLTTGW 186
STHGGFSVPRRAAEK+FPPLD+S QPP QEL+ RDLH N W FRHI+RGQPKRHLLTTGW
Sbjct: 168 STHGGFSVPRRAAEKLFPPLDYSAQPPTQELVVRDLHENTWTFRHIYRGQPKRHLLTTGW 227
Query: 187 SVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHLGLLAAAAHA 246
S+FV +KRL AGDSVLFI +EK+QL++G+RRA+R QT +PSSVLS+DSMH+G+LAAAAHA
Sbjct: 228 SLFVGSKRLRAGDSVLFIRDEKSQLMVGVRRANRQQTALPSSVLSADSMHIGVLAAAAHA 287
Query: 247 AATNSRFTIFYNPRACPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESSVRRYMGTI 306
A + F IFYNPRACP+EFVIPLAKY KA+ +++SVGMRF M+FETE+S RRYMGTI
Sbjct: 288 TANRTPFLIFYNPRACPAEFVIPLAKYRKAICGSQLSVGMRFGMMFETEDSGKRRYMGTI 347
Query: 307 TGICDLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEPLTTFPMYPSPFPLRLKRP 366
GI DLD +RWP S WR+++V WDE ++ RVS W+IE + ++PS LKR
Sbjct: 348 VGISDLDPLRWPGSKWRNLQVEWDEPGCNDKPTRVSPWDIETPESLFIFPS-LTSGLKRQ 406
Query: 367 WPPGLPSFHGMKDDDFGMSSPLMWLRDTDRGLQS-LNYQGIGVNPWMQPRFDPAMLNMQT 425
P F G + + PL+ + D+ G+ ++ + M+ + M+
Sbjct: 407 LHPSY--FAGETEWGSLIKRPLIRVPDSANGIMPYASFPSMASEQLMK-------MMMRP 457
Query: 426 DMYQAVAAAALQDMRTVVDPSKQLPGSLLQFQQPPNFPNRTAALMQAQMLQQSQPQQ--- 482
Q V + + + +V + L G ++ QQP LM Q + QPQ
Sbjct: 458 HNNQNVPSFMSEMQQNIVMGNGGLLGD-MKMQQP---------LMMNQKSEMVQPQNKLT 507
Query: 483 ---AFQNNNQENQNLSQSQPQAQTNPQQHPQHQHSFNNQLHHHSQQQQQTQQQVVDNNQQ 539
+ N + + QNLSQS A P+ S + ++ H +Q + QV +
Sbjct: 508 VNPSASNTSGQEQNLSQSM-SAPAKPENSTLSGCS-SGRVQHGLEQSMEQASQVTTSTVC 565
Query: 540 ISGSVSTMSQFVSATQP-QSPPPMQALSSLCHQQS-----FSDSNVNSSTTIVSPLHSIM 593
V+ + Q A+ P Q+ + + QS FS + T+ VS S+
Sbjct: 566 NEEKVNQLLQKPGASSPVQADQCLDITHQIYQPQSDPINGFSFLETDELTSQVSSFQSLA 625
Query: 594 GSSFPHDESSLLMSLPRTSSWVPVQNSTGWPSKRIAVDPLLSSGASQCILPQVEQ-LGQA 652
GS + + +L S + SS V + +ST P D L+ P ++Q L +
Sbjct: 626 GS---YKQPFILSS--QDSSAVVLPDSTNSPLFHDVWDTQLNGLKFDQFSPLMQQDLYAS 680
Query: 653 RNSMSQNAITL----PPFPGRE----CSI-DQEGSNDPQSNLLFGVNIDPSSLLLHNGMS 703
+N N+ T PP C+I D + N P L+ G +
Sbjct: 681 QNICMSNSTTSNILDPPLSNTVLDDFCAIKDTDFQNHPSGCLV--------------GNN 726
Query: 704 NFKGISGNNNDSSTMSYHQSSSYMNTAGADSSLNHGVTPSIGESGFLHTQENGEQGN--- 760
N + ++ S+ S ++ D+S G + S + ++N + +
Sbjct: 727 NTSFAQDVQSQITSASFADSQAFSRQDFPDNSGGTGTSSSNVDFDDCSLRQNSKGSSWQK 786
Query: 761 --NPLNKTFVKVYKSGSFGRSLDITKFSSYNELRSELARMFGLEGELEDPVRSGWQLVFV 818
P +T+ KV K+GS GRS+D+T F Y EL+S + MFGLEG L P SGW+LV+V
Sbjct: 787 IATPRVRTYTKVQKTGSVGRSIDVTSFKDYEELKSAIECMFGLEGLLTHPQSSGWKLVYV 846
Query: 819 DRENDVLLLGDGPWPEFVNSVWCIKILSPEEVQQMGNTGLGLLNSVPIQRLSNSI 873
D E+DVLL+GD PW EFV V CI+ILSP EVQQM G+ LLNS I L S+
Sbjct: 847 DYESDVLLVGDDPWEEFVGCVRCIRILSPTEVQQMSEEGMKLLNSAGINDLKTSV 901
>ARFS_ARATH (Q8RYC8) Auxin response factor 19 (Auxin-responsive
protein IAA22)
Length = 1086
Score = 568 bits (1465), Expect = e-161
Identities = 361/814 (44%), Positives = 471/814 (57%), Gaps = 77/814 (9%)
Query: 5 GEKRVLDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQ 64
GEK+ ++S+LWHACAGPLVSLP VGS VVYFPQGHSEQVA S K+ D IPNYP+LP +
Sbjct: 15 GEKKPINSQLWHACAGPLVSLPPVGSLVVYFPQGHSEQVAASMQKQTD-FIPNYPNLPSK 73
Query: 65 LICQLHNLTMHADVETDEVYAQMTLQPLNAQEQKEAYLPAELGTP-SKQPTNYFCKTLTA 123
LIC LH++T+HAD ETDEVYAQMTLQP+N + +EA L +++G ++QPT +FCKTLTA
Sbjct: 74 LICLLHSVTLHADTETDEVYAQMTLQPVNKYD-REALLASDMGLKLNRQPTEFFCKTLTA 132
Query: 124 SDTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFRGQPKRHLLT 183
SDTSTHGGFSVPRRAAEK+FPPLDFS QPPAQE++A+DLH W FRHI+RGQPKRHLLT
Sbjct: 133 SDTSTHGGFSVPRRAAEKIFPPLDFSMQPPAQEIVAKDLHDTTWTFRHIYRGQPKRHLLT 192
Query: 184 TGWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHLGLLAAA 243
TGWSVFVS KRL AGDSVLF+ +EK+QL+LGIRRA+R + SSV+SSDSMH+G+LAAA
Sbjct: 193 TGWSVFVSTKRLFAGDSVLFVRDEKSQLMLGIRRANRQTPTLSSSVISSDSMHIGILAAA 252
Query: 244 AHAAATNSRFTIFYNPRACPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESSVRRYM 303
AHA A +S FTIF+NPRA PSEFV+PLAKY KA+Y +VS+GMRFRM+FETE+ VRRYM
Sbjct: 253 AHANANSSPFTIFFNPRASPSEFVVPLAKYNKALY-AQVSLGMRFRMMFETEDCGVRRYM 311
Query: 304 GTITGICDLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEP-LTTFPMYPSPFPLR 362
GT+TGI DLD VRW S WR+++VGWDESTAG+R RVS+WEIEP +T F + P PF R
Sbjct: 312 GTVTGISDLDPVRWKGSQWRNLQVGWDESTAGDRPSRVSIWEIEPVITPFYICPPPF-FR 370
Query: 363 LKRPWPPGLPSFHGMKDDDFGMSSPLMWLRDTDRGLQSLNYQGIGVNPWM-QPRFDPAML 421
K P PG+P ++ F + P M + QS + G+ + WM + +P
Sbjct: 371 PKYPRQPGMPDDELDMENAFKRAMPWMGEDFGMKDAQSSMFPGLSLVQWMSMQQNNPLSG 430
Query: 422 NMQTDMYQAVAAAALQDMRTVVDPSKQLPGSLLQFQQP-----------PNFPNRTAALM 470
+ + A+++ L + DPSK LL FQ P PN N + M
Sbjct: 431 SATPQLPSALSSFNLPNNFASNDPSK-----LLNFQSPNLSSANSQFNKPNTVNHISQQM 485
Query: 471 QAQ--MLQQSQPQQAFQNNNQENQNLSQSQPQAQTNPQQHPQHQHSFNNQLHHHSQQ--- 525
QAQ M++ Q QQ Q +Q Q Q Q Q Q + QQ Q Q +NN + Q
Sbjct: 486 QAQPAMVKSQQQQQQQQQQHQHQQQQLQQQQQLQMS-QQQVQQQGIYNNGTIAVANQVSC 544
Query: 526 ----------QQQTQQQVV----------DNNQQISGSVSTMSQFVSATQPQSPPPMQAL 565
Q Q QQQ + + N + +S M+ F Q Q M
Sbjct: 545 QSPNQPTGFSQSQLQQQSMLPTGAKMTHQNINSMGNKGLSQMTSFAQEMQFQQQLEMHNS 604
Query: 566 SSLCHQQSFSDSNVNSSTTIVS----PLHSIMGSSFPHDESSLLMSLPRTSSWVPVQNST 621
S L Q S+++S +S L SS P L + L + Q S
Sbjct: 605 SQLLRNQQ-EQSSLHSLQQNLSQNPQQLQMQQQSSKPSPSQQLQLQLLQKLQQQQQQQSI 663
Query: 622 GWPSKRIAVDPLLS------SGASQCILPQVEQLGQARNSMSQNAITLPPFPGRECSIDQ 675
P ++ P LS S Q +L Q A + S A T P + S Q
Sbjct: 664 --PPVSSSLQPQLSALQQTQSHQLQQLLSSQNQQPLAHGNNSFPASTFMQPPQIQVSPQQ 721
Query: 676 EGSNDPQSNLLFGVNIDPSSLLLHNGMSNFKGISGNNNDSSTMSYHQSSSYMNTAGADSS 735
+G ++ + G + H+G ++ + S + + S+ + H + S N +
Sbjct: 722 QGQMSNKNLVAAGRS--------HSGHTDGEAPSCSTSPSANNTGHDNVSPTNFLSRNQQ 773
