

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143337.6 + phase: 0
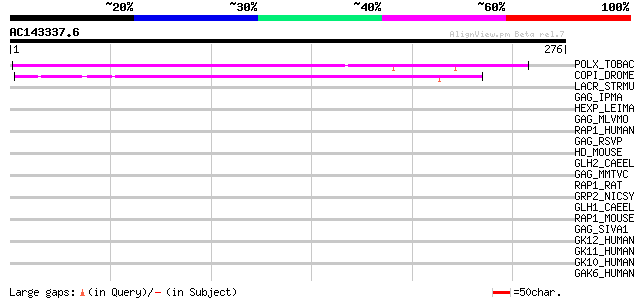
(276 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 176 6e-44
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 76 1e-13
LACR_STRMU (P26422) Lactose phosphotransferase system repressor 40 0.005
GAG_IPMA (P11365) Retrovirus-related Gag polyprotein [Contains: ... 38 0.031
HEXP_LEIMA (Q04832) DNA-binding protein HEXBP (Hexamer-binding p... 37 0.052
GAG_MLVMO (P03332) Gag polyprotein [Contains: Core protein p15; ... 37 0.052
RAP1_HUMAN (Q15276) Rab GTPase binding effector protein 1 (Rabap... 37 0.068
GAG_RSVP (P03322) Gag polyprotein [Contains: Core protein p19; C... 36 0.089
HD_MOUSE (P42859) Huntingtin (Huntington's disease protein homol... 35 0.15
GLH2_CAEEL (Q966L9) ATP-dependent RNA helicase glh-2 (EC 3.6.1.-... 35 0.20
GAG_MMTVC (P11284) Gag polyprotein [Contains: Protein p10; Phosp... 35 0.20
RAP1_RAT (O35550) Rab GTPase binding effector protein 1 (Rabapti... 35 0.26
GRP2_NICSY (P27484) Glycine-rich protein 2 35 0.26
GLH1_CAEEL (P34689) ATP-dependent RNA helicase glh-1 (EC 3.6.1.-... 35 0.26
RAP1_MOUSE (O35551) Rab GTPase binding effector protein 1 (Rabap... 34 0.34
GAG_SIVA1 (P27972) Gag polyprotein [Contains: Core protein p17; ... 34 0.34
GK12_HUMAN (P63130) HERV-K_1q22 provirus ancestral Gag polyprote... 34 0.44
GK11_HUMAN (P63145) HERV-K_22q11.21 provirus ancestral Gag polyp... 34 0.44
GK10_HUMAN (P87889) HERV-K_5q33.3 provirus ancestral Gag polypro... 34 0.44
GAK6_HUMAN (P62685) HERV-K_8p23.1 provirus ancestral Gag polypro... 34 0.44
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 176 bits (446), Expect = 6e-44
Identities = 97/267 (36%), Positives = 150/267 (55%), Gaps = 11/267 (4%)
Query: 2 GSKWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVDKARSA 61
G K+++ KF GDN F W+ +M +LIQQ K L + P TM + + ++A SA
Sbjct: 3 GVKYEVAKFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAEDWADLDERAASA 62
Query: 62 VVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQL 121
+ L L D V+ + E TA +W +LESLYM+K+L ++ +LK+QLY+ M E + L
Sbjct: 63 IRLHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKKQLYALHMSEGTNFLSHL 122
Query: 122 TEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGKEGTVTLEEVQAALRT 181
FN ++ L N+ V++E+EDKAILLL +LP S+++ T+L+GK T+ L++V +AL
Sbjct: 123 NVFNGLITQLANLGVKIEEEDKAILLLNSLPSSYDNLATTILHGKT-TIELKDVTSALLL 181
Query: 182 KDLTKSKD-------LTHEHGEGLSVTRGNGGGRGNRRKSGNKSRF---ECFNCHKMGHF 231
+ + K +T G + N G G R KS N+S+ C+NC++ GHF
Sbjct: 182 NEKMRKKPENQGQALITEGRGRSYQRSSNNYGRSGARGKSKNRSKSRVRNCYNCNQPGHF 241
Query: 232 KKDCPKINGNSAQIVSEGYEDAGALMV 258
K+DCP + + +D A MV
Sbjct: 242 KRDCPNPRKGKGETSGQKNDDNTAAMV 268
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 75.9 bits (185), Expect = 1e-13
Identities = 56/246 (22%), Positives = 112/246 (44%), Gaps = 17/246 (6%)
Query: 3 SKWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVDKARSAV 62
+K +I+ F G+ + +WK ++ A+L +Q K + +G +P + + K K A+S +
Sbjct: 4 AKRNIKPFDGEK-YAIWKFRIRALLAEQDVLKVV--DGLMPNEVDDSWK-KAERCAKSTI 59
Query: 63 VLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQLT 122
+ L D L + TA + L+++Y KSLA + L+++L S ++ +++
