

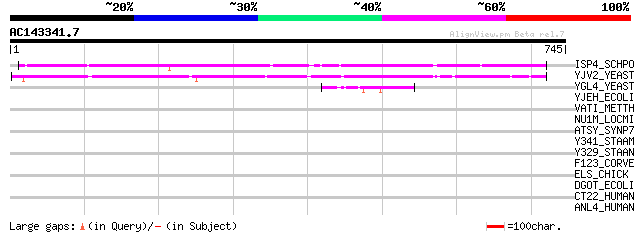
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143341.7 + phase: 0
(745 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ISP4_SCHPO (P40900) Sexual differentiation process protein isp4 509 e-143
YJV2_YEAST (P40897) Hypothetical 91.6 kDa protein in HXT8-CRT1 i... 450 e-126
YGL4_YEAST (P53134) Hypothetical 80.0 kDa protein in SNF4-TAF60 ... 49 4e-05
YJEH_ECOLI (P39277) Hypothetical protein yjeH 34 1.2
VATI_METTH (O27041) V-type ATP synthase subunit I (EC 3.6.3.14) ... 33 2.7
NU1M_LOCMI (P09045) NADH-ubiquinone oxidoreductase chain 1 (EC 1... 33 2.7
ATSY_SYNP7 (P37385) Probable copper-transporting ATPase synA (EC... 32 4.7
Y341_STAAM (Q99WN6) Hypothetical UPF0324 membrane protein SAV0341 32 6.1
Y329_STAAN (Q7A7M5) Hypothetical UPF0324 membrane protein SA0329 32 6.1
F123_CORVE (O74631) Protein FDD123 (CvHSP30/1) 32 6.1
ELS_CHICK (P07916) Elastin precursor (Tropoelastin) (Fragment) 32 6.1
DGOT_ECOLI (P31457) D-galactonate transporter 32 8.0
CT22_HUMAN (Q8N2K0) Protein C20orf22 32 8.0
ANL4_HUMAN (Q9BY76) Angiopoietin-related protein 4 precursor (An... 32 8.0
>ISP4_SCHPO (P40900) Sexual differentiation process protein isp4
Length = 785
Score = 509 bits (1310), Expect = e-143
Identities = 261/713 (36%), Positives = 404/713 (56%), Gaps = 23/713 (3%)
Query: 13 EDAEIDEYSINDSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCILLAFVNQFLGYRTN 72
E + +DE+ DSP +VR VP TDDPS P T R W +GL + A VN F R
Sbjct: 59 EISTVDEFE-EDSPYPEVRAAVPPTDDPSMPCNTIRMWTIGLIYSTVGAAVNMFFSLRNP 117
Query: 73 PLKITSVSAQIIALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSLKEHVLITIFASSG 132
+ ++ + ++++A P ++ P + ++ F+ PGPF++KEH LI + +S
Sbjct: 118 TVTLSVLISELLAYPALQIWDLIFPDREFRIG--RLKFNFKPGPFNVKEHALIVVMSSVS 175
Query: 133 SSGVYAINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRFLVDSPYMWWPESL 192
Y+ +II + Y + L L+TQ++GYG AG+ RR LV M WP +L
Sbjct: 176 FGNAYSTDIILAQRVHYKQRFGFGYEICLTLATQLIGYGLAGLSRRLLVRPASMLWPVNL 235
Query: 193 VQVSLFRAFHEKEKRPRGGT----SRLQFFFVVFVASFAYYIIPGYFFQAISTVSFVCLI 248
VQ +L + H K+ R S +FF VF+ASF + P Y FQA+S ++V I
Sbjct: 236 VQCTLIKTLHRKDLRNAVANGWRISPFRFFLYVFIASFIWNWFPSYIFQALSLFAWVTWI 295
Query: 249 WKESITAQQIGSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAGFFLYMYVLIP 308
S T QI G+ I DWN ++ ++ SPL P A++NI+ G L+ +++ P
Sbjct: 296 RPNSPTVNQIFGESTGISILPMTFDWNQISAYILSPLMAPADALMNILLGVILFFWIVTP 355
