

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144406.4 - phase: 1 /pseudo
(322 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
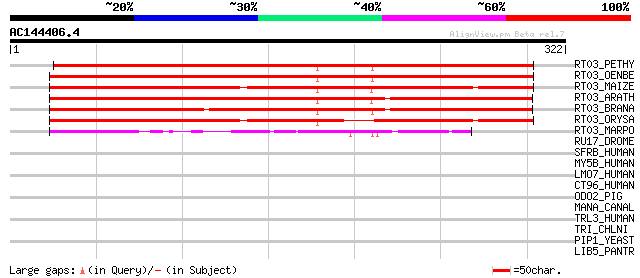
Score E
Sequences producing significant alignments: (bits) Value
RT03_PETHY (Q04716) Mitochondrial ribosomal protein S3 494 e-139
RT03_OENBE (P27754) Mitochondrial ribosomal protein S3 489 e-138
RT03_MAIZE (P27928) Mitochondrial ribosomal protein S3 479 e-135
RT03_ARATH (Q95749) Mitochondrial ribosomal protein S3 473 e-133
RT03_BRANA (P49386) Mitochondrial ribosomal protein S3 460 e-129
RT03_ORYSA (P46773) Mitochondrial ribosomal protein S3 452 e-127
RT03_MARPO (P26865) Mitochondrial ribosomal protein S3 132 2e-30
RU17_DROME (P17133) U1 small nuclear ribonucleoprotein 70 kDa (U... 35 0.33
SFRB_HUMAN (Q05519) Splicing factor arginine/serine-rich 11 (Arg... 34 0.56
MY5B_HUMAN (Q9ULV0) Myosin Vb (Myosin 5B) 32 1.6
LMO7_HUMAN (Q8WWI1) LIM domain only protein 7 (LOMP) (F-box only... 31 3.6
CT96_HUMAN (Q9NUD7) Protein C20orf96 31 3.6
ODO2_PIG (Q9N0F1) Dihydrolipoyllysine-residue succinyltransferas... 30 6.2
MANA_CANAL (P34948) Mannose-6-phosphate isomerase (EC 5.3.1.8) (... 30 6.2
TRL3_HUMAN (Q9HCF6) Long transient receptor potential channel 3 ... 30 8.1
TRI_CHLNI (Q7M3Y3) Troponin I (TnI) 30 8.1
PIP1_YEAST (P40020) Polymerase-interacting protein 1 (Factor int... 30 8.1
LIB5_PANTR (Q8MJZ7) Leukocyte immunoglobulin-like receptor subfa... 30 8.1
>RT03_PETHY (Q04716) Mitochondrial ribosomal protein S3
Length = 562
Score = 494 bits (1271), Expect = e-139
Identities = 247/284 (86%), Positives = 255/284 (88%), Gaps = 5/284 (1%)
Query: 26 YYYGKLVYQDVNLRSYFGSIRPPARLTFGFRLGRCILIHFPKRTFIHFFLPRRPRRLKRR 85
YYYGK VYQDVNLRSYFGSIRPP RLTFGFRLGRCI++HFPKRTFIHFFLPRRPRRLKRR
Sbjct: 25 YYYGKSVYQDVNLRSYFGSIRPPTRLTFGFRLGRCIILHFPKRTFIHFFLPRRPRRLKRR 84
Query: 86 DKSRPGKEKGRWWAFGKVGPIGCLHSSNKKDEERNEVRGRRAGKRVESIRLDDREKQNEI 145
+KSRP KEKGRW AFGKVGPIGCLHSS+ +EERNEVRGR AGKRVESIRLDDREKQNEI
Sbjct: 85 EKSRPVKEKGRWGAFGKVGPIGCLHSSDGTEEERNEVRGRGAGKRVESIRLDDREKQNEI 144
Query: 146 RIWPKKKQRYGYHDRSPSIKKNLSKSLRVSGA--HPKYAGAVNDIAFLIENYDSFRKTKF 203
RIWPKKKQ YGYHDRSPSIKKNLSKSLRVSGA HPKYAG NDIAFLIEN DSFRKT
Sbjct: 145 RIWPKKKQGYGYHDRSPSIKKNLSKSLRVSGAFKHPKYAGIENDIAFLIENDDSFRKTNL 204
