

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143341.10 + phase: 0
(633 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
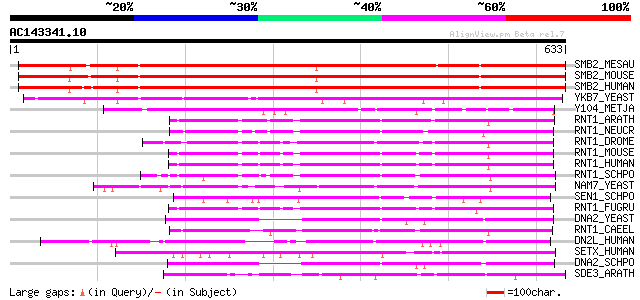
Score E
Sequences producing significant alignments: (bits) Value
SMB2_MESAU (Q60560) DNA-binding protein SMUBP-2 (Immunoglobulin ... 526 e-149
SMB2_MOUSE (P40694) DNA-binding protein SMUBP-2 (Immunoglobulin ... 524 e-148
SMB2_HUMAN (P38935) DNA-binding protein SMUBP-2 (Immunoglobulin ... 520 e-147
YKB7_YEAST (P34243) Hypothetical 78.3 kDa protein in RAM2-ATP7 i... 342 2e-93
Y104_METJA (Q57568) Hypothetical ATP-binding protein MJ0104 288 2e-77
RNT1_ARATH (Q9FJR0) Regulator of nonsense transcripts 1 homolog 216 2e-55
RNT1_NEUCR (Q9HEH1) Regulator of nonsense transcripts 1 homolog 214 4e-55
RNT1_DROME (Q9VYS3) Regulator of nonsense transcripts 1 homolog 209 2e-53
RNT1_MOUSE (Q9EPU0) Regulator of nonsense transcripts 1 (Nonsens... 203 1e-51
RNT1_HUMAN (Q92900) Regulator of nonsense transcripts 1 (Nonsens... 203 1e-51
RNT1_SCHPO (Q09820) Regulator of nonsense transcripts 1 homolog 194 8e-49
NAM7_YEAST (P30771) NAM7 protein ((Nuclear accomodation of mitoc... 194 8e-49
SEN1_SCHPO (Q92355) Helicase sen1 (EC 3.6.1.-) (Endonuclease sen1) 187 6e-47
RNT1_FUGRU (Q98TR3) Putative regulator of nonsense transcripts 1 178 4e-44
DNA2_YEAST (P38859) DNA replication helicase DNA2 178 4e-44
RNT1_CAEEL (O76512) Regulator of nonsense transcripts 1 (Nonsens... 175 3e-43
DN2L_HUMAN (P51530) DNA2-like homolog (DNA replication helicase-... 171 5e-42
SETX_HUMAN (Q7Z333) Probable helicase senataxin (EC 3.6.1.-) (SE... 163 1e-39
DNA2_SCHPO (Q9URU2) DNA replication helicase dna2 159 2e-38
SDE3_ARATH (Q8GYD9) Probable RNA helicase SDE3 (EC 3.6.1.-) (Sil... 150 1e-35
>SMB2_MESAU (Q60560) DNA-binding protein SMUBP-2 (Immunoglobulin MU
binding protein 2) (SMUBP-2) (Insulin II gene
enhancer-binding protein) (RIPE3B-binding complex 3B2
P110 subunit) (RIP-1)
Length = 989
Score = 526 bits (1356), Expect = e-149
Identities = 301/637 (47%), Positives = 406/637 (63%), Gaps = 22/637 (3%)
Query: 11 LFFFHQDAEISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQSTKAD-- 68
L +DAE+ + +L Q RG +L L+ TGL G+ L+ + K
Sbjct: 16 LLELERDAEVEERRSWQEHSSLKELQSRGVCLLKLQVSSQCTGLYGQRLVTFEPRKLGPV 75
Query: 69 -VLPAHKFGTHDVVVL-KLNKADLGSPALGQGVVYRLKDSSITVAFDDIPEDGLN----S 122
VLP++ F + D+V L N+ S L GV+ R+ S+TVAFD+ + LN +
Sbjct: 76 VVLPSNSFTSGDIVGLYDANE----SSQLATGVLTRITQKSVTVAFDESHDFQLNLDREN 131
Query: 123 PLRLEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSINKN 182
RL K+AN+VTY R+K AL+ L K H GPAS LI VL G P+ + + FT N
Sbjct: 132 TYRLLKLANDVTYKRLKKALMTLKK-YHSGPASSLIDVLLGGSSPSPTTEIPPFTFYNTA 190
Query: 183 LDYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNI 242
LD SQK+A+S AL+ K V ++HGPPGTGKTTTVVEIILQ VK+G KIL CA SN+AVDN+
Sbjct: 191 LDPSQKEAVSFALAQKEVAIIHGPPGTGKTTTVVEIILQAVKQGLKILCCAPSNVAVDNL 250
Query: 243 VERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTK 302
VERL + +++R+GHPARLL +LDA + R DN+ + DIRK++ + GK KT+
Sbjct: 251 VERLALCKKRILRLGHPARLLESAQQHSLDAVLARSDNAQIVADIRKDIDQVFGKNKKTQ 310
Query: 303 EKNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASS----KKLGNTSFDL 358
+K + + E++ L +E ++R++ A+ + +DV+L T GASS K L FD+
Sbjct: 311 DKREKSNFRNEIKLLRKELKEREEAAIVQSLTAADVVLATNTGASSDGPLKLLPENHFDV 370
Query: 359 VIIDEAAQALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYG 418
V++DE AQALE +CWIPLLK +CILAGDH QLPPT S +A GL R+L ERL E +G
Sbjct: 371 VVVDECAQALEASCWIPLLKAPKCILAGDHRQLPPTTISHKAALAGLSRSLMERLVEKHG 430
Query: 419 DEVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLI 478
MLTVQYRMHQ I W+S+ +Y+ ++ AH VA H+L DL GV T T LLLI
Sbjct: 431 AGAVRMLTVQYRMHQAITRWASEAMYHGQLTAHPSVAGHLLKDLPGVADTEETSVPLLLI 490
Query: 479 DTAGCDMEEKKDEEDSTL--NEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKM 536
DTAGC + E DEEDS N GE + H + LV +GV DI +I PY QV LL+
Sbjct: 491 DTAGCGLLE-LDEEDSQSKGNPGEVRLVTLHIQALVDAGVHAGDIAVIAPYNLQVDLLR- 548
Query: 537 LKNKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQC 596
++ N ++EI +VDGFQGREKEA+I++ VRSN K EVGFL++ RR+NVAVTRARR
Sbjct: 549 -QSLSNKHPELEIKSVDGFQGREKEAVILTFVRSNRKGEVGFLAEDRRINVAVTRARRHV 607
Query: 597 CIVCDTETVSSDGFLKRLIEYFEEHGEYQSASEYQNE 633
++CD+ TV++ FLK L++YF EHGE ++A EY ++
Sbjct: 608 AVICDSRTVNNHAFLKTLVDYFTEHGEVRTAFEYLDD 644
>SMB2_MOUSE (P40694) DNA-binding protein SMUBP-2 (Immunoglobulin MU
binding protein 2) (SMUBP-2) (Cardiac transcription
factor 1) (CATF1)
Length = 993
Score = 524 bits (1349), Expect = e-148
Identities = 293/635 (46%), Positives = 407/635 (63%), Gaps = 18/635 (2%)
Query: 11 LFFFHQDAEISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQSTK---A 67
L +DAE+ + +L Q RG +L L+ +TGL G+ L+ + K A
Sbjct: 16 LLELERDAEVEERRSWQEHSSLRELQSRGVCLLKLQVSSQRTGLYGQRLVTFEPRKFGPA 75
Query: 68 DVLPAHKFGTHDVVVLKLNKADLGSPALGQGVVYRLKDSSITVAFDDIPEDGLN----SP 123
VLP++ F + D+V L + L GV+ R+ S+TVAFD+ + LN +
Sbjct: 76 VVLPSNSFTSGDIVGLYDTNEN---SQLATGVLTRITQKSVTVAFDESHDLQLNLDRENT 132
Query: 124 LRLEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSINKNL 183
RL K+AN+VTY R+K AL+ L K H GPAS LI +L G P+ + + + N L
Sbjct: 133 YRLLKLANDVTYKRLKKALMTLKK-YHSGPASSLIDILLGSSTPSPAMEIPPLSFYNTTL 191
Query: 184 DYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIV 243
D SQK+A+S AL+ K + ++HGPPGTGKTTTVVEIILQ VK+G K+L CA SNIAVDN+V
Sbjct: 192 DLSQKEAVSFALAQKELAIIHGPPGTGKTTTVVEIILQAVKQGLKVLCCAPSNIAVDNLV 251