Query: 736 LNHGVTPSIGESGFLHTQENGEQGNNPLNKTFVK 769
+ S +S F E+ +NP+ + + K
Sbjct: 774 QGQAASVSASDSVF-------ERASNPVQELYTK 800
Score = 140 bits (353), Expect = 2e-32
Identities = 137/461 (29%), Positives = 208/461 (44%), Gaps = 54/461 (11%)
Query: 422 NMQTDMYQAVAAAALQDMRTVVDPSKQLPGSLLQF----QQPPNFPNRTAALM-QAQMLQ 476
++Q ++ Q +Q + PS+QL LLQ QQ + P +++L Q LQ
Sbjct: 619 SLQQNLSQNPQQLQMQQQSSKPSPSQQLQLQLLQKLQQQQQQQSIPPVSSSLQPQLSALQ 678
Query: 477 QSQPQQAFQNNNQENQNLSQSQPQAQTNPQQHPQHQHSFNNQLHHHSQQQ-QQTQQQVVD 535
Q+Q Q Q + +NQ QP A N P Q+ QQQ Q + + +V
Sbjct: 679 QTQSHQLQQLLSSQNQ-----QPLAHGN-NSFPASTFMQPPQIQVSPQQQGQMSNKNLVA 732
Query: 536 NNQQISGSVS------TMSQFVSATQPQSPPPMQALSSLCHQQSFSDSNVNSSTTIVSPL 589
+ SG + S + T + P LS +QQ ++V++S ++
Sbjct: 733 AGRSHSGHTDGEAPSCSTSPSANNTGHDNVSPTNFLSR--NQQQGQAASVSASDSVFERA 790
Query: 590 HSIMGSSFPHDESSLLMSLPRTSSWVPVQNSTGWPSKRIAVDPLLSSGASQCILPQVEQL 649
+ + + ES + + S + K D + S A P V +
Sbjct: 791 SNPVQELYTKTESRISQGMMNMKS-----AGEHFRFKSAVTDQIDVSTAGTTYCPDV--V 843
Query: 650 GQARNSMSQNAITLPPFP-GRECSIDQEGSNDPQSNLLFGVNIDPSSLLLHNGMSNFKGI 708
G + Q LP F +C S+ P++NL F N++ + +F+ +
Sbjct: 844 GPVQQ---QQTFPLPSFGFDGDCQ-----SHHPRNNLAFPGNLEAVTSDPLYSQKDFQNL 895
Query: 709 SGNNNDSSTMSYHQSSSYMNTAGADSSLNHGVTPSIGESGFLHTQENGEQGNNPLN---- 764
N ++ + SS A SS + G+ PSI + G + +N
Sbjct: 896 VPNYGNTPRDIETELSS-----AAISSQSFGI-PSIPFKPGCSNEVGGINDSGIMNGGGL 949
Query: 765 --------KTFVKVYKSGSFGRSLDITKFSSYNELRSELARMFGLEGELEDPVRSGWQLV 816
+T+ KV K GS GRS+D+T++S Y+ELR +LARMFG+EG+LEDP+ S W+LV
Sbjct: 950 WPNQTQRMRTYTKVQKRGSVGRSIDVTRYSGYDELRHDLARMFGIEGQLEDPLTSDWKLV 1009
Query: 817 FVDRENDVLLLGDGPWPEFVNSVWCIKILSPEEVQQMGNTG 857
+ D END+LL+GD PW EFVN V IKILS EVQQM G
Sbjct: 1010 YTDHENDILLVGDDPWEEFVNCVQNIKILSSVEVQQMSLDG 1050
>ARFG_ARATH (P93022) Auxin response factor 7 (Non-phototropic
hypocotyl 4) (BIPOSTO protein) (Auxin-responsive protein
IAA21/IAA23/IAA25)
Length = 1164
Score = 560 bits (1443), Expect = e-159
Identities = 337/662 (50%), Positives = 412/662 (61%), Gaps = 69/662 (10%)
Query: 5 GEKRVLDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQ 64
GE+R ++SELWHACAGPL+SLP GS VVYFPQGHSEQVA S K+ D IP+YP+LP +
Sbjct: 16 GERRNINSELWHACAGPLISLPPAGSLVVYFPQGHSEQVAASMQKQTD-FIPSYPNLPSK 74
Query: 65 LICQLHNLTMHADVETDEVYAQMTLQPLNAQEQKEAYLPAELGTP-SKQPTNYFCKTLTA 123
LIC LHN+T++AD ETDEVYAQMTLQP+N + ++A L +++G ++QP +FCKTLTA
Sbjct: 75 LICMLHNVTLNADPETDEVYAQMTLQPVNKYD-RDALLASDMGLKLNRQPNEFFCKTLTA 133
Query: 124 SDTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFRGQPKRHLLT 183
SDTSTHGGFSVPRRAAEK+FP LDFS QPP QEL+A+D+H N W FRHI+RGQPKRHLLT
Sbjct: 134 SDTSTHGGFSVPRRAAEKIFPALDFSMQPPCQELVAKDIHDNTWTFRHIYRGQPKRHLLT 193
Query: 184 TGWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHLGLLAAA 243
TGWSVFVS KRL AGDSVLFI + K QLLLGIRRA+R Q + SSV+SSDSMH+G+LAAA
Sbjct: 194 TGWSVFVSTKRLFAGDSVLFIRDGKAQLLLGIRRANRQQPALSSSVISSDSMHIGVLAAA 253
Query: 244 AHAAATNSRFTIFYNPRACPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESSVRRYM 303
AHA A NS FTIFYNPRA P+EFV+PLAKY KA+Y +VS+GMRFRM+FETEE VRRYM
Sbjct: 254 AHANANNSPFTIFYNPRAAPAEFVVPLAKYTKAMY-AQVSLGMRFRMIFETEECGVRRYM 312
Query: 304 GTITGICDLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEP-LTTFPMYPSPFPLR 362
GT+TGI DLD VRW NS WR++++GWDES AG+R RVS+W+IEP LT F + P PF
Sbjct: 313 GTVTGISDLDPVRWKNSQWRNLQIGWDESAAGDRPSRVSVWDIEPVLTPFYICPPPF--- 369
Query: 363 LKRPWPPGLPSFHGMKDDDFGMSSPLM----WLRDT--DRGLQSLNYQGIGVNPWM---- 412
RP G P GM DD+ M S L WL ++ + S + G+ + WM
Sbjct: 370 -FRPRFSGQP---GMPDDETDMESALKRAMPWLDNSLEMKDPSSTIFPGLSLVQWMNMQQ 425
Query: 413 ----------QPRFDPAMLNMQTDMYQAVAAAALQDMRTVVDPSKQLPGSLLQFQQPP-- 460
QP F P+ML + AA ++ DPSK LL FQ P
Sbjct: 426 QNGQLPSAAAQPGFFPSML--------SPTAALHNNLGGTDDPSK-----LLSFQTPHGG 472
Query: 461 --------NFPNRTAALMQ-----AQMLQQSQPQQAFQN--NNQENQNLSQSQPQAQTNP 505
N N+ A + Q + QQ Q QQ + N+Q+ Q+ SQ Q Q Q
Sbjct: 473 ISSSNLQFNKQNQQAPMSQLPQPPTTLSQQQQLQQLLHSSLNHQQQQSQSQQQQQQQQLL 532
Query: 506 QQHPQ---HQHSFNNQLHHHSQQ---QQQTQQQVVDNNQQISGSVSTMSQF-VSATQPQS 558
QQ Q QHS NNQ QQ QQQ QQQ+ +QQ + Q Q S
Sbjct: 533 QQQQQLQSQQHSNNNQSQSQQQQQLLQQQQQQQLQQQHQQPLQQQTQQQQLRTQPLQSHS 592
Query: 559 PPPMQALSSLCHQQSFSDSNVNSSTTIVSPLHSIMGSSFPHDESSLLMSLPRTSSWVPVQ 618
P Q L QQ N + + H +S H + L+ +S P
Sbjct: 593 HPQPQQLQQHKLQQLQVPQNQLYNGQQAAQQHQSQQASTHHLQPQLVSGSMASSVITPPS 652
Query: 619 NS 620
+S
Sbjct: 653 SS 654
Score = 146 bits (368), Expect = 3e-34
Identities = 147/451 (32%), Positives = 196/451 (42%), Gaps = 83/451 (18%)
Query: 457 QQPPNFPNRTAALMQAQMLQQSQPQQAFQNNNQENQNLSQSQPQAQTNPQQHPQHQH-SF 515
QQPP + + Q Q+Q QQ FQ + E ++ Q Q QQ Q Q S
Sbjct: 712 QQPPGLNGQNQQTLLQQKAHQAQAQQIFQQSLLEQPHI---QFQLLQRLQQQQQQQFLSP 768
Query: 516 NNQLHHHSQQQQQTQQ-QVVDNNQQISGS-----VSTM---SQFVSATQPQSPPP----M 562
+QL HH Q QQ QQ + Q S +ST+ VS Q + PP +
Sbjct: 769 QSQLPHHQLQSQQLQQLPTLSQGHQFPSSCTNNGLSTLQPPQMLVSRPQEKQNPPVGGGV 828
Query: 563 QALSSLCHQQSFSDSNVNSSTTIVSPLHSIMGSSFPHDESS----------------LLM 606
+A S + S+ + ST I S F + S L+
Sbjct: 829 KAYSGITDGGDAPSSSTSPSTNNC----QISSSGFLNRSQSGPAILIPDAAIDMSGNLVQ 884
Query: 607 SLPRTSSWVPVQNSTG-WPSKRIAVDPLLSSGASQCILPQVEQLGQARNSMSQNAITLPP 665
L S Q G SK D L + AS L N+ QN + P
Sbjct: 885 DLYSKSDMRLKQELVGQQKSKASLTDHQLEASASGTSY----GLDGGENNRQQNFLA-PT 939
Query: 666 FPGRECSIDQEGSNDPQSNLLFGVNID----PSSLL---------LHNGMSNFKGISGNN 712
F +D D +++LL G N+D P +LL L N +SN+ G++
Sbjct: 940 F-----GLD----GDSRNSLLGGANVDNGFVPDTLLSRGYDSQKDLQNMLSNYGGVT--- 987
Query: 713 NDSSTMSYHQSSSYMNTAGADSSLNHGVTPSIGESGFLHTQENGEQG------NNPLNKT 766
