

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144474.7 - phase: 0
(329 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
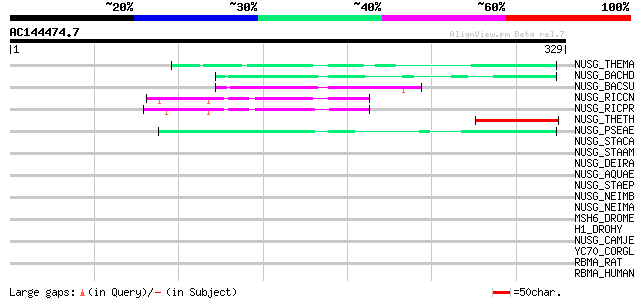
Score E
Sequences producing significant alignments: (bits) Value
NUSG_THEMA (P29397) Transcription antitermination protein nusG 58 3e-08
NUSG_BACHD (Q9KGE7) Transcription antitermination protein nusG 47 7e-05
NUSG_BACSU (Q06795) Transcription antitermination protein nusG 46 1e-04
NUSG_RICCN (Q92J91) Transcription antitermination protein nusG 45 3e-04
NUSG_RICPR (P50056) Transcription antitermination protein nusG 44 4e-04
NUSG_THETH (P35872) Transcription antitermination protein nusG 44 7e-04
NUSG_PSEAE (Q9HWC4) Transcription antitermination protein nusG 44 7e-04
NUSG_STACA (P36264) Transcription antitermination protein nusG 42 0.002
NUSG_STAAM (O08386) Transcription antitermination protein nusG 41 0.005
NUSG_DEIRA (Q9RSS6) Transcription antitermination protein nusG 40 0.010
NUSG_AQUAE (O67757) Transcription antitermination protein nusG 39 0.014
NUSG_STAEP (Q8CQ85) Transcription antitermination protein nusG 39 0.023
NUSG_NEIMB (P65592) Transcription antitermination protein nusG 38 0.040
NUSG_NEIMA (P65591) Transcription antitermination protein nusG 38 0.040
MSH6_DROME (Q9VUM0) Probable DNA mismatch repair protein MSH6 38 0.040
H1_DROHY (P17268) Histone H1 38 0.040
NUSG_CAMJE (Q9PI36) Transcription antitermination protein nusG 37 0.052
YC70_CORGL (P42531) Hypothetical protein Cgl1270/cg1434 36 0.12
RBMA_RAT (P70501) RNA-binding protein 10 (RNA binding motif prot... 36 0.12
RBMA_HUMAN (P98175) RNA-binding protein 10 (RNA binding motif pr... 36 0.12
>NUSG_THEMA (P29397) Transcription antitermination protein nusG
Length = 353
Score = 58.2 bits (139), Expect = 3e-08
Identities = 54/228 (23%), Positives = 92/228 (39%), Gaps = 61/228 (26%)
Query: 97 RVSRVKGQYTAEALARSLAKFFPDIEFKVYAPAIHEKKRLKSGSISVKSKPLFPGCIFLR 156
R+ + K E LA + KFF + +V +K ++ K + LFPG +F+
Sbjct: 183 RIKKGKEVKQGEMLAEA-RKFFAKVSGRVEVVDYSTRKEIRI--YKTKRRKLFPGYVFVE 239
Query: 157 CELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKPRPVAEDDMEAIFKQAKVEQENADKA 216
+N +++++ V GF+ S +P PV + +M I + A +E+
Sbjct: 240 MIMNDEAYNFVRSVPYVMGFVSSG--------GQPVPVKDREMRPILRLAGLEE------ 285
Query: 217 FEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKRGSRKTSNQLTITEEASSAKKKPKLVT 276
+EE++K V G
Sbjct: 286 YEEKKKPVKVELG--------------------------------------------FKV 301
Query: 277 GSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTMFGKENIVDLDVSEI 324
G V+IISG F FAG +K+++ + + V+ T+FG+E V L VSE+
Sbjct: 302 GDMVKIISGPFEDFAGVIKEIDPERQELKVNVTIFGRETPVVLHVSEV 349
>NUSG_BACHD (Q9KGE7) Transcription antitermination protein nusG
Length = 178
Score = 47.0 bits (110), Expect = 7e-05
Identities = 41/202 (20%), Positives = 80/202 (39%), Gaps = 62/202 (30%)
Query: 123 FKVYAPAIHEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVG 182