Query: 204 FKLFFP---RSDGPTGHLFKRTLPAVRPSLNYSVMQYFFCRKNQMHFDPVVVLNHFVAPG 260
FK FFP RSD PT HL KRTLPAVRPSLNYSVMQY KN++HFDPVVVLNHFVAPG
Sbjct: 205 FKFFFPKKSRSDRPTSHLLKRTLPAVRPSLNYSVMQYLLNTKNKIHFDPVVVLNHFVAPG 264
Query: 261 VAEPSTMGGTNAQGRSLDKRIRSRIAFFVESSTSEKKCLAEAKK 304
VAEPSTMGG NAQGRSLDKRI S IAFFVE+STSEKKCLAEAKK
Sbjct: 265 VAEPSTMGGANAQGRSLDKRIASCIAFFVENSTSEKKCLAEAKK 308
>RT03_OENBE (P27754) Mitochondrial ribosomal protein S3
Length = 550
Score = 489 bits (1260), Expect = e-138
Identities = 244/286 (85%), Positives = 253/286 (88%), Gaps = 5/286 (1%)
Query: 24 SIYYYGKLVYQDVNLRSYFGSIRPPARLTFGFRLGRCILIHFPKRTFIHFFLPRRPRRLK 83
S YYYGK VYQDVNLRSYFGSIRPP RLTFGFRLGRCI++HFPKRTFIHFFLPRRPRRLK
Sbjct: 24 SDYYYGKSVYQDVNLRSYFGSIRPPTRLTFGFRLGRCIILHFPKRTFIHFFLPRRPRRLK 83
Query: 84 RRDKSRPGKEKGRWWAFGKVGPIGCLHSSNKKDEERNEVRGRRAGKRVESIRLDDREKQN 143
R KSRPGKEK RWW FGKVGPIGCLHS++ +EERNEV GR AGKRVESIRLDDREKQN
Sbjct: 84 RGKKSRPGKEKARWWEFGKVGPIGCLHSNDDTEEERNEVGGRGAGKRVESIRLDDREKQN 143
Query: 144 EIRIWPKKKQRYGYHDRSPSIKKNLSKSLRVSGA--HPKYAGAVNDIAFLIENYDSFRKT 201
EIRIWPKKKQ YGYHDRSPSIKKNLSKSLRVSGA HPKYAG +NDIAFLIEN DSFRKT
Sbjct: 144 EIRIWPKKKQGYGYHDRSPSIKKNLSKSLRVSGAFKHPKYAGVLNDIAFLIENDDSFRKT 203
Query: 202 KFFKLFFP---RSDGPTGHLFKRTLPAVRPSLNYSVMQYFFCRKNQMHFDPVVVLNHFVA 258
K FK FFP RSDGPT HL KRT PAVRPSLNYSVMQY+ KNQMHFDP+VVLNHFVA
Sbjct: 204 KLFKFFFPKKSRSDGPTSHLLKRTPPAVRPSLNYSVMQYYLNTKNQMHFDPLVVLNHFVA 263
Query: 259 PGVAEPSTMGGTNAQGRSLDKRIRSRIAFFVESSTSEKKCLAEAKK 304
PGVAEPSTMGG N QGRS + RIRSRIAFFVESSTSEKK LAE KK
Sbjct: 264 PGVAEPSTMGGANRQGRSNELRIRSRIAFFVESSTSEKKSLAEDKK 309
>RT03_MAIZE (P27928) Mitochondrial ribosomal protein S3
Length = 559
Score = 479 bits (1234), Expect = e-135
Identities = 243/287 (84%), Positives = 252/287 (87%), Gaps = 11/287 (3%)
Query: 24 SIYYYGKLVYQDVNLRSYFGSIRPPARLTFGFRLGRCILIHFPKRTFIHFFLPRRPRRLK 83
S YYYGK +YQDVNLRSYF SIRPP RLTFGFRLGRCI++HFPKRTFIHFFLPRRP RLK
Sbjct: 24 SDYYYGKSLYQDVNLRSYFSSIRPPTRLTFGFRLGRCIILHFPKRTFIHFFLPRRPLRLK 83
Query: 84 RRDKSRPGKEKGRWWAFGKVGPIGCLHSSNKKDEERNEVRGRRAGKRVESIRLDDREKQN 143
RRDKSRPGK+KGRWWAFGKVGPIGCLHSS +EERNEVRGR AGKRVESI DREKQN
Sbjct: 84 RRDKSRPGKDKGRWWAFGKVGPIGCLHSSEGTEEERNEVRGRGAGKRVESI---DREKQN 140
Query: 144 EIRIWPKKKQRYGYHDRSPSIKKNLSKSLRVSGA--HPKYAGAVNDIAFLIENYDSFRKT 201
