

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144389.8 - phase: 0
(468 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
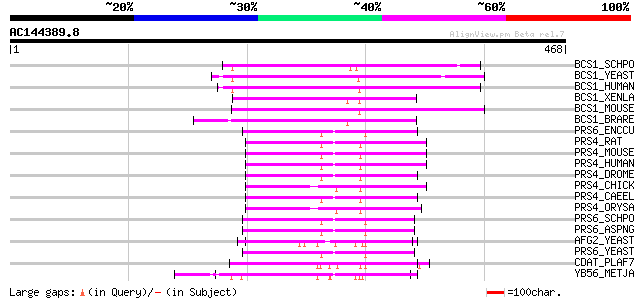
Score E
Sequences producing significant alignments: (bits) Value
BCS1_SCHPO (Q9P6Q3) Probable mitochondrial chaperone BCS1 (BCS1-... 122 2e-27
BCS1_YEAST (P32839) Mitochondrial chaperone BCS1 114 7e-25
BCS1_HUMAN (Q9Y276) Mitochondrial chaperone BCS1 (BCS1-like prot... 105 2e-22
BCS1_XENLA (Q7ZTL7) Mitochondrial chaperone BCS1 (BCS1-like prot... 105 2e-22
BCS1_MOUSE (Q9CZP5) Mitochondrial chaperone BCS1 (BCS1-like prot... 105 3e-22
BCS1_BRARE (Q7ZV60) Mitochondrial chaperone BCS1 (BCS1-like prot... 104 6e-22
PRS6_ENCCU (Q8SQI9) 26S protease regulatory subunit 6B homolog 62 2e-09
PRS4_RAT (P62193) 26S protease regulatory subunit 4 (P26s4) (Pro... 61 7e-09
PRS4_MOUSE (P62192) 26S protease regulatory subunit 4 (P26s4) (P... 61 7e-09
PRS4_HUMAN (P62191) 26S protease regulatory subunit 4 (P26s4) (P... 61 7e-09
PRS4_DROME (P48601) 26S protease regulatory subunit 4 (P26s4) 60 9e-09
PRS4_CHICK (Q90732) 26S protease regulatory subunit 4 (P26s4) (P... 60 9e-09
PRS4_CAEEL (O16368) Probable 26S protease regulatory subunit 4 60 9e-09
PRS4_ORYSA (P46466) 26S protease regulatory subunit 4 homolog (T... 57 1e-07
PRS6_SCHPO (O74894) 26S protease regulatory subunit 6B homolog 56 2e-07
PRS6_ASPNG (P78578) 26S protease regulatory subunit 6B homolog 56 2e-07
AFG2_YEAST (P32794) AFG2 protein 56 2e-07
PRS6_YEAST (P33298) 26S protease regulatory subunit 6B homolog (... 55 3e-07
CDAT_PLAF7 (P46468) Putative cell division cycle ATPase 55 3e-07
YB56_METJA (Q58556) Cell division cycle protein 48 homolog MJ1156 55 4e-07
>BCS1_SCHPO (Q9P6Q3) Probable mitochondrial chaperone BCS1
(BCS1-like protein)
Length = 449
Score = 122 bits (307), Expect = 2e-27
Identities = 71/229 (31%), Positives = 122/229 (53%), Gaps = 13/229 (5%)
Query: 180 PFTHPST---FETITMETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKS 236
PF HP + ++ +E+++K + D+ FL+ Q+Y G ++R +LLYG G+GK+
Sbjct: 197 PFGHPRSKRMLSSVVLESNVKKMITDDVHDFLRNSQWYDTRGIPYRRGYLLYGPPGSGKT 256
Query: 237 SFIAAMANFLSYDVYYIDLSRIS-TDSDLKSILLQTAPKSIIVVEDLDRYL--TEKSS-- 291
SF+ A+A L YD+ ++L+ TD L +L PK+++++ED+D E+S
Sbjct: 257 SFLYALAGELDYDICVLNLAEKGLTDDRLNHLLSNVPPKAVVLLEDVDSAFQGRERSGEV 316
Query: 292 ---TTVTSSGILNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSF 348
VT SG+LN +DG+ S +ER++ T N E +DP L+RPGRVDV +
Sbjct: 317 GFHANVTFSGLLNALDGVTSSDERIIFMTTNHPEKLDPALVRPGRVDVKAYLGNATPEQV 376
Query: 349 KTLASNYLGVKDHKLFPQVQEIFENGASLSPAEIGELMIANRNSPSRAI 397
+ + + + G + S+ A + L + N++SP+ A+