ND T S+S + T + GV S L + G G +T
Sbjct: 988 NDIGT---EMSTSAVRTQ------SFGVPNVPAISNDLAVNDAGVLGGGLWPAQTQRMRT 1038
Query: 767 FVKVYKSGSFGRSLDITKFSSYNELRSELARMFGLEGELEDPVRSGWQLVFVDRENDVLL 826
+ KV K GS GRS+D+ ++ Y+ELR +LARMFG+EG+LEDP S W+LV+VD END+LL
Sbjct: 1039 YTKVQKRGSVGRSIDVNRYRGYDELRHDLARMFGIEGQLEDPQTSDWKLVYVDHENDILL 1098
Query: 827 LGDGPWPEFVNSVWCIKILSPEEVQQMGNTG 857
+GD PW EFVN V IKILS EVQQM G
Sbjct: 1099 VGDDPWEEFVNCVQSIKILSSAEVQQMSLDG 1129
Score = 38.9 bits (89), Expect = 0.062
Identities = 85/371 (22%), Positives = 122/371 (31%), Gaps = 83/371 (22%)
Query: 386 SPLMWLRDTDRGLQSLNYQGIGVN---PWMQPRFDPAMLNMQTDMYQAVAAAALQDMRTV 442
S L+ + G+ S N Q N P Q P L+ Q + Q + ++ +
Sbjct: 461 SKLLSFQTPHGGISSSNLQFNKQNQQAPMSQLPQPPTTLSQQQQLQQLLHSSLNHQQQQS 520
Query: 443 VDPSKQLPGSLLQFQQPPNFP-----NRTAALMQAQMLQQSQPQQAFQNNNQENQNLSQS 497
+Q LLQ QQ N++ + Q Q+LQQ Q QQ Q + Q Q +Q
Sbjct: 521 QSQQQQQQQQLLQQQQQLQSQQHSNNNQSQSQQQQQLLQQQQQQQLQQQHQQPLQQQTQQ 580
Query: 498 QPQAQTNPQQ---HPQ----HQHSFN------NQLH----------------HH------ 522
Q Q +T P Q HPQ QH NQL+ HH
Sbjct: 581 Q-QLRTQPLQSHSHPQPQQLQQHKLQQLQVPQNQLYNGQQAAQQHQSQQASTHHLQPQLV 639
Query: 523 ----------------------SQQQQQTQQQVVDNNQQISGSVSTMSQFVSATQPQSPP 560
QQQ + QQ + + S + S++ S P
Sbjct: 640 SGSMASSVITPPSSSLNQSFQQQQQQSKQLQQAHHHLGASTSQSSVIETSKSSSNLMSAP 699
Query: 561 PMQALSS--LCHQQSFSDSNVNSSTTIVSPLHSIMGSSF--------PHDESSLLMSLPR 610
P + S + QQ + N T + H PH + LL L +
Sbjct: 700 PQETQFSRQVEQQQPPGLNGQNQQTLLQQKAHQAQAQQIFQQSLLEQPHIQFQLLQRLQQ 759
Query: 611 TSSWVPVQNSTGWPSKRIAVDPLLSSGASQCILPQVEQLGQARNSMSQNAITLPPFPGRE 670
Q P ++ L S Q LP + Q Q +S + N ++ P
Sbjct: 760 QQ-----QQQFLSPQSQLPHHQLQSQQLQQ--LPTLSQGHQFPSSCTNNGLSTLQPPQML 812
Query: 671 CSIDQEGSNDP 681
S QE N P
Sbjct: 813 VSRPQEKQNPP 823
>ARFB_ARATH (Q94JM3) Auxin response factor 2 (ARF1-binding protein)
(ARF1-BP)
Length = 859
Score = 396 bits (1017), Expect = e-109
Identities = 200/370 (54%), Positives = 260/370 (70%), Gaps = 10/370 (2%)
Query: 4 AGEKRVLDSE------LWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPN 57
A +R LD E LWHACAGPLV++P RV YFPQGH EQV STN+ + +P
Sbjct: 46 AAAERALDPEAALYRELWHACAGPLVTVPRQDDRVFYFPQGHIEQVEASTNQAAEQQMPL 105
Query: 58 YPSLPPQLICQLHNLTMHADVETDEVYAQMTLQPLNAQEQKEAYLPAELGTPSKQPTNYF 117
Y LP +L+C++ N+ + A+ +TDEVYAQ+TL P Q++ A L P + + F
Sbjct: 106 Y-DLPSKLLCRVINVDLKAEADTDEVYAQITLLPEANQDENAIEKEAPLPPPPRFQVHSF 164
Query: 118 CKTLTASDTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFRGQP 177
CKTLTASDTSTHGGFSV RR A++ PPLD S+QPP QEL+A+DLH NEW+FRHIFRGQP
Sbjct: 165 CKTLTASDTSTHGGFSVLRRHADECLPPLDMSRQPPTQELVAKDLHANEWRFRHIFRGQP 224
Query: 178 KRHLLTTGWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHL 237
+RHLL +GWSVFVS+KRLVAGD+ +F+ E +L +G+RRA R Q +PSSV+SS SMHL
Sbjct: 225 RRHLLQSGWSVFVSSKRLVAGDAFIFLRGENGELRVGVRRAMRQQGNVPSSVISSHSMHL 284
Query: 238 GLLAAAAHAAATNSRFTIFYNPRACPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEES 297
G+LA A HA +T + FT++Y PR PSEF++P +Y+++V S+GMRF+M FE EE+
Sbjct: 285 GVLATAWHAISTGTMFTVYYKPRTSPSEFIVPFDQYMESV-KNNYSIGMRFKMRFEGEEA 343
Query: 298 SVRRYMGTITGICDLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEPLTTFP-MYP 356
+R+ GTI GI + D RWP S WRS+KV WDE+++ R RVS W++EP P + P
Sbjct: 344 PEQRFTGTIVGIEESDPTRWPKSKWRSLKVRWDETSSIPRPDRVSPWKVEPALAPPALSP 403
Query: 357 SPFPLRLKRP 366
P P R KRP
Sbjct: 404 VPMP-RPKRP 412
Score = 85.1 bits (209), Expect = 8e-16
Identities = 62/213 (29%), Positives = 110/213 (51%), Gaps = 18/213 (8%)
Query: 647 EQLGQARNSMSQNAITLPPFPGRECSIDQEGSNDPQSNL-LFGVNIDPSSLLLHNGMSNF 705
E + + ++ +T PF +I +E + + N LFG+ + + NG +
Sbjct: 621 EVVNAQAQAQAREQVTKQPF-----TIQEETAKSREGNCRLFGIPLTNNM----NGTDST 671
Query: 706 KGISGNNNDSSTMSYHQSSSYMNTA----GADSSLNHGVTPSIGESGFLHTQENGEQGNN 761
N ND++ ++ S + + G+ S+ +H ++ H ++ + N+
Sbjct: 672 MSQRNNLNDAAGLTQIASPKVQDLSDQSKGSKSTNDHREQGRPFQTNNPHPKDAQTKTNS 731
Query: 762 PLNKTFVKVYKSG-SFGRSLDITKFSSYNELRSELARMFGLEGELEDPVRSGWQLVFVDR 820
+++ KV+K G + GRS+D++KF +Y EL +EL R+F GEL P + W +V+ D
Sbjct: 732 --SRSCTKVHKQGIALGRSVDLSKFQNYEELVAELDRLFEFNGELMAP-KKDWLIVYTDE 788
Query: 821 ENDVLLLGDGPWPEFVNSVWCIKILSPEEVQQM 853
END++L+GD PW EF V I I + EEV++M
Sbjct: 789 ENDMMLVGDDPWQEFCCMVRKIFIYTKEEVRKM 821
>ARFA_ARATH (Q8L7G0) Auxin response factor 1
Length = 665
Score = 388 bits (997), Expect = e-107
Identities = 200/365 (54%), Positives = 261/365 (70%), Gaps = 9/365 (2%)
Query: 13 ELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQLICQLHNL 72
ELWHACAGPLV+LP G RV YFP+GH EQ+ S ++ ++ +P++ +LP +++C++ N+
Sbjct: 22 ELWHACAGPLVTLPREGERVYYFPEGHMEQLEASMHQGLEQQMPSF-NLPSKILCKVINI 80
Query: 73 TMHADVETDEVYAQMTLQPLNAQEQKEAYLP-AELGTPSKQPTNYFCKTLTASDTSTHGG 131
A+ ETDEVYAQ+TL P +Q E P A + P K + FCKTLTASDTSTHGG
Sbjct: 81 QRRAEPETDEVYAQITLLP--ELDQSEPTSPDAPVQEPEKCTVHSFCKTLTASDTSTHGG 138
Query: 132 FSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFRGQPKRHLLTTGWSVFVS 191
FSV RR A+ PPLD SQQPP QEL+A DLH +EW FRHIFRGQP+RHLLTTGWSVFVS
Sbjct: 139 FSVLRRHADDCLPPLDMSQQPPWQELVATDLHNSEWHFRHIFRGQPRRHLLTTGWSVFVS 198
Query: 192 AKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHLGLLAAAAHAAATNS 251
+K+LVAGD+ +F+ E +L +G+RR R QT +PSSV+SS SMH+G+LA AAHA T +
Sbjct: 199 SKKLVAGDAFIFLRGENEELRVGVRRHMRQQTNIPSSVISSHSMHIGVLATAAHAITTGT 258
Query: 252 RFTIFYNPRACPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESSVRRYMGTITGICD 311
F++FY PR SEF++ + +Y++A ++SVGMRF+M FE EE+ +R+ GTI G+ +
Sbjct: 259 IFSVFYKPRTSRSEFIVSVNRYLEAKTQ-KLSVGMRFKMRFEGEEAPEKRFSGTIVGVQE 317
