

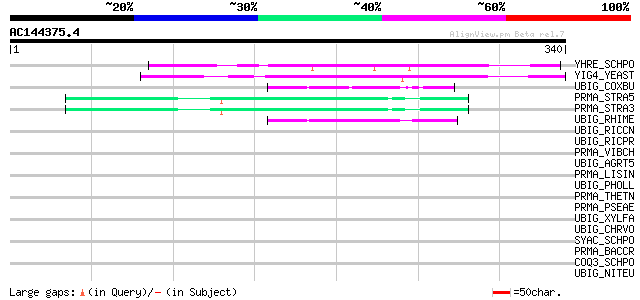
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144375.4 - phase: 0
(340 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YHRE_SCHPO (Q9P7Z3) Hypothetical protein C839.14c in chromosome II 112 2e-24
YIG4_YEAST (P40516) Hypothetical 28.7 kDa protein in RNR3-ARC15 ... 109 9e-24
UBIG_COXBU (Q820B5) 3-demethylubiquinone-9 3-methyltransferase (... 46 1e-04
PRMA_STRA5 (Q8DX85) Ribosomal protein L11 methyltransferase (EC ... 44 6e-04
PRMA_STRA3 (Q8E307) Ribosomal protein L11 methyltransferase (EC ... 44 6e-04
UBIG_RHIME (Q92MK1) 3-demethylubiquinone-9 3-methyltransferase (... 44 8e-04
UBIG_RICCN (Q92H07) 3-demethylubiquinone-9 3-methyltransferase (... 41 0.005
UBIG_RICPR (Q9ZCT9) 3-demethylubiquinone-9 3-methyltransferase (... 40 0.008
PRMA_VIBCH (Q9KV64) Ribosomal protein L11 methyltransferase (EC ... 40 0.008
UBIG_AGRT5 (Q8UA66) 3-demethylubiquinone-9 3-methyltransferase (... 40 0.011
PRMA_LISIN (Q92BP0) Ribosomal protein L11 methyltransferase (EC ... 39 0.014
UBIG_PHOLL (Q7N2M5) 3-demethylubiquinone-9 3-methyltransferase (... 39 0.019
PRMA_THETN (Q8RB66) Ribosomal protein L11 methyltransferase (EC ... 39 0.019
PRMA_PSEAE (Q9HUW3) Ribosomal protein L11 methyltransferase (EC ... 39 0.019
UBIG_XYLFA (Q9PAM5) 3-demethylubiquinone-9 3-methyltransferase (... 39 0.024
UBIG_CHRVO (Q7NZ91) 3-demethylubiquinone-9 3-methyltransferase (... 39 0.024
SYAC_SCHPO (O13914) Probable alanyl-tRNA synthetase, cytoplasmic... 39 0.024
PRMA_BACCR (Q818F1) Ribosomal protein L11 methyltransferase (EC ... 39 0.024
COQ3_SCHPO (O74421) Hexaprenyldihydroxybenzoate methyltransferas... 39 0.024
UBIG_NITEU (Q81ZZ2) 3-demethylubiquinone-9 3-methyltransferase (... 38 0.032
>YHRE_SCHPO (Q9P7Z3) Hypothetical protein C839.14c in chromosome II
Length = 238
Score = 112 bits (279), Expect = 2e-24
Identities = 83/264 (31%), Positives = 119/264 (44%), Gaps = 53/264 (20%)
Query: 86 SMLGFQSYWDAAYTDELTNFHEHGHAGEVWFGDNVMEVVASWTKTLCIDISQGRLPNHVD 145
S LG + YWD Y E++NF E GEVWFG+ E + W L +H+
Sbjct: 7 SKLGTKQYWDNVYEREVSNFTEFNDEGEVWFGEEAEERIVQW------------LEDHIS 54
Query: 146 DVKADAGELDDKLLSSWNVLDIGTGNGLLLQELAKQGFS------DLTGTDYSERAINLA 199
+ E + + VLD+GTGNG LL L ++ + L G DYSE AI LA
Sbjct: 55 TSFREVSE-----AAPFRVLDLGTGNGHLLFRLLEEEDTLLPSPCQLVGVDYSEAAIVLA 109
Query: 200 QSLANRDGFPN-IKFLVDDVLETK--LEQVFQLVMDKGTLDAIGLHP---DGPVKRMMYW 253
+++A F + +KF D+++ + + L++DKGT DAI L DG +Y
Sbjct: 110 KNIARHRQFSDKVKFQQLDIIKDSKFCSKDWDLILDKGTFDAISLSGELLDGRPLNSVYV 169
Query: 254 DSVSRLVAPGGILVVTSCNSTKDELVQEVESFNQRKSATAPEPEVAKDDESCRDPLFQYV 313
D V +++P GI ++TSCN T EL + +T P