Query: 309 ISYWNNLYDAKKFPLISSHTFDSTGATYNVTRILNTE-TFDIDMESYSNYSKIYLSVAFA 367
+ N + P+ SS D G +YNVTRIL + TFD+D +Y NYS I++S +A
Sbjct: 356 ALNFTNTWYGDYLPISSSGIIDHFGNSYNVTRILTKDATFDLD--AYQNYSPIFMSTTYA 413
Query: 368 FEYGFCFAALTATISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYEQVPEWWF 427
+G FA++T+ I HV+L+HG+ I + D+H ++MK Y++VP +W+
Sbjct: 414 LAFGLSFASITSVIFHVILYHGKEIYDRLRDPPAP------DIHEKLMKA-YDEVPFYWY 466
Query: 428 VTILILMVMMALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVIT 487
+++ + M + +G + + PWW I++ + + ++ +PIG+++A TNI+ GLNV T
Sbjct: 467 LSVFLAFFGMMMGTI--YGWKTETPWWVIIVGVIFSAVWFIPIGIVQAITNIQLGLNVFT 524
Query: 488 ELVIGFIYPGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKIAPKSMFIVQLVGTVVA 547
E ++G++YPG+PLA + FKT G+I+M Q L F D K GHYMK+ P+ MF Q++ T+ +
Sbjct: 525 EFIVGYMYPGRPLAMMIFKTVGYITMTQGLAFAADLKFGHYMKLPPRIMFYTQMIATIWS 584
Query: 548 SSVHFGTAWWLLTSIENICDESLLPKGSPWTCPGDDVFYNASIIWGVVGPKRMFTKDGVY 607
V G W L +I+N+C + +TCP VF+N+S+IWGV+GPKRMF+ Y
Sbjct: 585 CFVQIGVLDWALGNIDNVCQAD---QPDNYTCPNATVFFNSSVIWGVIGPKRMFSGKNTY 641
Query: 608 PELNWFFLIGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPVRSVNYITWGIVG 667
L +F+L G++ + W L K+P QKW +N P+I G IPP VNY+ W +G
Sbjct: 642 TGLQYFWLAGVLGTILFWALWKKWP-QKWWGQLNGPLIFGGTGYIPPATPVNYLAWSGIG 700
Query: 668 IFFNFYVYRKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQSYGIYGPTWWG 720
+FFN+Y+ + F WW ++ + LSA LD G ++L+F LQ + P WWG
Sbjct: 701 LFFNYYLKKIFADWWQKYNFTLSAALDTGTQLSVIILFFCLQLPMVNFPDWWG 753
>YJV2_YEAST (P40897) Hypothetical 91.6 kDa protein in HXT8-CRT1
intergenic region
Length = 799
Score = 450 bits (1158), Expect = e-126
Identities = 250/730 (34%), Positives = 411/730 (56%), Gaps = 32/730 (4%)
Query: 3 SSRVSQNTVIEDAEI---DEYSINDSPIEQVRLTVPITDDPSQPALTFRTWVLGLASCIL 59
S + ++++ E+ + D + +SP +VR V I DDP+ +RTW L ++
Sbjct: 60 SQKFDRHSIQEEGLVWKGDPTYLPNSPYPEVRSAVSIEDDPTIRLNHWRTWFLTTVFVVV 119
Query: 60 LAFVNQFLGYRTNPLKITSVSAQIIALPLGKLMAATLPTKPIQVPFTTWSFSLNPGPFSL 119
A VNQF R L+I + AQ++ P+G+++A K +VPF F LNPGPF+
Sbjct: 120 FAGVNQFFSLRYPSLEINFLVAQVVCYPIGRILALLPDWKCSKVPF----FDLNPGPFTK 175
Query: 120 KEHVLITIFASSGSSGVYAINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGIFRRF 179
KEH ++TI + SS YA+ I+ +FY+ ++ +LL ++QM+GYG AG+ RR+
Sbjct: 176 KEHAVVTIAVALTSSTAYAMYILNAQGSFYNMKLNVGYQFLLVWTSQMIGYGAAGLTRRW 235