Query: 312 LDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEPL---TTFPMYPSPFPLRLKRPWP 368
S W +S WRS+KV WDE ++ R RVS WE+EPL +T P P P R KRP P
Sbjct: 318 NKSSVWHDSEWRSLKVQWDEPSSVFRPERVSPWELEPLVANSTPSSQPQP-PQRNKRPRP 376
Query: 369 PGLPS 373
PGLPS
Sbjct: 377 PGLPS 381
Score = 71.6 bits (174), Expect = 9e-12
Identities = 35/90 (38%), Positives = 59/90 (64%), Gaps = 2/90 (2%)
Query: 765 KTFVKVYKSGS-FGRSLDITKFSSYNELRSELARMFGLEGELEDPVRSGWQLVFVDREND 823
++ KV+ GS GR++D+T+ Y +L +L MF ++GEL + + WQ+V+ D E+D
Sbjct: 542 RSCTKVHMQGSAVGRAIDLTRSECYEDLFKKLEEMFDIKGELLESTKK-WQVVYTDDEDD 600
Query: 824 VLLLGDGPWPEFVNSVWCIKILSPEEVQQM 853
++++GD PW EF V I I +PEEV+++
Sbjct: 601 MMMVGDDPWNEFCGMVRKIFIYTPEEVKKL 630
>ARFD_ARATH (Q9ZTX9) Auxin response factor 4
Length = 788
Score = 376 bits (966), Expect = e-103
Identities = 200/385 (51%), Positives = 254/385 (65%), Gaps = 21/385 (5%)
Query: 5 GEKRVLDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQ 64
G + SELWHACAGPL LP G+ VVYFPQGH EQ A+ + IP + L PQ
Sbjct: 57 GSASSIYSELWHACAGPLTCLPKKGNVVVYFPQGHLEQDAMVSYSS-PLEIPKF-DLNPQ 114
Query: 65 LICQLHNLTMHADVETDEVYAQMTLQPLN--------AQEQKEAYLPAELGTPS--KQPT 114
++C++ N+ + A+ +TDEVY Q+TL PL +E KE E S K+
Sbjct: 115 IVCRVVNVQLLANKDTDEVYTQVTLLPLQEFSMLNGEGKEVKELGGEEERNGSSSVKRTP 174
Query: 115 NYFCKTLTASDTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFR 174
+ FCKTLTASDTSTHGGFSVPRRAAE F PLD+ QQ P+QELIA+DLHG EWKFRHI+R
Sbjct: 175 HMFCKTLTASDTSTHGGFSVPRRAAEDCFAPLDYKQQRPSQELIAKDLHGVEWKFRHIYR 234
Query: 175 GQPKRHLLTTGWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDS 234
GQP+RHLLTTGWS+FVS K LV+GD+VLF+ +E +L LGIRRA+RP+ +P S++ +S
Sbjct: 235 GQPRRHLLTTGWSIFVSQKNLVSGDAVLFLRDEGGELRLGIRRAARPRNGLPDSIIEKNS 294
Query: 235 MHLGLLAAAAHAAATNSRFTIFYNPRACPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFET 294
+L+ A+A +T S F +FY+PRA +EFVIP KY+ ++ + V +G RFRM FE
Sbjct: 295 CS-NILSLVANAVSTKSMFHVFYSPRATHAEFVIPYEKYITSI-RSPVCIGTRFRMRFEM 352
Query: 295 EESSVRRYMGTITGICDLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEPLTTFPM 354
++S RR G +TG+CDLD RWPNS WR + V WDES + Q RVS WEI+P + P
Sbjct: 353 DDSPERRCAGVVTGVCDLDPYRWPNSKWRCLLVRWDESFVSDHQERVSPWEIDPSVSLPH 412
Query: 355 YPSPFPLRLKRPW-------PPGLP 372
R KRPW PPG P
Sbjct: 413 LSIQSSPRPKRPWAGLLDTTPPGNP 437
Score = 85.5 bits (210), Expect = 6e-16
Identities = 48/152 (31%), Positives = 82/152 (53%), Gaps = 5/152 (3%)
Query: 734 SSLNHGVTPSIGESGF-LHTQENGEQGNNPLNKTFVKVYKSGS-FGRSLDITKFSSYNEL 791
S LN + GF L + + + + KV+K GS GR++D+++ + Y++L
Sbjct: 633 SELNMNASSGCKLFGFSLPVETPASKPQSSSKRICTKVHKQGSQVGRAIDLSRLNGYDDL 692
Query: 792 RSELARMFGLEGELEDPVRSGWQLVFVDRENDVLLLGDGPWPEFVNSVWCIKILSPEEVQ 851
EL R+F +EG L DP + GW++++ D END++++GD PW +F N VW I + + EEV+
Sbjct: 693 LMELERLFNMEGLLRDPEK-GWRILYTDSENDMMVVGDDPWHDFCNVVWKIHLYTKEEVE 751
Query: 852 QMGNTGLGLLNSVPIQRLSNSICDDYVSRQDS 883
+ L + ++ VS+ DS
Sbjct: 752 NANDDNKSCLEQAALMMEASK--SSSVSQPDS 781
>ARFR_ARATH (Q9C5W9) Auxin response factor 18
Length = 602
Score = 359 bits (921), Expect = 2e-98
Identities = 185/346 (53%), Positives = 243/346 (69%), Gaps = 6/346 (1%)
Query: 10 LDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQLICQL 69
L +ELW CAGPLV +P RV YFPQGH EQ+ STN+ +++ LPP+++C++
Sbjct: 22 LYTELWKVCAGPLVEVPRAQERVFYFPQGHMEQLVASTNQGINSEEIPVFDLPPKILCRV 81
Query: 70 HNLTMHADVETDEVYAQMTLQPLNAQEQKEAYLPAELGTPSKQPTNYFCKTLTASDTSTH 129
++T+ A+ ETDEVYAQ+TLQP Q + + P +G P+KQ + F K LTASDTSTH
Sbjct: 82 LDVTLKAEHETDEVYAQITLQPEEDQSEPTSLDPPIVG-PTKQEFHSFVKILTASDTSTH 140
Query: 130 GGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFRGQPKRHLLTTGWSVF 189
GGFSV R+ A + P LD +Q P QEL+ RDLHG EW+F+HIFRGQP+RHLLTTGWS F
Sbjct: 141 GGFSVLRKHATECLPSLDMTQATPTQELVTRDLHGFEWRFKHIFRGQPRRHLLTTGWSTF 200
Query: 190 VSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHLGLLAAAAHAAAT 249
VS+KRLVAGD+ +F+ E L +G+RR +R Q+ MP+SV+SS SMHLG+LA A+HA T
Sbjct: 201 VSSKRLVAGDAFVFLRGENGDLRVGVRRLARHQSTMPTSVISSQSMHLGVLATASHAVRT 260
Query: 250 NSRFTIFYNPRACPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESSVRRYMGTITGI 309
+ F +FY PR S+F++ + KY++A+ H S+G RFRM FE EES R + GTI G
Sbjct: 261 TTIFVVFYKPRI--SQFIVGVNKYMEAIKH-GFSLGTRFRMRFEGEESPERIFTGTIVGS 317
Query: 310 CDLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEP-LTTFPM 354
DL S +WP S WRS++V WDE T +R +VS WEIEP L T P+
Sbjct: 318 GDLSS-QWPASKWRSLQVQWDEPTTVQRPDKVSPWEIEPFLATSPI 362
Score = 69.7 bits (169), Expect = 3e-11
Identities = 50/151 (33%), Positives = 75/151 (49%), Gaps = 15/151 (9%)
Query: 722 QSSSYMNTAGADSSLNHGVTPSIGESGFLHTQENGEQGNNPLNKTFVKVYKSG-SFGRSL 780
Q M+T GA + +TP+ EQ +++ KV G + GR++
Sbjct: 457 QDKQPMDTCGA-AKCQEPITPT----------SMSEQKKQQTSRSRTKVQMQGIAVGRAV 505
Query: 781 DITKFSSYNELRSELARMFGLEGELEDPVRSGWQLVFVDRENDVLLLGDGPWPEFVNSVW 840
D+T SY+EL EL MF ++G+L R W +VF D E D++L GD PW EF
Sbjct: 506 DLTLLKSYDELIDELEEMFEIQGQLL--ARDKWIVVFTDDEGDMMLAGDDPWNEFCKMAK 563
Query: 841 CIKILSPEEVQQMGNTGLGLLNSVPIQRLSN 871
I I S +EV++M T L + +S+ + N
Sbjct: 564 KIFIYSSDEVKKM-TTKLKISSSLENEEYGN 593
>ARFK_ARATH (Q9ZPY6) Auxin response factor 11
Length = 601
Score = 357 bits (917), Expect = 6e-98
Identities = 185/350 (52%), Positives = 247/350 (69%), Gaps = 7/350 (2%)
Query: 10 LDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEV-DAHIPNYPSLPPQLICQ 68
L +ELW ACAGPLV +P G RV YFPQGH EQ+ STN+ V D IP + +LPP+++C+
Sbjct: 18 LYTELWKACAGPLVEVPRYGERVFYFPQGHMEQLVASTNQGVVDQEIPVF-NLPPKILCR 76
Query: 69 LHNLTMHADVETDEVYAQMTLQPLNAQEQKEAYLPAELGTPSKQPTNYFCKTLTASDTST 128