Sbjct: 60 IEYLSDSFLNFATSDITARQILENLDAVYERKSLASQLALRKRLLSLKLSSEMSLLSHFH 119
Query: 123 EFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKDTMLYGKEGTVTLEEVQAALRTK 182
F++++ +L ++E+ DK LL LP ++ + E +TL V+ L +
Sbjct: 120 IFDELISELLAAGAKIEEMDKISHLLITLPSCYDGIITAIETLSEENLTLAFVKNRLLDQ 179
Query: 183 DLTKSKDLTHEHGEGLSVTRGNGGGRGNRR-------------KSGNKSRFECFNCHKMG 229
++ D + ++ N K +K + +C +C + G
Sbjct: 180 EIKIKNDHNDTSKKVMNAIVHNNNNTYKNNLFKNRVTKPKKIFKGNSKYKVKCHHCGREG 239
Query: 230 HFKKDC 235
H KKDC
Sbjct: 240 HIKKDC 245
>LACR_STRMU (P26422) Lactose phosphotransferase system repressor
Length = 251
Score = 40.4 bits (93), Expect = 0.005
Identities = 30/95 (31%), Positives = 44/95 (45%), Gaps = 17/95 (17%)
Query: 161 TMLYGKEGTVTLEEVQAALRTKDLTKSKDLTHEHGEGLSVTRGNGGGRGNRRKSGNKSRF 220
T L K GT+ + EV L+ D+T +DLT G G+ +TR +GG R N
Sbjct: 11 TKLINKRGTIRVTEVVERLKVSDMTVRRDLTELEGLGV-LTRIHGGARSN---------- 59
Query: 221 ECFNCHKMGHFKKDCPKIN------GNSAQIVSEG 249
F +M H +K +I +A++V EG
Sbjct: 60 NIFQYKEMSHEEKHSRQIEEKHYIAQKAAELVEEG 94
>GAG_IPMA (P11365) Retrovirus-related Gag polyprotein [Contains:
Protease (EC 3.4.23.-)]
Length = 827
Score = 37.7 bits (86), Expect = 0.031
Identities = 23/49 (46%), Positives = 26/49 (52%), Gaps = 6/49 (12%)
Query: 210 NRRKSGNKSRFECFNCHKMGHFKKDC--PKINGNSAQIVS---EGYEDA 253
NR S N R CFNC K GHFKKDC P G + + S +GY A
Sbjct: 449 NRSMSRNDQR-TCFNCGKPGHFKKDCRAPDKQGGTLTLCSKCGKGYHRA 496
>HEXP_LEIMA (Q04832) DNA-binding protein HEXBP (Hexamer-binding
protein)
Length = 271
Score = 37.0 bits (84), Expect = 0.052
Identities = 24/78 (30%), Positives = 33/78 (41%), Gaps = 18/78 (23%)
Query: 203 GNGGGRGNRR-KSGNKSRFE----CFNCHKMGHFKKDCPKINGNSAQIVSEGYEDAGALM 257
G+ GG G +R +SG + + C+ C GH +DCP G GY AG
Sbjct: 118 GSRGGYGQKRGRSGAQGGYSGDRTCYKCGDAGHISRDCPNGQG--------GYSGAGDRT 169
Query: 258 VWCCLEEEKGDVSHLGID 275
+ C GD H+ D
Sbjct: 170 CYKC-----GDAGHISRD 182
Score = 37.0 bits (84), Expect = 0.052
Identities = 14/33 (42%), Positives = 17/33 (51%)
Query: 210 NRRKSGNKSRFECFNCHKMGHFKKDCPKINGNS 242
N K G FEC+ C + GH +DCP G S
Sbjct: 87 NSAKPGAAKGFECYKCGQEGHLSRDCPSSQGGS 119
Score = 33.1 bits (74), Expect = 0.76
Identities = 19/69 (27%), Positives = 30/69 (42%), Gaps = 11/69 (15%)
Query: 195 GEGLSVTRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPKINGNSAQIVSEGYEDAG 254
G+ ++R G+G +G+++ C+ C GH +DCP G GY AG
Sbjct: 146 GDAGHISRDCPNGQGGYSGAGDRT---CYKCGDAGHISRDCPNGQG--------GYSGAG 194
Query: 255 ALMVWCCLE 263
+ C E
Sbjct: 195 DRKCYKCGE 203
Score = 32.7 bits (73), Expect = 0.99
Identities = 16/43 (37%), Positives = 23/43 (53%), Gaps = 5/43 (11%)
Query: 210 NRRKSGNKSRFECFNCHKMGHFKKDCPKINGNSAQI-VSEGYE 251
N +SG CF C + GH +DCP NSA+ ++G+E
Sbjct: 60 NEARSGAAGAMTCFRCGEAGHMSRDCP----NSAKPGAAKGFE 98
Score = 32.0 bits (71), Expect = 1.7
Identities = 12/32 (37%), Positives = 18/32 (55%), Gaps = 3/32 (9%)
Query: 205 GGGRGNRRKSGNKSRFECFNCHKMGHFKKDCP 236
GG G R G+++ C+ C + GH +DCP
Sbjct: 241 GGSYGGSRGGGDRT---CYKCGEAGHISRDCP 269
>GAG_MLVMO (P03332) Gag polyprotein [Contains: Core protein p15;
Inner coat protein p12; Core shell protein p30;
Nucleoprotein p10]
Length = 538
Score = 37.0 bits (84), Expect = 0.052
Identities = 23/71 (32%), Positives = 31/71 (43%), Gaps = 6/71 (8%)
Query: 173 EEVQAALRTKDLTKSKDLTHEHGEGLS-----VTRGNGGGR-GNRRKSGNKSRFECFNCH 226