EIRIWPKK QRYGYHDR+PS KKN SKSLRVSGA HPKYAG VNDIAFLIEN DSF KT
Sbjct: 141 EIRIWPKKMQRYGYHDRTPSRKKNFSKSLRVSGAFKHPKYAGVVNDIAFLIENDDSFIKT 200
Query: 202 KFFKLFF----PRSDGPTGHLFKRTLPAVRPSLNYSVMQYFFCRKNQMHFDPVVVLNHFV 257
K FK FF RSDGPT HL KRTLPAVRPSLNYSVMQYFF KN+MHFDPVVVLNHFV
Sbjct: 201 KLFKFFFLPKKSRSDGPTSHLLKRTLPAVRPSLNYSVMQYFFNTKNKMHFDPVVVLNHFV 260
Query: 258 APGVAEPSTMGGTNAQGRSLDKRIRSRIAFFVESSTSEKKCLAEAKK 304
APGVAEPSTMGG A+G SLDKRIRSRIAFFVESSTS+KKCLA AKK
Sbjct: 261 APGVAEPSTMGG--AKGGSLDKRIRSRIAFFVESSTSDKKCLARAKK 305
>RT03_ARATH (Q95749) Mitochondrial ribosomal protein S3
Length = 556
Score = 473 bits (1216), Expect = e-133
Identities = 239/286 (83%), Positives = 252/286 (87%), Gaps = 8/286 (2%)
Query: 24 SIYYYGKLVYQDVNLRSYFGSIRPPARLTFGFRLGRCILIHFPKRTFIHFFLPRRPRRLK 83
S YYYGK VYQDVNLRSYFGSIRPP RLTFGFRLGRCI++HFPKRTFIHFFLPRRPRRLK
Sbjct: 24 SEYYYGKFVYQDVNLRSYFGSIRPPTRLTFGFRLGRCIILHFPKRTFIHFFLPRRPRRLK 83
Query: 84 RRDKSRPGKEKGRWW-AFGKVGPIGCLHSSNKKDEERNEVRGRRAGKRVESIRLDDREKQ 142
RR+K+RPGKEKGRWW FGK GPI CLHSS+ +EERNEVRGR A KRVESIRLDDR+KQ
Sbjct: 84 RREKTRPGKEKGRWWTTFGKAGPIECLHSSDDTEEERNEVRGRGARKRVESIRLDDRKKQ 143
Query: 143 NEIRIWPKKKQRYGYHDRSPSIKKNLSKSLRVSGA--HPKYAGAVNDIAFLIENYDSFRK 200
NEIR WPKKKQRYGYHDR PSIKKNLSK LR+SGA HPKYAG VNDIAFLIEN DSF+K
Sbjct: 144 NEIRGWPKKKQRYGYHDRLPSIKKNLSKLLRISGAFKHPKYAGVVNDIAFLIENDDSFKK 203
Query: 201 TKFFKLFF---PRSDGPTGHLFKRTLPAVRPSLNYSVMQYFFCRKNQMHFDPVVVLNHFV 257
TK FKLFF RSDGPT +L RTLPAVRPSLN+ VMQYFF KNQ++FDPVVVLNHFV
Sbjct: 204 TKLFKLFFQNKSRSDGPTSYL--RTLPAVRPSLNFLVMQYFFNTKNQINFDPVVVLNHFV 261
Query: 258 APGVAEPSTMGGTNAQGRSLDKRIRSRIAFFVESSTSEKKCLAEAK 303
APG AEPSTMG NAQGRSL KRIRSRIAFFVES TSEKKCLAEAK
Sbjct: 262 APGAAEPSTMGRANAQGRSLQKRIRSRIAFFVESLTSEKKCLAEAK 307
>RT03_BRANA (P49386) Mitochondrial ribosomal protein S3
Length = 555
Score = 460 bits (1183), Expect = e-129
Identities = 237/287 (82%), Positives = 247/287 (85%), Gaps = 11/287 (3%)
Query: 24 SIYYYGKLVYQDVNLRSYFGSIRPPARLTFGFRLGRCILIHFPKRTFIHFFLPRRPRRLK 83
S YYYGK VYQDVNLRSYFGSIRPP RLTFGFRLGRCIL+HFPKRTFIHFFLPRRPRRLK
Sbjct: 24 SEYYYGKFVYQDVNLRSYFGSIRPPTRLTFGFRLGRCILLHFPKRTFIHFFLPRRPRRLK 83
Query: 84 RRDKSRPGKEKGRWWAF-GKVGPIGCLHSSNKKDEERNEVRGRRAGKRVESIRLDDREKQ 142
RR+K+RPGKEKGRWW GK GPIGCL +EERNEVRGR A KRVESIRLDDR+KQ