+ ++T+ A+ ETDEVYAQ+TLQP Q + + P L P+K + F K LTASDTST
Sbjct: 77 VLSVTLKAEHETDEVYAQITLQPEEDQSEPTSLDPP-LVEPAKPTVDSFVKILTASDTST 135
Query: 129 HGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFRGQPKRHLLTTGWSV 188
HGGFSV R+ A + P LD +Q P QEL+ARDLHG EW+F+HIFRGQP+RHLLTTGWS
Sbjct: 136 HGGFSVLRKHATECLPSLDMTQPTPTQELVARDLHGYEWRFKHIFRGQPRRHLLTTGWST 195
Query: 189 FVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHLGLLAAAAHAAA 248
FV++KRLVAGD+ +F+ E L +G+RR ++ Q+ MP+SV+SS SM LG+LA A+HA
Sbjct: 196 FVTSKRLVAGDAFVFLRGETGDLRVGVRRLAKQQSTMPASVISSQSMRLGVLATASHAVT 255
Query: 249 TNSRFTIFYNPRACPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESSVRRYMGTITG 308
T + F +FY PR S+F+I + KY+ A+ S+GMR+RM FE EES R + GTI G
Sbjct: 256 TTTIFVVFYKPRI--SQFIISVNKYMMAM-KNGFSLGMRYRMRFEGEESPERIFTGTIIG 312
Query: 309 ICDLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEPLTTFPMYPSP 358
DL S +WP S WRS+++ WDE ++ +R +VS WEIEP + + P+P
Sbjct: 313 SGDLSS-QWPASKWRSLQIQWDEPSSIQRPNKVSPWEIEPFSPSALTPTP 361
Score = 69.3 bits (168), Expect = 4e-11
Identities = 39/103 (37%), Positives = 58/103 (55%), Gaps = 3/103 (2%)
Query: 757 EQGNNPLNKTFVKVYKSGS-FGRSLDITKFSSYNELRSELARMFGLEGELEDPVRSGWQL 815
EQ ++ +KV G+ GR++D+T SY+EL EL +MF +EGEL + W +
Sbjct: 482 EQKQQTSTRSRIKVQMQGTAVGRAVDLTLLRSYDELIKELEKMFEIEGELSP--KDKWAI 539
Query: 816 VFVDRENDVLLLGDGPWPEFVNSVWCIKILSPEEVQQMGNTGL 858
VF D E D +L+GD PW EF + I +EV++M + L
Sbjct: 540 VFTDDEGDRMLVGDDPWNEFCKMAKKLFIYPSDEVKKMRSKSL 582
>ARFI_ARATH (Q9XED8) Auxin response factor 9
Length = 638
Score = 345 bits (885), Expect = 3e-94
Identities = 181/361 (50%), Positives = 241/361 (66%), Gaps = 12/361 (3%)
Query: 10 LDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNK-EVDAHIPNYPSLPPQLICQ 68
L ELW CAGPLV +P RV YFPQGH EQ+ ST + +++ P + LPP+++C
Sbjct: 9 LYDELWKLCAGPLVDVPQAQERVYYFPQGHMEQLEASTQQVDLNTMKPLFV-LPPKILCN 67
Query: 69 LHNLTMHADVETDEVYAQMTLQPLNAQEQKEAY---LPAELGTPSKQPTNYFCKTLTASD 125
+ N+++ A+ +TDEVYAQ+TL P+ + + P EL P + F K LTASD
Sbjct: 68 VMNVSLQAEKDTDEVYAQITLIPVGTEVDEPMSPDPSPPELQRPK---VHSFSKVLTASD 124
Query: 126 TSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFRGQPKRHLLTTG 185
TSTHGGFSV R+ A + PPLD +QQ P QEL+A D+HG +WKF+HIFRGQP+RHLLTTG
Sbjct: 125 TSTHGGFSVLRKHATECLPPLDMTQQTPTQELVAEDVHGYQWKFKHIFRGQPRRHLLTTG 184
Query: 186 WSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHLGLLAAAAH 245
WS FV++KRLVAGD+ +F+ E +L +G+RRA+ Q+ MPSSV+SS SMHLG+LA A H
Sbjct: 185 WSTFVTSKRLVAGDTFVFLRGENGELRVGVRRANLQQSSMPSSVISSHSMHLGVLATARH 244
Query: 246 AAATNSRFTIFYNPRACPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESSVRRYMGT 305
A T + F ++Y PR S+F+I L KY++A+ + SVGMRF+M FE E+S RRY GT
Sbjct: 245 ATQTKTMFIVYYKPRT--SQFIISLNKYLEAM-SNKFSVGMRFKMRFEGEDSPERRYSGT 301
Query: 306 ITGICDLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEPLTTFPMYPSPFPLRLKR 365
+ G+ D S W +S WR ++V WDE + R +VS WEIEP P L+ KR
Sbjct: 302 VIGVKDC-SPHWKDSKWRCLEVHWDEPASISRPNKVSPWEIEPFVNSENVPKSVMLKNKR 360
Query: 366 P 366
P
Sbjct: 361 P 361
Score = 77.4 bits (189), Expect = 2e-13
Identities = 42/103 (40%), Positives = 62/103 (59%), Gaps = 4/103 (3%)
Query: 777 GRSLDITKFSSYNELRSELARMFGLEGELEDPVRSGWQLVFVDRENDVLLLGDGPWPEFV 836
GR++D+ YNEL ++ ++F ++GEL R+ W++VF D E D++L+GD PWPEF
Sbjct: 536 GRAVDLNALKGYNELIDDIEKLFDIKGELRS--RNQWEIVFTDDEGDMMLVGDDPWPEFC 593
Query: 837 NSVWCIKILSPEEVQQM--GNTGLGLLNSVPIQRLSNSICDDY 877
N V I I S EEV++M GN LL V + S D++
Sbjct: 594 NMVKRIFIWSKEEVKKMTPGNQLRMLLREVETTLTTTSKTDNH 636
>ARFC_ARATH (O23661) Auxin response factor 3 (ETTIN protein)
Length = 608
Score = 337 bits (863), Expect = 1e-91
Identities = 182/385 (47%), Positives = 246/385 (63%), Gaps = 24/385 (6%)
Query: 13 ELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQLICQLHNL 72
ELWHACAGPL+SLP GS V+YFPQGH EQ D Y LPP + C++ ++
Sbjct: 54 ELWHACAGPLISLPKRGSLVLYFPQGHLEQAP-------DFSAAIY-GLPPHVFCRILDV 105
Query: 73 TMHADVETDEVYAQMTLQPLNA---QEQKEAYLPAELGTPSKQ------PTNYFCKTLTA 123
+HA+ TDEVYAQ++L P + ++ +E + + G + + FCKTLTA
Sbjct: 106 KLHAETTTDEVYAQVSLLPESEDIERKVREGIIDVDGGEEDYEVLKRSNTPHMFCKTLTA 165
Query: 124 SDTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFRGQPKRHLLT 183
SDTSTHGGFSVPRRAAE FPPLD+SQ P+QEL+ARDLHG EW+FRHI+RGQP+RHLLT
Sbjct: 166 SDTSTHGGFSVPRRAAEDCFPPLDYSQPRPSQELLARDLHGLEWRFRHIYRGQPRRHLLT 225
Query: 184 TGWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHLGLLAAA 243
TGWS FV+ K+LV+GD+VLF+ + +L LG+RRAS+ + S + +M+ +
Sbjct: 226 TGWSAFVNKKKLVSGDAVLFLRGDDGKLRLGVRRASQIEGTAALSAQYNQNMNHNNFSEV 285
Query: 244 AHAAATNSRFTIFYNPRACPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESSVRRYM 303
AHA +T+S F+I YNP+A S F+IP K++K V + +GMRF+ E+E++S RR
Sbjct: 286 AHAISTHSVFSISYNPKASWSNFIIPAPKFLKVVDYP-FCIGMRFKARVESEDASERRSP 344
Query: 304 GTITGICDLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEP---LTTFPMYPSPFP 360
G I+GI DLD +RWP S WR + V WD+ A Q RVS WEIEP ++ + + P
Sbjct: 345 GIISGISDLDPIRWPGSKWRCLLVRWDDIVANGHQQRVSPWEIEPSGSISNSGSFVTTGP 404
Query: 361 LRLK---RPWPPGLPSFHGMKDDDF 382
R + P +P G++ DF
Sbjct: 405 KRSRIGFSSGKPDIPVSEGIRATDF 429
>ARFL_ARATH (Q9XID4) Putative auxin response factor 12
Length = 593
Score = 289 bits (740), Expect = 2e-77
Identities = 156/361 (43%), Positives = 220/361 (60%), Gaps = 9/361 (2%)
Query: 5 GEKRVLDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQ 64
G K + +LW CAGPL +P +G +V YFPQGH E V ST +E++ P LP +
Sbjct: 19 GSKSYVYEQLWKLCAGPLCDIPKLGEKVYYFPQGHIELVETSTREELNELQP-ICDLPSK 77
