

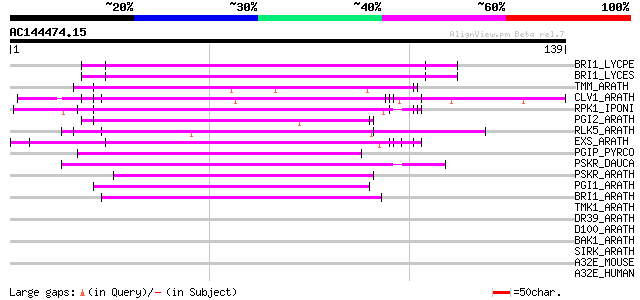
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144474.15 - phase: 0 /pseudo
(139 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 50 1e-06
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 50 1e-06
TMM_ARATH (Q9SSD1) TOO MANY MOUTHS protein precursor (TMM) 49 4e-06
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 49 4e-06
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 47 1e-05
PGI2_ARATH (Q9M5J8) Polygalacturonase inhibitor 2 precursor (Pol... 45 4e-05
RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC... 45 5e-05
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 45 5e-05
PGIP_PYRCO (Q05091) Polygalacturonase inhibitor precursor (Polyg... 44 8e-05
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 44 1e-04
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 43 2e-04
PGI1_ARATH (Q9M5J9) Polygalacturonase inhibitor 1 precursor (Pol... 42 4e-04
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 42 5e-04
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 41 0.001
DR39_ARATH (Q9LVT1) Putative disease resistance protein At5g47280 41 0.001
D100_ARATH (Q00874) DNA-damage-repair/toleration protein DRT100 ... 40 0.001
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 40 0.002
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 40 0.002
A32E_MOUSE (P97822) Acidic leucine-rich nuclear phosphoprotein 3... 40 0.002
A32E_HUMAN (Q9BTT0) Acidic leucine-rich nuclear phosphoprotein 3... 40 0.002
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 50.1 bits (118), Expect = 1e-06
Identities = 28/88 (31%), Positives = 46/88 (51%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L +S+S+ LSG + +SL RL NL ++ LG N+ S +P N ++L L+L L
Sbjct: 525 LNWISLSNNQLSGEIPASLGRLSNLAILKLGNNSISGNIPAELGNCQSLIWLDLNTNFLN 584
Query: 85 GTFPQNIFQIKSQLLTYLTTPTSMVFFQ 112
G+ P +F+ + L T V+ +
Sbjct: 585 GSIPPPLFKQSGNIAVALLTGKRYVYIK 612
Score = 42.7 bits (99), Expect = 2e-04
Identities = 25/86 (29%), Positives = 39/86 (45%)
Query: 19 LLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNL 78
L+ L+ L+ L + DL+GP+ +SL+ L I L N S +P + NL L L
Sbjct: 495 LMYLQALENLILDFNDLTGPIPASLSNCTKLNWISLSNNQLSGEIPASLGRLSNLAILKL 554
Query: 79 RKCGLIGTFPQNIFQIKSQLLTYLTT 104
+ G P + +S + L T
Sbjct: 555 GNNSISGNIPAELGNCQSLIWLDLNT 580
Score = 39.7 bits (91), Expect = 0.002
Identities = 22/68 (32%), Positives = 36/68 (52%), Gaps = 1/68 (1%)
Query: 25 LQKLSMSDCDLSGPLD-SSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGL 83
L+ + +S+ + SG L +L +L N+ +VL N F +P +F+N L TL++ L
Sbjct: 354 LELVDISNNNFSGKLPVDTLLKLSNIKTMVLSFNKFVGGLPDSFSNLPKLETLDMSSNNL 413
Query: 84 IGTFPQNI 91
G P I
Sbjct: 414 TGIIPSGI 421
Score = 36.6 bits (83), Expect = 0.017
Identities = 25/66 (37%), Positives = 36/66 (53%), Gaps = 4/66 (6%)
Query: 19 LLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTV--IVLGENNFSSPVP--QTFANFKNLT 74
LLPL +L+ L + + +LSG L S+ +T+ I L EN S P+ +F NL
Sbjct: 104 LLPLSNLESLVLKNANLSGSLTSAAKSQCGVTLDSIDLAENTISGPISDISSFGVCSNLK 163
Query: 75 TLNLRK 80
+LNL K
Sbjct: 164 SLNLSK 169
Score = 36.2 bits (82), Expect = 0.022
Identities = 19/64 (29%), Positives = 31/64 (47%)
Query: 28 LSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTF 87
L +S L G + L + L+++ LG N+ S +PQ KN+ L+L GT
Sbjct: 668 LDLSYNKLEGSIPKELGAMYYLSILNLGHNDLSGMIPQQLGGLKNVAILDLSYNRFNGTI 727
Query: 88 PQNI 91
P ++
Sbjct: 728 PNSL 731
Score = 35.4 bits (80), Expect = 0.038
Identities = 18/66 (27%), Positives = 33/66 (49%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L L++ DLSG + L L+N+ ++ L N F+ +P + + L ++L L
Sbjct: 689 LSILNLGHNDLSGMIPQQLGGLKNVAILDLSYNRFNGTIPNSLTSLTLLGEIDLSNNNLS 748
Query: 85 GTFPQN 90
G P++
Sbjct: 749 GMIPES 754
Score = 34.3 bits (77), Expect = 0.084
Identities = 25/84 (29%), Positives = 41/84 (48%), Gaps = 7/84 (8%)
Query: 6 ISITSRGHEWSNALLP-----LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFS 60
+SI + GH + ++P L+++ L +S +G + +SLT L L I L NN S
Sbjct: 689 LSILNLGHNDLSGMIPQQLGGLKNVAILDLSYNRFNGTIPNSLTSLTLLGEIDLSNNNLS 748
Query: 61 SPVPQT--FANFKNLTTLNLRKCG 82
+P++ F F + N CG
Sbjct: 749 GMIPESAPFDTFPDYRFANNSLCG 772
Score = 33.1 bits (74), Expect = 0.19
Identities = 20/80 (25%), Positives = 40/80 (50%)
Query: 17 NALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTL 76
++L L L+ L + LSG + L L+ L ++L N+ + P+P + +N L +
Sbjct: 469 SSLGSLSKLKDLILWLNQLSGEIPQELMYLQALENLILDFNDLTGPIPASLSNCTKLNWI 528
Query: 77 NLRKCGLIGTFPQNIFQIKS 96
+L L G P ++ ++ +
Sbjct: 529 SLSNNQLSGEIPASLGRLSN 548
Score = 30.4 bits (67), Expect = 1.2
Identities = 22/87 (25%), Positives = 39/87 (44%), Gaps = 5/87 (5%)
Query: 20 LPLRDLQKLSMSDCDLSGPLDSSLTRL-ENLTVIVLGENNFSSPVPQTFANFKNLTTLNL 78
LP LQ L + D G + L L + + + L NNFS VP++ +L +++
Sbjct: 300 LPSESLQYLYLRGNDFQGVYPNQLADLCKTVVELDLSYNNFSGMVPESLGECSSLELVDI 359
Query: 79 RKCGLIGTFPQN----IFQIKSQLLTY 101
G P + + IK+ +L++
Sbjct: 360 SNNNFSGKLPVDTLLKLSNIKTMVLSF 386
Score = 30.0 bits (66), Expect = 1.6
Identities = 21/67 (31%), Positives = 30/67 (44%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L L +S L+G + SSL L L ++L N S +PQ + L L L L
Sbjct: 453 LVSLDLSFNYLTGSIPSSLGSLSKLKDLILWLNQLSGEIPQELMYLQALENLILDFNDLT 512
Query: 85 GTFPQNI 91
G P ++
Sbjct: 513 GPIPASL 519
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor
(EC 2.7.1.37) (tBRI1) (Altered brassinolide sensitivity
1) (Systemin receptor SR160)
Length = 1207
Score = 50.1 bits (118), Expect = 1e-06
Identities = 28/88 (31%), Positives = 46/88 (51%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L +S+S+ LSG + +SL RL NL ++ LG N+ S +P N ++L L+L L
Sbjct: 525 LNWISLSNNQLSGEIPASLGRLSNLAILKLGNNSISGNIPAELGNCQSLIWLDLNTNFLN 584
Query: 85 GTFPQNIFQIKSQLLTYLTTPTSMVFFQ 112
G+ P +F+ + L T V+ +
Sbjct: 585 GSIPPPLFKQSGNIAVALLTGKRYVYIK 612
Score = 42.7 bits (99), Expect = 2e-04
Identities = 25/86 (29%), Positives = 39/86 (45%)
Query: 19 LLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNL 78
L+ L+ L+ L + DL+GP+ +SL+ L I L N S +P + NL L L
Sbjct: 495 LMYLQALENLILDFNDLTGPIPASLSNCTKLNWISLSNNQLSGEIPASLGRLSNLAILKL 554
Query: 79 RKCGLIGTFPQNIFQIKSQLLTYLTT 104
+ G P + +S + L T
Sbjct: 555 GNNSISGNIPAELGNCQSLIWLDLNT 580