Sbjct: 377 REMFTRFYGHSPEMADDLSDIVCPKNTSM--ASLQGLFVMNKSSPADAV 423
>BCS1_YEAST (P32839) Mitochondrial chaperone BCS1
Length = 456
Score = 114 bits (284), Expect = 7e-25
Identities = 76/244 (31%), Positives = 129/244 (52%), Gaps = 18/244 (7%)
Query: 171 SDFGP-WRFVPFTHPST---FETITMETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFL 226
+ FGP WR F P ++ +++ +K + D+ F+K ++Y G ++R +L
Sbjct: 207 TSFGPEWR--KFGQPKAKRMLPSVILDSGIKEGILDDVYDFMKNGKWYSDRGIPYRRGYL 264
Query: 227 LYGSSGTGKSSFIAAMANFLSYDVYYIDLSRIS-TDSDLKSILLQTAPKSIIVVEDLDRY 285
LYG G+GK+SFI A+A L Y++ ++LS + TD L ++ +SI+++ED+D
Sbjct: 265 LYGPPGSGKTSFIQALAGELDYNICILNLSENNLTDDRLNHLMNNMPERSILLLEDIDAA 324
Query: 286 LTEKSST-------TVTSSGILNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHI 338
++S T +VT SG+LN +DG+ S EE + T N E +D ++RPGR+D +
Sbjct: 325 FNKRSQTGEQGFHSSVTFSGLLNALDGVTSSEETITFMTTNHPEKLDAAIMRPGRIDYKV 384
Query: 339 HF-PLCDFSSFKTLASNYLGVKD-HKLFPQVQEIFENGASLSPAEIGELMIANRNSPSRA 396
+ K Y G D K F V + E ++S A++ L + N+++P A
Sbjct: 385 FVGNATPYQVEKMFMKFYPGETDICKKF--VNSVKELDITVSTAQLQGLFVMNKDAPHDA 442
Query: 397 IKTV 400
+K V
Sbjct: 443 LKMV 446
>BCS1_HUMAN (Q9Y276) Mitochondrial chaperone BCS1 (BCS1-like
protein) (H-BCS1)
Length = 419
Score = 105 bits (263), Expect = 2e-22
Identities = 68/238 (28%), Positives = 116/238 (48%), Gaps = 18/238 (7%)
Query: 176 WRFVPFTHPST---FETITMETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSG 232
WR PF +P ++ ++ L +R+ D++ F+ ++Y G ++R +LLYG G
Sbjct: 176 WR--PFGYPRRRRPLNSVVLQQGLADRIVRDVQEFIDNPKWYTDRGIPYRRGYLLYGPPG 233
Query: 233 TGKSSFIAAMANFLSYDVYYIDLSRISTDSDLKSILLQTAP-KSIIVVEDLDRYLTEKSS 291
GKSSFI A+A L + + + L+ S D + LL AP +S++++ED+D +
Sbjct: 234 CGKSSFITALAGELEHSICLLSLTDSSLSDDRLNHLLSVAPQQSLVLLEDVDAAFLSRDL 293
Query: 292 TT-----------VTSSGILNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHF 340
+T SG+LN +DG+ S E R++ T N + +DP L+RPGRVD+ +
Sbjct: 294 AVENPVKYQGLGRLTFSGLLNALDGVASTEARIVFMTTNHVDRLDPALIRPGRVDLKEYV 353
Query: 341 PLCDFSSFKTLASNYLGVKDHKLFPQVQE-IFENGASLSPAEIGELMIANRNSPSRAI 397
C + + + L E + +SPA++ + +N P AI
Sbjct: 354 GYCSHWQLTQMFQRFYPGQAPSLAENFAEHVLRATNQISPAQVQGYFMLYKNDPVGAI 411
>BCS1_XENLA (Q7ZTL7) Mitochondrial chaperone BCS1 (BCS1-like
protein)
Length = 419
Score = 105 bits (262), Expect = 2e-22
Identities = 61/167 (36%), Positives = 97/167 (57%), Gaps = 12/167 (7%)
Query: 189 TITMETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSY 248
++ +E + ++ D++ F++ ++Y G ++R +LLYG G GKSSFI A+A L Y
Sbjct: 190 SVVLEQGISEKIVQDVKGFIENPKWYSDRGIPYRRGYLLYGPPGCGKSSFITALAGELEY 249
Query: 249 DVYYIDLSRISTDSDLKSILLQTAP-KSIIVVEDLD-----RYLTEKSSTT------VTS 296
+ + LS S D + LL AP +SII++ED+D R L +++ T +T