Query: 244 ERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKE 303
ERL + +++R+GHPARLL V +LDA + R DN+ + DIR+++ + GK KT++
Sbjct: 252 ERLALCKKRILRLGHPARLLESVQHHSLDAVLARSDNAQIVADIRRDIDQVFGKNKKTQD 311
Query: 304 KNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASS----KKLGNTSFDLV 359
K + + E++ L +E ++R++ A+ + +DV+L T GASS K L FD+V
Sbjct: 312 KREKGNFRSEIKLLRKELKEREEAAIVQSLTAADVVLATNTGASSDGPLKLLPEDYFDVV 371
Query: 360 IIDEAAQALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGD 419
++DE AQALE +CWIPLLK +CILAGDH QLPPT S A GL R+L ERLAE +G
Sbjct: 372 VVDECAQALEASCWIPLLKAPKCILAGDHRQLPPTTVSHRAALAGLSRSLMERLAEKHGA 431
Query: 420 EVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLID 479
V MLTVQYRMHQ IM W+S+ +Y+ + +H VA H+L DL GV T T LLLID
Sbjct: 432 GVVRMLTVQYRMHQAIMCWASEAMYHGQFTSHPSVAGHLLKDLPGVTDTEETRVPLLLID 491
Query: 480 TAGCDMEEKKDEE-DSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLK 538
TAGC + E ++E+ S N GE + H + LV +GV DI +I PY QV LL+ +
Sbjct: 492 TAGCGLLELEEEDSQSKGNPGEVRLVTLHIQALVDAGVQAGDIAVIAPYNLQVDLLR--Q 549
Query: 539 NKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCI 598
+ N ++EI +VDGFQGREKEA++++ VRSN K EVGFL++ RR+NVAVTRARR +
Sbjct: 550 SLSNKHPELEIKSVDGFQGREKEAVLLTFVRSNRKGEVGFLAEDRRINVAVTRARRHVAV 609
Query: 599 VCDTETVSSDGFLKRLIEYFEEHGEYQSASEYQNE 633
+CD+ TV++ FL+ L++YF EHGE ++A EY ++
Sbjct: 610 ICDSHTVNNHAFLETLVDYFTEHGEVRTAFEYLDD 644
>SMB2_HUMAN (P38935) DNA-binding protein SMUBP-2 (Immunoglobulin MU
binding protein 2) (SMUBP-2) (Glial factor-1) (GF-1)
Length = 993
Score = 520 bits (1339), Expect = e-147
Identities = 296/635 (46%), Positives = 414/635 (64%), Gaps = 17/635 (2%)
Query: 11 LFFFHQDAEISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQSTK---A 67
L +DAE+ + + +L Q RG +L L+ +TGL G+ L+ + + A
Sbjct: 16 LLELERDAEVEERRSWQENISLKELQSRGVCLLKLQVSSQRTGLYGRLLVTFEPRRYGSA 75
Query: 68 DVLPAHKFGTHDVVVLKLNKADLGSPALGQGVVYRLKDSSITVAFDDIPEDGLN----SP 123
LP++ F + D+V L + A+ GS L G++ R+ S+TVAFD+ + L+ +
Sbjct: 76 AALPSNSFTSGDIVGL-YDAANEGSQ-LATGILTRVTQKSVTVAFDESHDFQLSLDRENS 133
Query: 124 LRLEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSINKNL 183
RL K+AN+VTY R+K ALI L K H GPAS LI VLFG P+ + + T N L
Sbjct: 134 YRLLKLANDVTYRRLKKALIALKK-YHSGPASSLIEVLFGRSAPSPASEIHPLTFFNTCL 192
Query: 184 DYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIV 243
D SQK+A+S ALS K + ++HGPPGTGKTTTVVEIILQ VK+G K+L CA SNIAVDN+V
Sbjct: 193 DTSQKEAVSFALSQKELAIIHGPPGTGKTTTVVEIILQAVKQGLKVLCCAPSNIAVDNLV 252
Query: 244 ERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKE 303
ERL + +++R+GHPARLL V +LDA + R D++ DIRK++ + K KT++
Sbjct: 253 ERLALCKQRILRLGHPARLLESVQQHSLDAVLARSDSAQNVADIRKDIDQVFVKNKKTQD 312
Query: 304 KNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASS----KKLGNTSFDLV 359
K + + E++ L +E ++R++ A+ + + +++V+L T GAS+ K L + FD+V
Sbjct: 313 KREKSNFRNEIKLLRKELKEREEAAMLESLTSANVVLATNTGASADGPLKLLPESYFDVV 372
Query: 360 IIDEAAQALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGD 419
+IDE AQALE +CWIPLLK +CILAGDH QLPPT S +A GL +L ERLAE YG
Sbjct: 373 VIDECAQALEASCWIPLLKARKCILAGDHKQLPPTTVSHKAALAGLSLSLMERLAEEYGA 432
Query: 420 EVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLID 479
V LTVQYRMHQ IM W+S +Y +V AH+ VA H+L DL GV T T LLL+D
Sbjct: 433 RVVRTLTVQYRMHQAIMRWASDTMYLGQVTAHSSVARHLLRDLPGVAATEETGVPLLLVD 492
Query: 480 TAGCDM-EEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLK 538
TAGC + E ++++E S N GE + H + LV +GV DI +++PY QV LL+ +
Sbjct: 493 TAGCGLFELEEEDEQSKGNPGEVRLVSLHIQALVDAGVPARDIAVVSPYNLQVDLLR--Q 550
Query: 539 NKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCI 598
+ + ++EI +VDGFQGREKEA+I+S VRSN K EVGFL++ RR+NVAVTRARR +
Sbjct: 551 SLVHRHPELEIKSVDGFQGREKEAVILSFVRSNRKGEVGFLAEDRRINVAVTRARRHVAV 610
Query: 599 VCDTETVSSDGFLKRLIEYFEEHGEYQSASEYQNE 633
+CD+ TV++ FLK L+EYF +HGE ++A EY ++
Sbjct: 611 ICDSRTVNNHAFLKTLVEYFTQHGEVRTAFEYLDD 645
>YKB7_YEAST (P34243) Hypothetical 78.3 kDa protein in RAM2-ATP7
intergenic region
Length = 683
Score = 342 bits (876), Expect = 2e-93
Identities = 247/672 (36%), Positives = 375/672 (55%), Gaps = 65/672 (9%)
Query: 16 QDAEISSSIATGASRNLDTAQKRGSTILNLKCVDVQTGLMGKSLIELQSTKA--DVLPAH 73
QD + +S + T S + + G I N+ ++++GL+GK +EL A D +
Sbjct: 19 QDIQTTSRLLTTLS--IQQLVQNGLAINNIHLENIRSGLIGKLYMELGPNLAVNDKIQRG 76
Query: 74 KFGTHDVVVL----------------KLNKADLGSPALGQGVVYRLKDSSITVAFDDIPE 117
D+V++ K+++ G A GVVY++ D+ IT+A ++ +
Sbjct: 77 DIKVGDIVLVRPAKTKVNTKTKPKVKKVSEDSNGEQAECSGVVYKMSDTQITIALEESQD 136
Query: 118 DGLN-----SPLRLEKVANEVTYHRMKDALIQLSKGVHKGPASD-LIPVLFGERQ--PTV 169
S L + K N VTY+RM+ + +LS+ P D +I L ER P
Sbjct: 137 VIATTFYSYSKLYILKTTNVVTYNRMESTMRKLSE--ISSPIQDKIIQYLVNERPFIPNT 194
Query: 170 SKKDVVFTSINKNLDYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRG--S 227
+ + + +N NL+ SQK AI+ A+++ ++ ++HGPPGTGKT T++E+I Q + +
Sbjct: 195 NSFQNIKSFLNPNLNDSQKTAINFAINN-DLTIIHGPPGTGKTFTLIELIQQLLIKNPEE 253
Query: 228 KILACAASNIAVDNIVERLVP--HRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLAN 285
+IL C SNI+VD I+ERL P L+RIGHPARLL +LD +L N+ +
Sbjct: 254 RILICGPSNISVDTILERLTPLVPNNLLLRIGHPARLLDSNKRHSLD--ILSKKNT-IVK 310
Query: 286 DIRKEMKVLNGKLLKTKEKNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIG 345
DI +E+ L + K K R+E E++ L ++ +KR+ + D+I S +++TTL G
Sbjct: 311 DISQEIDKLIQENKKLKNYKQRKENWNEIKLLRKDLKKREFKTIKDLIIQSRIVVTTLHG 370