Score = 38.9 bits (89), Expect = 0.003
Identities = 22/68 (32%), Positives = 36/68 (52%), Gaps = 1/68 (1%)
Query: 25 LQKLSMSDCDLSGPLD-SSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGL 83
L+ + +S + SG L +L++L N+ +VL N F +P +F+N L TL++ L
Sbjct: 354 LELVDISYNNFSGKLPVDTLSKLSNIKTMVLSFNKFVGGLPDSFSNLLKLETLDMSSNNL 413
Query: 84 IGTFPQNI 91
G P I
Sbjct: 414 TGVIPSGI 421
Score = 36.6 bits (83), Expect = 0.017
Identities = 25/66 (37%), Positives = 36/66 (53%), Gaps = 4/66 (6%)
Query: 19 LLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTV--IVLGENNFSSPVP--QTFANFKNLT 74
LLPL +L+ L + + +LSG L S+ +T+ I L EN S P+ +F NL
Sbjct: 104 LLPLSNLESLVLKNANLSGSLTSAAKSQCGVTLDSIDLAENTISGPISDISSFGVCSNLK 163
Query: 75 TLNLRK 80
+LNL K
Sbjct: 164 SLNLSK 169
Score = 36.2 bits (82), Expect = 0.022
Identities = 19/64 (29%), Positives = 31/64 (47%)
Query: 28 LSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTF 87
L +S L G + L + L+++ LG N+ S +PQ KN+ L+L GT
Sbjct: 668 LDLSYNKLEGSIPKELGAMYYLSILNLGHNDLSGMIPQQLGGLKNVAILDLSYNRFNGTI 727
Query: 88 PQNI 91
P ++
Sbjct: 728 PNSL 731
Score = 35.4 bits (80), Expect = 0.038
Identities = 18/66 (27%), Positives = 33/66 (49%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L L++ DLSG + L L+N+ ++ L N F+ +P + + L ++L L
Sbjct: 689 LSILNLGHNDLSGMIPQQLGGLKNVAILDLSYNRFNGTIPNSLTSLTLLGEIDLSNNNLS 748
Query: 85 GTFPQN 90
G P++
Sbjct: 749 GMIPES 754
Score = 34.3 bits (77), Expect = 0.084
Identities = 25/84 (29%), Positives = 41/84 (48%), Gaps = 7/84 (8%)
Query: 6 ISITSRGHEWSNALLP-----LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFS 60
+SI + GH + ++P L+++ L +S +G + +SLT L L I L NN S
Sbjct: 689 LSILNLGHNDLSGMIPQQLGGLKNVAILDLSYNRFNGTIPNSLTSLTLLGEIDLSNNNLS 748
Query: 61 SPVPQT--FANFKNLTTLNLRKCG 82
+P++ F F + N CG
Sbjct: 749 GMIPESAPFDTFPDYRFANNSLCG 772
Score = 33.1 bits (74), Expect = 0.19
Identities = 20/80 (25%), Positives = 40/80 (50%)
Query: 17 NALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTL 76
++L L L+ L + LSG + L L+ L ++L N+ + P+P + +N L +
Sbjct: 469 SSLGSLSKLKDLILWLNQLSGEIPQELMYLQALENLILDFNDLTGPIPASLSNCTKLNWI 528
Query: 77 NLRKCGLIGTFPQNIFQIKS 96
+L L G P ++ ++ +
Sbjct: 529 SLSNNQLSGEIPASLGRLSN 548
Score = 30.0 bits (66), Expect = 1.6
Identities = 22/87 (25%), Positives = 38/87 (43%), Gaps = 5/87 (5%)
Query: 20 LPLRDLQKLSMSDCDLSGPLDSSLTRL-ENLTVIVLGENNFSSPVPQTFANFKNLTTLNL 78
LP LQ L + D G + L L + + + L NNFS VP++ +L +++
Sbjct: 300 LPSESLQYLYLRGNDFQGVYPNQLADLCKTVVELDLSYNNFSGMVPESLGECSSLELVDI 359
Query: 79 RKCGLIGTFP----QNIFQIKSQLLTY 101
G P + IK+ +L++
Sbjct: 360 SYNNFSGKLPVDTLSKLSNIKTMVLSF 386
Score = 30.0 bits (66), Expect = 1.6
Identities = 21/67 (31%), Positives = 30/67 (44%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L L +S L+G + SSL L L ++L N S +PQ + L L L L
Sbjct: 453 LVSLDLSFNYLTGSIPSSLGSLSKLKDLILWLNQLSGEIPQELMYLQALENLILDFNDLT 512
Query: 85 GTFPQNI 91
G P ++
Sbjct: 513 GPIPASL 519
>TMM_ARATH (Q9SSD1) TOO MANY MOUTHS protein precursor (TMM)
Length = 496
Score = 48.5 bits (114), Expect = 4e-06
Identities = 28/88 (31%), Positives = 48/88 (53%), Gaps = 3/88 (3%)
Query: 17 NALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTL 76
NA L++L L +S+ ++ G + SLTRL +L V+ L NN + +P F + K+L+ L
Sbjct: 321 NAFKGLKNLMILVLSNTNIQGSIPKSLTRLNSLRVLHLEGNNLTGEIPLEFRDVKHLSEL 380
Query: 77 NLRKCGLIGTFP---QNIFQIKSQLLTY 101
L L G P +++++ +L Y
Sbjct: 381 RLNDNSLTGPVPFERDTVWRMRRKLRLY 408
Score = 42.7 bits (99), Expect = 2e-04
Identities = 28/83 (33%), Positives = 44/83 (52%), Gaps = 2/83 (2%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVL-GENNFSSPVPQ-TFANFKNLTTLNLR 79
L L L +S LSGP SSL L +L ++L G FS+ +P+ F KNL L L
Sbjct: 276 LNQLVLLDLSYNRLSGPFPSSLQGLNSLQALMLKGNTKFSTTIPENAFKGLKNLMILVLS 335
Query: 80 KCGLIGTFPQNIFQIKSQLLTYL 102
+ G+ P+++ ++ S + +L
Sbjct: 336 NTNIQGSIPKSLTRLNSLRVLHL 358
Score = 38.1 bits (87), Expect = 0.006
Identities = 27/76 (35%), Positives = 37/76 (48%), Gaps = 3/76 (3%)
Query: 16 SNALLPLRDLQKLSMSDCDLSGP--LDSSLTRL-ENLTVIVLGENNFSSPVPQTFANFKN 72
S +L L+ L+ L C P + + L RL +L +VL EN F P+P N N
Sbjct: 125 SESLTRLKHLKALFFYRCLGRAPQRIPAFLGRLGSSLQTLVLRENGFLGPIPDELGNLTN 184
Query: 73 LTTLNLRKCGLIGTFP 88
L L+L K L G+ P
Sbjct: 185 LKVLDLHKNHLNGSIP 200
Score = 38.1 bits (87), Expect = 0.006
Identities = 29/99 (29%), Positives = 43/99 (43%), Gaps = 3/99 (3%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L L ++ L+GP+ +LT +L I L N + P+P++ L L+L L
Sbjct: 231 LSVLDLNQNLLTGPVPPTLTSCGSLIKIDLSRNRVTGPIPESINRLNQLVLLDLSYNRLS 290
Query: 85 GTFPQNIFQIKSQLLTYLTTPTSMVFFQTTHLVNLFIAL 123
G FP ++ + S L T F TT N F L
Sbjct: 291 GPFPSSLQGLNSLQALMLKGNTK---FSTTIPENAFKGL 326
Score = 32.7 bits (73), Expect = 0.25
Identities = 21/79 (26%), Positives = 42/79 (52%), Gaps = 2/79 (2%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L+ L +S L+G + + L L+V+ L +N + PVP T + +L ++L + +
Sbjct: 209 LRSLDLSGNRLTGSIPGFV--LPALSVLDLNQNLLTGPVPPTLTSCGSLIKIDLSRNRVT 266
Query: 85 GTFPQNIFQIKSQLLTYLT 103
G P++I ++ +L L+
Sbjct: 267 GPIPESINRLNQLVLLDLS 285
Score = 32.0 bits (71), Expect = 0.42
Identities = 14/56 (25%), Positives = 26/56 (46%)
Query: 40 DSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQIK 95
+++ L+NL ++VL N +P++ +L L+L L G P +K
Sbjct: 320 ENAFKGLKNLMILVLSNTNIQGSIPKSLTRLNSLRVLHLEGNNLTGEIPLEFRDVK 375
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 48.5 bits (114), Expect = 4e-06
Identities = 27/80 (33%), Positives = 42/80 (51%)
Query: 24 DLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGL 83
+L KL +SD L+G + L R E L +++L N F P+P+ K+LT + + K L
Sbjct: 362 NLIKLDVSDNHLTGLIPKDLCRGEKLEMLILSNNFFFGPIPEELGKCKSLTKIRIVKNLL 421
Query: 84 IGTFPQNIFQIKSQLLTYLT 103
GT P +F + + LT
Sbjct: 422 NGTVPAGLFNLPLVTIIELT 441
Score = 46.2 bits (108), Expect = 2e-05
Identities = 36/128 (28%), Positives = 63/128 (49%), Gaps = 12/128 (9%)
Query: 24 DLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGL 83
DL+ L + + +G L ++ L+ L + G N FS +P+++ + ++L L L GL
Sbjct: 145 DLEVLDTYNNNFNGKLPPEMSELKKLKYLSFGGNFFSGEIPESYGDIQSLEYLGLNGAGL 204
Query: 84 IGTFPQNIFQIKS---QLLTYLTTPTSMV---FFQTTHLVNLFIALYCVT------LSSL 131
G P + ++K+ + Y + T V F T L L +A +T LS+L