Sbjct: 250 SICLMSLSDSSLSDDRLNHLLSVAPQQSIILLEDVDAAFVSRDLNKQNPTAYQGMGRLTF 309
Query: 297 SGILNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLC 343
SG+LN +DG+ S E R++ T N + +DP L+RPGRVDV + C
Sbjct: 310 SGLLNALDGVASTEARIVFMTTNHIDRLDPALIRPGRVDVKQYVGHC 356
>BCS1_MOUSE (Q9CZP5) Mitochondrial chaperone BCS1 (BCS1-like
protein)
Length = 418
Score = 105 bits (261), Expect = 3e-22
Identities = 62/226 (27%), Positives = 113/226 (49%), Gaps = 13/226 (5%)
Query: 188 ETITMETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLS 247
+++ ++ L +R+ D+ F+ ++Y G ++R +LLYG G GKSSFI A+A L
Sbjct: 189 DSVVLQQGLADRIVKDIREFIDNPKWYIDRGIPYRRGYLLYGPPGCGKSSFITALAGELE 248
Query: 248 YDVYYIDLSRISTDSDLKSILLQTAP-KSIIVVEDLDRYLTEKSSTT-----------VT 295
+ + + L+ S D + LL AP +S++++ED+D + +T
Sbjct: 249 HSICLLSLTDSSLSDDRLNHLLSVAPQQSLVLLEDVDAAFLSRDLAVENPIKYQGLGRLT 308
Query: 296 SSGILNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNY 355
SG+LN +DG+ S E R++ T N + +DP L+RPGRVD+ + C + +
Sbjct: 309 FSGLLNALDGVASTEARIVFMTTNYIDRLDPALIRPGRVDLKEYVGYCSHWQLTQMFQRF 368
Query: 356 LGVKDHKLFPQVQE-IFENGASLSPAEIGELMIANRNSPSRAIKTV 400
+ L E + + + +SPA++ + +N P A+ +
Sbjct: 369 YPGQAPSLAENFAEHVLKATSEISPAQVQGYFMLYKNDPMGAVHNI 414
>BCS1_BRARE (Q7ZV60) Mitochondrial chaperone BCS1 (BCS1-like
protein)
Length = 420
Score = 104 bits (259), Expect = 6e-22
Identities = 64/201 (31%), Positives = 107/201 (52%), Gaps = 15/201 (7%)
Query: 156 IEKQGNRDLRFYMNASDFGPWRFVPFTHPSTFETITMETDLKNRVKSDLESFLKGKQYYH 215
++++ R + + +++ P+ F P ++ +E+ + R+ D++ F+ ++Y
Sbjct: 159 LKQEEGRTVMYTAMGAEWRPFGFPRRRRP--LSSVVLESGVAERIVDDVKEFIGNPKWYT 216
Query: 216 RLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTDSDLKSILLQTAP-K 274
G ++R +LLYG G GKSSFI A+A L Y + + LS S D + LL AP +
Sbjct: 217 DRGIPYRRGYLLYGPPGCGKSSFITALAGELGYSICLMSLSDRSLSDDRLNHLLSVAPQQ 276
Query: 275 SIIVVEDLD------------RYLTEKSSTTVTSSGILNFMDGIWSGEERVMVFTMNSKE 322
SII++ED+D L + +T SG+LN +DG+ S E R++ T N E
Sbjct: 277 SIILLEDVDAAFVSRELLPTENPLAYQGMGRLTFSGLLNALDGVASSEARIVFMTTNFIE 336
Query: 323 NVDPNLLRPGRVDVHIHFPLC 343
+DP L+RPGRVD+ + C
Sbjct: 337 RLDPALVRPGRVDLKQYVGHC 357
>PRS6_ENCCU (Q8SQI9) 26S protease regulatory subunit 6B homolog
Length = 387
Score = 62.4 bits (150), Expect = 2e-09
Identities = 45/165 (27%), Positives = 74/165 (44%), Gaps = 18/165 (10%)
Query: 197 KNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLS 256
K +K +E L Y ++G + LLYG GTGK+ + A+AN ++ S
Sbjct: 143 KQEIKETVELPLLQSDLYRQIGIDPPQGVLLYGPPGTGKTMLVKAVANHTKATFIRVNGS 202
Query: 257 RISTD---------SDLKSILLQTAPKSIIVVEDLDRYLTEKSSTTVTSSG--------I 299
D+ + + AP SI+ ++++D T++ + ++ +
Sbjct: 203 EFVQKYLGEGPRMVRDVFRLAREKAP-SIVFIDEVDSIATKRFDASTSADREVQRVLIEL 261