Sbjct: 84 RREKTRPGKEKGRWWTTPGKAGPIGCLRDDT--EEERNEVRGRGARKRVESIRLDDRKKQ 141
Query: 143 NEIRIWPKKKQRYGYHDRSPSIKKNLSKSLRVSGA--HPKYAGAVNDIAFLIENYDSFRK 200
NEIR WPKKKQRYGYHDR+PSIKKNLSKSLR+SGA HPKY G VNDIAFLIEN DSFRK
Sbjct: 142 NEIRGWPKKKQRYGYHDRTPSIKKNLSKSLRISGAFKHPKYGGVVNDIAFLIENDDSFRK 201
Query: 201 TKFFKLFFP---RSDGPTGHLFKRTLPAVRPSLNYSVMQYFFCRKNQMHFDPVVVLNHFV 257
TK FK FFP RSDGPT +L RTLPAV PSLN+ VMQYFF KNQM+FDPVVVLNHFV
Sbjct: 202 TKLFKFFFPKKSRSDGPTSYL--RTLPAVGPSLNFLVMQYFFNTKNQMNFDPVVVLNHFV 259
Query: 258 APGVAEPSTMGGTNAQG-RSLDKRIRSRIAFFVESSTSEKKCLAEAK 303
APG AEPSTMG N G RSL KRIRSRIAFFVESSTSEKKCLAEAK
Sbjct: 260 APGAAEPSTMGRANGTGDRSLQKRIRSRIAFFVESSTSEKKCLAEAK 306
>RT03_ORYSA (P46773) Mitochondrial ribosomal protein S3
Length = 538
Score = 452 bits (1163), Expect = e-127
Identities = 232/283 (81%), Positives = 239/283 (83%), Gaps = 24/283 (8%)
Query: 24 SIYYYGKLVYQDVNLRSYFGSIRPPARLTFGFRLGRCILIHFPKRTFIHFFLPRRPRRLK 83
S YYYGK +YQDVNLRSYF SIRPP LTFGFRLGRCI++HFPKRTFIHFFLPRRP RLK
Sbjct: 24 SDYYYGKSLYQDVNLRSYFSSIRPPTILTFGFRLGRCIILHFPKRTFIHFFLPRRPLRLK 83
Query: 84 RRDKSRPGKEKGRWWAFGKVGPIGCLHSSNKKDEERNEVRGRRAGKRVESIRLDDREKQN 143
RRDKSRPGK+KGRWWAFGKVGPIGCLHSS +EERNEVRGR AGKRVESI DREKQN
Sbjct: 84 RRDKSRPGKDKGRWWAFGKVGPIGCLHSSEGTEEERNEVRGRGAGKRVESI---DREKQN 140
Query: 144 EIRIWPKKKQRYGYHDRSPSIKKNLSKSLRVSGA--HPKYAGAVNDIAFLIENYDSFRKT 201
EIRIWPKK QRYGYHDRSPS KKN SKSLRVSGA HPKYAG VNDIAFLIEN
Sbjct: 141 EIRIWPKKMQRYGYHDRSPSRKKNFSKSLRVSGAFKHPKYAGVVNDIAFLIEN------- 193
Query: 202 KFFKLFFPRSDGPTGHLFKRTLPAVRPSLNYSVMQYFFCRKNQMHFDPVVVLNHFVAPGV 261
DGPT HL KRTLPAVRPSLNYSVMQYFF KN+MHFDPVVVLNHFVAPGV
Sbjct: 194 ----------DGPTSHLLKRTLPAVRPSLNYSVMQYFFNTKNKMHFDPVVVLNHFVAPGV 243
Query: 262 AEPSTMGGTNAQGRSLDKRIRSRIAFFVESSTSEKKCLAEAKK 304
AEPSTMGG A+G SLDKRIRSRIAFFVESSTSEKKCLA AKK
Sbjct: 244 AEPSTMGG--AKGGSLDKRIRSRIAFFVESSTSEKKCLARAKK 284
>RT03_MARPO (P26865) Mitochondrial ribosomal protein S3
Length = 430
Score = 132 bits (331), Expect = 2e-30
Identities = 105/259 (40%), Positives = 131/259 (50%), Gaps = 56/259 (21%)
Query: 24 SIYYYGKLVYQDVNLRSYFGSIRPPARLTFGFRLGRCILIHFPKRTFIHFFLPRRPRRLK 83
S YYYGKL+YQD+N R YFGSIRPP TFGFRLGRCI+ HFPKRTFIH F L
Sbjct: 24 SDYYYGKLLYQDLNFRDYFGSIRPPTGNTFGFRLGRCIIHHFPKRTFIHVFF------LD 77
Query: 84 RRDKSRPGKEKGRWWAFGKVGPIGCLHSSNKKDEERNEVRGRRAGKRVESIRLDDREKQN 143