Sbjct: 205 SGKSPAFLSRLKNLREMYIGYYNSYTGGVPPEFGGLTKLEILDMASCTLTGEIPTSLSNL 264
Query: 132 EHVHTLLV 139
+H+HTL +
Sbjct: 265 KHLHTLFL 272
Score = 43.1 bits (100), Expect = 2e-04
Identities = 29/92 (31%), Positives = 47/92 (50%), Gaps = 1/92 (1%)
Query: 3 LDGISITSRGHEWSNALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSP 62
LD S T G E +L L+ L L + +L+G + L+ L +L + L N +
Sbjct: 246 LDMASCTLTG-EIPTSLSNLKHLHTLFLHINNLTGHIPPELSGLVSLKSLDLSINQLTGE 304
Query: 63 VPQTFANFKNLTTLNLRKCGLIGTFPQNIFQI 94
+PQ+F N N+T +NL + L G P+ I ++
Sbjct: 305 IPQSFINLGNITLINLFRNNLYGQIPEAIGEL 336
Score = 42.7 bits (99), Expect = 2e-04
Identities = 22/78 (28%), Positives = 40/78 (51%)
Query: 19 LLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNL 78
+ L+ L +++ S +++G + S++R L + L N + +P+ N KNL TLN+
Sbjct: 500 IFELKHLSRINTSANNITGGIPDSISRCSTLISVDLSRNRINGEIPKGINNVKNLGTLNI 559
Query: 79 RKCGLIGTFPQNIFQIKS 96
L G+ P I + S
Sbjct: 560 SGNQLTGSIPTGIGNMTS 577
Score = 42.4 bits (98), Expect = 3e-04
Identities = 22/73 (30%), Positives = 39/73 (53%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L L+ L M+ C L+G + +SL+ L++L + L NN + +P + +L +L+L
Sbjct: 240 LTKLEILDMASCTLTGEIPTSLSNLKHLHTLFLHINNLTGHIPPELSGLVSLKSLDLSIN 299
Query: 82 GLIGTFPQNIFQI 94
L G PQ+ +
Sbjct: 300 QLTGEIPQSFINL 312
Score = 42.0 bits (97), Expect = 4e-04
Identities = 22/75 (29%), Positives = 40/75 (53%), Gaps = 1/75 (1%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLG-ENNFSSPVPQTFANFKNLTTLNLRK 80
++ L+ L ++ LSG + L+RL+NL + +G N+++ VP F L L++
Sbjct: 191 IQSLEYLGLNGAGLSGKSPAFLSRLKNLREMYIGYYNSYTGGVPPEFGGLTKLEILDMAS 250
Query: 81 CGLIGTFPQNIFQIK 95
C L G P ++ +K
Sbjct: 251 CTLTGEIPTSLSNLK 265
Score = 38.1 bits (87), Expect = 0.006
Identities = 23/68 (33%), Positives = 35/68 (50%), Gaps = 1/68 (1%)
Query: 28 LSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTF 87
+ ++D SG L +++ + L I L N FS +P NF NL TL L + G
Sbjct: 438 IELTDNFFSGELPVTMSG-DVLDQIYLSNNWFSGEIPPAIGNFPNLQTLFLDRNRFRGNI 496
Query: 88 PQNIFQIK 95
P+ IF++K
Sbjct: 497 PREIFELK 504
Score = 35.4 bits (80), Expect = 0.038
Identities = 21/67 (31%), Positives = 33/67 (48%), Gaps = 1/67 (1%)
Query: 28 LSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCG-LIGT 86
L++S L G + + L +L + L NNF+ +P + +L LN+ G L GT
Sbjct: 75 LNVSFTPLFGTISPEIGMLTHLVNLTLAANNFTGELPLEMKSLTSLKVLNISNNGNLTGT 134
Query: 87 FPQNIFQ 93
FP I +
Sbjct: 135 FPGEILK 141
Score = 32.7 bits (73), Expect = 0.25
Identities = 16/67 (23%), Positives = 31/67 (45%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L ++ +S+ SG + ++ NL + L N F +P+ K+L+ +N +
Sbjct: 458 LDQIYLSNNWFSGEIPPAIGNFPNLQTLFLDRNRFRGNIPREIFELKHLSRINTSANNIT 517
Query: 85 GTFPQNI 91
G P +I
Sbjct: 518 GGIPDSI 524
Score = 31.2 bits (69), Expect = 0.72
Identities = 18/70 (25%), Positives = 29/70 (40%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L L+ L +S L+G + S L N+T+I L NN +P+ L + +
Sbjct: 288 LVSLKSLDLSINQLTGEIPQSFINLGNITLINLFRNNLYGQIPEAIGELPKLEVFEVWEN 347
Query: 82 GLIGTFPQNI 91
P N+
Sbjct: 348 NFTLQLPANL 357
Score = 30.8 bits (68), Expect = 0.93
Identities = 16/64 (25%), Positives = 30/64 (46%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L + +S ++G + + ++NL + + N + +P N +LTTL+L L
Sbjct: 530 LISVDLSRNRINGEIPKGINNVKNLGTLNISGNQLTGSIPTGIGNMTSLTTLDLSFNDLS 589
Query: 85 GTFP 88
G P
Sbjct: 590 GRVP 593
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 47.0 bits (110), Expect = 1e-05
Identities = 24/74 (32%), Positives = 37/74 (49%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
+ +L L + D SGP+ SSL + L + L +NN +P T N +NL L++R
Sbjct: 187 MSELTTLWLDDNQFSGPVPSSLGNITTLQELYLNDNNLVGTLPVTLNNLENLVYLDVRNN 246
Query: 82 GLIGTFPQNIFQIK 95
L+G P + K
Sbjct: 247 SLVGAIPLDFVSCK 260
Score = 47.0 bits (110), Expect = 1e-05
Identities = 26/85 (30%), Positives = 44/85 (51%), Gaps = 2/85 (2%)
Query: 18 ALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLN 77
+LL + L+ + + L+G + S++ + LT + L +N FS PVP + N L L
Sbjct: 159 SLLSIPHLETVYFTGNGLNGSIPSNIGNMSELTTLWLDDNQFSGPVPSSLGNITTLQELY 218
Query: 78 LRKCGLIGTFPQNIFQIKSQLLTYL 102
L L+GT P + +++ L YL
Sbjct: 219 LNDNNLVGTLPVTLNNLEN--LVYL 241
Score = 46.2 bits (108), Expect = 2e-05
Identities = 30/104 (28%), Positives = 51/104 (48%), Gaps = 2/104 (1%)
Query: 2 YLDGISITSRG--HEWSNALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNF 59
++D ++++S G E+ + L+ L+K+ +S G + S L L I L N+F
Sbjct: 69 FVDTLNLSSYGISGEFGPEISHLKHLKKVVLSGNGFFGSIPSQLGNCSLLEHIDLSSNSF 128
Query: 60 SSPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQIKSQLLTYLT 103
+ +P T +NL L+L LIG FP+++ I Y T
Sbjct: 129 TGNIPDTLGALQNLRNLSLFFNSLIGPFPESLLSIPHLETVYFT 172
Score = 42.4 bits (98), Expect = 3e-04
Identities = 26/83 (31%), Positives = 42/83 (50%), Gaps = 3/83 (3%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
++ LQ L + +LSG L +T L+ L + L EN+F+ +PQ +L L+L +
Sbjct: 379 IQSLQSLQLYQNNLSGELPVDMTELKQLVSLALYENHFTGVIPQDLGANSSLEVLDLTRN 438
Query: 82 GLIGTFPQNIF---QIKSQLLTY 101
G P N+ ++K LL Y
Sbjct: 439 MFTGHIPPNLCSQKKLKRLLLGY 461
Score = 39.7 bits (91), Expect = 0.002
Identities = 27/106 (25%), Positives = 49/106 (45%), Gaps = 6/106 (5%)
Query: 3 LDGISITSRGHEWSNALLP----LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENN 58
+D IS+++ ++++ L P L++ C LSGP+ S +L L + L N+
Sbjct: 262 IDTISLSN--NQFTGGLPPGLGNCTSLREFGAFSCALSGPIPSCFGQLTKLDTLYLAGNH 319
Query: 59 FSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQIKSQLLTYLTT 104
FS +P K++ L L++ L G P + + +L T
Sbjct: 320 FSGRIPPELGKCKSMIDLQLQQNQLEGEIPGELGMLSQLQYLHLYT 365
Score = 37.7 bits (86), Expect = 0.008
Identities = 29/113 (25%), Positives = 46/113 (40%), Gaps = 13/113 (11%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L++L+ LS+ L GP SL + +L + N + +P N LTTL L
Sbjct: 139 LQNLRNLSLFFNSLIGPFPESLLSIPHLETVYFTGNGLNGSIPSNIGNMSELTTLWLDDN 198
Query: 82 GLIGTFPQNIFQIKSQLLTYLTTPTSMVFFQTTHLVNLFIALYCVTLSSLEHV 134
G P ++ I + YL N + VTL++LE++
Sbjct: 199 QFSGPVPSSLGNITTLQELYLND-------------NNLVGTLPVTLNNLENL 238
Score = 37.7 bits (86), Expect = 0.008
Identities = 21/70 (30%), Positives = 36/70 (51%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L LQ L + +LSG + S+ ++++L + L +NN S +P K L +L L +