Query: 180 LVDSPYMWWPESLVQVSLFRAFHEK--EKRPRGGTS--RLQFFFVVFVASFAYYIIPGYF 235
+V+ WP++L+ VSLF + H + EK G + R +FF +V + SF +Y +PG+
Sbjct: 236 VVNPASSIWPQTLISVSLFDSLHSRKVEKTVANGWTMPRYRFFLIVLIGSFIWYWVPGFL 295
Query: 236 FQAISTVSFVCLIW----KESITAQQIGSGMKGLGIGSFGLDWNTVAGFL-GSPLAVPGF 290
F +S F ++W + + A I GLG D+ V+ + GS A P +
Sbjct: 296 FTGLSY--FNVILWGSKTRHNFIANTIFGTQSGLGALPITFDYTQVSQAMSGSVFATPFY 353
Query: 291 AIINIMAGFFLYMYVLIPISYWNNLYDAKKFPLISSHTFDSTGATYNVTRILNTETFDID 350
N A ++ +++P Y+ N + AK P+IS T+D+T YNVT+ILN E + I+
Sbjct: 354 VSANTYASVLIFFVIVLPCLYFTNTWYAKYMPVISGSTYDNTQNKYNVTKILN-EDYSIN 412
Query: 351 MESYSNYSKIYLSVAFAFEYGFCFAALTATISHVVLFHGEMIVQMWKKTTTSLKNQLGDV 410
+E Y YS +++ ++ Y FAA+ A H +L+HG+ IV +K KN D+
Sbjct: 413 LEKYKEYSPVFVPFSYLLSYALNFAAVIAVFVHCILYHGKDIVAKFKDR----KNGGTDI 468
Query: 411 HTRIMKKNYEQVPEWWFVTILILMVMMALLACEGFGKQLQLPWWGILLSLTIALIFTLPI 470
H RI KNY+ P+WW++ + I+M+ + +A F + P W ++++ I+L+ +P
Sbjct: 469 HMRIYSKNYKDCPDWWYLLLQIVMIGLGFVAVCCF--DTKFPAWAFVIAILISLVNFIPQ 526
Query: 471 GVIEATTNIRSGLNVITELVIGFIYPGKPLANVAFKTYGHISMVQALGFLGDFKLGHYMK 530
G++EA TN GLN+ITEL+ G++ P +P+AN+ FK YG I M Q L D KL YMK
Sbjct: 527 GILEAMTNQHVGLNIITELICGYMLPLRPMANLLFKLYGFIVMRQGLNLSRDLKLAMYMK 586
Query: 531 IAPKSMFIVQLVGTVVASSVHFGTAWWLLTSIENICDESLLPKGSPWTCPGDDVFYNASI 590
++P+ +F VQ+ T+++ V+ G W++ +I+ +C P G +TC +NASI
Sbjct: 587 VSPRLIFAVQIYATIISGMVNVGVQEWMMHNIDGLCTTD-QPNG--FTCANGRTVFNASI 643
Query: 591 IWGVVGPKRMFTKDGVYPELNWFFLIGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAGAS 650
IW + PK +F+ +Y L WFFLIGL+ P+ V+ + KFP K+ + I+ P+ G
Sbjct: 644 IWSL--PKYLFSSGRIYNPLMWFFLIGLLFPLAVYAVQWKFPKFKFAKHIHTPVFFTGPG 701
Query: 651 DIPPVRSVNYITWGIVGIFFNFYVYRKFKAWWARHTYILSAGLDAGVAFIGLLLYFSLQS 710
+IPP NY + + N + ++++AW+ ++ +++ AG++AGVA I +++ F
Sbjct: 702 NIPPSTPYNYSLFFAMSFCLNL-IRKRWRAWFNKYNFVMGAGVEAGVA-ISVVIIFLCVQ 759
Query: 711 YGIYGPTWWG 720
Y +WWG
Sbjct: 760 YPGGKLSWWG 769
>YGL4_YEAST (P53134) Hypothetical 80.0 kDa protein in SNF4-TAF60
intergenic region
Length = 725
Score = 49.3 bits (116), Expect = 4e-05
Identities = 40/132 (30%), Positives = 64/132 (48%), Gaps = 13/132 (9%)
Query: 419 YEQVPEWWFVTILILMVMMALLACEGFGKQLQLPWWGILLSLTIALIFTLPIGVI---EA 475
Y V V+ +I +V + L FG Q+ +P + I+ +L +AL ++ +G+ E
Sbjct: 445 YTTVISGCLVSSIICIVSIIYL----FGIQV-IPLYAIITALILALFLSI-LGIRALGET 498
Query: 476 TTNIRSGLNVITELVIGFIYP----GKPLANVAFKTYGHISMVQALGFLGDFKLGHYMKI 531