F+V P + E+ +K+G S+ +FPG + + + ++ GV GF+GS
Sbjct: 36 FRVMVP-VEEETEVKNGKSKQVSRKVFPGYVLVEMVMTDDSWYVVRNTPGVTGFVGSAGA 94
Query: 183 NTKKQINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKA 242
+KP P+ ++++AI +Q +++E +++E D
Sbjct: 95 G-----SKPTPLLPEEVDAILRQMGMQEE---------------------RQVEIDFQ-- 126
Query: 243 IVDSKPKRGSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTK 302
+E+ K+ P F F GT++++ +
Sbjct: 127 -------------------LKESVKVKEGP--------------FANFIGTIEEIQLDKR 153
Query: 303 MATVHFTMFGKENIVDLDVSEI 324
VH MFG+E V+L+ ++I
Sbjct: 154 KLKVHVNMFGRETPVELEFTQI 175
>NUSG_BACSU (Q06795) Transcription antitermination protein nusG
Length = 177
Score = 45.8 bits (107), Expect = 1e-04
Identities = 33/127 (25%), Positives = 57/127 (43%), Gaps = 11/127 (8%)
Query: 123 FKVYAPAIHEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVG 182
F+V P E+ +K+G V K +FPG + + + ++ GV GF+GS
Sbjct: 36 FRVVVPE-EEETDIKNGKKKVVKKKVFPGYVLVEIVMTDDSWYVVRNTPGVTGFVGSAGS 94
Query: 183 NTKKQINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPN-----KELES 237
+KP P+ + E I K+ +++ D FE +E ++ N +E++
Sbjct: 95 G-----SKPTPLLPGEAETILKRMGMDERKTDIDFELKETVKVIDGPFANFTGSIEEIDY 149
Query: 238 DVSKAIV 244
D SK V
Sbjct: 150 DKSKVKV 156
>NUSG_RICCN (Q92J91) Transcription antitermination protein nusG
Length = 192
Score = 44.7 bits (104), Expect = 3e-04
Identities = 37/141 (26%), Positives = 62/141 (43%), Gaps = 22/141 (15%)
Query: 82 NLDNLM----KLGPQWWGVRVSRVKGQYTAEALARSLAK-----FFPDIEFKVYAPAIHE 132
++DN++ K QW+ V + + E + R +AK FF DI V+ + E
Sbjct: 5 SIDNILSSSEKNVKQWYVVHTASGAEKRIKEDMLRKIAKQNMTHFFEDILIPVFG--VSE 62
Query: 133 KKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKPR 192
KR K+ + K L P I ++ + +K GV GF+GSK P+
Sbjct: 63 VKRGKNVKVE---KKLMPSYILIKMNMTDKSWHLVKNISGVTGFLGSK--------TTPK 111
Query: 193 PVAEDDMEAIFKQAKVEQENA 213
+ E +++ IF + E + A
Sbjct: 112 ALTESEIQNIFNNLEAEAKEA 132
Score = 36.6 bits (83), Expect = 0.089
Identities = 22/84 (26%), Positives = 38/84 (45%), Gaps = 10/84 (11%)
Query: 251 GSRKTSNQLTITE----------EASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSK 300
GS+ T LT +E EA AK G V + G F F GT+++++ +
Sbjct: 105 GSKTTPKALTESEIQNIFNNLEAEAKEAKNSKLYEVGEIVTVTDGPFETFMGTVEEIDQE 164
Query: 301 TKMATVHFTMFGKENIVDLDVSEI 324
V +FGK ++L+ +++
Sbjct: 165 KNRLKVSVAIFGKATPIELNFNQV 188
>NUSG_RICPR (P50056) Transcription antitermination protein nusG
Length = 192
Score = 44.3 bits (103), Expect = 4e-04
Identities = 37/143 (25%), Positives = 62/143 (42%), Gaps = 22/143 (15%)
Query: 80 ELNLDNLMKLGP----QWWGVRVSRVKGQYTAEALARSLAK-----FFPDIEFKVYAPAI 130
E +DN++ QW+ V + + E + R +AK FF DI V+ +
Sbjct: 3 EQTIDNILPASKNNVKQWYVVHTASGAEKRIKEDILRKIAKQKMTDFFEDILIPVFG--V 60
Query: 131 HEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINK 190
E KR K+ + K L P I ++ + +K GV GF+GSK+
Sbjct: 61 SEVKRGKNVKVE---KKLMPSYILIKMNMTDKSWHLVKNIPGVTGFLGSKI--------V 109
Query: 191 PRPVAEDDMEAIFKQAKVEQENA 213
P+ + E +++ IF + E + A