Sbjct: 355 LSQLQYLHLYTNNLSGEVPLSIWKIQSLQSLQLYQNNLSGELPVDMTELKQLVSLALYEN 414
Query: 82 GLIGTFPQNI 91
G PQ++
Sbjct: 415 HFTGVIPQDL 424
Score = 37.4 bits (85), Expect = 0.010
Identities = 21/69 (30%), Positives = 32/69 (45%)
Query: 23 RDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCG 82
++L +S + +GP+ SL L+N+T I L N S +P + L LNL
Sbjct: 499 QNLLFFDLSGNNFTGPIPPSLGNLKNVTAIYLSSNQLSGSIPPELGSLVKLEHLNLSHNI 558
Query: 83 LIGTFPQNI 91
L G P +
Sbjct: 559 LKGILPSEL 567
Score = 37.4 bits (85), Expect = 0.010
Identities = 28/81 (34%), Positives = 37/81 (45%), Gaps = 5/81 (6%)
Query: 13 HEWSNALLP--LRDLQKLSMSDCD---LSGPLDSSLTRLENLTVIVLGENNFSSPVPQTF 67
H +LP L + KLS D L+G + S+L L LT + LGEN+FS +P +
Sbjct: 556 HNILKGILPSELSNCHKLSELDASHNLLNGSIPSTLGSLTELTKLSLGENSFSGGIPTSL 615
Query: 68 ANFKNLTTLNLRKCGLIGTFP 88
L L L L G P
Sbjct: 616 FQSNKLLNLQLGGNLLAGDIP 636
Score = 35.8 bits (81), Expect = 0.029
Identities = 20/69 (28%), Positives = 33/69 (46%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L+ L++S L G L S L+ L+ + N + +P T + LT L+L +
Sbjct: 549 LEHLNLSHNILKGILPSELSNCHKLSELDASHNLLNGSIPSTLGSLTELTKLSLGENSFS 608
Query: 85 GTFPQNIFQ 93
G P ++FQ
Sbjct: 609 GGIPTSLFQ 617
Score = 30.0 bits (66), Expect = 1.6
Identities = 26/102 (25%), Positives = 35/102 (33%), Gaps = 24/102 (23%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLT------------------------VIVLGENNFS 60
LQ+L ++D +L G L +L LENL I L N F+
Sbjct: 214 LQELYLNDNNLVGTLPVTLNNLENLVYLDVRNNSLVGAIPLDFVSCKQIDTISLSNNQFT 273
Query: 61 SPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQIKSQLLTYL 102
+P N +L C L G P Q+ YL
Sbjct: 274 GGLPPGLGNCTSLREFGAFSCALSGPIPSCFGQLTKLDTLYL 315
Score = 28.1 bits (61), Expect = 6.1
Identities = 18/70 (25%), Positives = 36/70 (50%), Gaps = 1/70 (1%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L+ L+ L++S L+G L L +L+ L + + NN S + + + ++LT +N+
Sbjct: 641 LQALRSLNLSSNKLNGQLPIDLGKLKMLEELDVSHNNLSGTL-RVLSTIQSLTFINISHN 699
Query: 82 GLIGTFPQNI 91
G P ++
Sbjct: 700 LFSGPVPPSL 709
>PGI2_ARATH (Q9M5J8) Polygalacturonase inhibitor 2 precursor
(Polygalacturonase-inhibiting protein) (PGIP-2)
Length = 330
Score = 45.4 bits (106), Expect = 4e-05
Identities = 25/74 (33%), Positives = 42/74 (55%), Gaps = 1/74 (1%)
Query: 19 LLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFK-NLTTLN 77
L L++L+ + +S DLSG + SSL+ L L + L N + P+P++F F + +L
Sbjct: 139 LSQLKNLEYIDLSFNDLSGSIPSSLSSLRKLEYLELSRNKLTGPIPESFGTFSGKVPSLF 198
Query: 78 LRKCGLIGTFPQNI 91
L L GT P+++
Sbjct: 199 LSHNQLSGTIPKSL 212
Score = 42.0 bits (97), Expect = 4e-04
Identities = 22/69 (31%), Positives = 40/69 (57%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L++L L +S +L+GP+ L++L+NL I L N+ S +P + ++ + L L L +
Sbjct: 118 LKNLTFLRLSWTNLTGPVPEFLSQLKNLEYIDLSFNDLSGSIPSSLSSLRKLEYLELSRN 177
Query: 82 GLIGTFPQN 90
L G P++
Sbjct: 178 KLTGPIPES 186
Score = 40.8 bits (94), Expect = 0.001
Identities = 21/69 (30%), Positives = 39/69 (56%), Gaps = 2/69 (2%)
Query: 34 DLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQ 93
+L+G + ++ +L+NLT + L N + PVP+ + KNL ++L L G+ P ++
Sbjct: 106 NLTGHIQPTIAKLKNLTFLRLSWTNLTGPVPEFLSQLKNLEYIDLSFNDLSGSIPSSLSS 165
Query: 94 IKSQLLTYL 102
++ L YL
Sbjct: 166 LRK--LEYL 172
>RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC
2.7.1.37)
Length = 999
Score = 45.1 bits (105), Expect = 5e-05
Identities = 24/78 (30%), Positives = 41/78 (51%)
Query: 14 EWSNALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNL 73
E SN L + L ++ +S+ LSG + L L+++ L +N+F+ +P+T KNL
Sbjct: 394 EISNNLGKCKSLTRVRLSNNKLSGQIPHGFWGLPRLSLLELSDNSFTGSIPKTIIGAKNL 453
Query: 74 TTLNLRKCGLIGTFPQNI 91
+ L + K G+ P I
Sbjct: 454 SNLRISKNRFSGSIPNEI 471
Score = 44.7 bits (104), Expect = 6e-05
Identities = 23/75 (30%), Positives = 40/75 (52%)
Query: 17 NALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTL 76
N + L + ++S ++ D SG + SL +L+ L+ + L +N S +P+ +KNL L
Sbjct: 469 NEIGSLNGIIEISGAENDFSGEIPESLVKLKQLSRLDLSKNQLSGEIPRELRGWKNLNEL 528
Query: 77 NLRKCGLIGTFPQNI 91
NL L G P+ +
Sbjct: 529 NLANNHLSGEIPKEV 543
Score = 42.7 bits (99), Expect = 2e-04
Identities = 32/100 (32%), Positives = 46/100 (46%), Gaps = 4/100 (4%)
Query: 24 DLQKLSMSDCDLSGPLDSSLT-RLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCG 82
+L L +S+ L G + SL L NL + + NN S +P +F F+ L +LNL
Sbjct: 115 NLISLDLSENLLVGSIPKSLPFNLPNLKFLEISGNNLSDTIPSSFGEFRKLESLNLAGNF 174
Query: 83 LIGTFPQ---NIFQIKSQLLTYLTTPTSMVFFQTTHLVNL 119
L GT P N+ +K L Y S + Q +L L
Sbjct: 175 LSGTIPASLGNVTTLKELKLAYNLFSPSQIPSQLGNLTEL 214
Score = 38.9 bits (89), Expect = 0.003
Identities = 20/73 (27%), Positives = 37/73 (50%)
Query: 19 LLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNL 78
++ ++L L +S SG + + + L + I EN+FS +P++ K L+ L+L
Sbjct: 447 IIGAKNLSNLRISKNRFSGSIPNEIGSLNGIIEISGAENDFSGEIPESLVKLKQLSRLDL 506
Query: 79 RKCGLIGTFPQNI 91
K L G P+ +
Sbjct: 507 SKNQLSGEIPREL 519
Score = 38.9 bits (89), Expect = 0.003
Identities = 20/70 (28%), Positives = 35/70 (49%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L +LQ L ++ C+L GP+ SL+RL +L + L N + +P K + + L
Sbjct: 211 LTELQVLWLAGCNLVGPIPPSLSRLTSLVNLDLTFNQLTGSIPSWITQLKTVEQIELFNN 270
Query: 82 GLIGTFPQNI 91
G P+++
Sbjct: 271 SFSGELPESM 280
Score = 36.6 bits (83), Expect = 0.017
Identities = 21/74 (28%), Positives = 36/74 (48%)
Query: 18 ALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLN 77
+L L L L ++ L+G + S +T+L+ + I L N+FS +P++ N L +
Sbjct: 231 SLSRLTSLVNLDLTFNQLTGSIPSWITQLKTVEQIELFNNSFSGELPESMGNMTTLKRFD 290
Query: 78 LRKCGLIGTFPQNI 91
L G P N+
Sbjct: 291 ASMNKLTGKIPDNL 304
Score = 36.2 bits (82), Expect = 0.022
Identities = 23/86 (26%), Positives = 38/86 (43%)
Query: 14 EWSNALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNL 73
E +L+ L+ L +L +S LSG + L +NL + L N+ S +P+ L
Sbjct: 490 EIPESLVKLKQLSRLDLSKNQLSGEIPRELRGWKNLNELNLANNHLSGEIPKEVGILPVL 549
Query: 74 TTLNLRKCGLIGTFPQNIFQIKSQLL 99
L+L G P + +K +L
Sbjct: 550 NYLDLSSNQFSGEIPLELQNLKLNVL 575
Score = 33.1 bits (74), Expect = 0.19
Identities = 14/71 (19%), Positives = 35/71 (48%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L L +SD +G + ++ +NL+ + + +N FS +P + + ++ +