N SG+ I++L+ FI P G L NV S QA + D K GH +
Sbjct: 499 DLNPVSGIGKISQLIFAFIIPRDRPGSVLMNVVSGGIAEASAQQAGDLMQDLKTGHLLGA 558
Query: 532 APKSMFIVQLVG 543
+P++ F QL+G
Sbjct: 559 SPRAQFCAQLIG 570
>YJEH_ECOLI (P39277) Hypothetical protein yjeH
Length = 418
Score = 34.3 bits (77), Expect = 1.2
Identities = 28/101 (27%), Positives = 54/101 (52%), Gaps = 9/101 (8%)
Query: 420 EQVPEWWFVTILILMVMMALLACEGFGKQLQLPW--WGILLSL--TIALIFTLPIGVIEA 475
E+V W F++++ + + AL GFG Q W W +LL+ T+AL++ + +
Sbjct: 85 ERVTGWLFLSVIPVGLPAALQIAAGFG-QAMFGWHSWQLLLAELGTLALVWYIGTRGASS 143
Query: 476 TTNIRSGL-NVITELVIGFIYPG--KPLANVAFKTYGHISM 513
+ N+++ + +I L++ + G KP AN+ F G+I +
Sbjct: 144 SANLQTVIAGLIVALIVAIWWAGDIKP-ANIPFPAPGNIEL 183
>VATI_METTH (O27041) V-type ATP synthase subunit I (EC 3.6.3.14)
(V-type ATPase subunit I)
Length = 658
Score = 33.1 bits (74), Expect = 2.7
Identities = 36/104 (34%), Positives = 50/104 (47%), Gaps = 11/104 (10%)
Query: 431 LILMVMMALLACEG-FGKQLQLPWWGILLSLTIALIFTLPIGVIEATTNIRSGLNVITEL 489
LI++ LL G FG + G LLS L L G I T NI +GL+
Sbjct: 535 LIILAFAMLLYYNGLFGLMDVSGFLGTLLSYARLLALCLSTGGIAMTVNILTGLSYEMIP 594
Query: 490 VIGFIYPGKPLANVAFKTYGHI--SMVQALG-FLGDFKLGHYMK 530
VIG + LA + F +GHI + Q+LG F+ +L HY++
Sbjct: 595 VIGVV-----LAPIIF-VFGHIANNAFQSLGAFINSLRL-HYVE 631
>NU1M_LOCMI (P09045) NADH-ubiquinone oxidoreductase chain 1 (EC
1.6.5.3)
Length = 313
Score = 33.1 bits (74), Expect = 2.7
Identities = 48/197 (24%), Positives = 81/197 (40%), Gaps = 30/197 (15%)
Query: 361 YLSVAFAFEYGFCFAALTATISHVVLFHGEMIVQMWKKTTTSLKNQLGDVHTRIMKKNYE 420
YL+ +F YGF F T+ + + G W ++ S LG + + +YE
Sbjct: 97 YLTYMCSFPYGFYFLCCTSLSVYTFMIAG------W--SSNSNYALLGSLRSVAQTISYE 148
Query: 421 QVPEWWFVTILILMVMMALLACEGFGKQLQLPWWGILLSLTIALIF---------TLPIG 471
++++IL+ +L F QL W I+ S +AL F P
Sbjct: 149 VSLALILLSLIILIGSFNMLYFMNF----QLYCWFIVFSFPLALSFFGSSLAETNRTPFD 204
Query: 472 VIEATTNIRSGLNVITELVIGFIYPGKPLANVA-FKTYGHISMVQALGFLGD--FKLGHY 528
E + + SG N I + G L +A + + +SM+ +L FLG + +
Sbjct: 205 FAEGESELVSGFN------IEYSTGGFTLIFLAEYSSIIFMSMLFSLIFLGGDFYSFYFF 258
Query: 529 MKIAPKSMFIVQLVGTV 545
K++ S F V + GT+
Sbjct: 259 FKLSLVSFFFVWVRGTL 275
>ATSY_SYNP7 (P37385) Probable copper-transporting ATPase synA (EC
3.6.3.4)
Length = 790
Score = 32.3 bits (72), Expect = 4.7
Identities = 19/77 (24%), Positives = 37/77 (47%), Gaps = 8/77 (10%)
Query: 580 PGDDVFYNASII--WGVVGPKRMFTKDGVY------PELNWFFLIGLIAPVPVWLLSLKF 631