Query: 346 ASSKKLGNTS--------FDLVIIDEAAQALEVACWIPLLKGT----RCILAGDHLQLPP 393
+SS++L + FD +IIDE +QA+E CWIPL+ + +LAGD+ QLPP
Sbjct: 371 SSSRELCSLYRDDPNFQLFDTLIIDEVSQAMEPQCWIPLIAHQNQFHKLVLAGDNKQLPP 430
Query: 394 TIQSVEAEK--KGLGRTLFERLAELYGD-EVTSMLTVQYRMHQLIMDWSSKELYNSKVKA 450
TI++ + + L TLF+R+ +++ ++ L VQYRM+Q IM++ S +YN K+ A
Sbjct: 431 TIKTEDDKNVIHNLETTLFDRIIKIFPKRDMVKFLNVQYRMNQKIMEFPSHSMYNGKLLA 490
Query: 451 HACVASHMLYDLEGVKKTSS-----TEPTLLLIDTAGCDMEEKKDEED---STLNEGESE 502
A VA+ +L DL V T S T+ L+ DT G + +E DE S NEGE
Sbjct: 491 DATVANRLLIDLPTVDATPSEDDDDTKIPLIWYDTQGDEFQETADEATILGSKYNEGEIA 550
Query: 503 VAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKNKENSLKDIEISTVDGFQGREKEA 562
+ H + L V + IG+I+PY AQV LK L + E L DIEISTVDGFQGREK+
Sbjct: 551 IVKEHIENLRSFNVPENSIGVISPYNAQVSHLKKLIHDELKLTDIEISTVDGFQGREKDV 610
Query: 563 IIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDTETVSSDG--FLKRLIEYFEE 620
II+S+VRSN K EVGFL + RR+NVA+TR RRQ +V + E + G +LK E+ EE
Sbjct: 611 IILSLVRSNEKFEVGFLKEERRLNVAMTRPRRQLVVVGNIEVLQRCGNKYLKSWSEWCEE 670
Query: 621 HGE--YQSASEY 630
+ + Y + +Y
Sbjct: 671 NADVRYPNIDDY 682
>Y104_METJA (Q57568) Hypothetical ATP-binding protein MJ0104
Length = 663
Score = 288 bits (738), Expect = 2e-77
Identities = 202/563 (35%), Positives = 311/563 (54%), Gaps = 68/563 (12%)
Query: 108 ITVAFD-DIPEDGLNSPLRLEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQ 166
I VAFD D+P+ +R++ N++T+ RMK+AL + ++ K L ++ G
Sbjct: 113 IDVAFDVDVPKWVYKERVRVDLYVNDITFKRMKEALREFARKRDK-----LAYIILGIEH 167
Query: 167 PTVS-KKDVVFTSINKNLDYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKR 225
P ++D+ +KNL+ SQK A+ KA+ S++++L+HGPPGTGKT T+ E+I+QEVK
Sbjct: 168 PEKPLREDIKLEFYDKNLNESQKLAVKKAVLSRDLYLIHGPPGTGKTRTITEVIVQEVKF 227
Query: 226 GS-KILACAASNIAVDNIVERLVPHR--VKLVRIGHPARLLPQVVDSALDAQVLRGDNSG 282
K+LA A SNIA DNI+E L+ +K+VR+GHP R+ ++ +L + +
Sbjct: 228 NKHKVLATADSNIAADNILEYLIKKYPDLKVVRVGHPTRISKDLIQHSLPYLIENHEKYQ 287
Query: 283 LANDIR---KEMKVLNGKLLK-----------------TKEKNTRREIQKE--------- 313
+R KE+K K LK K K + R I KE
Sbjct: 288 EILALREKIKEIKEQRDKFLKPSPRWRRGMSDEQILKVAKRKKSYRGIPKEKIVSMAEWI 347
Query: 314 -----LRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFDLVIIDEAAQAL 368
++ + + + + +++ +DVI+ T A S+ L FD+++IDE +QA+
Sbjct: 348 IRNKKIKRIINNLDEITEKIMNEILAEADVIVATNSMAGSEILKGWEFDVIVIDEGSQAM 407
Query: 369 EVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDEVTSMLTVQ 428
E +C IP++KG + I+AGDH QLPPT+ S E + L +TLFERL + Y E +S+L +Q
Sbjct: 408 EPSCLIPIVKGRKLIMAGDHKQLPPTVLS---ENEELKKTLFERLIKKY-PEFSSILEIQ 463
Query: 429 YRMHQLIMDWSSKELYNSKVKAHACVASHMLYDL------EGVKKTSSTEPTLLLIDTAG 482
YRM++ IM++ +K YN+K+KA V + L DL + V + E + I+ G
Sbjct: 464 YRMNEKIMEFPNKMFYNNKLKADESVKNITLLDLVKEEEIDEVDRDIINEIPVQFINVEG 523
Query: 483 CDMEEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKNKEN 542
+E K E S N E+E + K+LV+ + +ITPY AQV L+ L + N
Sbjct: 524 --IERKDKESPSYYNIEEAEKVLEIVKKLVKYKI---PTNVITPYDAQVRYLRRLFEEHN 578
Query: 543 SLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDT 602
DIE++TVDGFQGRE EAI+IS VR+ K GFL D RR+NVA+TRA+R+ ++ +
Sbjct: 579 I--DIEVNTVDGFQGRENEAIVISFVRT---KNFGFLKDLRRLNVAITRAKRKLILIGNE 633
Query: 603 ETVSSDGFLKRLIEYF----EEH 621
+ D +I++ EEH
Sbjct: 634 NLLKQDKVYNEMIKWAKSVEEEH 656
>RNT1_ARATH (Q9FJR0) Regulator of nonsense transcripts 1 homolog
Length = 1235
Score = 216 bits (549), Expect = 2e-55
Identities = 147/446 (32%), Positives = 232/446 (51%), Gaps = 26/446 (5%)
Query: 183 LDYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGS-KILACAASNIAVDN 241
L+ SQ +A+ K++ K + L+ GPPGTGKT T I+ K+G ++L CA SN+AVD
Sbjct: 486 LNASQVNAV-KSVLQKPISLIQGPPGTGKTVTSAAIVYHMAKQGQGQVLVCAPSNVAVDQ 544
Query: 242 IVERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKT 301
+ E++ +K+VR+ +R + V S ++ L L + E+ L + LK
Sbjct: 545 LAEKISATGLKVVRLCAKSR---EAVSSPVEYLTLHYQVRHLDTSEKSELHKL--QQLKD 599
Query: 302 KEKNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFDLVII 361
++ +K+ + L R + ++ +++DVI T +GA+ +L N F V+I
Sbjct: 600 EQGELSSSDEKKYKNLKRATER-------EITQSADVICCTCVGAADLRLSNFRFRQVLI 652
Query: 362 DEAAQALEVACWIPLLKGTR-CILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDE 420
DE+ QA E C IPL+ G + +L GDH QL P I +A + GL ++LFERL L
Sbjct: 653 DESTQATEPECLIPLVLGVKQVVLVGDHCQLGPVIMCKKAARAGLAQSLFERLVTLGIKP 712
Query: 421 VTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLIDT 480
+ L VQYRMH + ++ S Y ++ + ++ + P +
Sbjct: 713 IR--LQVQYRMHPALSEFPSNSFYEGTLQNGVTIIERQTTGIDFPWPVPN-RPMFFYVQL 769
Query: 481 AGCDMEEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKNK 540
EE S LN E+ ++SGV+PS IG+ITPY Q + +
Sbjct: 770 G---QEEISASGTSYLNRTEAANVEKLVTAFLKSGVVPSQIGVITPYEGQRAYIVNYMAR 826
Query: 541 ENSL-----KDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQ 595
SL K+IE+++VD FQGREK+ II+S VRSN + +GFL+D RR+NVA+TRAR
Sbjct: 827 NGSLRQQLYKEIEVASVDSFQGREKDYIILSCVRSNEHQGIGFLNDPRRLNVALTRARYG 886
Query: 596 CCIVCDTETVSSDGFLKRLIEYFEEH 621
I+ + + +S L+ +++EH
Sbjct: 887 IVILGNPKVLSKQPLWNGLLTHYKEH 912
>RNT1_NEUCR (Q9HEH1) Regulator of nonsense transcripts 1 homolog
Length = 1093
Score = 214 bits (546), Expect = 4e-55
Identities = 148/445 (33%), Positives = 237/445 (53%), Gaps = 28/445 (6%)
Query: 183 LDYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRG-SKILACAASNIAVDN 241
L+ SQ AI + LS+ + L+ GPPGTGKT T II K S++L CA SN+AVD
Sbjct: 456 LNASQIAAIKQVLSNP-LSLIQGPPGTGKTVTSATIIYHLAKMSNSQVLVCAPSNVAVDQ 514