R +SR +G +G + S K + I + +++N
Sbjct: 78 RLSQSR---HQG----------LGAIPSV----------------KLIRRINDNTVKQRN 108
Query: 144 EIRIWPKKKQRYGYHDRSPSIKKNLSKSLRVSGAHPKYAGAVNDIAFLIENYD--SFRKT 201
E+ IWPKK RY YHDR PSI+K + + LRVS I EN D + +
Sbjct: 109 EVGIWPKK--RYEYHDRLPSIQK-IDQLLRVSDWMADIHSTFQSIWPKDENDDRRASEER 165
Query: 202 KFFKLFFP------RSD------GPTGHLFKRTLPAVRPSLNYSVMQYFFCRKNQMHFDP 249
F F P R++ G G F T R L+Y VMQYFF KNQ+ FDP
Sbjct: 166 YAFSRFAPSILVAVRAEKKKAIFGSEGDFFGFT---GRAFLDYFVMQYFFNLKNQIQFDP 222
Query: 250 VVVLNHFVAPGVAEPSTMG 268
+V + VA GVA+ S +G
Sbjct: 223 MVNRSP-VAQGVAKTSMIG 240
>RU17_DROME (P17133) U1 small nuclear ribonucleoprotein 70 kDa (U1
snRNP 70 kDa) (snRNP70)
Length = 448
Score = 34.7 bits (78), Expect = 0.33
Identities = 27/97 (27%), Positives = 44/97 (44%), Gaps = 13/97 (13%)
Query: 77 RRPRRLKRRDKSRPGKEKGRWWAFGKVGPIGCLHSSNKKDEERNEVRGRRAGKRVESIRL 136
RR R +RRD+ R ++GR G + S+ + E R RRAG R
Sbjct: 256 RRSRSRERRDRER---DRGR----------GAVASNGRSRSRSRERRKRRAGSRERYDEF 302
Query: 137 DDREKQNEIRIWPKKKQRYGYHDRSPSIKKNLSKSLR 173
D R++++ R + ++R RS S ++ S+ R
Sbjct: 303 DRRDRRDRERERDRDREREKKKKRSKSRERESSRERR 339
>SFRB_HUMAN (Q05519) Splicing factor arginine/serine-rich 11
(Arginine-rich 54 kDa nuclear protein) (p54)
Length = 484
Score = 33.9 bits (76), Expect = 0.56
Identities = 31/116 (26%), Positives = 49/116 (41%), Gaps = 14/116 (12%)
Query: 66 PKRTFIHFFLPRRPRRLKRRDKSRPGKEKGRWWAFGKVGPIGCLHSSNKKDEERNEVRGR 125
PKR PRR ++ K++DK + R + K KD+E++ R
Sbjct: 361 PKRKLSRSPSPRRHKKEKKKDKDKERSRDERERSTSK--------KKKSKDKEKDRERKS 412
Query: 126 RAGKRVESIRLD-DREKQNEIRIWPKKKQRYGYHDRSP-----SIKKNLSKSLRVS 175
+ K V+ + D D E+Q KK+++ SP S++K SLR S
Sbjct: 413 ESDKDVKQVTRDYDEEEQGYDSEKEKKEEKKPIETGSPKTKECSVEKGTGDSLRES 468
>MY5B_HUMAN (Q9ULV0) Myosin Vb (Myosin 5B)
Length = 1849
Score = 32.3 bits (72), Expect = 1.6
Identities = 21/82 (25%), Positives = 39/82 (46%), Gaps = 4/82 (4%)
Query: 118 ERNEVRGRRAGKRVESIRLDDREKQNEIRIWPKKKQRYGYHDRSPSIKKNLSKSLRVSGA 177
E++E+R R A E+ L D ++Q +I + K + + S+K+NL +
Sbjct: 1009 EKDELRKRVADLEQENALLKDEKEQLNNQILCQSKDEFAQN----SVKENLLMKKELEEE 1064
Query: 178 HPKYAGAVNDIAFLIENYDSFR 199
+Y V + + L + YD+ R
Sbjct: 1065 RSRYQNLVKEYSQLEQRYDNLR 1086
>LMO7_HUMAN (Q8WWI1) LIM domain only protein 7 (LOMP) (F-box only