Sbjct: 170 DRVRGMLSPNGIFLITSCNWTIQELEERFTKNGFIVHSTVP------------------- 210
Query: 314 SHVRTYPTFMFGGSVGSRVATVAF 337
P F F GS GS + +AF
Sbjct: 211 -----VPVFEFQGSTGSSTSVIAF 229
>YIG4_YEAST (P40516) Hypothetical 28.7 kDa protein in RNR3-ARC15
intergenic region
Length = 257
Score = 109 bits (273), Expect = 9e-24
Identities = 84/268 (31%), Positives = 130/268 (48%), Gaps = 52/268 (19%)
Query: 81 AEAVSSMLGFQSYWDAAYTDELTNFHEHGH-AGEVWFGDNVMEVVASWTKTLCIDISQGR 139
A+ +S LG + YWD Y EL NF + G+ WF D+ D Q
Sbjct: 33 ADLSTSKLGTKKYWDELYALELENFRRNPQDTGDCWFSDS--------------DAEQKM 78
Query: 140 LPNHVDDVKADAGELDDKLLSSWNVLDIGTGNGLLLQELAKQGFSD-LTGTDYSERAINL 198
+ VD++ A ++ + +V+D+GTGNG +L EL + F L G DYSE ++ L
Sbjct: 79 IDFLVDNIGAY------RISENASVVDLGTGNGHMLFELHQTEFQGKLVGIDYSEESVKL 132
Query: 199 AQSLANRDGFPN-IKFLVDDVLETKLEQ-VFQLVMDKGTLDAI---GLHPDGPVKRM-MY 252
A ++A G N I F D+ + + +V+DKGTLDAI G+ +G + + +Y
Sbjct: 133 ASNIAEATGVDNFISFQQADIFSGDWKPGKYDIVLDKGTLDAISLSGMKINGKLDVVDVY 192
Query: 253 WDSVSRLVAPGGILVVTSCNSTKDELVQEVESFNQRKSATAPEPEVAKDDESCRDPLFQY 312
V R++ GI ++TSCN T+DELV+ +E+ N + T
Sbjct: 193 AGVVERILKKDGIFLITSCNFTQDELVKIIETDNLKMWKTI------------------- 233
Query: 313 VSHVRTYPTFMFGGSVGSRVATVAFLRK 340
YP F FGG G+ + +VAF+++
Sbjct: 234 -----KYPVFQFGGVQGATICSVAFVKQ 256
>UBIG_COXBU (Q820B5) 3-demethylubiquinone-9 3-methyltransferase (EC
2.1.1.64) (3,4-dihydroxy-5-hexaprenylbenzoate
methyltransferase) (DHHB methyltransferase)
Length = 234
Score = 46.2 bits (108), Expect = 1e-04
Identities = 39/115 (33%), Positives = 63/115 (53%), Gaps = 10/115 (8%)
Query: 159 LSSWNVLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFPNIKFLVDDV 218
L +VLD+G G GLL + LAK G + +TG D SE I++A++ A + NI + D+
Sbjct: 50 LKGKHVLDVGCGGGLLSEALAKHG-AIVTGVDMSESLIDVAKNHAEQQQL-NINYQCQDI 107
Query: 219 -LETKLEQVFQLVMDKGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGGILVVTSCN 272
+ TK Q F ++ L+ + PD +RM+ + + L+ PGG L ++ N
Sbjct: 108 EILTKDAQRFDIITCMELLEHV---PD--PQRMI--KNCAALIKPGGKLFFSTIN 155
>PRMA_STRA5 (Q8DX85) Ribosomal protein L11 methyltransferase (EC
2.1.1.-) (L11 Mtase)
Length = 317
Score = 43.9 bits (102), Expect = 6e-04
Identities = 60/257 (23%), Positives = 102/257 (39%), Gaps = 40/257 (15%)
Query: 35 ADSWSIKSEYGSTLDDDQRHADAAEALSNVNLRAASDYSSDKDEPDAEAVSSMLGFQSYW 94
+D +I + Y TLD + AD A+ L+N + S + D + ++ Y+
Sbjct: 56 SDMIAITAYYPDTLDIEAVKADLADRLANFEGFGLATGSVNLDSQELVEEDWADNWKKYY 115
Query: 95 DAA-YTDELTNFHEHGHAGEVWFGDNVMEVVASWT--------KTLCIDISQGRLPNHVD 145
+ A T +LT +V SWT K + +D
Sbjct: 116 EPARITHDLT-------------------IVPSWTDYEAKAGEKIIKMDPGMAFGTGTHP 156