Query: 242 IVERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKT 301
+ ER+ +K+VR+ +R + V+S++ L N KE+ L LKT
Sbjct: 515 LCERIHRTGLKVVRLTAKSR---EDVESSVSFLALH--EQVRMNTTNKELDGL--VKLKT 567
Query: 302 KEKNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFDLVII 361
+ + +K + L+R+ + ++++ +DV+ T +GA +L F V+I
Sbjct: 568 ETGELSSQDEKRFKQLTRQAER-------EILQNADVVCCTCVGAGDPRLSKMKFRNVLI 620
Query: 362 DEAAQALEVACWIPLLKGTR-CILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDE 420
DE+ Q+ E C IPL+ G + +L GDH QL P I + +A K GL ++LFERL +L
Sbjct: 621 DESTQSAEPECMIPLVLGCKQVVLVGDHKQLGPVIMNKKAAKAGLNQSLFERLVKLQFTP 680
Query: 421 VTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLIDT 480
+ L VQYRMH + ++ S Y ++ A + D++ T ++
Sbjct: 681 IR--LKVQYRMHPCLSEFPSNMFYEGSLQNGVTAAERLRKDVDFPWPVPETP----MMFW 734
Query: 481 AGCDMEEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQ-VVLLKMLKN 539
+ EE S LN E+ R ++GV P+DIG+ITPY Q ++ ++N
Sbjct: 735 SNLGNEEISASGTSYLNRTEAANVEKIVTRFFKAGVKPADIGVITPYEGQRSYIVNTMQN 794
Query: 540 ----KENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQ 595
K+ S +++E+++VD FQGREK+ I++S VRSN + +GFLSD RR+NVA+TRA+
Sbjct: 795 TGTFKKESYREVEVASVDAFQGREKDFIVLSCVRSNENQGIGFLSDPRRLNVALTRAKYG 854
Query: 596 CCIVCDTETVSSDGFLKRLIEYFEE 620
I+ + + + L+ +F++
Sbjct: 855 LVIIGNPKVLCKHELWHHLLVHFKD 879
>RNT1_DROME (Q9VYS3) Regulator of nonsense transcripts 1 homolog
Length = 1180
Score = 209 bits (532), Expect = 2e-53
Identities = 156/477 (32%), Positives = 237/477 (48%), Gaps = 36/477 (7%)
Query: 152 GPASDLIPVLFGERQPTVSKKDVVFTSINKNLDYSQKDAISKALSSKNVFLLHGPPGTGK 211
G A VLF QP + + +N++ Y+ K A+ + LS L+ GPPGTGK
Sbjct: 426 GRADSNDEVLFRGPQPKLFSAPHL-PDLNRSQVYAVKHALQRPLS-----LIQGPPGTGK 479
Query: 212 TTTVVEIILQEVK-RGSKILACAASNIAVDNIVERLVPHRVKLVRIGHPARLLPQVVDSA 270
T T I+ Q VK G +L CA SN AVD + E++ +K+VR+ +R + +DS
Sbjct: 480 TVTSATIVYQLVKLHGGTVLVCAPSNTAVDQLTEKIHRTNLKVVRVCAKSR---EAIDSP 536
Query: 271 LDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKEKNTRREIQKELRTLSREERKRQQLAVT 330
+ L + + E+K L +T E ++ E K R L R A
Sbjct: 537 VSFLALHNQIRNM--ETNSELKKLQQLKDETGELSSADE--KRYRNLKRA-------AEN 585
Query: 331 DVIKTSDVILTTLIGASSKKLGNTSFDLVIIDEAAQALEVACWIPLLKGTR-CILAGDHL 389
+++ +DVI T +GA +L F ++IDE+ Q+ E C +P++ G + IL GDH
Sbjct: 586 QLLEAADVICCTCVGAGDGRLSRVKFTSILIDESMQSTEPECMVPVVLGAKQLILVGDHC 645
Query: 390 QLPPTIQSVEAEKKGLGRTLFERLAELYGDEVTSMLTVQYRMHQLIMDWSSKELYNSKVK 449
QL P + +A + GL ++LFERL L L VQYRMH + + S Y ++
Sbjct: 646 QLGPVVMCKKAARAGLSQSLFERLVVL--GIRPFRLEVQYRMHPELSQFPSNFFYEGSLQ 703
Query: 450 AHACVASHML-YDLEGVKKTSSTEPTLLLIDTAGCDMEEKKDEEDSTLNEGESEVAMAHA 508
C L D + E + + T G EE S LN E+
Sbjct: 704 NGVCAEDRRLKLDFPWPQP----ERPMFFLVTQG--QEEIAGSGTSFLNRTEAANVEKIT 757
Query: 509 KRLVQSGVLPSDIGIITPYAAQVVLLKMLKNKENSL-----KDIEISTVDGFQGREKEAI 563
R +++G+ P IGIITPY Q L + SL ++IEI++VD FQGREK+ I
Sbjct: 758 TRFLKAGIKPEQIGIITPYEGQRAYLVQYMQYQGSLHSRLYQEIEIASVDAFQGREKDII 817
Query: 564 IISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDTETVSSDGFLKRLIEYFEE 620
I+S VRSN ++ +GFL+D RR+NVA+TRA+ IV + + ++ L+ ++++
Sbjct: 818 IMSCVRSNERQGIGFLNDPRRLNVALTRAKFGIIIVGNPKVLAKQQLWNHLLNFYKD 874
>RNT1_MOUSE (Q9EPU0) Regulator of nonsense transcripts 1 (Nonsense
mRNA reducing factor 1) (NORF1) (Up-frameshift
suppressor 1 homolog)
Length = 1113
Score = 203 bits (517), Expect = 1e-51
Identities = 146/446 (32%), Positives = 227/446 (50%), Gaps = 28/446 (6%)
Query: 182 NLDYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGS-KILACAASNIAVD 240
+L++SQ A+ K + + + L+ GPPGTGKT T I+ ++G+ +L CA SNIAVD
Sbjct: 465 DLNHSQVYAV-KTVLQRPLSLIQGPPGTGKTVTSATIVYHLARQGNGPVLVCAPSNIAVD 523
Query: 241 NIVERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLK 300
+ E++ +K+VR+ +R + +DS + L + D E++ L +
Sbjct: 524 QLTEKIHQTGLKVVRLCAKSR---EAIDSPVSFLALHNQIRNM--DSMPELQKLQQLKDE 578
Query: 301 TKEKNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFDLVI 360
T E ++ E K R L R + +++ +DVI T +GA +L F ++
Sbjct: 579 TGELSSADE--KRYRALKRTAER-------ELLMNADVICCTCVGAGDPRLAKMQFRSIL 629
Query: 361 IDEAAQALEVACWIPLLKGTR-CILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGD 419
IDE+ QA E C +P++ G + IL GDH QL P + +A K GL ++LFERL L
Sbjct: 630 IDESTQATEPECMVPVVLGAKQLILVGDHCQLGPVVMCKKAAKAGLSQSLFERLVVLGIR 689
Query: 420 EVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLID 479
+ L VQYRMH + + S Y ++ A + + + + +
Sbjct: 690 PIR--LQVQYRMHPALSAFPSNIFYEGSLQNGVTAADRVKKGFDF--QWPQPDKPMFFYV 745
Query: 480 TAGCDMEEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKN 539
T G EE S LN E+ +L+++G P IGIITPY Q L
Sbjct: 746 TQG--QEEIASSGTSYLNRTEAANVEKITTKLLKAGAKPDQIGIITPYEGQRSYLVQYMQ 803
Query: 540 KENSL-----KDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARR 594
SL +++EI++VD FQGREK+ II+S VR+N + +GFL+D RR+NVA+TRAR
Sbjct: 804 FSGSLHTKLYQEVEIASVDAFQGREKDFIILSCVRANEHQGIGFLNDPRRLNVALTRARY 863
Query: 595 QCCIVCDTETVSSDGFLKRLIEYFEE 620
IV + + +S L+ Y++E
Sbjct: 864 GVIIVGNPKALSKQPLWNHLLSYYKE 889
>RNT1_HUMAN (Q92900) Regulator of nonsense transcripts 1 (Nonsense
mRNA reducing factor 1) (NORF1) (Up-frameshift
suppressor 1 homolog)
Length = 1129
Score = 203 bits (517), Expect = 1e-51
Identities = 146/446 (32%), Positives = 227/446 (50%), Gaps = 28/446 (6%)
Query: 182 NLDYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGS-KILACAASNIAVD 240
+L++SQ A+ K + + + L+ GPPGTGKT T I+ ++G+ +L CA SNIAVD