Query: 65 LICQLHNLTMHADVETDEVYAQMTLQPLNAQEQKEAYLPAELGTPSKQPTNYFCKTLTAS 124
L C++ + + + +DE YA++TL P + + +P + + N F K LTAS
Sbjct: 78 LQCRVIAIHLKVENNSDETYAEITLMP----DTTQVVIPTQNENQFRPLVNSFTKVLTAS 133
Query: 125 DTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFRGQPKRHLLTT 184
DTS HGGF VP++ A + P LD SQ PAQEL+A DLHGN+W+F H +RG P+RHLLTT
Sbjct: 134 DTSAHGGFFVPKKHAIECLPSLDMSQPLPAQELLAIDLHGNQWRFNHNYRGTPQRHLLTT 193
Query: 185 GWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHLGLLAAAA 244
GW+ F ++K+LVAGD ++F+ E +L +GIRRA Q +PSS++S D M G++A+A
Sbjct: 194 GWNAFTTSKKLVAGDVIVFVRGETGELRVGIRRARHQQGNIPSSIVSIDCMRHGVVASAK 253
Query: 245 HAAATNSRFTIFYNPRACPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESSVRRYMG 304
HA FT+ Y PR+ S+F++ K++ AV + + +VG RF M E ++ S RR G
Sbjct: 254 HAFDNQCMFTVVYKPRS--SKFIVSYDKFLDAV-NNKFNVGSRFTMRLEGDDFSERRCFG 310
Query: 305 TITGICDLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEPLTTFPMYPSPFPLRLK 364
TI G+ D S W S WRS++V WDE T+ +VS W+IE L P F L+ K
Sbjct: 311 TIIGVSDF-SPHWKCSEWRSLEVQWDEFTSFPGPKKVSPWDIEHLMPAINVPRSFLLKNK 369
Query: 365 R 365
R
Sbjct: 370 R 370
Score = 65.1 bits (157), Expect = 8e-10
Identities = 29/77 (37%), Positives = 51/77 (65%), Gaps = 3/77 (3%)
Query: 764 NKTFVKVYKSG-SFGRSLDITKFSSYNELRSELARMFGLEGELEDPVRSGWQLVFVDREN 822
++T KV G + GR++D++ + Y++L EL ++F ++G+L+ R+ W++ F D +
Sbjct: 510 SRTCTKVQMQGVTIGRAVDLSVLNGYDQLILELEKLFDIKGQLQ--TRNQWEIAFTDSDE 567
Query: 823 DVLLLGDGPWPEFVNSV 839
D +L+GD PWPEF N V
Sbjct: 568 DKMLVGDDPWPEFCNMV 584
>ARFU_ARATH (Q9C8N9) Putative auxin response factor 21
Length = 606
Score = 288 bits (736), Expect = 6e-77
Identities = 154/361 (42%), Positives = 221/361 (60%), Gaps = 9/361 (2%)
Query: 5 GEKRVLDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQ 64
G K + +LW CAGPL +P +G V YFPQG+ E V ST +E++ P LP +
Sbjct: 19 GSKSYMYEQLWKLCAGPLCDIPKLGENVYYFPQGNIELVQASTREELNELQP-ICDLPSK 77
Query: 65 LICQLHNLTMHADVETDEVYAQMTLQPLNAQEQKEAYLPAELGTPSKQPTNYFCKTLTAS 124
L C++ + + + +DE+YA++TL P + + +P + + N F K LTAS
Sbjct: 78 LQCRVIAIHLKVENNSDEIYAEITLMP----DTTQVVIPTQSENRFRPLVNSFTKVLTAS 133
Query: 125 DTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFRGQPKRHLLTT 184
DTS +GGFSVP++ A + PPLD SQ PAQE++A DLH N+W+FRH +RG P+RH LTT
Sbjct: 134 DTSAYGGFSVPKKHAIECLPPLDMSQPLPAQEILAIDLHDNQWRFRHNYRGTPQRHSLTT 193
Query: 185 GWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHLGLLAAAA 244
GW+ F+++K+LV GD ++F+ E +L +GIRRA Q +PSS++S D M G++A+A
Sbjct: 194 GWNEFITSKKLVKGDVIVFVRGETGELRVGIRRARHQQGNIPSSIVSIDCMRHGVIASAK 253
Query: 245 HAAATNSRFTIFYNPRACPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESSVRRYMG 304
HA F + Y PR+ S+F++ K++ AV + + +VG RF M FE ++ S RRY G
Sbjct: 254 HAFDNQCIFIVVYKPRS--SQFIVSYDKFLDAV-NNKFNVGSRFTMRFEGDDFSERRYFG 310
Query: 305 TITGICDLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEPLTTFPMYPSPFPLRLK 364
TI G+ D S W S WRS++V WDE + R +VS WEIE L P L+ K
Sbjct: 311 TIIGVSDF-SPHWKCSEWRSLEVQWDEFASFSRPNKVSPWEIEHLVPALNVPRSSLLKNK 369
Query: 365 R 365
R
Sbjct: 370 R 370
Score = 66.2 bits (160), Expect = 4e-10
Identities = 33/91 (36%), Positives = 57/91 (62%), Gaps = 3/91 (3%)
Query: 764 NKTFVKVYKSG-SFGRSLDITKFSSYNELRSELARMFGLEGELEDPVRSGWQLVFVDREN 822
++T KV G + GR++D++ + Y++L EL ++F ++G+L+ R+ W++ F D +
Sbjct: 510 SRTCTKVQMQGVTIGRAVDLSVLNGYDQLILELEKLFDIKGQLQ--TRNQWKIAFTDSDG 567
Query: 823 DVLLLGDGPWPEFVNSVWCIKILSPEEVQQM 853
+L+GD PWPEF V I I S EEV+ +
Sbjct: 568 YEMLVGDDPWPEFCKMVKKILIYSKEEVKNL 598
>ARFV_ARATH (Q9C8N7) Putative auxin response factor 22
Length = 598
Score = 287 bits (735), Expect = 8e-77
Identities = 154/361 (42%), Positives = 221/361 (60%), Gaps = 11/361 (3%)
Query: 5 GEKRVLDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQ 64
G K + +LW CAGPL +P +G ++ YFPQG+ E V ST +E++ P LP +
Sbjct: 19 GSKSYMYEQLWKLCAGPLCDIPKLGEKIYYFPQGNIELVEASTREELNELKP-ICDLPSK 77
Query: 65 LICQLHNLTMHADVETDEVYAQMTLQPLNAQEQKEAYLPAELGTPSKQPTNYFCKTLTAS 124
L C++ + + + +DE YA++TL P + + +P + + N F K LTAS
Sbjct: 78 LQCRVIAIQLKVENNSDETYAEITLMP----DTTQVVIPTQNENQFRPLVNSFTKVLTAS 133
Query: 125 DTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFRGQPKRHLLTT 184
DTS GGF VP++ A + PPLD SQ P QEL+A DLHGN+W+F H +RG P+RHLLTT
Sbjct: 134 DTS--GGFFVPKKHAIECLPPLDMSQPLPTQELLATDLHGNQWRFNHNYRGTPQRHLLTT 191
Query: 185 GWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHLGLLAAAA 244
GW+ F ++K+LVAGD ++F+ E +L +GIRRA Q +PSS++S +SM G++A+A
Sbjct: 192 GWNAFTTSKKLVAGDVIVFVRGETGELRVGIRRAGHQQGNIPSSIISIESMRHGVIASAK 251
Query: 245 HAAATNSRFTIFYNPRACPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESSVRRYMG 304
HA F + Y PR+ S+F++ K++ AV + + +VG RF M FE ++ S RRY G
Sbjct: 252 HAFDNQCMFIVVYKPRS--SQFIVSYDKFLDAV-NNKFNVGSRFTMRFEGDDFSERRYFG 308
Query: 305 TITGICDLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEPLTTFPMYPSPFPLRLK 364
TI G+ D S W S WR+++V WDE + R +VS WEIE L P P L+ K
Sbjct: 309 TIIGVSDF-SPHWKCSEWRNLEVQWDEFASFSRPNKVSPWEIEHLMPALNVPRPSLLKNK 367
Query: 365 R 365
R
Sbjct: 368 R 368
Score = 62.4 bits (150), Expect = 5e-09
Identities = 32/96 (33%), Positives = 55/96 (56%), Gaps = 3/96 (3%)
Query: 745 GESGFLHTQENGEQGNNPLNKTFVKVYKSG-SFGRSLDITKFSSYNELRSELARMFGLEG 803
G+S L + + +T KV G + R++D++ + Y++L EL +F L+G
Sbjct: 489 GQSQTLRSPTEIQSKQFSSTRTCTKVQMQGVTIERAVDLSVLNGYDQLILELEELFDLKG 548
Query: 804 ELEDPVRSGWQLVFVDRENDVLLLGDGPWPEFVNSV 839
+L+ R+ W++ F D ++D +L+GD PWPEF N V
Sbjct: 549 QLQ--TRNQWEIAFTDSDDDKMLVGDDPWPEFCNMV 582
>ARFN_ARATH (Q9LQE8) Putative auxin response factor 14
Length = 605