Query: 146 DVKADAGELDDKLLSSWNVLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQ-SLAN 204
K L+ L V+D+GTG+G+L + G D+ D + A+ +AQ ++
Sbjct: 157 TTKMSLFALEQVLRGGETVIDVGTGSGVLSIASSLLGAKDIYAYDLDDVAVRVAQENIDM 216
Query: 205 RDGFPNIKFLVDDVLETKLEQVFQLVMDKGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGG 264
G NI D+L+ ++V +V + L I +H D RLV G
Sbjct: 217 NPGTENIHVAAGDLLKGVQQEVDVIVAN--ILADILIH---------LTDDAYRLVKDEG 265
Query: 265 ILVVTSCNSTKDELVQE 281
L+++ S K ++V+E
Sbjct: 266 YLIMSGIISEKWDMVRE 282
>PRMA_STRA3 (Q8E307) Ribosomal protein L11 methyltransferase (EC
2.1.1.-) (L11 Mtase)
Length = 317
Score = 43.9 bits (102), Expect = 6e-04
Identities = 60/257 (23%), Positives = 102/257 (39%), Gaps = 40/257 (15%)
Query: 35 ADSWSIKSEYGSTLDDDQRHADAAEALSNVNLRAASDYSSDKDEPDAEAVSSMLGFQSYW 94
+D +I + Y TLD + AD A+ L+N + S + D + ++ Y+
Sbjct: 56 SDMIAITAYYPDTLDIEAVKADLADRLANFEGFGLATGSVNLDSQELVEEDWADNWKKYY 115
Query: 95 DAA-YTDELTNFHEHGHAGEVWFGDNVMEVVASWT--------KTLCIDISQGRLPNHVD 145
+ A T +LT +V SWT K + +D
Sbjct: 116 EPARITHDLT-------------------IVPSWTDYEAKAGEKIIKMDPGMAFGTGTHP 156
Query: 146 DVKADAGELDDKLLSSWNVLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQ-SLAN 204
K L+ L V+D+GTG+G+L + G D+ D + A+ +AQ ++
Sbjct: 157 TTKMSLFALEQVLRGGETVIDVGTGSGVLSIASSLLGAKDIYAYDLDDVAVRVAQENIDM 216
Query: 205 RDGFPNIKFLVDDVLETKLEQVFQLVMDKGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGG 264
G NI D+L+ ++V +V + L I +H D RLV G
Sbjct: 217 NPGTENIHVAAGDLLKGVQQEVDVIVAN--ILADILIH---------LTDDAYRLVKDEG 265
Query: 265 ILVVTSCNSTKDELVQE 281
L+++ S K ++V+E
Sbjct: 266 YLIMSGIISEKWDMVRE 282
>UBIG_RHIME (Q92MK1) 3-demethylubiquinone-9 3-methyltransferase (EC
2.1.1.64) (3,4-dihydroxy-5-hexaprenylbenzoate
methyltransferase) (DHHB methyltransferase)
Length = 248
Score = 43.5 bits (101), Expect = 8e-04
Identities = 31/116 (26%), Positives = 55/116 (46%), Gaps = 8/116 (6%)
Query: 159 LSSWNVLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFPNIKFLVDDV 218
L VLDIG G GLL + +A+ G +D+ G D SE+ I +A++ A G V
Sbjct: 63 LEGLRVLDIGCGGGLLSEPMARMG-ADVVGADASEKNIGIARTHAAGSGVSVDYRAVTAE 121
Query: 219 LETKLEQVFQLVMDKGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGGILVVTSCNST 274
+ + F +V++ ++ + + + + +V PGG++ V + N T
Sbjct: 122 ALAEAGESFDVVLNMEVVEHV-------ADVEFFMTTCAHMVRPGGLMFVATINRT 170
>UBIG_RICCN (Q92H07) 3-demethylubiquinone-9 3-methyltransferase (EC
2.1.1.64) (3,4-dihydroxy-5-hexaprenylbenzoate
methyltransferase) (DHHB methyltransferase)
Length = 289
Score = 40.8 bits (94), Expect = 0.005
Identities = 28/117 (23%), Positives = 60/117 (50%), Gaps = 9/117 (7%)