Query: 300 LNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCD 344
LN MDG ++ N + +DP LLRPGR+D I FPL D
Sbjct: 262 LNQMDGFDPAANVKVIMATNRADTIDPALLRPGRLDRKIEFPLPD 306
>PRS4_RAT (P62193) 26S protease regulatory subunit 4 (P26s4)
(Proteasome 26S subunit ATPase 1)
Length = 440
Score = 60.8 bits (146), Expect = 7e-09
Identities = 47/169 (27%), Positives = 78/169 (45%), Gaps = 18/169 (10%)
Query: 200 VKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRIS 259
+K +E L +YY +G + +LYG GTGK+ A+AN S + S +
Sbjct: 197 IKESVELPLTHPEYYEEMGIKPPKGVILYGPPGTGKTLLAKAVANQTSATFLRVVGSELI 256
Query: 260 TD---------SDLKSILLQTAPKSIIVVEDLDRYLTEKSSTTV--------TSSGILNF 302
+L + + AP SI+ ++++D T++ + T +LN
Sbjct: 257 QKYLGDGPKLVRELFRVAEEHAP-SIVFIDEIDAIGTKRYDSNSGGEREIQRTMLELLNQ 315
Query: 303 MDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTL 351
+DG S + ++ N E +DP L+RPGR+D I FPL D + K +
Sbjct: 316 LDGFDSRGDVKVIMATNRIETLDPALIRPGRIDRKIEFPLPDEKTKKRI 364
>PRS4_MOUSE (P62192) 26S protease regulatory subunit 4 (P26s4)
(Proteasome 26S subunit ATPase 1)
Length = 440
Score = 60.8 bits (146), Expect = 7e-09
Identities = 47/169 (27%), Positives = 78/169 (45%), Gaps = 18/169 (10%)
Query: 200 VKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRIS 259
+K +E L +YY +G + +LYG GTGK+ A+AN S + S +
Sbjct: 197 IKESVELPLTHPEYYEEMGIKPPKGVILYGPPGTGKTLLAKAVANQTSATFLRVVGSELI 256
Query: 260 TD---------SDLKSILLQTAPKSIIVVEDLDRYLTEKSSTTV--------TSSGILNF 302
+L + + AP SI+ ++++D T++ + T +LN
Sbjct: 257 QKYLGDGPKLVRELFRVAEEHAP-SIVFIDEIDAIGTKRYDSNSGGEREIQRTMLELLNQ 315
Query: 303 MDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTL 351
+DG S + ++ N E +DP L+RPGR+D I FPL D + K +
Sbjct: 316 LDGFDSRGDVKVIMATNRIETLDPALIRPGRIDRKIEFPLPDEKTKKRI 364
>PRS4_HUMAN (P62191) 26S protease regulatory subunit 4 (P26s4)
(Proteasome 26S subunit ATPase 1)
Length = 440
Score = 60.8 bits (146), Expect = 7e-09
Identities = 47/169 (27%), Positives = 78/169 (45%), Gaps = 18/169 (10%)
Query: 200 VKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRIS 259
+K +E L +YY +G + +LYG GTGK+ A+AN S + S +
Sbjct: 197 IKESVELPLTHPEYYEEMGIKPPKGVILYGPPGTGKTLLAKAVANQTSATFLRVVGSELI 256
Query: 260 TD---------SDLKSILLQTAPKSIIVVEDLDRYLTEKSSTTV--------TSSGILNF 302
+L + + AP SI+ ++++D T++ + T +LN
Sbjct: 257 QKYLGDGPKLVRELFRVAEEHAP-SIVFIDEIDAIGTKRYDSNSGGEREIQRTMLELLNQ 315
Query: 303 MDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTL 351
+DG S + ++ N E +DP L+RPGR+D I FPL D + K +
Sbjct: 316 LDGFDSRGDVKVIMATNRIETLDPALIRPGRIDRKIEFPLPDEKTKKRI 364
>PRS4_DROME (P48601) 26S protease regulatory subunit 4 (P26s4)
Length = 439
Score = 60.5 bits (145), Expect = 9e-09