Sbjct: 429 LSLLELSDNSFTGSIPKTIIGAKNLSNLRISKNRFSGSIPNEIGSLNGIIEISGAENDFS 488
Query: 85 GTFPQNIFQIK 95
G P+++ ++K
Sbjct: 489 GEIPESLVKLK 499
Score = 32.7 bits (73), Expect = 0.25
Identities = 20/72 (27%), Positives = 34/72 (46%)
Query: 20 LPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLR 79
L L +L+ L++ + L GPL S+TR + L+ + L N + +P L ++L
Sbjct: 304 LNLLNLESLNLFENMLEGPLPESITRSKTLSELKLFNNRLTGVLPSQLGANSPLQYVDLS 363
Query: 80 KCGLIGTFPQNI 91
G P N+
Sbjct: 364 YNRFSGEIPANV 375
Score = 31.2 bits (69), Expect = 0.72
Identities = 18/58 (31%), Positives = 28/58 (48%)
Query: 39 LDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQIKS 96
+ S L L L V+ L N P+P + + +L L+L L G+ P I Q+K+
Sbjct: 204 IPSQLGNLTELQVLWLAGCNLVGPIPPSLSRLTSLVNLDLTFNQLTGSIPSWITQLKT 261
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells
protein) (EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 45.1 bits (105), Expect = 5e-05
Identities = 27/79 (34%), Positives = 43/79 (54%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L ++S+S+ LSG + +SL+RL NLT++ L N + +P+ N L LNL L
Sbjct: 606 LVEISLSNNHLSGEIPASLSRLTNLTILDLSGNALTGSIPKEMGNSLKLQGLNLANNQLN 665
Query: 85 GTFPQNIFQIKSQLLTYLT 103
G P++ + S + LT
Sbjct: 666 GHIPESFGLLGSLVKLNLT 684
Score = 45.1 bits (105), Expect = 5e-05
Identities = 22/72 (30%), Positives = 36/72 (49%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L+ + C +GPL +++L++L + L N +P++F NL+ LNL LI
Sbjct: 212 LKNFAAPSCFFNGPLPKEISKLKHLAKLDLSYNPLKCSIPKSFGELHNLSILNLVSAELI 271
Query: 85 GTFPQNIFQIKS 96
G P + KS
Sbjct: 272 GLIPPELGNCKS 283
Score = 44.3 bits (103), Expect = 8e-05
Identities = 25/93 (26%), Positives = 43/93 (45%)
Query: 6 ISITSRGHEWSNALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQ 65
+S ++ +W L + LS+ L G + ++ L+NL + L N FS +P
Sbjct: 48 VSSSASHCDWVGVTCLLGRVNSLSLPSLSLRGQIPKEISSLKNLRELCLAGNQFSGKIPP 107
Query: 66 TFANFKNLTTLNLRKCGLIGTFPQNIFQIKSQL 98
N K+L TL+L L G P+ + ++ L
Sbjct: 108 EIWNLKHLQTLDLSGNSLTGLLPRLLSELPQLL 140
Score = 42.4 bits (98), Expect = 3e-04
Identities = 26/95 (27%), Positives = 42/95 (43%)
Query: 1 MYLDGISITSRGHEWSNALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFS 60
+YLD G + + L L L +S+ LSG + + +L NL+ + +G N+FS
Sbjct: 140 LYLDLSDNHFSGSLPPSFFISLPALSSLDVSNNSLSGEIPPEIGKLSNLSNLYMGLNSFS 199
Query: 61 SPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQIK 95
+P N L C G P+ I ++K
Sbjct: 200 GQIPSEIGNISLLKNFAAPSCFFNGPLPKEISKLK 234
Score = 42.0 bits (97), Expect = 4e-04
Identities = 25/80 (31%), Positives = 43/80 (53%), Gaps = 3/80 (3%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L++L +SD L+G + + +L +L+V+ L N F +P + +LTTL+L L
Sbjct: 474 LKRLVLSDNQLTGEIPREIGKLTSLSVLNLNANMFQGKIPVELGDCTSLTTLDLGSNNLQ 533
Query: 85 GTFPQNI---FQIKSQLLTY 101
G P I Q++ +L+Y
Sbjct: 534 GQIPDKITALAQLQCLVLSY 553
Score = 40.0 bits (92), Expect = 0.002
Identities = 21/70 (30%), Positives = 36/70 (51%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L++L + +S +LSG L S L+ +E L + + +N F+ +P N L L++ +
Sbjct: 699 LKELTHMDLSFNNLSGELSSELSTMEKLVGLYIEQNKFTGEIPSELGNLTQLEYLDVSEN 758
Query: 82 GLIGTFPQNI 91
L G P I
Sbjct: 759 LLSGEIPTKI 768
Score = 38.5 bits (88), Expect = 0.004
Identities = 27/96 (28%), Positives = 49/96 (50%), Gaps = 5/96 (5%)
Query: 12 GHEWSNALLP----LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQT- 66
G+++S + P L+ LQ L +S L+G L L+ L L + L +N+FS +P +
Sbjct: 98 GNQFSGKIPPEIWNLKHLQTLDLSGNSLTGLLPRLLSELPQLLYLDLSDNHFSGSLPPSF 157
Query: 67 FANFKNLTTLNLRKCGLIGTFPQNIFQIKSQLLTYL 102
F + L++L++ L G P I ++ + Y+
Sbjct: 158 FISLPALSSLDVSNNSLSGEIPPEIGKLSNLSNLYM 193
Score = 36.2 bits (82), Expect = 0.022
Identities = 24/85 (28%), Positives = 35/85 (40%)
Query: 6 ISITSRGHEWSNALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQ 65
+S + E S+ L + L L + +G + S L L L + + EN S +P
Sbjct: 707 LSFNNLSGELSSELSTMEKLVGLYIEQNKFTGEIPSELGNLTQLEYLDVSENLLSGEIPT 766
Query: 66 TFANFKNLTTLNLRKCGLIGTFPQN 90
NL LNL K L G P +
Sbjct: 767 KICGLPNLEFLNLAKNNLRGEVPSD 791
Score = 36.2 bits (82), Expect = 0.022
Identities = 21/78 (26%), Positives = 38/78 (47%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
LQ L++++ L+G + S L +L + L +N PVP + N K LT ++L L
Sbjct: 654 LQGLNLANNQLNGHIPESFGLLGSLVKLNLTKNKLDGPVPASLGNLKELTHMDLSFNNLS 713
Query: 85 GTFPQNIFQIKSQLLTYL 102
G + ++ + Y+
Sbjct: 714 GELSSELSTMEKLVGLYI 731
Score = 35.8 bits (81), Expect = 0.029
Identities = 19/57 (33%), Positives = 28/57 (48%)
Query: 35 LSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNI 91
LSGP+ L L I L N+ S +P + + NLT L+L L G+ P+ +
Sbjct: 592 LSGPIPEELGECLVLVEISLSNNHLSGEIPASLSRLTNLTILDLSGNALTGSIPKEM 648
Score = 34.3 bits (77), Expect = 0.084
Identities = 20/70 (28%), Positives = 33/70 (46%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L L KL+++ L GP+ +SL L+ LT + L NN S + + + L L + +
Sbjct: 675 LGSLVKLNLTKNKLDGPVPASLGNLKELTHMDLSFNNLSGELSSELSTMEKLVGLYIEQN 734
Query: 82 GLIGTFPQNI 91
G P +
Sbjct: 735 KFTGEIPSEL 744
Score = 31.6 bits (70), Expect = 0.55
Identities = 25/106 (23%), Positives = 44/106 (40%), Gaps = 2/106 (1%)
Query: 24 DLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGL 83
+L + + S L G L + + +L +VL +N + +P+ +L+ LNL
Sbjct: 449 NLMEFTASYNRLEGYLPAEIGNAASLKRLVLSDNQLTGEIPREIGKLTSLSVLNLNANMF 508
Query: 84 IGTFPQNIFQIKSQLLTYLTTPTSMVFFQTTHLVNLFIALYCVTLS 129
G P + S LT L ++ + Q + L C+ LS
Sbjct: 509 QGKIPVELGDCTS--LTTLDLGSNNLQGQIPDKITALAQLQCLVLS 552
Score = 28.9 bits (63), Expect = 3.6
Identities = 17/57 (29%), Positives = 28/57 (48%)
Query: 35 LSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNI 91
LSG L S + + + L ++L N FS +P + L L+L L G+ P+ +
Sbjct: 317 LSGSLPSWMGKWKVLDSLLLANNRFSGEIPHEIEDCPMLKHLSLASNLLSGSIPREL 373
>PGIP_PYRCO (Q05091) Polygalacturonase inhibitor precursor
(Polygalacturonase-inhibiting protein)
Length = 330
Score = 44.3 bits (103), Expect = 8e-05
Identities = 24/71 (33%), Positives = 39/71 (54%)
Query: 18 ALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLN 77
A+ L+ L+ L +S +LSG + L++L+NLT + L NN + +P + + NL L
Sbjct: 114 AIAKLKGLKSLRLSWTNLSGSVPDFLSQLKNLTFLDLSFNNLTGAIPSSLSELPNLGALR 173
Query: 78 LRKCGLIGTFP 88
L + L G P
Sbjct: 174 LDRNKLTGHIP 184
Score = 39.7 bits (91), Expect = 0.002