Query: 159 LSSWNVLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFPNIKFLVDDV 218
+S +LD+G G GL+ LA QGF ++T D + I A + A +G I +L +
Sbjct: 100 ISKLEILDVGCGGGLIATPLAAQGF-NVTAIDALQSNIETATAYAKENGV-KINYLQSTI 157
Query: 219 LETKLEQVFQLVMDKGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGGILVVTSCNSTK 275
E ++++ +V+ ++ H + + ++ ++ + + P G+ ++++ N TK
Sbjct: 158 EELDSDKLYDVVICLEVIE----HVENVQQFIL---NLVKHIKPNGMAIISTINRTK 207
>UBIG_RICPR (Q9ZCT9) 3-demethylubiquinone-9 3-methyltransferase (EC
2.1.1.64) (3,4-dihydroxy-5-hexaprenylbenzoate
methyltransferase) (DHHB methyltransferase)
Length = 252
Score = 40.0 bits (92), Expect = 0.008
Identities = 27/117 (23%), Positives = 58/117 (49%), Gaps = 9/117 (7%)
Query: 159 LSSWNVLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFPNIKFLVDDV 218
+S +LD+G G GL+ LA QGF ++T D + + A A ++G I +L +
Sbjct: 51 ISKLQILDVGCGGGLIAAPLALQGF-NVTAIDALKSNVETATIYAQKNGL-KINYLQATI 108
Query: 219 LETKLEQVFQLVMDKGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGGILVVTSCNSTK 275
E + ++++ +V+ ++ + + ++ + + P GI ++++ N TK
Sbjct: 109 EELENDKLYDVVICLEVIEHV-------ANIQQFILNLVQHIKPNGIAIISTMNRTK 158
>PRMA_VIBCH (Q9KV64) Ribosomal protein L11 methyltransferase (EC
2.1.1.-) (L11 Mtase)
Length = 295
Score = 40.0 bits (92), Expect = 0.008
Identities = 37/130 (28%), Positives = 61/130 (46%), Gaps = 15/130 (11%)
Query: 154 LDDKLLSSWNVLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFPNIKF 213
LD+ LS V+D G G+G+L K G + + G D +A+ ++ A R+G
Sbjct: 153 LDNLDLSGKTVIDFGCGSGILAIAAIKLGAAKVIGIDIDPQALLASKDNAARNG------ 206
Query: 214 LVDDVLETKL--EQVFQLVMDKGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGGILVVTSC 271
V+D +E L +Q LV D + + GP++ + + L+ PGG L ++
Sbjct: 207 -VEDQIEVYLPKDQPEGLVADVVVANILA----GPLRELS--PIIKGLLKPGGQLAMSGI 259
Query: 272 NSTKDELVQE 281
T+ E V E
Sbjct: 260 LDTQAESVAE 269
>UBIG_AGRT5 (Q8UA66) 3-demethylubiquinone-9 3-methyltransferase (EC
2.1.1.64) (3,4-dihydroxy-5-hexaprenylbenzoate
methyltransferase) (DHHB methyltransferase)
Length = 250
Score = 39.7 bits (91), Expect = 0.011
Identities = 29/117 (24%), Positives = 58/117 (48%), Gaps = 10/117 (8%)
Query: 159 LSSWNVLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFP-NIKFLVDD 217
L VLDIG G GLL + +A+ G + + G D SE+ I +A + A G + + + +
Sbjct: 63 LEGLRVLDIGCGGGLLSEPVARMG-ATVVGADPSEKNIGIASTHARESGVSVDYRAVTAE 121
Query: 218 VLETKLEQVFQLVMDKGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGGILVVTSCNST 274
L+ E F ++++ ++ + ++ + +++V PGG++ + N T
Sbjct: 122 QLQEAGES-FDVILNMEVVEHV-------ANVDLFVTTCAKMVRPGGLMFAATINRT 170
>PRMA_LISIN (Q92BP0) Ribosomal protein L11 methyltransferase (EC
2.1.1.-) (L11 Mtase)