Sbjct: 481 DLNHSQVYAV-KTVLQRPLSLIQGPPGTGKTVTSATIVYHLARQGNGPVLVCAPSNIAVD 539
Query: 241 NIVERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLK 300
+ E++ +K+VR+ +R + +DS + L + D E++ L +
Sbjct: 540 QLTEKIHQTGLKVVRLCAKSR---EAIDSPVSFLALHNQIRNM--DSMPELQKLQQLKDE 594
Query: 301 TKEKNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFDLVI 360
T E ++ E K R L R + +++ +DVI T +GA +L F ++
Sbjct: 595 TGELSSADE--KRYRALKRTAER-------ELLMNADVICCTCVGAGDPRLAKMQFRSIL 645
Query: 361 IDEAAQALEVACWIPLLKGTR-CILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGD 419
IDE+ QA E C +P++ G + IL GDH QL P + +A K GL ++LFERL L
Sbjct: 646 IDESTQATEPECMVPVVLGAKQLILVGDHCQLGPVVMCKKAAKAGLSQSLFERLVVLGIR 705
Query: 420 EVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLID 479
+ L VQYRMH + + S Y ++ A + + + + +
Sbjct: 706 PIR--LQVQYRMHPALSAFPSNIFYEGSLQNGVTAADRVKKGFDF--QWPQPDKPMFFYV 761
Query: 480 TAGCDMEEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKN 539
T G EE S LN E+ +L+++G P IGIITPY Q L
Sbjct: 762 TQG--QEEIASSGTSYLNRTEAANVEKITTKLLKAGAKPDQIGIITPYEGQRSYLVQYMQ 819
Query: 540 KENSL-----KDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARR 594
SL +++EI++VD FQGREK+ II+S VR+N + +GFL+D RR+NVA+TRAR
Sbjct: 820 FSGSLHTKLYQEVEIASVDAFQGREKDFIILSCVRANEHQGIGFLNDPRRLNVALTRARY 879
Query: 595 QCCIVCDTETVSSDGFLKRLIEYFEE 620
IV + + +S L+ Y++E
Sbjct: 880 GVIIVGNPKALSKQPLWNHLLNYYKE 905
>RNT1_SCHPO (Q09820) Regulator of nonsense transcripts 1 homolog
Length = 925
Score = 194 bits (492), Expect = 8e-49
Identities = 147/488 (30%), Positives = 246/488 (50%), Gaps = 43/488 (8%)
Query: 150 HKGPASDLIPVLFGERQPTVSKKDVVFTSINKNLDYSQKDAISKALSSKNVFLLHGPPGT 209
HK D+ P + P+ D+ ++ K L+ SQ +A+ +A+ SK + L+ GPPGT
Sbjct: 365 HKLLGHDIPPSFLKPKLPS----DLSVPNLPK-LNASQSEAV-RAVLSKPLSLIQGPPGT 418
Query: 210 GKTTTVVEII-----LQEVKRGSK--ILACAASNIAVDNIVERLVPHRVKLVRIGHPARL 262
GKT T ++ +Q KR S +L CA SN+AVD + E++ +++VR+ +R
Sbjct: 419 GKTVTSASVVYHLATMQSRKRKSHSPVLVCAPSNVAVDQLAEKIHRTGLRVVRVAAKSR- 477
Query: 263 LPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKEKNTRREIQ--KELRTLSRE 320
+ ++S++ L E++ +LLK + +N IQ K+LR L
Sbjct: 478 --EDIESSVSFLSLHEQIKNYK--FNPELQ----RLLKLRSENNELSIQDEKKLRILVAA 529
Query: 321 ERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFDLVIIDEAAQALEVACWIPLLKGT 380
K ++++ + VI T +GA +++ F V+IDEA QA E C IPL+ G
Sbjct: 530 AEK-------ELLRAAHVICCTCVGAGDRRISKYKFRSVLIDEATQASEPECMIPLVLGA 582
Query: 381 R-CILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDEVTSMLTVQYRMHQLIMDWS 439
+ +L GDH QL P + + + L ++LFERL L L VQYRMH + ++
Sbjct: 583 KQVVLVGDHQQLGPVVMNKKVALASLSQSLFERLIILGNSPFR--LVVQYRMHPCLSEFP 640
Query: 440 SKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLIDTAGCDMEEKKDEEDSTLNEG 499
S Y ++ + + ++ +P L+ A EE S LN
Sbjct: 641 SNTFYEGTLQNGVTTSERIARHVD----FPWIQPDSPLMFYANFGQEELSASGTSFLNRT 696
Query: 500 ESEVAMAHAKRLVQSGVLPSDIGIITPYAAQ-VVLLKMLKNKENSLKD----IEISTVDG 554
E+ ++S VLP IGI+TPY Q +++ ++N + KD +E+++VD
Sbjct: 697 EASTCEKIVTTFLRSNVLPEQIGIVTPYDGQRSYIVQYMQNNGSMQKDLYKAVEVASVDA 756
Query: 555 FQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDTETVSSDGFLKRL 614
FQGREK+ II+S VRS+ + +GF++D RR+NVA+TRA+ ++ + + ++
Sbjct: 757 FQGREKDFIILSCVRSSEHQGIGFVNDPRRLNVALTRAKYGVIVLGNPKVLAKHALWYHF 816
Query: 615 IEYFEEHG 622
+ + +E G
Sbjct: 817 VLHCKEKG 824
>NAM7_YEAST (P30771) NAM7 protein ((Nuclear accomodation of
mitochondria 7 protein) Nonsense-mediated mRNA decay
protein 1) (Up-frameshift suppressor 1)
Length = 971
Score = 194 bits (492), Expect = 8e-49
Identities = 170/549 (30%), Positives = 267/549 (47%), Gaps = 51/549 (9%)
Query: 96 GQGVVYRLKDS---SITVAFDDI---PEDGLNSPLRLEKVANEVTYHRMKDALIQLSKGV 149
G+G + RL +S + T+ P L + E + +Y RM+DAL + +
Sbjct: 313 GRGYIVRLPNSFQDTFTLELKPSKTPPPTHLTTGFTAEFIWKGTSYDRMQDALKKFAID- 371
Query: 150 HKGPASDLIPVLFGERQPTVS-----KKDVVFTSINKNLDYSQKDAISKALSSKNVFLLH 204
K + L + G + +S K+ + + L+ SQ +A+S L + + L+
Sbjct: 372 KKSISGYLYYKILGHQVVDISFDVPLPKEFSIPNFAQ-LNSSQSNAVSHVLQ-RPLSLIQ 429
Query: 205 GPPGTGKTTTVVEIILQEVK-RGSKILACAASNIAVDNIVERLVPHRVKLVRIGHPARLL 263
GPPGTGKT T I+ K +IL CA SN+AVD++ +L +K+VR+ +R
Sbjct: 430 GPPGTGKTVTSATIVYHLSKIHKDRILVCAPSNVAVDHLAAKLRDLGLKVVRLTAKSR-- 487
Query: 264 PQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKEKNTRREIQKELRTLSREERK 323
+ V+S++ N L N + + K LLK K+ E+ LS + K
Sbjct: 488 -EDVESSVS-------NLALHNLVGRGAKGELKNLLKLKD---------EVGELSASDTK 530
Query: 324 RQQLAV----TDVIKTSDVILTTLIGASSKKLGNTSFDLVIIDEAAQALEVACWIPLLKG 379
R V +++ +DV+ T +GA K+L +T F V+IDE+ QA E C IP++KG
Sbjct: 531 RFVKLVRKTEAEILNKADVVCCTCVGAGDKRL-DTKFRTVLIDESTQASEPECLIPIVKG 589
Query: 380 TR-CILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDEVTSMLTVQYRMHQLIMDW 438
+ IL GDH QL P I +A GL ++LFERL L V L VQYRM+ + ++
Sbjct: 590 AKQVILVGDHQQLGPVILERKAADAGLKQSLFERLISL--GHVPIRLEVQYRMNPYLSEF 647
Query: 439 SSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLIDTAGCDMEEKKDEEDSTLNE 498
S Y ++ + + + K + ++ A EE S LN
Sbjct: 648 PSNMFYEGSLQNGVTIEQRTVPN----SKFPWPIRGIPMMFWANYGREEISANGTSFLNR 703
Query: 499 GESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLKNKENSL-KD----IEISTVD 553
E+ +L + GV P IG+ITPY Q + SL KD +E+++VD
Sbjct: 704 IEAMNCERIITKLFRDGVKPEQIGVITPYEGQRAYILQYMQMNGSLDKDLYIKVEVASVD 763
Query: 554 GFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQCCIVCDTETVSSDGFLKR 613
FQGREK+ II+S VR+N ++ +GFL D RR+NV +TRA+ I+ + +++ +
Sbjct: 764 AFQGREKDYIILSCVRANEQQAIGFLRDPRRLNVGLTRAKYGLVILGNPRSLARNTLWNH 823