Identities = 19/61 (31%), Positives = 33/61 (53%)
Query: 34 DLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNIFQ 93
+L+GP+ ++ +L+ L + L N S VP + KNLT L+L L G P ++ +
Sbjct: 106 NLTGPIQPAIAKLKGLKSLRLSWTNLSGSVPDFLSQLKNLTFLDLSFNNLTGAIPSSLSE 165
Query: 94 I 94
+
Sbjct: 166 L 166
Score = 38.5 bits (88), Expect = 0.004
Identities = 36/121 (29%), Positives = 57/121 (46%), Gaps = 7/121 (5%)
Query: 19 LLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANF-KNLTTLN 77
L L++L L +S +L+G + SSL+ L NL + L N + +P +F F N+ L
Sbjct: 139 LSQLKNLTFLDLSFNNLTGAIPSSLSELPNLGALRLDRNKLTGHIPISFGQFIGNVPDLY 198
Query: 78 LRKCGLIGTFPQNIFQIKSQLL----TYLTTPTSMVF--FQTTHLVNLFIALYCVTLSSL 131
L L G P + Q+ + L S++F +TT +V+L L LS +
Sbjct: 199 LSHNQLSGNIPTSFAQMDFTSIDLSRNKLEGDASVIFGLNKTTQIVDLSRNLLEFNLSKV 258
Query: 132 E 132
E
Sbjct: 259 E 259
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 43.5 bits (101), Expect = 1e-04
Identities = 28/96 (29%), Positives = 45/96 (46%), Gaps = 2/96 (2%)
Query: 14 EWSNALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNL 73
E +L R + LS+ + LSG + + + + NLT + L N+FS +P N L
Sbjct: 293 EMPRSLSNSRSISLLSLRNNTLSGQIYLNCSAMTNLTSLDLASNSFSGSIPSNLPNCLRL 352
Query: 74 TTLNLRKCGLIGTFPQNIFQIKSQLLTYLTTPTSMV 109
T+N K I P++ +S LT L+ S +
Sbjct: 353 KTINFAKIKFIAQIPESFKNFQS--LTSLSFSNSSI 386
Score = 40.0 bits (92), Expect = 0.002
Identities = 27/103 (26%), Positives = 46/103 (44%), Gaps = 2/103 (1%)
Query: 19 LLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNL 78
L L +L L++ + LSG L S L +L NL + + N FS +P F L +
Sbjct: 226 LFQLSNLSVLALQNNRLSGALSSKLGKLSNLGRLDISSNKFSGKIPDVFLELNKLWYFSA 285
Query: 79 RKCGLIGTFPQNIFQIKSQLLTYL--TTPTSMVFFQTTHLVNL 119
+ G P+++ +S L L T + ++ + + NL
Sbjct: 286 QSNLFNGEMPRSLSNSRSISLLSLRNNTLSGQIYLNCSAMTNL 328
Score = 38.9 bits (89), Expect = 0.003
Identities = 24/76 (31%), Positives = 40/76 (52%), Gaps = 1/76 (1%)
Query: 16 SNALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTT 75
S ++ L L+ L+++ LSG + +SL L NL V+ L N+FS P + N +L
Sbjct: 103 SESVAKLDQLKVLNLTHNSLSGSIAASLLNLSNLEVLDLSSNDFSGLFP-SLINLPSLRV 161
Query: 76 LNLRKCGLIGTFPQNI 91
LN+ + G P ++
Sbjct: 162 LNVYENSFHGLIPASL 177
Score = 38.5 bits (88), Expect = 0.004
Identities = 20/70 (28%), Positives = 37/70 (52%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
LR L L++ + +LSG + ++L+ + +L V+ L NN S +P + L+T ++
Sbjct: 556 LRQLHVLNLKNNNLSGNIPANLSGMTSLEVLDLSHNNLSGNIPPSLVKLSFLSTFSVAYN 615
Query: 82 GLIGTFPQNI 91
L G P +
Sbjct: 616 KLSGPIPTGV 625
Score = 34.3 bits (77), Expect = 0.084
Identities = 22/80 (27%), Positives = 38/80 (47%), Gaps = 1/80 (1%)
Query: 18 ALLPLRDLQKLSMSDCDLSGPLDSSL-TRLENLTVIVLGENNFSSPVPQTFANFKNLTTL 76
+L+ L L+ L++ + G + +SL L + I L N F +P N ++ L
Sbjct: 152 SLINLPSLRVLNVYENSFHGLIPASLCNNLPRIREIDLAMNYFDGSIPVGIGNCSSVEYL 211
Query: 77 NLRKCGLIGTFPQNIFQIKS 96
L L G+ PQ +FQ+ +
Sbjct: 212 GLASNNLSGSIPQELFQLSN 231
Score = 32.7 bits (73), Expect = 0.25
Identities = 17/70 (24%), Positives = 31/70 (44%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
++ L ++ +LSG + L +L NL+V+ L N S + NL L++
Sbjct: 208 VEYLGLASNNLSGSIPQELFQLSNLSVLALQNNRLSGALSSKLGKLSNLGRLDISSNKFS 267
Query: 85 GTFPQNIFQI 94
G P ++
Sbjct: 268 GKIPDVFLEL 277
Score = 30.4 bits (67), Expect = 1.2
Identities = 18/75 (24%), Positives = 33/75 (44%)
Query: 17 NALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTL 76
+ L L L S +G + SL+ +++++ L N S + + NLT+L
Sbjct: 272 DVFLELNKLWYFSAQSNLFNGEMPRSLSNSRSISLLSLRNNTLSGQIYLNCSAMTNLTSL 331
Query: 77 NLRKCGLIGTFPQNI 91
+L G+ P N+
Sbjct: 332 DLASNSFSGSIPSNL 346
Score = 30.0 bits (66), Expect = 1.6
Identities = 17/67 (25%), Positives = 30/67 (44%)
Query: 28 LSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTF 87
+ +S L+G + L L V+ L NN S +P + +L L+L L G
Sbjct: 538 IDLSYNSLNGSIWPEFGDLRQLHVLNLKNNNLSGNIPANLSGMTSLEVLDLSHNNLSGNI 597
Query: 88 PQNIFQI 94
P ++ ++
Sbjct: 598 PPSLVKL 604
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 42.7 bits (99), Expect = 2e-04
Identities = 23/65 (35%), Positives = 34/65 (51%)
Query: 27 KLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGT 86
+L + + LSG L SL +L+ + V+ L N +P + N KNL TL+L L G
Sbjct: 80 RLELGNKKLSGKLSESLGKLDEIRVLNLSRNFIKDSIPLSIFNLKNLQTLDLSSNDLSGG 139
Query: 87 FPQNI 91
P +I
Sbjct: 140 IPTSI 144
Score = 35.0 bits (79), Expect = 0.050
Identities = 19/77 (24%), Positives = 38/77 (48%)
Query: 20 LPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLR 79
L L+ L +++C L+G + L+ L ++ L N + +P +FK L L+L
Sbjct: 412 LHFEKLKVLVVANCRLTGSMPRWLSSSNELQLLDLSWNRLTGAIPSWIGDFKALFYLDLS 471
Query: 80 KCGLIGTFPQNIFQIKS 96
G P+++ +++S
Sbjct: 472 NNSFTGEIPKSLTKLES 488
Score = 34.7 bits (78), Expect = 0.065
Identities = 30/111 (27%), Positives = 51/111 (45%), Gaps = 13/111 (11%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L L++ + LSG L + T + L + LG N F+ +P+ + K L +NL +
Sbjct: 294 LNLLNLRNNSLSGRLMLNCTAMIALNSLDLGTNRFNGRLPENLPDCKRLKNVNLARNTFH 353
Query: 85 GTFPQNIFQIKSQLLTYLTTPTSMVFFQTTHLVNLFIAL----YCVTLSSL 131
G P++ +S L+Y + S L N+ AL +C L++L
Sbjct: 354 GQVPESFKNFES--LSYFSLSNS-------SLANISSALGILQHCKNLTTL 395
Score = 33.9 bits (76), Expect = 0.11
Identities = 25/100 (25%), Positives = 38/100 (38%), Gaps = 36/100 (36%)
Query: 28 LSMSDCDLSGPLDSSLTRLENLT------------------------------------V 51
L +S+ +G + SLT+LE+LT
Sbjct: 468 LDLSNNSFTGEIPKSLTKLESLTSRNISVNEPSPDFPFFMKRNESARALQYNQIFGFPPT 527
Query: 52 IVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNI 91
I LG NN S P+ + F N K L +L+ L G+ P ++
Sbjct: 528 IELGHNNLSGPIWEEFGNLKKLHVFDLKWNALSGSIPSSL 567
Score = 30.8 bits (68), Expect = 0.93
Identities = 22/75 (29%), Positives = 31/75 (41%)
Query: 16 SNALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTT 75
S + L L +L +S SG + L L + N F +P++ AN +L
Sbjct: 237 SREIRNLSSLVRLDVSWNLFSGEIPDVFDELPQLKFFLGQTNGFIGGIPKSLANSPSLNL 296
Query: 76 LNLRKCGLIGTFPQN 90
LNLR L G N
Sbjct: 297 LNLRNNSLSGRLMLN 311
Score = 30.0 bits (66), Expect = 1.6
Identities = 20/68 (29%), Positives = 33/68 (48%), Gaps = 2/68 (2%)
Query: 17 NALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQ--TFANFKNLT 74
++L + L+ L +S+ LSG + SL +L L+ + NN S +P F F N +