Length = 314
Score = 39.3 bits (90), Expect = 0.014
Identities = 32/129 (24%), Positives = 61/129 (46%), Gaps = 11/129 (8%)
Query: 154 LDDKLLSSWNVLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFPNIKF 213
L D L ++D+GTG+G+L AK G + TD E +A R NI+
Sbjct: 168 LSDYLQPGDELIDVGTGSGVLSIASAKLGAKSILATDLDE--------IATRAAEENIRL 219
Query: 214 -LVDDVLETKLEQVFQLVMDKGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGGILVVTSCN 272
++++ K + Q + +K +D + + V +++ + V + + PGGI + +
Sbjct: 220 NKTENIITVKQNNLLQDI-NKSDVDIVVANILAEV-ILLFPEDVYKALKPGGIFIASGII 277
Query: 273 STKDELVQE 281
K ++V+E
Sbjct: 278 EDKAKVVEE 286
>UBIG_PHOLL (Q7N2M5) 3-demethylubiquinone-9 3-methyltransferase (EC
2.1.1.64) (3,4-dihydroxy-5-hexaprenylbenzoate
methyltransferase) (DHHB methyltransferase)
Length = 240
Score = 38.9 bits (89), Expect = 0.019
Identities = 30/112 (26%), Positives = 54/112 (47%), Gaps = 7/112 (6%)
Query: 164 VLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFPNIKFLVDDVLETKL 223
VLD+G G G+L + +A++G +D+TG D + +A+ A G P V + +E+
Sbjct: 60 VLDVGCGGGILSESMAREG-ADVTGLDMGTEPLQVARLHALETGIP--VTYVQETVESHA 116
Query: 224 EQVFQLVMDKGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGGILVVTSCNSTK 275
E+ Q ++ + PD + ++LV PGG + ++ N K
Sbjct: 117 EKYPQAYDIVTCMEMLEHVPD----PQSVVHACAQLVKPGGHVFFSTINRNK 164
>PRMA_THETN (Q8RB66) Ribosomal protein L11 methyltransferase (EC
2.1.1.-) (L11 Mtase)
Length = 309
Score = 38.9 bits (89), Expect = 0.019
Identities = 20/67 (29%), Positives = 35/67 (51%)
Query: 154 LDDKLLSSWNVLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFPNIKF 213
L+D + V D+G G+G+L +K G S + G D E A+ +A+ +G N++
Sbjct: 167 LEDIVKPGAIVFDVGCGSGILSIAASKLGASYVYGADVDEMAVKIARENVKLNGLENVEI 226
Query: 214 LVDDVLE 220
D+L+
Sbjct: 227 FQSDLLK 233
>PRMA_PSEAE (Q9HUW3) Ribosomal protein L11 methyltransferase (EC
2.1.1.-) (L11 Mtase)
Length = 294
Score = 38.9 bits (89), Expect = 0.019
Identities = 34/131 (25%), Positives = 56/131 (41%), Gaps = 11/131 (8%)
Query: 154 LDDKLLSSWNVLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFPNIKF 213
LD + L+ VLD G G+G+L G GTD +A+ ++ A+R+G +F
Sbjct: 151 LDGQELAGRQVLDFGCGSGILAIAALLLGAERAVGTDIDPQALEASRDNASRNGIEPARF 210
Query: 214 LVDDVLETKLEQVFQLVMDKGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGGILVVTSCNS 273
V + Q LV + + L P ++ LV PGG+L ++ +
Sbjct: 211 PVYLPADLPQRQADVLVANILAGPLVSLAP-----------QLTGLVRPGGLLALSGILA 259
Query: 274 TKDELVQEVES 284
+ E V+ S
Sbjct: 260 EQAEEVRAAYS 270
>UBIG_XYLFA (Q9PAM5) 3-demethylubiquinone-9 3-methyltransferase (EC
2.1.1.64) (3,4-dihydroxy-5-hexaprenylbenzoate
methyltransferase) (DHHB methyltransferase)