protein 20)
Length = 1683
Score = 31.2 bits (69), Expect = 3.6
Identities = 17/52 (32%), Positives = 28/52 (53%), Gaps = 7/52 (13%)
Query: 114 KKDEERNEV------RGRRAGKRVESIRLDDREKQNEIRIWPKKKQRYGYHD 159
KK EER E+ + +R+ K + + L DRE QN+ P +++ Y + D
Sbjct: 758 KKKEEREEIEKQALEKSKRSSKTFKEM-LQDRESQNQKSTVPSRRRMYSFDD 808
>CT96_HUMAN (Q9NUD7) Protein C20orf96
Length = 363
Score = 31.2 bits (69), Expect = 3.6
Identities = 14/37 (37%), Positives = 18/37 (47%)
Query: 62 LIHFPKRTFIHFFLPRRPRRLKRRDKSRPGKEKGRWW 98
+ HF T + P+ PR L RR K PGK + W
Sbjct: 63 VFHFKPTTVVTSCQPKNPRELHRRRKLDPGKMHAKIW 99
>ODO2_PIG (Q9N0F1) Dihydrolipoyllysine-residue succinyltransferase
component of 2-oxoglutarate dehydrogenase complex,
mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide
succinyltransferase component of 2-oxoglutarate
dehydrogenase complex) (E2) (E2K)
Length = 455
Score = 30.4 bits (67), Expect = 6.2
Identities = 16/38 (42%), Positives = 20/38 (52%)
Query: 249 PVVVLNHFVAPGVAEPSTMGGTNAQGRSLDKRIRSRIA 286
PV + AP VAEP + G A+ R R+R RIA
Sbjct: 200 PVSAVKPTAAPPVAEPGAVKGLRAEHREKMNRMRQRIA 237
>MANA_CANAL (P34948) Mannose-6-phosphate isomerase (EC 5.3.1.8)
(Phosphomannose isomerase) (PMI) (Phosphohexomutase)
Length = 440
Score = 30.4 bits (67), Expect = 6.2
Identities = 32/127 (25%), Positives = 50/127 (39%), Gaps = 20/127 (15%)
Query: 175 SGAHPKYAGAVNDIAFLIENYDSFRKTKFFKLFFPRSDGPTGHLFKRTLPAVRPSLNYSV 234
+G PK+ N + L +Y+S K K FPRS G +++ P +SV
Sbjct: 304 AGFTPKFKDVKNLVEMLTYSYESVEKQKMPLQEFPRSKGDA----VKSVLYDPPIAEFSV 359
Query: 235 MQYFFCRKNQMHFDPVVVLNHFVAPGVAEPSTMGGTNAQGR-------SLDKRIRSRIAF 287
+Q F + V G+ PS + TN +G S ++I + F
Sbjct: 360 LQTIFDKSKG---------GKQVIEGLNGPSIVIATNGKGTIQITGDDSTKQKIDTGYVF 410
Query: 288 FVESSTS 294
FV +S
Sbjct: 411 FVAPGSS 417
>TRL3_HUMAN (Q9HCF6) Long transient receptor potential channel 3
(LTrpC3) (Fragment)
Length = 1017
Score = 30.0 bits (66), Expect = 8.1
Identities = 23/71 (32%), Positives = 31/71 (43%), Gaps = 10/71 (14%)
Query: 102 KVGPIGCLH-------SSNKKDEERNEVRGRRAGKRVESIRLDDREK--QNEIRIWPKKK 152
KV + C H S D E NE +GRRA + S D+ E+ N I + PK +
Sbjct: 885 KVEDLTCCHPEREAELSHPSSDSEENEAKGRRATIAISSQEGDNSERTLSNNITV-PKIE 943
Query: 153 QRYGYHDRSPS 163
+ Y PS
Sbjct: 944 RANSYSAEEPS 954