Identities = 46/162 (28%), Positives = 75/162 (45%), Gaps = 18/162 (11%)
Query: 200 VKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRIS 259
+K +E L +YY +G + +LYG GTGK+ A+AN S + S +
Sbjct: 196 IKESVELPLTHPEYYEEMGIKPPKGVILYGPPGTGKTLLAKAVANQTSATFLRVVGSELI 255
Query: 260 TD---------SDLKSILLQTAPKSIIVVEDLDRYLTEKSSTTV--------TSSGILNF 302
+L + + AP SI+ ++++D T++ + T +LN
Sbjct: 256 QKYLGDGPKLVRELFRVAEEHAP-SIVFIDEIDAVGTKRYDSNSGGEREIQRTMLELLNQ 314
Query: 303 MDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCD 344
+DG S + ++ N E +DP L+RPGR+D I FPL D
Sbjct: 315 LDGFDSRGDVKVIMATNRIETLDPALIRPGRIDRKIEFPLPD 356
>PRS4_CHICK (Q90732) 26S protease regulatory subunit 4 (P26s4)
(Proteasome 26S subunit ATPase 1)
Length = 440
Score = 60.5 bits (145), Expect = 9e-09
Identities = 48/174 (27%), Positives = 76/174 (43%), Gaps = 28/174 (16%)
Query: 200 VKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRIS 259
+K +E L +YY +G + +LYG GTGK+ A+AN S +
Sbjct: 197 IKESVELPLTHPEYYEEMGIKPPKGVILYGPPGTGKTLLAKAVANQTSATFLRV------ 250
Query: 260 TDSDLKSILLQTAPK--------------SIIVVEDLDRYLTEKSSTTV--------TSS 297
S+L L PK SI+ ++++D T++ + T
Sbjct: 251 VGSELIQKYLGDGPKLVRELFRVAEEHGPSIVFIDEIDAIGTKRYDSNSGGEREIQRTML 310
Query: 298 GILNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTL 351
+LN +DG S + ++ N E +DP L+RPGR+D I FPL D + K +
Sbjct: 311 ELLNQLDGFDSRGDVKVIMATNRIETLDPALIRPGRIDRKIEFPLPDEKTKKRI 364
>PRS4_CAEEL (O16368) Probable 26S protease regulatory subunit 4
Length = 443
Score = 60.5 bits (145), Expect = 9e-09
Identities = 47/162 (29%), Positives = 76/162 (46%), Gaps = 18/162 (11%)
Query: 200 VKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRIS 259
+K +E L +YY +G + +LYG GTGK+ A+AN S I S +
Sbjct: 200 IKEAVELPLTHPEYYEEMGIRPPKGVILYGCPGTGKTLLAKAVANQTSATFLRIVGSELI 259
Query: 260 TD---------SDLKSILLQTAPKSIIVVEDLDRYLTEKSSTTV--------TSSGILNF 302
+L + + AP SI+ ++++D T++ + T +LN
Sbjct: 260 QKYLGDGPKMVRELFRVAEENAP-SIVFIDEIDAVGTKRYDSNSGGEREIQRTMLELLNQ 318
Query: 303 MDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCD 344
+DG S + ++ N E++DP L+RPGR+D I FPL D
Sbjct: 319 LDGFDSRGDVKVLMATNRIESLDPALIRPGRIDRKIEFPLPD 360
>PRS4_ORYSA (P46466) 26S protease regulatory subunit 4 homolog
(TAT-binding protein homolog 2)
Length = 448
Score = 56.6 bits (135), Expect = 1e-07
Identities = 47/170 (27%), Positives = 73/170 (42%), Gaps = 28/170 (16%)
Query: 200 VKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRIS 259
+K +E L + Y +G + +LYG GTGK+ A+AN S +
Sbjct: 205 IKEAVELPLTHPELYEDIGIRPPKGVILYGEPGTGKTLLAKAVANSTSATFLRV------ 258
Query: 260 TDSDLKSILLQTAPK--------------SIIVVEDLDRYLTEKSSTTV--------TSS 297
S+L L PK SI+ ++++D T++ T