Sbjct: 110 PKALTESEIQNIFNNLEAEAKVA 132
Score = 32.7 bits (73), Expect = 1.3
Identities = 16/61 (26%), Positives = 29/61 (47%)
Query: 264 EASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTMFGKENIVDLDVSE 323
EA AK G V + G F F GT++ ++ V ++FGK ++L+ ++
Sbjct: 128 EAKVAKNSKLYEVGEIVTVTDGPFETFMGTVEAIDKARNRLKVSVSIFGKATPIELNFNQ 187
Query: 324 I 324
+
Sbjct: 188 V 188
>NUSG_THETH (P35872) Transcription antitermination protein nusG
Length = 184
Score = 43.5 bits (101), Expect = 7e-04
Identities = 19/49 (38%), Positives = 31/49 (62%)
Query: 277 GSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTMFGKENIVDLDVSEIV 325
G V+++SG F F GT+ ++N + V T+FG+E V+LD S++V
Sbjct: 134 GDQVRVVSGPFADFTGTVTEINPERGKVKVMVTIFGRETPVELDFSQVV 182
Score = 38.5 bits (88), Expect = 0.023
Identities = 37/173 (21%), Positives = 72/173 (41%), Gaps = 24/173 (13%)
Query: 92 QWWGVRVSRVKGQYTAEALARSLAKF-FPDIEFKVYAPAIHEKKRLKSGSISVKSKPLFP 150
+W+ V + + L + + F D F+V P + + G V K LFP
Sbjct: 4 EWYAVHTLVGQEEKAKANLEKRIKAFGLQDKIFQVLIPTEEVVELREGGKKEVVRKKLFP 63
Query: 151 GCIFLRCELN-----KPLHDYIKEYEGVGGFIGSKVGNTKKQINKPRPVAEDDMEAIFKQ 205
G +F++ +L + ++ G+ GF+G+ + +P P++ D++ I +
Sbjct: 64 GYLFIQMDLGDEEEPNEAWEVVRGTPGITGFVGAGM--------RPVPLSPDEVRHILEV 115
Query: 206 A----KVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKRGSRK 254
+ K E A AF E ++ V+ +D + + + P+RG K
Sbjct: 116 SGLLGKKEAPKAQVAFREGDQVRVVSGP------FADFTGTVTEINPERGKVK 162
>NUSG_PSEAE (Q9HWC4) Transcription antitermination protein nusG
Length = 177
Score = 43.5 bits (101), Expect = 7e-04
Identities = 43/236 (18%), Positives = 83/236 (34%), Gaps = 62/236 (26%)
Query: 89 LGPQWWGVRVSRVKGQYTAEALARSLAKFFPDIEFKVYAPAIHEKKRLKSGSISVKSKPL 148
+ +W+ V ++ +L + + EF E +++G +
Sbjct: 1 MAKRWYVVHAYSGYEKHVMRSLIERVKLAGMEEEFGEILVPTEEVVEMRNGQKRKSERKF 60
Query: 149 FPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKPRPVAEDDMEAIFKQAKV 208
FPG + ++ E+N+ +K+ V GFIG +KP P+ + + +AI ++
Sbjct: 61 FPGYVLVQMEMNEGTWHLVKDTPRVMGFIGGTA-------DKPAPITDREADAILRR--- 110
Query: 209 EQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKRGSRKTSNQLTITEEASSA 268
+ DS K
Sbjct: 111 ----------------------------------VADSGDK------------------P 118
Query: 269 KKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTMFGKENIVDLDVSEI 324
K K G TV++I G F F G ++++N + V +FG+ V+L+ S++
Sbjct: 119 KPKTLFEPGETVRVIDGPFADFNGVVEEVNYEKSRIQVAVLIFGRSTPVELEFSQV 174
>NUSG_STACA (P36264) Transcription antitermination protein nusG
Length = 182
Score = 42.4 bits (98), Expect = 0.002
Identities = 32/127 (25%), Positives = 55/127 (43%), Gaps = 11/127 (8%)
Query: 123 FKVYAPAIHEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVG 182
F+V P E+ ++K G +K FPG + + + ++ GV GF+GS
Sbjct: 41 FRVVIPE-EEETQVKDGKAKKLTKKTFPGYVLVELVMTDESWYVVRNTPGVTGFVGSAGA 99
Query: 183 NTKKQINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAA-----VNSGNPNKELES 237
+KP P+ D++ I KQ ++++ D E E+ N +E+E+
Sbjct: 100 G-----SKPNPLLPDEVRFILKQMGMKEKTIDVEVEVGEQVRIKSGPFANQVGEVQEIEA 154