Score = 287 bits (734), Expect = 1e-76
Identities = 149/346 (43%), Positives = 214/346 (61%), Gaps = 9/346 (2%)
Query: 5 GEKRVLDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQ 64
G K + +LW CAGPL +P +G +V YFPQGH E V ST +E++ P P +
Sbjct: 19 GSKSYMYEQLWKLCAGPLCDIPKLGEKVYYFPQGHIELVEASTREELNELQP-ICDFPSK 77
Query: 65 LICQLHNLTMHADVETDEVYAQMTLQPLNAQEQKEAYLPAELGTPSKQPTNYFCKTLTAS 124
L C++ + + + +DE YA++TL P + + +P + + N F K LTAS
Sbjct: 78 LQCRVIAIQLKVENNSDETYAEITLMP----DTTQVVIPTQNQNQFRPLVNSFTKVLTAS 133
Query: 125 DTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFRGQPKRHLLTT 184
DTS HGGFSVP++ A + PPLD SQ P QE++A DLHGN+W+FRHI+RG +RHLLT
Sbjct: 134 DTSVHGGFSVPKKHAIECLPPLDMSQPLPTQEILAIDLHGNQWRFRHIYRGTAQRHLLTI 193
Query: 185 GWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHLGLLAAAA 244
GW+ F ++K+LV GD ++F+ E +L +GIRRA Q +PSS++S +SM G++A+A
Sbjct: 194 GWNAFTTSKKLVEGDVIVFVRGETGELRVGIRRAGHQQGNIPSSIVSIESMRHGIIASAK 253
Query: 245 HAAATNSRFTIFYNPRACPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESSVRRYMG 304
HA F + Y PR+ S+F++ K++ V + + +VG RF M FE ++ S RR G
Sbjct: 254 HAFDNQCMFIVVYKPRS--SQFIVSYDKFLDVV-NNKFNVGSRFTMRFEGDDFSERRSFG 310
Query: 305 TITGICDLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEPLT 350
TI G+ D S W S WRS++V WDE + R +VS W+IE LT
Sbjct: 311 TIIGVSDF-SPHWKCSEWRSLEVQWDEFASFPRPNQVSPWDIEHLT 355
Score = 73.2 bits (178), Expect = 3e-12
Identities = 69/269 (25%), Positives = 117/269 (42%), Gaps = 19/269 (7%)
Query: 595 SSFPHDESSLLMSLPRTSSWVPVQNSTGWPSKRIAVDPLLSSGASQCILPQVEQLGQARN 654
+SFP + + W V S+ +KR + S +S + P + Q GQ
Sbjct: 338 ASFPRPNQVSPWDIEHLTPWSNVSRSSFLKNKRSREVNEIGSSSSHLLPPTLTQ-GQ--- 393
Query: 655 SMSQNAITLPPFPGRECSIDQEGSNDPQSNLLFGVNIDPSSLLLHNGMSNFKGISGNNND 714
+ Q ++ P E + P S LL + P + L +N + I N
Sbjct: 394 EIGQQSMATPMNISLRYRDITEDAMTP-SRLLMSYPVQPMAKLNYNNVVT--PIEENITT 450
Query: 715 SSTMSYHQSSSYMNTAGA--DSSLNHGVTPS-------IGESGFLHTQENGEQGNNPLNK 765
++ S+ + T D G+ S G+S L + + +
Sbjct: 451 NAVASFRLFGVSLATPSVIKDPVEQIGLEISRLTQEKKFGQSQILRSPTEIQSKQFSSTR 510
Query: 766 TFVKVYKSG-SFGRSLDITKFSSYNELRSELARMFGLEGELEDPVRSGWQLVFVDRENDV 824
T KV G + GR++D++ + Y++L EL ++F L+G+L+ R+ W++ F + E D
Sbjct: 511 TCTKVQMQGVTIGRAVDLSVLNGYDQLILELEKLFDLKGQLQ--ARNQWEIAFTNNEEDK 568
Query: 825 LLLGDGPWPEFVNSVWCIKILSPEEVQQM 853
+L+G+ PWPEF N V I I S EEV+ +
Sbjct: 569 MLVGEDPWPEFCNMVKKIFIYSKEEVKNL 597
>ARFT_ARATH (Q9C7I9) Putative auxin response factor 20
Length = 606
Score = 286 bits (733), Expect = 1e-76
Identities = 154/361 (42%), Positives = 220/361 (60%), Gaps = 9/361 (2%)
Query: 5 GEKRVLDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQ 64
G K + +LW CAGPL +P +G V YFPQG+ E V ST +E++ P LP +
Sbjct: 19 GSKSYMYEQLWKLCAGPLCDIPKLGENVYYFPQGNIELVDASTREELNELQP-ICDLPSK 77
Query: 65 LICQLHNLTMHADVETDEVYAQMTLQPLNAQEQKEAYLPAELGTPSKQPTNYFCKTLTAS 124
L C++ + + + +DE YA++TL P + + +P + + N F K LTAS
Sbjct: 78 LQCRVIAIHLKVENNSDETYAEITLMP----DTTQVVIPTQSENQFRPLVNSFTKVLTAS 133
Query: 125 DTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFRGQPKRHLLTT 184
DTS +GGF VP++ A + PPLD SQ PAQEL+A+DLHGN+W+FRH +RG P+RH LTT
Sbjct: 134 DTSAYGGFFVPKKHAIECLPPLDMSQPLPAQELLAKDLHGNQWRFRHSYRGTPQRHSLTT 193
Query: 185 GWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHLGLLAAAA 244
GW+ F ++K+LV GD ++F+ E +L +GIRRA Q +PSS++S D M G++A+A
Sbjct: 194 GWNEFTTSKKLVKGDVIVFVRGETGELRVGIRRARHQQGNIPSSIVSIDCMRHGVIASAK 253
Query: 245 HAAATNSRFTIFYNPRACPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESSVRRYMG 304
HA F + Y PR+ S+F++ K++ A+ + + VG RF M FE ++ S RRY G
Sbjct: 254 HALDNQCIFIVVYKPRS--SQFIVSYDKFLDAM-NNKFIVGSRFTMRFEGDDFSERRYFG 310
Query: 305 TITGICDLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEPLTTFPMYPSPFPLRLK 364
TI G+ D S W S WRS++V WDE + R +VS WEIE L + P L+ K
Sbjct: 311 TIIGVNDF-SPHWKCSEWRSLEVQWDEFASFSRPNKVSPWEIEHLMSALNVPRSSLLKNK 369
Query: 365 R 365
R
Sbjct: 370 R 370
Score = 66.6 bits (161), Expect = 3e-10
Identities = 34/90 (37%), Positives = 56/90 (61%), Gaps = 3/90 (3%)
Query: 765 KTFVKVYKSG-SFGRSLDITKFSSYNELRSELARMFGLEGELEDPVRSGWQLVFVDREND 823
+T KV G + GR++D++ + Y++L EL ++F L+G+L+ R+ W++ F D +
Sbjct: 511 RTCTKVQMQGVTIGRAVDLSVLNGYDQLILELEKLFDLKGQLQ--TRNQWKIAFTDSDGY 568
Query: 824 VLLLGDGPWPEFVNSVWCIKILSPEEVQQM 853
+L+GD PWPEF V I I S EEV+ +
Sbjct: 569 EMLVGDDPWPEFCKMVKKILIYSKEEVKNL 598
>ARFJ_ARATH (Q9SKN5) Auxin response factor 10
Length = 693
Score = 280 bits (717), Expect = 9e-75
Identities = 166/424 (39%), Positives = 238/424 (55%), Gaps = 63/424 (14%)
Query: 6 EKRVLDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIP---NYPSLP 62
+++ LD +LWHACAG +V +P++ S V YF QGH+E AH P + P +P
Sbjct: 3 QEKSLDPQLWHACAGSMVQIPSLNSTVFYFAQGHTEH----------AHAPPDFHAPRVP 52
Query: 63 PQLICQLHNLTMHADVETDEVYAQMTLQPLNAQE---QKEAYL----PAELGTPS-KQPT 114
P ++C++ ++ AD ETDEV+A++TL PL + + +A L P+ G + K+
Sbjct: 53 PLILCRVVSVKFLADAETDEVFAKITLLPLPGNDLDLENDAVLGLTPPSSDGNGNGKEKP 112
Query: 115 NYFCKTLTASDTSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFR 174
F KTLT SD + GGFSVPR AE +FP LD+S +PP Q +IA+D+HG WKFRHI+R
Sbjct: 113 ASFAKTLTQSDANNGGGFSVPRYCAETIFPRLDYSAEPPVQTVIAKDIHGETWKFRHIYR 172
Query: 175 GQPKRHLLTTGWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASR-------------- 220
G P+RHLLTTGWS FV+ K+L+AGDS++F+ +E L +GIRRA R
Sbjct: 173 GTPRRHLLTTGWSTFVNQKKLIAGDSIVFLRSESGDLCVGIRRAKRGGLGSNAGSDNPYP 232
Query: 221 ---------PQTVMPSSVL-------------SSDSMHLGLLAAAAHAAATNSRFTIFYN 258
T S ++ ++ + + +A A AA F + Y