PG D + +++ W ++GP R + G P +N L+G + L++L +
Sbjct: 130 PGTDQLWFHALLATWALLGPGRSILQAGWQGLRCGAPNMNSLVLLGTGSAYLASLVALLW 189
Query: 632 PNQKWIQFINIPIIIAG 648
P W+ F + P+++ G
Sbjct: 190 PQLGWVCFFDEPVMLLG 206
>Y341_STAAM (Q99WN6) Hypothetical UPF0324 membrane protein SAV0341
Length = 331
Score = 32.0 bits (71), Expect = 6.1
Identities = 43/198 (21%), Positives = 78/198 (38%), Gaps = 21/198 (10%)
Query: 56 SCILLAFVNQFLGYRTNPLKITSVSAQIIALPLGKLMAATLPTKPIQVPFTTWSFSLNPG 115
S +++ FVN+ L N + V + +A ++ + +L
Sbjct: 104 SIVMMLFVNKLLHGDKNIALLLGVGTGVCGAAAIAAVAPIFKSREKDTAISIGIIALIGT 163
Query: 116 PFSLKEHVLITIFASSGSSGVYAINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGI 175
FSL + IF S ++ VY A+ S+H +A +LA G+G +
Sbjct: 164 IFSLIYTAIYAIF--SMTTNVYG--------AWSGVSLHEIAHVVLAG-----GFGGSDA 208
Query: 176 FRRFLVDS---PYMWWPESLVQVSLFRAFHEKEKRPRGGTSRLQFF--FVVFVASFAYYI 230
+ L+ ++ P ++V + + R F E +G S F FV+ Y
Sbjct: 209 LKIALLGKLGRVFLLIPLTIVLILIMR-FRSSESSSKGRISIPYFLIGFVIMALVNTYVT 267
Query: 231 IPGYFFQAISTVSFVCLI 248
IP ++T+S +CL+
Sbjct: 268 IPSVLLNILNTISTICLL 285
>Y329_STAAN (Q7A7M5) Hypothetical UPF0324 membrane protein SA0329
Length = 331
Score = 32.0 bits (71), Expect = 6.1
Identities = 43/198 (21%), Positives = 78/198 (38%), Gaps = 21/198 (10%)
Query: 56 SCILLAFVNQFLGYRTNPLKITSVSAQIIALPLGKLMAATLPTKPIQVPFTTWSFSLNPG 115
S +++ FVN+ L N + V + +A ++ + +L
Sbjct: 104 SIVMMLFVNKLLHGDKNIALLLGVGTGVCGAAAIAAVAPIFKSREKDTAISIGIIALIGT 163
Query: 116 PFSLKEHVLITIFASSGSSGVYAINIITIVKAFYHRSIHPVAAYLLALSTQMLGYGWAGI 175
FSL + IF S ++ VY A+ S+H +A +LA G+G +
Sbjct: 164 IFSLIYTAIYAIF--SMTTNVYG--------AWSGVSLHEIAHVVLAG-----GFGGSDA 208
Query: 176 FRRFLVDS---PYMWWPESLVQVSLFRAFHEKEKRPRGGTSRLQFF--FVVFVASFAYYI 230
+ L+ ++ P ++V + + R F E +G S F FV+ Y
Sbjct: 209 LKIALLGKLGRVFLLIPLTIVLILIMR-FRSSESSSKGRISIPYFLIGFVIMALVNTYVT 267
Query: 231 IPGYFFQAISTVSFVCLI 248
IP ++T+S +CL+
Sbjct: 268 IPSVLLNILNTISTICLL 285
>F123_CORVE (O74631) Protein FDD123 (CvHSP30/1)
Length = 283
Score = 32.0 bits (71), Expect = 6.1
Identities = 27/105 (25%), Positives = 43/105 (40%), Gaps = 13/105 (12%)
Query: 610 LNWFFLIGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAG--ASDIPPVRSVNYITWGIVG 667
+ WF L+ + L + F++I ++I G A+ +P Y T+G+
Sbjct: 98 IQWFITFPLLLLELLLATGLSLSDIFTTLFMSIVLVITGLVAALVPSTYKWGYYTFGVSA 157
Query: 668 IFFNFYVYRKFKAWWARHTYILSAG-------LDAGVAFIGLLLY 705