Query: 238 DVSKAIV 244
D K V
Sbjct: 155 DKFKLTV 161
>NUSG_STAAM (O08386) Transcription antitermination protein nusG
Length = 182
Score = 40.8 bits (94), Expect = 0.005
Identities = 31/127 (24%), Positives = 54/127 (42%), Gaps = 11/127 (8%)
Query: 123 FKVYAPAIHEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVG 182
F+V P E+ ++K G K FPG + + + ++ GV GF+GS
Sbjct: 41 FRVVIPE-EEETQVKDGKAKTTVKKTFPGYVLVELIMTDESWYVVRNTPGVTGFVGSAGA 99
Query: 183 NTKKQINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAA-----VNSGNPNKELES 237
+KP P+ +++ I KQ ++++ D E E+ N +E+E+
Sbjct: 100 G-----SKPNPLLPEEVRFILKQMGLKEKTIDVELEVGEQVRIKSGPFANQVGEVQEIET 154
Query: 238 DVSKAIV 244
D K V
Sbjct: 155 DKFKLTV 161
Score = 30.8 bits (68), Expect = 4.9
Identities = 16/52 (30%), Positives = 28/52 (53%)
Query: 273 KLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTMFGKENIVDLDVSEI 324
+L G V+I SG F G ++++ + TV MFG+E V+++ +I
Sbjct: 128 ELEVGEQVRIKSGPFANQVGEVQEIETDKFKLTVLVDMFGRETPVEVEFDQI 179
>NUSG_DEIRA (Q9RSS6) Transcription antitermination protein nusG
Length = 190
Score = 39.7 bits (91), Expect = 0.010
Identities = 21/64 (32%), Positives = 36/64 (55%), Gaps = 2/64 (3%)
Query: 261 ITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTMFGKENIVDLD 320
+ EEA + K L G V++ SG F F+G + ++N+ V ++FG+E V+LD
Sbjct: 127 VVEEAP--RIKVDLKAGDMVRVTSGPFADFSGVVSEVNAPQAKVKVLVSIFGRETPVELD 184
Query: 321 VSEI 324
S++
Sbjct: 185 FSQV 188
>NUSG_AQUAE (O67757) Transcription antitermination protein nusG
Length = 248
Score = 39.3 bits (90), Expect = 0.014
Identities = 16/54 (29%), Positives = 33/54 (60%)
Query: 271 KPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTMFGKENIVDLDVSEI 324
K + G V++I G F+ F GT+++++ + + TV ++FG+ V+LD ++
Sbjct: 192 KVEFEKGDQVRVIEGPFMNFTGTVEEVHPEKRKLTVMISIFGRMTPVELDFDQV 245
>NUSG_STAEP (Q8CQ85) Transcription antitermination protein nusG
Length = 182
Score = 38.5 bits (88), Expect = 0.023
Identities = 31/128 (24%), Positives = 56/128 (43%), Gaps = 13/128 (10%)
Query: 123 FKVYAPAIHEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVG 182
F+V P E+ ++K G K FPG + + + ++ GV GF+GS
Sbjct: 41 FRVVIPE-EEETQVKDGKAKKLVKKTFPGYVLVELIMTDESWYVVRNTPGVTGFVGSAGA 99
Query: 183 NTKKQINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSG------NPNKELE 236
+KP P+ +++ I KQ ++++ D + E + + SG +E+E
Sbjct: 100 G-----SKPNPLLPEEVRFILKQMGLKEKTIDVELDVGE-QVRIQSGPFANQIGEVQEIE 153
Query: 237 SDVSKAIV 244
+D K V
Sbjct: 154 ADKFKLTV 161
Score = 30.0 bits (66), Expect = 8.3
Identities = 16/52 (30%), Positives = 28/52 (53%)
Query: 273 KLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTMFGKENIVDLDVSEI 324
+L G V+I SG F G ++++ + TV MFG+E V+++ +I
Sbjct: 128 ELDVGEQVRIQSGPFANQIGEVQEIEADKFKLTVLVDMFGRETPVEVEFDQI 179
>NUSG_NEIMB (P65592) Transcription antitermination protein nusG
Length = 178
Score = 37.7 bits (86), Expect = 0.040
Identities = 22/95 (23%), Positives = 43/95 (45%), Gaps = 8/95 (8%)