Sbjct: 233 GFSGFLRDDESTTTTSKLMMMKRNGNNDGNAAATGRVRVEAVAEAVARAACGQAFEVVYY 292
Query: 259 PRACPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESS-VRRYMGTITGICDLDSVRW 317
PRA EF + A V++ R GMRF+M FETE+SS + +MGT++ + D +RW
Sbjct: 293 PRASTPEFCVKAAD-VRSAMRIRWCSGMRFKMAFETEDSSRISWFMGTVSAVQVADPIRW 351
Query: 318 PNSHWRSVKVGWDESTAGERQPRVSLWEIEPLTTFP-MYPSPFPLR--LKRPWPPGLPSF 374
PNS WR ++V WDE + RVS W +E ++ P ++ SPF R ++ P P P F
Sbjct: 352 PNSPWRLLQVAWDEPDLLQNVKRVSPWLVELVSNMPTIHLSPFSPRKKIRIPQPFEFP-F 410
Query: 375 HGMK 378
HG K
Sbjct: 411 HGTK 414
Score = 36.2 bits (82), Expect = 0.40
Identities = 22/67 (32%), Positives = 37/67 (54%), Gaps = 4/67 (5%)
Query: 772 KSGSFGRSLDITKFSSYNELRSELARMFGLEGELEDPVRSGWQLVFVDRENDVLLLGDGP 831
+S GR+LD++ SY EL +LA MF +E E D + +V+ D + +GD P
Sbjct: 588 ESEDVGRTLDLSVIGSYQELYRKLAEMFHIE-ERSDLLT---HVVYRDANGVIKRIGDEP 643
Query: 832 WPEFVNS 838
+ +F+ +
Sbjct: 644 FSDFMKA 650
>ARFO_ARATH (Q9LQE3) Putative auxin response factor 15
Length = 593
Score = 279 bits (714), Expect = 2e-74
Identities = 153/359 (42%), Positives = 218/359 (60%), Gaps = 9/359 (2%)
Query: 7 KRVLDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQLI 66
K + +LW CAGPL +P +G +V YFPQG+ E V ST +E++ P LP +L
Sbjct: 21 KSYMYEQLWKLCAGPLCDIPKLGEKVYYFPQGNIELVEASTREELNELQP-ICDLPSKLQ 79
Query: 67 CQLHNLTMHADVETDEVYAQMTLQPLNAQEQKEAYLPAELGTPSKQPTNYFCKTLTASDT 126
C++ + + + +DE YA++TL P + + +P + + N F K LTASD
Sbjct: 80 CRVIAIHLKVENNSDETYAKITLMP----DTTQVVIPTQNENQFRPLVNSFTKVLTASDI 135
Query: 127 STHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFRGQPKRHLLTTGW 186
S +G FSVP++ A + PPLD SQ PAQEL+A DLHGN+W FRH +RG P+RHLLTTGW
Sbjct: 136 SANGVFSVPKKHAIECLPPLDMSQPLPAQELLAIDLHGNQWSFRHSYRGTPQRHLLTTGW 195
Query: 187 SVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHLGLLAAAAHA 246
+ F ++K+LV GD ++F+ E +L +GIRRA Q +PSS++S D M G++A+A HA
Sbjct: 196 NEFTTSKKLVKGDVIVFVRGETGELRVGIRRARHQQGNIPSSIVSIDCMRHGVIASAKHA 255
Query: 247 AATNSRFTIFYNPRACPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESSVRRYMGTI 306
F + Y PR+ S+F++ K++ AV + + +VG RF M FE ++ S RRY GTI
Sbjct: 256 FDNQCMFIVVYKPRS--SQFIVSYDKFLDAV-NNKFNVGSRFTMRFEGDDLSERRYFGTI 312
Query: 307 TGICDLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEPLTTFPMYPSPFPLRLKR 365
G+ + S W S WRS++V WDE + R +VS WEIE L P L+ KR
Sbjct: 313 IGVSNF-SPHWKCSDWRSLEVQWDEFASFLRPNKVSPWEIEHLMPALNVPRSSFLKNKR 370
Score = 64.3 bits (155), Expect = 1e-09
Identities = 37/111 (33%), Positives = 62/111 (55%), Gaps = 6/111 (5%)
Query: 731 GAD-SSLNHGVTPSIGESGFLHTQENGEQGNNPLNKTFVKVYKSG-SFGRSLDITKFSSY 788
G+D S L G G+S L + + +T KV G + GR++D++ + Y
Sbjct: 478 GSDISKLTEG--KKFGQSQTLRSPTKIQSKQFSSTRTCTKVQMQGVTIGRAVDLSVLNGY 535
Query: 789 NELRSELARMFGLEGELEDPVRSGWQLVFVDRENDVLLLGDGPWPEFVNSV 839
++L EL ++F L+G+L+ R+ W+++F + D +L+GD PWPEF N V
Sbjct: 536 DQLILELEKLFDLKGQLQ--TRNQWKIIFTGSDEDEMLVGDDPWPEFCNMV 584
>ARFM_ARATH (Q9FX25) Putative auxin response factor 13
Length = 623
Score = 266 bits (679), Expect = 2e-70
Identities = 149/346 (43%), Positives = 208/346 (60%), Gaps = 13/346 (3%)
Query: 7 KRVLDSELWHACAGPLVSLPAVGSRVVYFPQGHSEQVAVSTNKEVDAHIPNYPSLPPQLI 66
K + +LW+ CAGPL LP G +V YFPQGH E + ST E+D HI LP +L
Sbjct: 21 KTYMYEKLWNICAGPLCVLPKPGEKVYYFPQGHIELIENSTRDELD-HIRPIFDLPSKLR 79
Query: 67 CQLHNLTMHADVETDEVYAQMTLQPLNAQEQKEAYLPAELGTPSKQPTNYF-CKTLTASD 125
C++ + D TDEVYAQ++L P + + +++P YF K LTASD
Sbjct: 80 CRVVAIDRKVDKNTDEVYAQISLMPDTTE-----VMTHNTTMDTRRPIVYFFSKILTASD 134
Query: 126 TSTHGGFSVPRRAAEKVFPPLDFSQQPPAQELIARDLHGNEWKFRHIFRGQPKRHLLTT- 184
S GG +P++ A + FPPLD SQ Q L+A+DL+G EW F+H+FRG P+RH+ T+
Sbjct: 135 VSLSGGLIIPKQYAIECFPPLDMSQPISTQNLVAKDLYGQEWSFKHVFRGTPQRHMFTSG 194
Query: 185 -GWSVFVSAKRLVAGDSVLFIWNEKNQLLLGIRRASRPQTVMPSSVLSSDSMHLGLLAAA 243
GWSVF + KRL+ GD + + E +L GIRRA Q +PSSV+S++ M G++A+
Sbjct: 195 GGWSVFATTKRLIVGDIFVLLRGENGELRFGIRRAKHQQGHIPSSVISANCMQHGVIASV 254
Query: 244 AHAAATNSRFTIFYNPRACPSEFVIPLAKYVKAVYHTRVSVGMRFRMLFETEESSVRRYM 303
+A T F + Y P + S+FVI K+V A+ + + VG RFRM FE ++ S +RY
Sbjct: 255 VNAFKTKCMFNVVYKPSS--SQFVISYDKFVDAMNNNYI-VGSRFRMQFEGKDFSEKRYD 311
Query: 304 GTITGICDLDSVRWPNSHWRSVKVGWDESTAGERQPRVSLWEIEPL 349
GTI G+ D+ S W +S WRS+KV WDE + R +VS W+IE L
Sbjct: 312 GTIIGVNDM-SPHWKDSEWRSLKVQWDELSPFLRPNQVSPWDIEHL 356
Score = 67.4 bits (163), Expect = 2e-10
Identities = 34/87 (39%), Positives = 53/87 (60%), Gaps = 3/87 (3%)
Query: 768 VKVYKSG-SFGRSLDITKFSSYNELRSELARMFGLEGELEDPVRSGWQLVFVDRENDVLL 826
+KV+ G + R++D+T YN+L +L +F L+ EL R+ W++VF + E +L
Sbjct: 513 IKVHMQGVAISRAVDLTAMHGYNQLIQKLEELFDLKDELR--TRNQWEIVFTNNEGAEML 570
Query: 827 LGDGPWPEFVNSVWCIKILSPEEVQQM 853
+GD PWPEF N I I S EE+++M
Sbjct: 571 VGDDPWPEFCNMAKRIFICSKEEIKKM 597
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.130 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 108,299,652
Number of Sequences: 164201
Number of extensions: 4816451
Number of successful extensions: 24229
Number of sequences better than 10.0: 529
Number of HSP's better than 10.0 without gapping: 237
Number of HSP's successfully gapped in prelim test: 317
Number of HSP's that attempted gapping in prelim test: 15802
Number of HSP's gapped (non-prelim): 2813
length of query: 898
length of database: 59,974,054
effective HSP length: 119
effective length of query: 779
effective length of database: 40,434,135
effective search space: 31498191165
effective search space used: 31498191165
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 70 (31.6 bits)
Medicago: description of AC144478.5