Query: 614 LIEYFEEHG 622
L+ +F E G
Sbjct: 824 LLIHFREKG 832
>SEN1_SCHPO (Q92355) Helicase sen1 (EC 3.6.1.-) (Endonuclease sen1)
Length = 1687
Score = 187 bits (476), Expect = 6e-47
Identities = 159/506 (31%), Positives = 243/506 (47%), Gaps = 84/506 (16%)
Query: 187 QKDAISKALSSKNVFLLHGPPGTGKTTTVVEII------------------LQEVKRGSK 228
Q AI AL + L+ GPPGTGKT T++ II + + +
Sbjct: 1134 QAKAIMCALDNNGFTLIQGPPGTGKTKTIIGIISALLVDLSRYHITRPNQQSKSTESKQQ 1193
Query: 229 ILACAASNIAVDNIVERL---------VPHRVKLVRIGHPARLLPQVVDSALDAQV---L 276
IL CA SN AVD ++ RL + ++VRIG+P + V D +L+ Q L
Sbjct: 1194 ILLCAPSNAAVDEVLLRLKRGFLLENGEKYIPRVVRIGNPETINVSVRDLSLEYQTEKQL 1253
Query: 277 RGDNSG--------------------------------LANDIRKEMKVLNGKLL-KTKE 303
N G +A D+ ++ K L +L K E
Sbjct: 1254 LEVNQGAIDLGSLQELTRWRDTFYDCIQKIEELEKQIDVARDVAEDTKSLGKELQNKINE 1313
Query: 304 KNTRREIQKELRTLSREERKRQQL----AVTDVIKTSDVILTTLIGASSKKLGNTS--FD 357
KN + +EL++ S + K L A ++K +DV+ TL G+ + ++S F
Sbjct: 1314 KNLAEQKVEELQSQSFTKNKEVDLLRKKAQKAILKQADVVCATLSGSGHDLVAHSSLNFS 1373
Query: 358 LVIIDEAAQALEVACWIPLLKGT-RCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAEL 416
VIIDEAAQA+E+ IPL G +CIL GD QLPPT+ S +A ++LF R+ +
Sbjct: 1374 TVIIDEAAQAVELDTIIPLRYGAKKCILVGDPNQLPPTVLSKKAASLNYSQSLFVRIQKN 1433
Query: 417 YGDEVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLL 476
+ +++ +L++QYRMH I + SK+ Y+S+++ +M + V +
Sbjct: 1434 FSNQM-CLLSIQYRMHPDISHFPSKKFYDSRLED----GDNMAEKTQQVWHVNPKFTQYR 1488
Query: 477 LIDTAGCDMEEKKDEEDSTLNEGESEVAMAHAKRLVQSGV---LPSDIGIITPYAAQVVL 533
L D G +E+ ST N E E + L+ IG+ITPY +Q+
Sbjct: 1489 LFDVRG---KERTSNTMSTYNLEEVEYLVNMVDELLNKFPDVNFTGRIGVITPYRSQLHE 1545
Query: 534 LK---MLKNKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVT 590
L+ +K ++ + I+I TVDGFQG+EK+ I S V+S SK +GFL D RR+NVA+T
Sbjct: 1546 LRRAFKVKYGKSFMSTIDIQTVDGFQGQEKDIIFFSCVKSYSKHGIGFLRDFRRLNVALT 1605
Query: 591 RARRQCCIVCDTETVSSDGFLKRLIE 616
RAR I+ + ET+ +D L++
Sbjct: 1606 RARSSLLIIGNMETLKTDDLWGSLVD 1631
>RNT1_FUGRU (Q98TR3) Putative regulator of nonsense transcripts 1
Length = 1097
Score = 178 bits (451), Expect = 4e-44
Identities = 136/444 (30%), Positives = 219/444 (48%), Gaps = 26/444 (5%)
Query: 182 NLDYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGS-KILACAASNIAVD 240
+L++SQ A+ K + + + L+ GPPGTGKT T I+ ++G+ +L CA SNIAVD
Sbjct: 452 DLNHSQVYAV-KTVLQRPLSLIQGPPGTGKTVTSATIVYHLSRQGNGPVLVCAPSNIAVD 510
Query: 241 NIVERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLK 300
+ E++ +K+VR+ +R + ++S + L S + D E++ L +
Sbjct: 511 QLTEKIDKTGLKVVRLCAKSR---EAIESPVSFLALHNQISNM--DSMPELQKLQQLKDE 565
Query: 301 TKEKNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFDLVI 360
T E ++ E K R L R + +++ +DVI T + A +L F ++
Sbjct: 566 TGELSSADE--KRYRALKRTAER-------ELLMNADVIWCTCVRAGDPRLAKMQFRSIL 616
Query: 361 IDEAAQALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDE 420
IDE+ QA E C P+ G + ++ G+ + +A K GL ++LFERL L
Sbjct: 617 IDESTQATEPKCIGPVELGAKQLILGEITASWSCVMCKKAAKAGLSQSLFERLVVLGIRP 676
Query: 421 VTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLLLIDT 480
+ L VQYRMH + + S Y ++ + + + E + T
Sbjct: 677 IR--LQVQYRMHPALSAFPSNIFYEGSLQNGVTAGDRIKKGFDF--QWPQPEKPMFFYVT 732
Query: 481 AGCDMEEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQ----VVLLKM 536
G EE S LN E+ RL+++G P IGIITPY Q V ++
Sbjct: 733 QG--QEEIASSGTSYLNRTEAANVEKITTRLLKAGAKPDQIGIITPYEGQRSYLVQYMQF 790
Query: 537 LKNKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTRARRQC 596
+ L +EI++VD FQGREK+ II+S VR+N + +GFL+D RR+NVA+TRA+
Sbjct: 791 SGSLHTKLYQVEIASVDAFQGREKDFIILSCVRANEHQGIGFLNDPRRLNVALTRAKYGV 850
Query: 597 CIVCDTETVSSDGFLKRLIEYFEE 620
IV + + +S L+ ++E
Sbjct: 851 IIVGNPKALSKQPLWNNLLNNYKE 874
>DNA2_YEAST (P38859) DNA replication helicase DNA2
Length = 1522
Score = 178 bits (451), Expect = 4e-44
Identities = 140/463 (30%), Positives = 223/463 (47%), Gaps = 69/463 (14%)
Query: 178 SINKNLDYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNI 237
S + L+ +QK+AI K + +++ L+ G PGTGKTT + EII V G ++L + ++
Sbjct: 1047 SKDTTLNLNQKEAIDKVMRAEDYALILGMPGTGKTTVIAEIIKILVSEGKRVLLTSYTHS 1106
Query: 238 AVDNIVERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGK 297
AVDNI+ +L + ++R+G ++ P + ++ N L+
Sbjct: 1107 AVDNILIKLRNTNISIMRLGMKHKVHPDTQKYVPNYASVKSYNDYLSK------------ 1154
Query: 298 LLKTKEKNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSK--KLGNTS 355
I ++ V+ TT +G + L
Sbjct: 1155 -----------------------------------INSTSVVATTCLGINDILFTLNEKD 1179
Query: 356 FDLVIIDEAAQALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAE 415
FD VI+DEA+Q PL G R I+ GDH QLPP +++ A GL +LF+ E
Sbjct: 1180 FDYVILDEASQISMPVALGPLRYGNRFIMVGDHYQLPPLVKNDAARLGGLEESLFKTFCE 1239
Query: 416 LYGDEVTSMLTVQYRMHQLIMDWSSKELYNSKVKAH------ACVASHMLYDLEGVKKTS 469
+ + V LT+QYRM I+ S+ +Y++K+K + M L + S
Sbjct: 1240 KHPESVAE-LTLQYRMCGDIVTLSNFLIYDNKLKCGNNEVFAQSLELPMPEALSRYRNES 1298
Query: 470 ST---------EPT--LLLIDTAGCDMEEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLP 518
+ EPT ++ ++ C ++ E+D+ N GE+E+ + + ++ SGV
Sbjct: 1299 ANSKQWLEDILEPTRKVVFLNYDNCPDIIEQSEKDNITNHGEAELTLQCVEGMLLSGVPC 1358
Query: 519 SDIGIITPYAAQVVLLKMLKNKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVG- 577
DIG++T Y AQ+ LLK + NK N +EI T D FQGR+K+ IIISMVR NS+ G
Sbjct: 1359 EDIGVMTLYRAQLRLLKKIFNK-NVYDGLEILTADQFQGRDKKCIIISMVRRNSQLNGGA 1417
Query: 578 FLSDRRRMNVAVTRARRQCCIVCDTETVSSDGFLKRLIEYFEE 620