Query: 136 LKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKPRPVA 195
+++G ++ + +PG + + E+ +K V GFIG + N+P P++
Sbjct: 48 IRNGRKTISERKSYPGYVLVEMEMTDDSWHLVKSTPRVSGFIGGRA-------NRPTPIS 100
Query: 196 EDDMEAIFKQAKVEQENADKAFE-EEEKKAAVNSG 229
+ + E I +Q + E E E ++ VN G
Sbjct: 101 QREAEIILQQVQTGIEKPKPKVEFEVGQQVRVNEG 135
Score = 35.4 bits (80), Expect = 0.20
Identities = 20/81 (24%), Positives = 41/81 (49%), Gaps = 2/81 (2%)
Query: 246 SKPKRGSRKTSNQLTITEEASSAKKKPKLV--TGSTVQIISGSFLGFAGTLKKLNSKTKM 303
++P S++ + + + K KPK+ G V++ G F F G ++++N +
Sbjct: 94 NRPTPISQREAEIILQQVQTGIEKPKPKVEFEVGQQVRVNEGPFADFNGVVEEVNYERNK 153
Query: 304 ATVHFTMFGKENIVDLDVSEI 324
V +FG+E V+L+ S++
Sbjct: 154 LRVSVQIFGRETPVELEFSQV 174
>NUSG_NEIMA (P65591) Transcription antitermination protein nusG
Length = 178
Score = 37.7 bits (86), Expect = 0.040
Identities = 22/95 (23%), Positives = 43/95 (45%), Gaps = 8/95 (8%)
Query: 136 LKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKPRPVA 195
+++G ++ + +PG + + E+ +K V GFIG + N+P P++
Sbjct: 48 IRNGRKTISERKSYPGYVLVEMEMTDDSWHLVKSTPRVSGFIGGRA-------NRPTPIS 100
Query: 196 EDDMEAIFKQAKVEQENADKAFE-EEEKKAAVNSG 229
+ + E I +Q + E E E ++ VN G
Sbjct: 101 QREAEIILQQVQTGIEKPKPKVEFEVGQQVRVNEG 135
Score = 35.4 bits (80), Expect = 0.20
Identities = 20/81 (24%), Positives = 41/81 (49%), Gaps = 2/81 (2%)
Query: 246 SKPKRGSRKTSNQLTITEEASSAKKKPKLV--TGSTVQIISGSFLGFAGTLKKLNSKTKM 303
++P S++ + + + K KPK+ G V++ G F F G ++++N +
Sbjct: 94 NRPTPISQREAEIILQQVQTGIEKPKPKVEFEVGQQVRVNEGPFADFNGVVEEVNYERNK 153
Query: 304 ATVHFTMFGKENIVDLDVSEI 324
V +FG+E V+L+ S++
Sbjct: 154 LRVSVQIFGRETPVELEFSQV 174
>MSH6_DROME (Q9VUM0) Probable DNA mismatch repair protein MSH6
Length = 1190
Score = 37.7 bits (86), Expect = 0.040
Identities = 40/124 (32%), Positives = 60/124 (48%), Gaps = 17/124 (13%)
Query: 185 KKQINKPRPVAEDDMEAIFKQAKVEQENADKA--FEEEEKKAAVNSGNPNKE----LESD 238
+K+I +P +E +ME K E + +D A +E +E +A+ +S + E E+D
Sbjct: 87 RKRIVQPESDSEPEMEV----TKSEDDFSDCASDYEPDENEASDDSVSSGAEEVSPSEND 142
Query: 239 VSKAIVDSKPKRGSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLN 298
+S VDS + SRK S L + KK KL ST+Q+ G+ F LK L
Sbjct: 143 MS---VDSPTPKKSRKKSKILNNNNNNEPSSKKVKL--ESTIQLAEGA--TFQEKLKNLQ 195
Query: 299 SKTK 302
S K
Sbjct: 196 SNAK 199
>H1_DROHY (P17268) Histone H1
Length = 249
Score = 37.7 bits (86), Expect = 0.040
Identities = 24/93 (25%), Positives = 40/93 (42%), Gaps = 1/93 (1%)
Query: 190 KPRPVAEDDM-EAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKP 248
KP+ VA+ + +A+ + V+++ A+KA ++ KK P + KP
Sbjct: 156 KPKGVADKKLSKAVVTKKSVDKKKAEKAKAKDAKKVGTIKAKPTTAKAKSSAAKPKTPKP 215
Query: 249 KRGSRKTSNQLTITEEASSAKKKPKLVTGSTVQ 281
K S K + +A KKPK T S +
Sbjct: 216 KTTSAKPKKVVAAASPKKAAAKKPKAKTASATK 248
>NUSG_CAMJE (Q9PI36) Transcription antitermination protein nusG
Length = 177
Score = 37.4 bits (85), Expect = 0.052
Identities = 28/88 (31%), Positives = 39/88 (43%), Gaps = 13/88 (14%)