Sbjct: 565 SSLSGMTSLEALDLSNNRLSGSIPVSLQQLSFLSKFSVAYNNLSGVIPSGGQFQTFPNSS 624
Query: 75 TLNLRKCG 82
+ CG
Sbjct: 625 FESNHLCG 632
Score = 30.0 bits (66), Expect = 1.6
Identities = 15/55 (27%), Positives = 31/55 (56%)
Query: 24 DLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNL 78
+LQ L +S L+G + S + + L + L N+F+ +P++ ++LT+ N+
Sbjct: 440 ELQLLDLSWNRLTGAIPSWIGDFKALFYLDLSNNSFTGEIPKSLTKLESLTSRNI 494
Score = 29.6 bits (65), Expect = 2.1
Identities = 19/68 (27%), Positives = 32/68 (46%), Gaps = 3/68 (4%)
Query: 24 DLQKLSMSDCD---LSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRK 80
+L+KL + D LSG + SSL+ + +L + L N S +P + L+ ++
Sbjct: 545 NLKKLHVFDLKWNALSGSIPSSLSGMTSLEALDLSNNRLSGSIPVSLQQLSFLSKFSVAY 604
Query: 81 CGLIGTFP 88
L G P
Sbjct: 605 NNLSGVIP 612
Score = 28.9 bits (63), Expect = 3.6
Identities = 20/78 (25%), Positives = 36/78 (45%), Gaps = 1/78 (1%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L+ L + DL+G + L L+ L ++ + EN S + + N +L L++
Sbjct: 198 LEHLCLGMNDLTGNIPEDLFHLKRLNLLGIQENRLSGSLSREIRNLSSLVRLDVSWNLFS 257
Query: 85 GTFPQNIFQIKSQLLTYL 102
G P ++F QL +L
Sbjct: 258 GEIP-DVFDELPQLKFFL 274
>PGI1_ARATH (Q9M5J9) Polygalacturonase inhibitor 1 precursor
(Polygalacturonase-inhibiting protein) (PGIP-1)
Length = 330
Score = 42.0 bits (97), Expect = 4e-04
Identities = 19/69 (27%), Positives = 40/69 (57%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L++L+ L +S +L+GP+ +++L+NL + L N+ S +P + + + L L +
Sbjct: 118 LKNLRMLRLSWTNLTGPIPDFISQLKNLEFLELSFNDLSGSIPSSLSTLPKILALELSRN 177
Query: 82 GLIGTFPQN 90
L G+ P++
Sbjct: 178 KLTGSIPES 186
Score = 40.4 bits (93), Expect = 0.001
Identities = 26/88 (29%), Positives = 44/88 (49%), Gaps = 8/88 (9%)
Query: 15 WSNALLP-------LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTF 67
W+N P L++L+ L +S DLSG + SSL+ L + + L N + +P++F
Sbjct: 128 WTNLTGPIPDFISQLKNLEFLELSFNDLSGSIPSSLSTLPKILALELSRNKLTGSIPESF 187
Query: 68 ANFK-NLTTLNLRKCGLIGTFPQNIFQI 94
+F + L L L G P+++ I
Sbjct: 188 GSFPGTVPDLRLSHNQLSGPIPKSLGNI 215
Score = 37.7 bits (86), Expect = 0.008
Identities = 17/58 (29%), Positives = 32/58 (54%)
Query: 34 DLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTFPQNI 91
+L+G + ++ +L+NL ++ L N + P+P + KNL L L L G+ P ++
Sbjct: 106 NLTGTIQPTIAKLKNLRMLRLSWTNLTGPIPDFISQLKNLEFLELSFNDLSGSIPSSL 163
Score = 30.8 bits (68), Expect = 0.93
Identities = 20/70 (28%), Positives = 33/70 (46%), Gaps = 1/70 (1%)
Query: 28 LSMSDCDLSGPLDSSLTRLENLTVIVLGE-NNFSSPVPQTFANFKNLTTLNLRKCGLIGT 86
L++ +SG + + + L L +V + +N + + T A KNL L L L G
Sbjct: 75 LTIFSGQISGQIPAEVGDLPYLETLVFRKLSNLTGTIQPTIAKLKNLRMLRLSWTNLTGP 134
Query: 87 FPQNIFQIKS 96
P I Q+K+
Sbjct: 135 IPDFISQLKN 144
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 41.6 bits (96), Expect = 5e-04
Identities = 22/70 (31%), Positives = 39/70 (55%)
Query: 24 DLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGL 83
+L +S+S+ L+G + + RLENL ++ L N+FS +P + ++L L+L
Sbjct: 513 NLNWISLSNNRLTGEIPKWIGRLENLAILKLSNNSFSGNIPAELGDCRSLIWLDLNTNLF 572
Query: 84 IGTFPQNIFQ 93
GT P +F+
Sbjct: 573 NGTIPAAMFK 582
Score = 39.7 bits (91), Expect = 0.002
Identities = 23/86 (26%), Positives = 38/86 (43%)
Query: 19 LLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNL 78
L+ ++ L+ L + DL+G + S L+ NL I L N + +P+ +NL L L
Sbjct: 484 LMYVKTLETLILDFNDLTGEIPSGLSNCTNLNWISLSNNRLTGEIPKWIGRLENLAILKL 543
Query: 79 RKCGLIGTFPQNIFQIKSQLLTYLTT 104
G P + +S + L T
Sbjct: 544 SNNSFSGNIPAELGDCRSLIWLDLNT 569
Score = 37.4 bits (85), Expect = 0.010
Identities = 30/102 (29%), Positives = 43/102 (41%), Gaps = 27/102 (26%)
Query: 17 NALLPLRDLQKLSMSDCDLSGPLDSSLTRLE-NLTVIVLGENNFSSPV------------ 63
+ LL +R L+ L +S + SG L SLT L +L + L NNFS P+
Sbjct: 359 DTLLKMRGLKVLDLSFNEFSGELPESLTNLSASLLTLDLSSNNFSGPILPNLCQNPKNTL 418
Query: 64 --------------PQTFANFKNLTTLNLRKCGLIGTFPQNI 91
P T +N L +L+L L GT P ++
Sbjct: 419 QELYLQNNGFTGKIPPTLSNCSELVSLHLSFNYLSGTIPSSL 460
Score = 34.3 bits (77), Expect = 0.084
Identities = 21/63 (33%), Positives = 31/63 (48%), Gaps = 5/63 (7%)
Query: 8 ITSRGHEWSNALLP-----LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSP 62
I + GH + +P LR L L +S L G + +++ L LT I L NN S P
Sbjct: 682 ILNLGHNDISGSIPDEVGDLRGLNILDLSSNKLDGRIPQAMSALTMLTEIDLSNNNLSGP 741
Query: 63 VPQ 65
+P+
Sbjct: 742 IPE 744
Score = 33.1 bits (74), Expect = 0.19
Identities = 16/62 (25%), Positives = 28/62 (44%)
Query: 28 LSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTF 87
L++ D+SG + + L L ++ L N +PQ + LT ++L L G
Sbjct: 683 LNLGHNDISGSIPDEVGDLRGLNILDLSSNKLDGRIPQAMSALTMLTEIDLSNNNLSGPI 742
Query: 88 PQ 89
P+
Sbjct: 743 PE 744
Score = 32.7 bits (73), Expect = 0.25
Identities = 19/62 (30%), Positives = 29/62 (46%)
Query: 28 LSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTF 87
L MS LSG + + + L ++ LG N+ S +P + + L L+L L G
Sbjct: 659 LDMSYNMLSGYIPKEIGSMPYLFILNLGHNDISGSIPDEVGDLRGLNILDLSSNKLDGRI 718
Query: 88 PQ 89
PQ
Sbjct: 719 PQ 720
Score = 31.2 bits (69), Expect = 0.72
Identities = 23/68 (33%), Positives = 29/68 (41%)
Query: 24 DLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGL 83
+L L +S LSG + SSL L L + L N +PQ K L TL L L
Sbjct: 441 ELVSLHLSFNYLSGTIPSSLGSLSKLRDLKLWLNMLEGEIPQELMYVKTLETLILDFNDL 500
Query: 84 IGTFPQNI 91
G P +
Sbjct: 501 TGEIPSGL 508
Score = 28.5 bits (62), Expect = 4.6
Identities = 18/64 (28%), Positives = 29/64 (45%), Gaps = 2/64 (3%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
LQ L +S LSG +++ L ++ + N F P+P K+L L+L +
Sbjct: 247 LQHLDISGNKLSGDFSRAISTCTELKLLNISSNQFVGPIPP--LPLKSLQYLSLAENKFT 304
Query: 85 GTFP 88
G P
Sbjct: 305 GEIP 308
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 40.8 bits (94), Expect = 0.001
Identities = 22/59 (37%), Positives = 36/59 (60%), Gaps = 4/59 (6%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRK 80
L++L+ LS+ D +GP+ +SL LE+L V+ L N+ PVP FK+ +++L K
Sbjct: 254 LKELESLSLRDNSFTGPVPASLLSLESLKVVNLTNNHLQGPVPV----FKSSVSVDLDK 308
Score = 38.1 bits (87), Expect = 0.006
Identities = 18/61 (29%), Positives = 31/61 (50%)
Query: 28 LSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTF 87
+S+ +L+G + +++L I+LG NN + +PQ NL TL++ L G
Sbjct: 367 ISLEKMELTGTISPEFGAIKSLQRIILGINNLTGMIPQELTTLPNLKTLDVSSNKLFGKV 426