Length = 246
Score = 38.5 bits (88), Expect = 0.024
Identities = 35/121 (28%), Positives = 54/121 (43%), Gaps = 17/121 (14%)
Query: 159 LSSWNVLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFPNIKFLVDDV 218
LS VLD+G G GLL + LA+QG + +T D + I +A+ G + + + +
Sbjct: 54 LSGARVLDVGCGGGLLSEALARQG-AHVTAIDLAPELIKVARLHGLESGI-QVDYRIQAI 111
Query: 219 LETKLEQVFQLVMDKGTLDAIGL-----HPDGPVKRMMYWDSVSRLVAPGGILVVTSCNS 273
+ EQ DAI H P + D+ + L+ PGG L V++ N
Sbjct: 112 EDLLAEQ-------PAPFDAIACMEMLEHVPDPAAIV---DACAHLLKPGGRLFVSTINR 161
Query: 274 T 274
T
Sbjct: 162 T 162
>UBIG_CHRVO (Q7NZ91) 3-demethylubiquinone-9 3-methyltransferase (EC
2.1.1.64) (3,4-dihydroxy-5-hexaprenylbenzoate
methyltransferase) (DHHB methyltransferase)
Length = 232
Score = 38.5 bits (88), Expect = 0.024
Identities = 29/110 (26%), Positives = 57/110 (51%), Gaps = 9/110 (8%)
Query: 164 VLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFP-NIKFLVDDVLETK 222
VLD+G G G+L + +A +G + +TG D +++++ +AQ + G P + + + + L +
Sbjct: 51 VLDVGCGGGILAESMALRG-AQVTGIDLAKKSLKVAQLHSLESGVPIDYRCVAVEDLAAE 109
Query: 223 LEQVFQLVMDKGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGGILVVTSCN 272
+ F V L+ + P+ V+ + S LV PGG + ++ N
Sbjct: 110 MPGAFDAVTCMEMLEHVP-DPESVVR------ACSTLVKPGGWVFFSTLN 152
>SYAC_SCHPO (O13914) Probable alanyl-tRNA synthetase, cytoplasmic
(EC 6.1.1.7) (Alanine--tRNA ligase) (AlaRS)
Length = 959
Score = 38.5 bits (88), Expect = 0.024
Identities = 50/229 (21%), Positives = 86/229 (36%), Gaps = 58/229 (25%)
Query: 24 DLVSDDDRSIAADSWSI------KSEYGSTLDDDQRHADAA---EALSNVNLRAASDYSS 74
+LV+ DD ++ + W+I + + GS RH D E L +V S+Y +
Sbjct: 207 ELVNMDDPNVL-EIWNIVFIQFNREKDGSLRPLPNRHVDTGMGFERLVSVIQNKTSNYDT 265
Query: 75 DKDEPDAEAVSSMLGFQSYWDAAYTDELTNFHEHGHAGEVWFGDNVMEVVASWTKTLCID 134
D P + + + Y +++ D VVA +TL
Sbjct: 266 DVFSPIFAKIQELTNARPYTGKMGDEDVDGI------------DTAYRVVADHVRTLTFA 313
Query: 135 ISQGRLPNHVDDVKADAGELDDKLLSSWNVLDIGTGNGLLLQELAKQG-------FSDLT 187
IS G +PN+ G G +L+ + ++G F
Sbjct: 314 ISDGGVPNN-------------------------EGRGYVLRRILRRGARYVRKKFGVPI 348
Query: 188 GTDYSERAINLAQSLANRDGFPNIKFLVDDVLE--TKLEQVFQLVMDKG 234
G +S ++ + + + D FP +K VDDV E + E+ F +D+G
Sbjct: 349 GNFFSRLSLTVVEQMG--DFFPELKRKVDDVRELLDEEEESFSRTLDRG 395
>PRMA_BACCR (Q818F1) Ribosomal protein L11 methyltransferase (EC
2.1.1.-) (L11 Mtase)
Length = 312
Score = 38.5 bits (88), Expect = 0.024
Identities = 27/119 (22%), Positives = 54/119 (44%), Gaps = 10/119 (8%)
Query: 163 NVLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFPNIKFLVDDVLETK 222
N++D+GTG+G+L AK G S + D A+ A+ + +I + + L