L + RR+NVA+TRA+ + I+ T+ S +K + EE
Sbjct: 1418 LLKELRRVNVAMTRAKSKLIIIGSKSTIGSVPEIKSFVNLLEE 1460
>RNT1_CAEEL (O76512) Regulator of nonsense transcripts 1 (Nonsense
mRNA reducing factor 1) (Up-frameshift suppressor 1
homolog)
Length = 1069
Score = 175 bits (444), Expect = 3e-43
Identities = 134/448 (29%), Positives = 221/448 (48%), Gaps = 37/448 (8%)
Query: 183 LDYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEV-KRGSKILACAASNIAVDN 241
L+ SQ A+ + L+ + + L+ GPPGTGKT I+ V K +L C+ SNIAVD+
Sbjct: 446 LNSSQMQAVKQVLT-RPLSLIQGPPGTGKTVVSATIVYHLVQKTEGNVLVCSPSNIAVDH 504
Query: 242 IVERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNG----K 297
+ E++ +K+VR+ +R + L Q ++KV+ G K
Sbjct: 505 LAEKIHKTGLKVVRLCARSREHSETTVPYLTLQ--------------HQLKVMGGAELQK 550
Query: 298 LLKTKEKNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFD 357
L++ K++ E + +LR + + K +L + +DVI T A+ +L
Sbjct: 551 LIQLKDEAGELEFKDDLRYMQLKRVKEHEL-----LAAADVICCTCSSAADARLSKIRTR 605
Query: 358 LVIIDEAAQALEVACWIPLLKGTR-CILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAEL 416
V+IDE+ QA E + +++G R +L GDH QL P + +A GL ++LFERL L
Sbjct: 606 TVLIDESTQATEPEILVSIMRGVRQLVLVGDHCQLGPVVICKKAAIAGLSQSLFERLV-L 664
Query: 417 YGDEVTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDLEGVKKTSSTEPTLL 476
G L VQYRMH ++ ++ S Y+ ++ + ++ +P
Sbjct: 665 LGIR-PFRLQVQYRMHPVLSEFPSNVFYDGSLQNGVTENDRHMTGVD----WHWPKPNKP 719
Query: 477 LIDTAGCDMEEKKDEEDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKM 536
EE S LN E+ +L+++GV P IG+IT Y Q +
Sbjct: 720 AFFWHCSGSEELSASGTSFLNRTEAANVEKLVSKLIKAGVQPHQIGVITSYEGQRSFIVN 779
Query: 537 LKNKENSL-----KDIEISTVDGFQGREKEAIIISMVRSNSKKEVGFLSDRRRMNVAVTR 591
+ + +L +++EI++VD FQGREK+ II++ VRSN +GFLSD RR+NVA+TR
Sbjct: 780 YMHTQGTLNSKLYENVEIASVDAFQGREKDYIIVTCVRSNDILGIGFLSDPRRLNVAITR 839
Query: 592 ARRQCCIVCDTETVSSDGFLKRLIEYFE 619
A+ +V + + ++ LI +++
Sbjct: 840 AKYGLVLVGNAKVLARHDLWHELINHYK 867
>DN2L_HUMAN (P51530) DNA2-like homolog (DNA replication helicase-like
homolog) (Fragment)
Length = 1077
Score = 171 bits (433), Expect = 5e-42
Identities = 168/620 (27%), Positives = 277/620 (44%), Gaps = 98/620 (15%)
Query: 36 QKRGSTILNL-KCVDVQTGLMGKSLIELQSTKADVLPAHKFGTHDVVVLKLNKADLGSPA 94
+K GS I NL + V+ G+ L Q K +P D V++ + L A
Sbjct: 490 EKSGSCIGNLIRMEHVKIVCDGQYLHNFQC-KHGAIPVTNLMAGDRVIVSGEERSLF--A 546
Query: 95 LGQGVVYRLKDSSITVAFDD----IPEDGL------------NSPL-RLEKVA-NEVTYH 136
L +G V + +++T D +PE L ++PL L K+ N
Sbjct: 547 LSRGYVKEINMTTVTCLLDRNLSVLPESTLFRLDQEEKNCDIDTPLGNLSKLMENTFVSK 606
Query: 137 RMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSINKNLDYSQKDAISKALS 196
+++D +I + S ++P KD V I K L+ Q+ A+ K L
Sbjct: 607 KLRDLIIDFREPQFISYLSSVLPH---------DAKDTV-ACILKGLNKPQRQAMKKVLL 656
Query: 197 SKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVDNIVERLVPHRVKLVRI 256
SK+ L+ G PGTGKTTT+ ++ G +L + ++ AVDNI+ +L ++ +R+
Sbjct: 657 SKDYTLIVGMPGTGKTTTICTLVRILYACGFSVLLTSYTHSAVDNILLKLAKFKIGFLRL 716
Query: 257 GHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLKTKEKNTRREIQKELRT 316
G ++ P + T+++ R + K L
Sbjct: 717 GQIQKVHPAIQQF-------------------------------TEQEICRSKSIKSLAL 745
Query: 317 LSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFDLVIIDEAAQALEVACWIPL 376
L EE QL ++ TT +G + FD I+DEA+Q + C PL
Sbjct: 746 L--EELYNSQL----------IVATTCMGINHPIFSRKIFDFCIVDEASQISQPICLGPL 793
Query: 377 LKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDEVTSMLTVQYRMHQLIM 436
R +L GDH QLPP + + EA G+ +LF+RL + LTVQYRM+ IM
Sbjct: 794 FFSRRFVLVGDHQQLPPLVLNREARALGMSESLFKRLEQ--NKSAVVQLTVQYRMNSKIM 851
Query: 437 DWSSKELYNSKVKAHACVASHMLYDLEGVKKTS---------STEPTLLLI---DTAGCD 484
S+K Y K++ + ++ + +L K S P L+ + + C
Sbjct: 852 SLSNKLTYEGKLECGSDKVANAVINLRHFKDVKLELEFYADYSDNPWLMGVFEPNNPVCF 911
Query: 485 MEEKK------DEEDSTLNEGESEVAMAHAKRLVQSGVLPSDIGIITPYAAQVVLLKMLK 538
+ K E+ N E+++ + V++G PSDIGII PY Q+ ++ L
Sbjct: 912 LNTDKVPAPEQVEKGGVSNVTEAKLIVFLTSIFVKAGCSPSDIGIIAPYRQQLKIINDLL 971
Query: 539 NKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVG-FLSDRRRMNVAVTRARRQCC 597
+ S+ +E++TVD +QGR+K +++S VRSN VG L D RR+NVA+TRA+ +
Sbjct: 972 AR--SIGMVEVNTVDKYQGRDKSIVLVSFVRSNKDGTVGELLKDWRRLNVAITRAKHKLI 1029
Query: 598 IVCDTETVSSDGFLKRLIEY 617
++ +++ L++L+ +
Sbjct: 1030 LLGCVPSLNCYPPLEKLLNH 1049
>SETX_HUMAN (Q7Z333) Probable helicase senataxin (EC 3.6.1.-) (SEN1
homolog)
Length = 2677
Score = 163 bits (413), Expect = 1e-39
Identities = 156/590 (26%), Positives = 271/590 (45%), Gaps = 98/590 (16%)
Query: 121 NSPLRLEKVANEVTYHRMKDALIQLSKGVHKGPASDLIPVLFGERQPTVSKKDVVFTSIN 180
N P L ++ N + + +L G + L + KD++ T+
Sbjct: 1866 NFPANLNELVNCIVISSLVTTQRKLKAMSLLGSRNQLARAVLNPNPMDFCTKDLLTTTSE 1925
Query: 181 KNLDY------SQKDAISKAL-------SSKNVFLLHGPPGTGKTTTVV----EIILQEV 223
+ + Y QK AI A S + L+HGPPGTGK+ T+V ++ +
Sbjct: 1926 RIIAYLRDFNEDQKKAIETAYAMVKHSPSVAKICLIHGPPGTGKSKTIVGLLYRLLTENQ 1985
Query: 224 KRG------------SKILACAASNIAVDNIVERLVPH----------------RVKLVR 255
++G +++L CA SN AVD ++++++ + LVR
Sbjct: 1986 RKGHSDENSNAKIKQNRVLVCAPSNAAVDELMKKIILEFKEKCKDKKNPLGNCGDINLVR 2045
Query: 256 IGHPARLLPQVVDSALDAQVLRGDNSGLANDIR---KEMKVLNGKLLKTKEKNTR----R 308
+G + +V+ +LD+QV L + ++ K + L+ +L + + R
Sbjct: 2046 LGPEKSINSEVLKFSLDSQVNHRMKKELPSHVQAMHKRKEFLDYQLDELSRQRALCRGGR 2105
Query: 309 EIQKEL--RTLSREERKRQQLAV-------------TDVIKTSDVILTTL-------IGA 346
EIQ++ +S+ ++RQ+LA + +I S +I TL + +