+F+ +YV W HT + G L AG LLLY
Sbjct: 158 LFYIWYVL----LWHGPHTTFAAGGVLRRGYILAAGYLSFLLLLY 198
>ELS_CHICK (P07916) Elastin precursor (Tropoelastin) (Fragment)
Length = 750
Score = 32.0 bits (71), Expect = 6.1
Identities = 17/40 (42%), Positives = 22/40 (54%), Gaps = 6/40 (15%)
Query: 259 GSGMKGLGIGSFGLDWNTVAGFLGSPLAVPGFAIINIMAG 298
G G+ GLG+G G AG LG+ + VPGF + I G
Sbjct: 675 GLGVGGLGVGGLG------AGGLGAGVGVPGFGVSPIFPG 708
>DGOT_ECOLI (P31457) D-galactonate transporter
Length = 430
Score = 31.6 bits (70), Expect = 8.0
Identities = 34/119 (28%), Positives = 50/119 (41%), Gaps = 19/119 (15%)
Query: 410 VHTRIMKKNYEQVPEWWFVTILILMVMMALLACEGFGKQLQLPWWGILLSLTIALIFTLP 469
+ T IM NY P M++M L+A FG W ++ SL + L
Sbjct: 324 ISTCIMGANYTNDP----------MMIMCLMALAFFGNGFASITWSLVSSLAPMRLIGLT 373
Query: 470 IGVIEATTNIRSGLNVIT-ELVIGFIYPGKPLANVAFKTYGHISMVQALGFL---GDFK 524
GV N GL IT LV+G++ G A A ++++ AL ++ GD K
Sbjct: 374 GGVF----NFAGGLGGITVPLVVGYLAQGYGFA-PALVYISAVALIGALSYILLVGDVK 427
>CT22_HUMAN (Q8N2K0) Protein C20orf22
Length = 398
Score = 31.6 bits (70), Expect = 8.0
Identities = 16/49 (32%), Positives = 26/49 (52%), Gaps = 6/49 (12%)
Query: 607 YPELNWFFLIGLIAPVPVWLLSLKFPNQKWIQFINIPIIIAGASDIPPV 655
+P +WFFL P+ +KF N + ++ I+ P++I A D P V
Sbjct: 294 FPGFDWFFLD------PITSSGIKFANDENVKHISCPLLILHAEDDPVV 336
>ANL4_HUMAN (Q9BY76) Angiopoietin-related protein 4 precursor
(Angiopoietin-like 4) (Hepatic
fibrinogen/angiopoietin-related protein) (HFARP)
(Angiopoietin-like protein PP1158)
Length = 406
Score = 31.6 bits (70), Expect = 8.0
Identities = 14/32 (43%), Positives = 20/32 (61%)
Query: 77 TSVSAQIIALPLGKLMAATLPTKPIQVPFTTW 108
T+ S Q+ A G+L A T+P + VPF+TW
Sbjct: 298 TAYSLQLTAPVAGQLGATTVPPSGLSVPFSTW 329
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.326 0.141 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 88,350,681
Number of Sequences: 164201
Number of extensions: 3824997
Number of successful extensions: 10656
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 10626
Number of HSP's gapped (non-prelim): 23
length of query: 745
length of database: 59,974,054
effective HSP length: 118
effective length of query: 627
effective length of database: 40,598,336
effective search space: 25455156672
effective search space used: 25455156672
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 70 (31.6 bits)
Medicago: description of AC143341.7