Sbjct: 178 NIIDVGTGSGVLSIAAAKLGASSVQAYDLDPVAVESAEMNVRLNKTDDIVSVGQNSLLEG 237
Query: 223 LEQVFQLVMDKGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGGILVVTSCNSTKDELVQE 281
+E L++ + I L P + +R+V GG+ + + + K++++ E
Sbjct: 238 IEGPVDLIVANLLAEIILLFP----------EDAARVVKSGGLFITSGIIAAKEKVISE 286
>COQ3_SCHPO (O74421) Hexaprenyldihydroxybenzoate methyltransferase,
mitochondrial precursor (EC 2.1.1.114)
(Dihydroxyhexaprenylbenzoate methyltransferase)
(3,4-dihydroxy-5-hexaprenylbenzoate methyltransferase)
(DHHB methyltransferase) (DHHB-MT) (DHHB-MTas
Length = 271
Score = 38.5 bits (88), Expect = 0.024
Identities = 33/117 (28%), Positives = 57/117 (48%), Gaps = 10/117 (8%)
Query: 160 SSWNVLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQSLANRDGFPN--IKFLVDD 217
S +LDIG G G+L + +A+ G S +T D S AI +A+ A+ D N ++++
Sbjct: 77 SGKKILDIGCGGGILSESMARLGAS-VTAVDASPMAIEVAKKHASLDPVLNGRLEYIHGS 135
Query: 218 VLETKLEQVFQLVMDKGTLDAIGLHPDGPVKRMMYWDSVSRLVAPGGILVVTSCNST 274
V ++L F +V L+ + D + S+ V P G LV+++ + T
Sbjct: 136 VEGSQLPTTFDVVTCMEVLEHVEQPRD-------FLFSLMEKVKPNGRLVLSTISRT 185
>UBIG_NITEU (Q81ZZ2) 3-demethylubiquinone-9 3-methyltransferase (EC
2.1.1.64) (3,4-dihydroxy-5-hexaprenylbenzoate
methyltransferase) (DHHB methyltransferase)
Length = 235
Score = 38.1 bits (87), Expect = 0.032
Identities = 34/132 (25%), Positives = 65/132 (48%), Gaps = 17/132 (12%)
Query: 142 NHVDDVKADAGELDDKLLSSWNVLDIGTGNGLLLQELAKQGFSDLTGTDYSERAINLAQ- 200
N++D++ G L +K V+D+G G G+L + +A +G S +TG D S++A+ +A+
Sbjct: 42 NYIDEI---IGGLSEK-----TVIDVGCGGGILSESMAARGAS-VTGIDLSDKALKVAKL 92
Query: 201 SLANRDGFPNIKFLVDDVLETKLEQVFQLVMDKGTLDAIGLHPDGPVKRMMYWDSVSRLV 260
L + + + + L T+ + + +V L+ H P + S +RLV
Sbjct: 93 HLLESGNQVDYRKITVEALATERPRYYDVVTCMEMLE----HVPDPASVI---QSCARLV 145
Query: 261 APGGILVVTSCN 272
GG + ++ N
Sbjct: 146 KSGGWVFFSTLN 157
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.132 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,459,287
Number of Sequences: 164201
Number of extensions: 1711795
Number of successful extensions: 3773
Number of sequences better than 10.0: 158
Number of HSP's better than 10.0 without gapping: 51
Number of HSP's successfully gapped in prelim test: 107
Number of HSP's that attempted gapping in prelim test: 3690
Number of HSP's gapped (non-prelim): 165
length of query: 340
length of database: 59,974,054
effective HSP length: 111
effective length of query: 229
effective length of database: 41,747,743
effective search space: 9560233147
effective search space used: 9560233147
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 66 (30.0 bits)
Medicago: description of AC144375.4