>TRI_CHLNI (Q7M3Y3) Troponin I (TnI)
Length = 292
Score = 30.0 bits (66), Expect = 8.1
Identities = 21/69 (30%), Positives = 35/69 (50%), Gaps = 5/69 (7%)
Query: 114 KKDEERNEVRGRRAGKRVESIRLD--DREKQNEIRIWP---KKKQRYGYHDRSPSIKKNL 168
+++ +R E RR + E +RL+ ++E++ E R KKK++ G SP KK L
Sbjct: 93 EEERQRREEEQRRQQEEEERLRLEREEQEREEEARRMAEEQKKKKKKGLGGLSPEKKKML 152
Query: 169 SKSLRVSGA 177
K + A
Sbjct: 153 KKLIMQKAA 161
>PIP1_YEAST (P40020) Polymerase-interacting protein 1 (Factor
interacting with REF)
Length = 925
Score = 30.0 bits (66), Expect = 8.1
Identities = 33/132 (25%), Positives = 54/132 (40%), Gaps = 14/132 (10%)
Query: 67 KRTFIHFFLPRRPRRLKRRDKSRPGKEKGRWWAFGKVGPIGCLHSSNKKDEERNEVRGRR 126
+ T H F+ + R +K P ++ + K P SN+ D + N + +R
Sbjct: 749 RSTLEHNFIEEK-RSIKNLGHGPPSQKNNYSFPRNKNTP------SNRHDLDFNTIYEKR 801
Query: 127 AGKRVESIRLDDRE----KQNEIRIWPKKKQRYGYHDRSPSIKKNLSKSLRVSGAHPKYA 182
GK VE I LD+ E K N+I + + + ++ KK LS + G K A
Sbjct: 802 DGKMVEVILLDEDEDVGLKNNDI---SRTRVCHAQKAKNEQQKKRLSHCNEILGMCDKTA 858
Query: 183 GAVNDIAFLIEN 194
I + + N
Sbjct: 859 DDAKRIIYQLVN 870
>LIB5_PANTR (Q8MJZ7) Leukocyte immunoglobulin-like receptor
subfamily B member 5 precursor (Leukocyte
immunoglobulin-like receptor 8) (LIR-8)
Length = 643
Score = 30.0 bits (66), Expect = 8.1
Identities = 24/79 (30%), Positives = 38/79 (47%), Gaps = 6/79 (7%)
Query: 213 GPTGHLFKRTLPAVRPSLNYSVMQYFFCRKNQMHFDPVVVLNHFVAPGVA-EPSTM---G 268
GP+ LF + V PS + Y++ RKN + L + PGV+ +PS + G
Sbjct: 174 GPSRALFP--VGPVTPSFRWRFRCYYYYRKNPQVWSHPSDLLEILVPGVSRKPSLLIPQG 231
Query: 269 GTNAQGRSLDKRIRSRIAF 287
A+G SL + RS + +
Sbjct: 232 SVVARGGSLTLQCRSDVGY 250
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.140 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,220,708
Number of Sequences: 164201
Number of extensions: 1687286
Number of successful extensions: 3848
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 3811
Number of HSP's gapped (non-prelim): 21
length of query: 322
length of database: 59,974,054
effective HSP length: 110
effective length of query: 212
effective length of database: 41,911,944
effective search space: 8885332128
effective search space used: 8885332128
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 66 (30.0 bits)
Medicago: description of AC144406.4