Sbjct: 259 VGSELIQKYLGDGPKLVRELFRVADELSPSIVFIDEIDAVGTKRYDAHSGGEREIQRTML 318
Query: 298 GILNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSS 347
+LN +DG S + ++ N E++DP LLRPGR+D I FPL D +
Sbjct: 319 ELLNQLDGFDSRGDVKVILATNRIESLDPALLRPGRIDRKIEFPLPDIKT 368
>PRS6_SCHPO (O74894) 26S protease regulatory subunit 6B homolog
Length = 389
Score = 56.2 bits (134), Expect = 2e-07
Identities = 44/162 (27%), Positives = 71/162 (43%), Gaps = 18/162 (11%)
Query: 197 KNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLS 256
K V+ +E L Y ++G R LLYG GTGK+ + A+AN + + + S
Sbjct: 143 KQEVREAVELPLTQGDLYRQIGIDPPRGVLLYGPPGTGKTMLVKAVANSTAANFIRVVGS 202
Query: 257 RISTD---------SDLKSILLQTAPKSIIVVEDLDRYLTEKSSTTVTSSG--------I 299
D+ + + AP +II ++++D T++ + +
Sbjct: 203 EFVQKYLGEGPRMVRDVFRMARENAP-AIIFIDEIDAIATKRFDAQTGADREVQRILIEL 261
Query: 300 LNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFP 341
L MDG G ++ N + +DP LLRPGR+D I FP
Sbjct: 262 LTQMDGFDQGANVKVIMATNRADTLDPALLRPGRLDRKIEFP 303
>PRS6_ASPNG (P78578) 26S protease regulatory subunit 6B homolog
Length = 423
Score = 55.8 bits (133), Expect = 2e-07
Identities = 42/162 (25%), Positives = 71/162 (42%), Gaps = 18/162 (11%)
Query: 197 KNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLS 256
K ++ +E L Y ++G R LLYG GTGK+ + A+AN + ++ S
Sbjct: 175 KQEIREAVELPLTQFDLYKQIGIDPPRGVLLYGPPGTGKTMLVKAVANSTTASFIRVNGS 234
Query: 257 RISTD---------SDLKSILLQTAPKSIIVVEDLDRYLTEKSSTTVTSSG--------I 299
D+ + + +P +II ++++D T++ + +
Sbjct: 235 EFVQKYLGEGPRMVRDVFRMARENSP-AIIFIDEIDAIATKRFDAQTGADREVQRILLEL 293
Query: 300 LNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFP 341
LN MDG ++ N + +DP LLRPGR+D I FP
Sbjct: 294 LNQMDGFEQSSNVKVIMATNRADTLDPALLRPGRLDRKIEFP 335
>AFG2_YEAST (P32794) AFG2 protein
Length = 780
Score = 55.8 bits (133), Expect = 2e-07
Identities = 43/163 (26%), Positives = 82/163 (49%), Gaps = 19/163 (11%)
Query: 193 ETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMA-----NFLS 247
+ +LK ++K ++ L+ + + RLG + LLYG G K+ A+A NFL+
Sbjct: 521 QEELKTKMKEMIQLPLEASETFARLGISAPKGVLLYGPPGCSKTLTAKALATESGINFLA 580
Query: 248 ------YDVYYIDLSRISTDSDLKSILLQTAPKSIIVVEDLDRYLTEKSSTTVTSSG--- 298
++ Y + R + K+ ++A SII +++D ++ ++ +++
Sbjct: 581 VKGPEIFNKYVGESERAIREIFRKA---RSAAPSIIFFDEIDALSPDRDGSSTSAANHVL 637
Query: 299 --ILNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIH 339
+LN +DG+ + V+V N + +D LLRPGR+D HI+
Sbjct: 638 TSLLNEIDGVEELKGVVIVAATNRPDEIDAALLRPGRLDRHIY 680
Score = 50.4 bits (119), Expect = 1e-05
Identities = 44/159 (27%), Positives = 73/159 (45%), Gaps = 14/159 (8%)
Query: 200 VKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRI- 258
+KS +E L + G R LL+G GTGK+ + +AN + V I+ I