Query: 88 P 88
P
Sbjct: 427 P 427
Score = 33.1 bits (74), Expect = 0.19
Identities = 22/66 (33%), Positives = 36/66 (54%), Gaps = 5/66 (7%)
Query: 25 LQKLSMSDCDLSGPLD--SSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCG 82
L+++ + SGPL S L LE+L+ L +N+F+ PVP + + ++L +NL
Sbjct: 234 LKEVWLHSNKFSGPLPDFSGLKELESLS---LRDNSFTGPVPASLLSLESLKVVNLTNNH 290
Query: 83 LIGTFP 88
L G P
Sbjct: 291 LQGPVP 296
>DR39_ARATH (Q9LVT1) Putative disease resistance protein At5g47280
Length = 623
Score = 40.8 bits (94), Expect = 0.001
Identities = 32/127 (25%), Positives = 64/127 (50%), Gaps = 10/127 (7%)
Query: 14 EWSNALLPLRDLQKLSMSDCDLSGPLDSSLTRLEN----LTVIVLGENNFSSPVPQTFAN 69
E S++++PL++L KL + C ++ D + + LT I + + + +P T
Sbjct: 426 ELSSSMIPLKNLHKLYLIICKINNSFDQTAIDIAQIFPKLTDITIDYCDDLAELPSTICG 485
Query: 70 FKNLTTLNLRKCGLIGTFPQNIFQIKS-QLLTYLTTP----TSMVFFQTTHLVNLFIALY 124
+L ++++ C I P+NI ++++ QLL P + + LV + I+ +
Sbjct: 486 ITSLNSISITNCPNIKELPKNISKLQALQLLRLYACPELKSLPVEICELPRLVYVDIS-H 544
Query: 125 CVTLSSL 131
C++LSSL
Sbjct: 545 CLSLSSL 551
>D100_ARATH (Q00874) DNA-damage-repair/toleration protein DRT100
precursor
Length = 372
Score = 40.4 bits (93), Expect = 0.001
Identities = 20/70 (28%), Positives = 38/70 (53%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L+ L ++ + +L+G + S++ +E L + L +N+ P+P+ N K L+ LNL
Sbjct: 206 LKMLSRVLLGRNELTGSIPESISGMERLADLDLSKNHIEGPIPEWMGNMKVLSLLNLDCN 265
Query: 82 GLIGTFPQNI 91
L G P ++
Sbjct: 266 SLTGPIPGSL 275
Score = 39.7 bits (91), Expect = 0.002
Identities = 22/70 (31%), Positives = 39/70 (55%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L L L++++ +SG + +SLT L L + L EN + +P F + K L+ + L +
Sbjct: 158 LSKLAVLNLAENQMSGEIPASLTSLIELKHLELTENGITGVIPADFGSLKMLSRVLLGRN 217
Query: 82 GLIGTFPQNI 91
L G+ P++I
Sbjct: 218 ELTGSIPESI 227
Score = 37.4 bits (85), Expect = 0.010
Identities = 17/72 (23%), Positives = 40/72 (54%)
Query: 18 ALLPLRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLN 77
+L L +L+ L +++ ++G + + L+ L+ ++LG N + +P++ + + L L+
Sbjct: 178 SLTSLIELKHLELTENGITGVIPADFGSLKMLSRVLLGRNELTGSIPESISGMERLADLD 237
Query: 78 LRKCGLIGTFPQ 89
L K + G P+
Sbjct: 238 LSKNHIEGPIPE 249
Score = 35.0 bits (79), Expect = 0.050
Identities = 20/76 (26%), Positives = 40/76 (52%), Gaps = 1/76 (1%)
Query: 25 LQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLI 84
L L +S + GP+ + ++ L+++ L N+ + P+P + + L NL + L
Sbjct: 233 LADLDLSKNHIEGPIPEWMGNMKVLSLLNLDCNSLTGPIPGSLLSNSGLDVANLSRNALE 292
Query: 85 GTFPQNIFQIKSQLLT 100
GT P ++F K+ L++
Sbjct: 293 GTIP-DVFGSKTYLVS 307
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 40.0 bits (92), Expect = 0.002
Identities = 22/70 (31%), Positives = 33/70 (46%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L +LQ L + +++G + L L L + L NN S P+P T K L L L
Sbjct: 91 LPNLQYLELYSNNITGTIPEQLGNLTELVSLDLYLNNLSGPIPSTLGRLKKLRFLRLNNN 150
Query: 82 GLIGTFPQNI 91
L G P+++
Sbjct: 151 SLSGEIPRSL 160
Score = 39.7 bits (91), Expect = 0.002
Identities = 23/69 (33%), Positives = 35/69 (50%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKC 81
L +L L + +LSGP+ S+L RL+ L + L N+ S +P++ L L+L
Sbjct: 115 LTELVSLDLYLNNLSGPIPSTLGRLKKLRFLRLNNNSLSGEIPRSLTAVLTLQVLDLSNN 174
Query: 82 GLIGTFPQN 90
L G P N
Sbjct: 175 PLTGDIPVN 183
Score = 39.7 bits (91), Expect = 0.002
Identities = 20/69 (28%), Positives = 35/69 (49%)
Query: 27 KLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGT 86
++ + + +LSG L L +L NL + L NN + +P+ N L +L+L L G
Sbjct: 72 RVDLGNANLSGQLVMQLGQLPNLQYLELYSNNITGTIPEQLGNLTELVSLDLYLNNLSGP 131
Query: 87 FPQNIFQIK 95
P + ++K
Sbjct: 132 IPSTLGRLK 140
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 39.7 bits (91), Expect = 0.002
Identities = 20/66 (30%), Positives = 34/66 (51%)
Query: 28 LSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLRKCGLIGTF 87
L++S +L G +D + + L ++ + L N + +P AN NLT LN+ L G
Sbjct: 419 LNISFSELRGQIDPAFSNLTSIRKLDLSGNTLTGEIPAFLANLPNLTELNVEGNKLTGIV 478
Query: 88 PQNIFQ 93
PQ + +
Sbjct: 479 PQRLHE 484
Score = 30.0 bits (66), Expect = 1.6
Identities = 18/58 (31%), Positives = 29/58 (49%)
Query: 22 LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSSPVPQTFANFKNLTTLNLR 79
L ++KL +S L+G + + L L NLT + + N + VPQ +L+LR
Sbjct: 437 LTSIRKLDLSGNTLTGEIPAFLANLPNLTELNVEGNKLTGIVPQRLHERSKNGSLSLR 494
>A32E_MOUSE (P97822) Acidic leucine-rich nuclear phosphoprotein 32
family member E (LANP-like protein) (LANP-L) (Cerebellar
postnatal development protein-1)
Length = 260
Score = 39.7 bits (91), Expect = 0.002
Identities = 31/91 (34%), Positives = 48/91 (52%), Gaps = 5/91 (5%)
Query: 16 SNALLP-LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSS-PVPQTFANFKNL 73
S A LP L L+KL +SD +SG L+ + NLT + L N + N KNL
Sbjct: 56 SLARLPSLNKLRKLELSDNIISGGLEVLAEKCPNLTYLNLSGNKIKDLSTVEALQNLKNL 115
Query: 74 TTLNLRKCGL--IGTFPQNIFQIKSQLLTYL 102
+L+L C + + + ++IF++ Q +TYL
Sbjct: 116 KSLDLFNCEITNLEDYRESIFELLQQ-ITYL 145
>A32E_HUMAN (Q9BTT0) Acidic leucine-rich nuclear phosphoprotein 32
family member E (LANP-like protein) (LANP-L)
Length = 268
Score = 39.7 bits (91), Expect = 0.002
Identities = 31/91 (34%), Positives = 48/91 (52%), Gaps = 5/91 (5%)
Query: 16 SNALLP-LRDLQKLSMSDCDLSGPLDSSLTRLENLTVIVLGENNFSS-PVPQTFANFKNL 73
S A LP L L+KL +SD +SG L+ + NLT + L N + N KNL
Sbjct: 56 SLARLPSLNKLRKLELSDNIISGGLEVLAEKCPNLTYLNLSGNKIKDLSTVEALQNLKNL 115
Query: 74 TTLNLRKCGL--IGTFPQNIFQIKSQLLTYL 102
+L+L C + + + ++IF++ Q +TYL
Sbjct: 116 KSLDLFNCEITNLEDYRESIFELLQQ-ITYL 145
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,268,892
Number of Sequences: 164201
Number of extensions: 544393
Number of successful extensions: 1536
Number of sequences better than 10.0: 89
Number of HSP's better than 10.0 without gapping: 37
Number of HSP's successfully gapped in prelim test: 52
Number of HSP's that attempted gapping in prelim test: 1291
Number of HSP's gapped (non-prelim): 280
length of query: 139
length of database: 59,974,054
effective HSP length: 99
effective length of query: 40
effective length of database: 43,718,155
effective search space: 1748726200
effective search space used: 1748726200
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144474.15