Sbjct: 2106 EIQRQELDENISKVSKERQELASKIKEVQGRPQKTQSIIILESHIICCTLSTSGGLLLES 2165
Query: 347 SSKKLGNTSFDLVIIDEAAQALEVACWIPLL-KGTRCILAGDHLQLPPTIQSVEAEKKGL 405
+ + G F VI+DEA Q+ E+ PL+ + + IL GD QLPPT+ S++A++ G
Sbjct: 2166 AFRGQGGVPFSCVIVDEAGQSCEIETLTPLIHRCNKLILVGDPKQLPPTVISMKAQEYGY 2225
Query: 406 GRTLFERLAELYGDEVTS---------MLTVQYRMHQLIMDWSSKELYNSKVKAHACVAS 456
+++ R L + V LTVQYRMH I + S +YN +K +
Sbjct: 2226 DQSMMARFCRLLEENVEHNMISRLPILQLTVQYRMHPDICLFPSNYVYNRNLKTNR---- 2281
Query: 457 HMLYDLEGVKKTSSTEPTLLLIDTAGCDMEEKKDEEDSTLNEGESEVAMAHAKRLV--QS 514
E ++ +S L+ G D E++D DS +N E ++ M K + +
Sbjct: 2282 ----QTEAIRCSSDWPFQPYLVFDVG-DGSERRDN-DSYINVQEIKLVMEIIKLIKDKRK 2335
Query: 515 GVLPSDIGIITPYAAQVVLLKMLKNKENSLKD-IEISTVDGFQGREKEAIIISMVRSNS- 572
V +IGIIT Y AQ +++ +KE K E+ TVD FQGR+K+ +I++ VR+NS
Sbjct: 2336 DVSFRNIGIITHYKAQKTMIQKDLDKEFDRKGPAEVDTVDAFQGRQKDCVIVTCVRANSI 2395
Query: 573 KKEVGFLSDRRRMNVAVTRARRQCCIVCDTETVSSDGFLKRLIEYFEEHG 622
+ +GFL+ +R+NV +TRA+ I+ T+ + +LI+ ++ G
Sbjct: 2396 QGSIGFLASLQRLNVTITRAKYSLFILGHLRTLMENQHWNQLIQDAQKRG 2445
>DNA2_SCHPO (Q9URU2) DNA replication helicase dna2
Length = 1398
Score = 159 bits (403), Expect = 2e-38
Identities = 134/458 (29%), Positives = 217/458 (47%), Gaps = 68/458 (14%)
Query: 181 KNLDYSQKDAISKALSSKNVFLLHGPPGTGKTTTVVEIILQEVKRGSKILACAASNIAVD 240
K L+ Q A+ K ++++ L+ G PGTGKTTT+ +I + + KIL + +++AVD
Sbjct: 931 KCLNEDQITALKKCHAAEHYSLILGMPGTGKTTTISSLIRSLLAKKKKILLTSFTHLAVD 990
Query: 241 NIVERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEMKVLNGKLLK 300
NI+ +L +VR+G P ++ P V + L D + L +
Sbjct: 991 NILIKLKGCDSTIVRLGSPHKIHPLVKEFCLTEGTTFDDLASLKH--------------- 1035
Query: 301 TKEKNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKLGNTSFDLVI 360
+ ++ + +G FD I
Sbjct: 1036 -------------------------------FYEDPQIVACSSLGVYHSIFNKRKFDYCI 1064
Query: 361 IDEAAQALEVACWIPLLKGTRCILAGDHLQLPPTIQSVEAEKKGLGRTLFERLAELYGDE 420
IDEA+Q C PL + +L GDH QLPP +++ K GL +LF+ L+E + +
Sbjct: 1065 IDEASQIPLPICLGPLQLAEKFVLVGDHYQLPPLVKNSRTSKDGLSLSLFKLLSEKHPEA 1124
Query: 421 VTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDL------EGVKKTSST--- 471
VT+ L +QYRM++ I SS+ +Y + + S L +G+ +SS+
Sbjct: 1125 VTT-LRLQYRMNEDINSLSSELIYGGNLVCGSKTISQKKLILPKAHLSDGLPDSSSSLHW 1183
Query: 472 -----EPTLLLIDTAGCDMEEKKDEEDSTL-NEGESEVAMAHAKRLVQSGVLPSDIGIIT 525
P+ +I D+ + + ++ L N E+ + ++ GV S IGII+
Sbjct: 1184 VNKLINPSHSVIFFNTDDILGVESKTNNILENHTEAFLIEQAVSSFLERGVKQSSIGIIS 1243
Query: 526 PYAAQVVLL-KMLKNKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKEVG-FLSDRR 583
Y +QV LL K LK S +IEI+TVD +QGR+K+ I+IS VRSNSK VG L D
Sbjct: 1244 IYKSQVELLSKNLK----SFTEIEINTVDRYQGRDKDIILISFVRSNSKNLVGELLRDWH 1299
Query: 584 RMNVAVTRARRQCCIVCDTETVSSDGFLKRLIEYFEEH 621
R+NVA++RA+ +C + T+SS + L++ E++
Sbjct: 1300 RLNVALSRAKVKCIMFGSLSTLSSSNIVSHLLKLLEKN 1337
>SDE3_ARATH (Q8GYD9) Probable RNA helicase SDE3 (EC 3.6.1.-)
(Silencing defective protein 3)
Length = 1002
Score = 150 bits (379), Expect = 1e-35
Identities = 142/479 (29%), Positives = 227/479 (46%), Gaps = 62/479 (12%)
Query: 176 FTSINKNLDYSQKDAISKALSSKNV--FLLHGPPGTGKTTTVVEIILQ--EVKRGSKILA 231
F I+ L+ Q +I L K +++HGPPGTGKT T+VE I+Q +R +++L
Sbjct: 390 FVPISPALNAEQICSIEMVLGCKGAPPYVIHGPPGTGKTMTLVEAIVQLYTTQRNARVLV 449
Query: 232 CAASNIAVDNIVERLVPHRVKLVRIGHPARLLPQVVDSALDAQVLRGDNSGLANDIRKEM 291
CA SN A D+I+E+L+ ++ VRI D ++ R N +
Sbjct: 450 CAPSNSAADHILEKLLC--LEGVRIK--------------DNEIFR------LNAATRSY 487
Query: 292 KVLNGKLLKTKEKNTRREIQKELRTLSREERKRQQLAVTDVIKTSDVILTTLIGASSKKL 351
+ + ++++ + L+ L+R + + S + +L+ A
Sbjct: 488 EEIKPEIIRFCFFDELIFKCPPLKALTRYK-----------LVVSTYMSASLLNAEGVNR 536
Query: 352 GNTSFDLVIIDEAAQALEVACWIPL----LKGTRCILAGDHLQLPPTIQSVEAEKKGLGR 407
G+ F +++DEA QA E I + L T +LAGD QL P I S +AE GLG+
Sbjct: 537 GH--FTHILLDEAGQASEPENMIAVSNLCLTETVVVLAGDPRQLGPVIYSRDAESLGLGK 594
Query: 408 TLFERLAE----LYGDE-VTSMLTVQYRMHQLIMDWSSKELYNSKVKAHACVASHMLYDL 462
+ ERL E GDE + L YR H I+D SK Y+ ++ A +L L
Sbjct: 595 SYLERLFECDYYCEGDENYVTKLVKNYRCHPEILDLPSKLFYDGELVASKEDTDSVLASL 654
Query: 463 EGVKKTSSTEPTLLLIDTAGCDMEEKKDEEDSTLNEGESEVAMAHAKRLVQSG-VLPSDI 521
+ + E ++ GCD E++ S N E + KRL + V DI
Sbjct: 655 NFL---PNKEFPMVFYGIQGCD--EREGNNPSWFNRIEISKVIETIKRLTANDCVQEEDI 709
Query: 522 GIITPYAAQVVLLKMLKNKENSLKDIEISTVDGFQGREKEAIIISMVRSNSKKE------ 575
G+ITPY QV+ +K + ++ + + ++++ +V+ FQG+EK+ IIIS VRS K
Sbjct: 710 GVITPYRQQVMKIKEVLDRLD-MTEVKVGSVEQFQGQEKQVIIISTVRSTIKHNEFDRAY 768
Query: 576 -VGFLSDRRRMNVAVTRARRQCCIVCDTETVSSDGFLKRLIEYFEEHGEYQSASEYQNE 633
+GFLS+ RR NVA+TRA I+ + + D +L+ ++ YQ + E
Sbjct: 769 CLGFLSNPRRFNVAITRAISLLVIIGNPHIICKDMNWNKLLWRCVDNNAYQGCGLPEQE 827
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.133 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 67,803,896
Number of Sequences: 164201
Number of extensions: 2784240
Number of successful extensions: 12760
Number of sequences better than 10.0: 138
Number of HSP's better than 10.0 without gapping: 47
Number of HSP's successfully gapped in prelim test: 92
Number of HSP's that attempted gapping in prelim test: 12479
Number of HSP's gapped (non-prelim): 214
length of query: 633
length of database: 59,974,054
effective HSP length: 116
effective length of query: 517
effective length of database: 40,926,738
effective search space: 21159123546
effective search space used: 21159123546
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 69 (31.2 bits)
Medicago: description of AC143341.10