Sbjct: 257 LKSAIEIPLHQPTLFSSFGVSPPRGILLHGPPGTGKTMLLRVVANTSNAHVLTINGPSIV 316
Query: 259 -----STDSDLKSILLQTAP--KSIIVVEDLDRYLTEKS---STTVTS---SGILNFMDG 305
T++ L+ I + SII ++++D ++ S V S + +L MDG
Sbjct: 317 SKYLGETEAALRDIFNEARKYQPSIIFIDEIDSIAPNRANDDSGEVESRVVATLLTLMDG 376
Query: 306 IWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCD 344
+ + + V++ N +VDP L RPGR D + + D
Sbjct: 377 MGAAGKVVVIAATNRPNSVDPALRRPGRFDQEVEIGIPD 415
>PRS6_YEAST (P33298) 26S protease regulatory subunit 6B homolog
(YNT1 protein) (TAT-binding homolog 2)
Length = 428
Score = 55.5 bits (132), Expect = 3e-07
Identities = 44/162 (27%), Positives = 69/162 (42%), Gaps = 18/162 (11%)
Query: 197 KNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLS 256
K ++ +E L Y ++G R LLYG GTGK+ + A+AN ++ S
Sbjct: 181 KQEIREAVELPLVQADLYEQIGIDPPRGVLLYGPPGTGKTMLVKAVANSTKAAFIRVNGS 240
Query: 257 RISTD---------SDLKSILLQTAPKSIIVVEDLDRYLTEKSSTTVTSSG--------I 299
D+ + + AP SII ++++D T++ S +
Sbjct: 241 EFVHKYLGEGPRMVRDVFRLARENAP-SIIFIDEVDSIATKRFDAQTGSDREVQRILIEL 299
Query: 300 LNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFP 341
L MDG ++ N + +DP LLRPGR+D I FP
Sbjct: 300 LTQMDGFDQSTNVKVIMATNRADTLDPALLRPGRLDRKIEFP 341
>CDAT_PLAF7 (P46468) Putative cell division cycle ATPase
Length = 1229
Score = 55.5 bits (132), Expect = 3e-07
Identities = 43/172 (25%), Positives = 80/172 (46%), Gaps = 13/172 (7%)
Query: 186 TFETITMETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANF 245
T+E + N+++ +E LK + + +G + L++G GTGK+S A+AN
Sbjct: 525 TYEDLGGMKKQLNKIRELIELPLKYPEIFMSIGISAPKGVLMHGIPGTGKTSIAKAIANE 584
Query: 246 LSYDVYYIDLSRI------STDSDLKSILLQTAPKS--IIVVEDLDRYLTEKSSTTV--- 294
+ Y I+ I ++ L+ I + + K+ II ++++D ++S +
Sbjct: 585 SNAYCYIINGPEIMSKHIGESEQKLRKIFKKASEKTPCIIFIDEIDSIANKRSKSNNELE 644
Query: 295 --TSSGILNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCD 344
S +L MDG+ +++ N ++DP L R GR D I P+ D
Sbjct: 645 KRVVSQLLTLMDGLKKNNNVLVLAATNRPNSIDPALRRFGRFDREIEIPVPD 696
Score = 52.8 bits (125), Expect = 2e-06
Identities = 45/187 (24%), Positives = 84/187 (44%), Gaps = 18/187 (9%)
Query: 186 TFETITMETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANF 245
T+ I ++K ++K + L+ K Y++ + + LLYG G GK+ A+AN
Sbjct: 932 TWNDIGGMNEVKEQLKETILYPLEYKHLYNKFNSNYNKGILLYGPPGCGKTLLAKAIANE 991
Query: 246 LSYDVYYIDLSRIST------DSDLKSIL--LQTAPKSIIVVEDLDRYLTEKSSTTVTSS 297
+ + + T +++++ + + A II +++D E++S T +
Sbjct: 992 CKANFISVKGPELLTMWFGESEANVRDLFDKARAASPCIIFFDEIDSLAKERNSNTNNDA 1051
Query: 298 G------ILNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCD----FSS 347
IL +DGI + ++ N + +D L RPGR+D I+ L D +S
Sbjct: 1052 SDRVINQILTEIDGINEKKTIFIIAATNRPDILDKALTRPGRLDKLIYISLPDLKSRYSI 1111