Query: 137 KSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKPRPVAE 196
K+G + + L+ G +F +LN L I+ VG FIG KP P+ E
Sbjct: 50 KNGKEKISERSLYSGYVFALLDLNTELWHRIQSLPKVGRFIGES--------KKPTPLTE 101
Query: 197 DDMEAIFKQAKVEQENADK---AFEEEE 221
D+ I + KV A K +FEE E
Sbjct: 102 KDINLILE--KVHNRAAPKPKISFEEGE 127
>YC70_CORGL (P42531) Hypothetical protein Cgl1270/cg1434
Length = 491
Score = 36.2 bits (82), Expect = 0.12
Identities = 20/81 (24%), Positives = 41/81 (49%), Gaps = 8/81 (9%)
Query: 179 SKVGNTKKQINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESD 238
S + + + + +P+P A + ++ + + +NAD + +N+GNP KEL+SD
Sbjct: 263 SLIDASPQALKEPKPEAAATVASLAASSNDDADNADAS--------VINAGNPEKELDSD 314
Query: 239 VSKAIVDSKPKRGSRKTSNQL 259
V + + S+ + K + L
Sbjct: 315 VLEQELSSEEPEETAKPDHSL 335
>RBMA_RAT (P70501) RNA-binding protein 10 (RNA binding motif protein
10) (S1-1 protein)
Length = 852
Score = 36.2 bits (82), Expect = 0.12
Identities = 35/139 (25%), Positives = 62/139 (44%), Gaps = 10/139 (7%)
Query: 187 QINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVD- 245
+I++ ++E+++EA+ K + + D+A E EK P + +S A VD
Sbjct: 711 EIHRRAHLSENELEALEKNDMEQMKYRDRAAERREKYGIPEPPEPKRRKYGGISTASVDF 770
Query: 246 SKPKRGSRKTSN-------QLTITEEASSAKKKPKLVTGSTVQI-ISGSFLGFAGTLKKL 297
+P R + N + E + +KK +VT Q + GS LG G+ +
Sbjct: 771 EQPTRDGLGSDNIGSRMLQAMGWKEGSGLGRKKQGIVTPIEAQTRVRGSGLGARGSSYGV 830
Query: 298 NS-KTKMATVHFTMFGKEN 315
S ++ T+H TM + N
Sbjct: 831 TSTESYKETLHKTMVTRFN 849
>RBMA_HUMAN (P98175) RNA-binding protein 10 (RNA binding motif
protein 10) (DXS8237E)
Length = 929
Score = 36.2 bits (82), Expect = 0.12
Identities = 35/139 (25%), Positives = 62/139 (44%), Gaps = 10/139 (7%)
Query: 187 QINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVD- 245
+I++ ++E+++EA+ K + + D+A E EK P + +S A VD
Sbjct: 788 EIHRRAHLSENELEALEKNDMEQMKYRDRAAERREKYGIPEPPEPKRRKYGGISTASVDF 847
Query: 246 SKPKRGSRKTSN-------QLTITEEASSAKKKPKLVTGSTVQI-ISGSFLGFAGTLKKL 297
+P R + N + E + +KK +VT Q + GS LG G+ +
Sbjct: 848 EQPTRDGLGSDNIGSRMLQAMGWKEGSGLGRKKQGIVTPIEAQTRVRGSGLGARGSSYGV 907
Query: 298 NS-KTKMATVHFTMFGKEN 315
S ++ T+H TM + N
Sbjct: 908 TSTESYKETLHKTMVTRFN 926
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.311 0.129 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 37,914,234
Number of Sequences: 164201
Number of extensions: 1577159
Number of successful extensions: 6434
Number of sequences better than 10.0: 163
Number of HSP's better than 10.0 without gapping: 52
Number of HSP's successfully gapped in prelim test: 113
Number of HSP's that attempted gapping in prelim test: 6223
Number of HSP's gapped (non-prelim): 302
length of query: 329
length of database: 59,974,054
effective HSP length: 111
effective length of query: 218
effective length of database: 41,747,743
effective search space: 9101007974
effective search space used: 9101007974
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC144474.7