Query: 348 FKTLASN 354
FK + N
Sbjct: 1112 FKAILKN 1118
>YB56_METJA (Q58556) Cell division cycle protein 48 homolog MJ1156
Length = 903
Score = 55.1 bits (131), Expect = 4e-07
Identities = 58/232 (25%), Positives = 105/232 (45%), Gaps = 32/232 (13%)
Query: 140 RILRAYIQHIHAVVDEIEKQGNRDLRFYMNASDFGPWRFVPFTHPSTFETITMET----- 194
R LR + I +EI K+ +L+ M+ DF + PS + +E
Sbjct: 396 RALRRVLPSIDLEAEEIPKEVLDNLKVTMD--DFK--EALKDVEPSAMREVLVEVPNVKW 451
Query: 195 -------DLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLS 247
++K ++ +E LK K+ + ++G + LL+G GTGK+ A+AN
Sbjct: 452 EDIGGLEEVKQELREAVEWPLKAKEVFEKIGVRPPKGVLLFGPPGTGKTLLAKAVANESG 511
Query: 248 YDVYYIDLSRI------STDSDLKSILL---QTAPKSIIVVEDLDRYLTEKS---STTVT 295
+ + I ++ ++ I Q+AP II +++D ++ S+ VT
Sbjct: 512 ANFISVKGPEIFSKWVGESEKAIREIFRKARQSAP-CIIFFDEIDAIAPKRGRDLSSAVT 570
Query: 296 S---SGILNFMDGIWSGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCD 344
+ +L +DG+ ++ V++ N + +DP LLRPGR+D I P+ D
Sbjct: 571 DKVVNQLLTELDGMEEPKDVVVIAATNRPDIIDPALLRPGRLDRVILVPVPD 622
Score = 53.1 bits (126), Expect = 1e-06
Identities = 51/196 (26%), Positives = 83/196 (42%), Gaps = 32/196 (16%)
Query: 174 GPWRFVPFTHPS-----------------TFETITMETDLKNRVKSDLESFLKGKQYYHR 216
GP R FTH T+E I + +V+ +E ++ + + +
Sbjct: 148 GPVRVTDFTHVELKEEPVSEIKETKVPDVTYEDIGGLKEEVKKVREMIELPMRHPELFEK 207
Query: 217 LGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRI------STDSDLKSIL-- 268
LG + LL G GTGK+ A+AN + Y I+ I T+ +L+ I
Sbjct: 208 LGIEPPKGVLLVGPPGTGKTLLAKAVANEAGANFYVINGPEIMSKYVGETEENLRKIFEE 267
Query: 269 -LQTAPKSIIVVEDLDRYLTEKSSTT-----VTSSGILNFMDGIWSGEERVMVFTMNSKE 322
+ AP SII ++++D ++ T + +L MDG+ + V++ N
Sbjct: 268 AEENAP-SIIFIDEIDAIAPKRDEATGEVERRLVAQLLTLMDGLKGRGQVVVIGATNRPN 326
Query: 323 NVDPNLLRPGRVDVHI 338
+DP L RPGR D I
Sbjct: 327 ALDPALRRPGRFDREI 342
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.138 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 55,031,038
Number of Sequences: 164201
Number of extensions: 2372397
Number of successful extensions: 7398
Number of sequences better than 10.0: 270
Number of HSP's better than 10.0 without gapping: 167
Number of HSP's successfully gapped in prelim test: 103
Number of HSP's that attempted gapping in prelim test: 6964
Number of HSP's gapped (non-prelim): 335
length of query: 468
length of database: 59,974,054
effective HSP length: 114
effective length of query: 354
effective length of database: 41,255,140
effective search space: 14604319560
effective search space used: 14604319560
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 68 (30.8 bits)
Medicago: description of AC144389.8