EE + RT+D K K+ +S V G R G R+ R +C C
Sbjct: 449 EEKEERRRTEDEQKEKERDRRRHREMSKLLATVVSGQKQDRQGGERRRSQLDRDQCAYCK 508
Query: 227 KMGHFKKDCPK 237
+ GH+ KDCPK
Sbjct: 509 EKGHWAKDCPK 519
>RAP1_HUMAN (Q15276) Rab GTPase binding effector protein 1
(Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4)
Length = 862
Score = 36.6 bits (83), Expect = 0.068
Identities = 30/117 (25%), Positives = 56/117 (47%), Gaps = 5/117 (4%)
Query: 24 EAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVDKARSAVVLCLGDKVLREVAKEATAASM 83
E ++ QK +L+G+ SL V++ +AE + D + L L K ++ TAA
Sbjct: 640 EELVRLQKDNDSLQGKHSLHVSLQQAEDFILPDTTEALRELVL--KYREDIINVRTAADH 697
Query: 84 WA---KLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQLTEFNKILDDLENIEVQ 137
K E L++ + + Q LK+ L +E + E++ + + +LE I+V+
Sbjct: 698 VEEKLKAEILFLKEQIQAEQCLKENLEETLQLEIENCKEEIASISSLKAELERIKVE 754
>GAG_RSVP (P03322) Gag polyprotein [Contains: Core protein p19; Core
protein p2A; Core protein p2B; Core protein p10; Capsid
protein p27; Inner coat protein p12; Protease p15 (EC
3.4.23.-)]
Length = 701
Score = 36.2 bits (82), Expect = 0.089
Identities = 19/39 (48%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query: 211 RRKSGNKSRFECFNCHKMGHFKKDCPKINGNSAQIVSEG 249
+RKSGN SR C C+ MGH K C K +GN Q +G
Sbjct: 525 KRKSGN-SRERCQLCNGMGHNAKQCRKRDGNQGQRPGKG 562
Score = 32.0 bits (71), Expect = 1.7
Identities = 14/39 (35%), Positives = 22/39 (55%), Gaps = 2/39 (5%)
Query: 208 RGNRRKSGNKSRFECFNCHKMGHFKKDCPK--INGNSAQ 244
R + SG ++R C+ C GH++ CPK +GNS +
Sbjct: 495 RDGQTGSGGRARGLCYTCGSPGHYQAQCPKKRKSGNSRE 533
>HD_MOUSE (P42859) Huntingtin (Huntington's disease protein homolog)
(HD protein)
Length = 3119
Score = 35.4 bits (80), Expect = 0.15
Identities = 24/78 (30%), Positives = 41/78 (51%), Gaps = 8/78 (10%)
Query: 124 FNKILDDLENIEVQLEDEDKAILLL-------CALPKSFESFKDTMLYGKEGTVTLEEVQ 176
+N +L+ L + V +E+E +L+L C +P + KDT L G G VT +E++
Sbjct: 267 YNWLLNVLLGLLVPMEEEHSTLLILGVLLTLRCLVPLLQQQVKDTSLKGSFG-VTRKEME 325
Query: 177 AALRTKDLTKSKDLTHEH 194
+ T+ L + +LT H
Sbjct: 326 VSPSTEQLVQVYELTLHH 343
>GLH2_CAEEL (Q966L9) ATP-dependent RNA helicase glh-2 (EC 3.6.1.-)
(Germline helicase-2)
Length = 974
Score = 35.0 bits (79), Expect = 0.20
Identities = 15/35 (42%), Positives = 20/35 (56%), Gaps = 4/35 (11%)
Query: 203 GNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPK 237
G+GGG +R + N CFNC + GH DCP+
Sbjct: 244 GSGGGGQDRGERNNN----CFNCQQPGHRSNDCPE 274
Score = 35.0 bits (79), Expect = 0.20
Identities = 15/35 (42%), Positives = 20/35 (56%), Gaps = 4/35 (11%)
Query: 203 GNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPK 237
G+GGG +R + N CFNC + GH DCP+
Sbjct: 358 GSGGGGQDRGERNNN----CFNCQQPGHRSNDCPE 388
Score = 31.6 bits (70), Expect = 2.2
Identities = 14/35 (40%), Positives = 18/35 (51%), Gaps = 3/35 (8%)
Query: 203 GNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPK 237
GN G GN + G +CFNC GH +CP+
Sbjct: 439 GNAEGFGNNEERGP---MKCFNCKGEGHRSAECPE 470
Score = 30.4 bits (67), Expect = 4.9
Identities = 20/69 (28%), Positives = 31/69 (43%), Gaps = 5/69 (7%)
Query: 204 NGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPKINGNSAQIVSEGYEDAGALMVWCCLE 263
N G G+R + CFNC + GH +CP N A+ EG E G + +E
Sbjct: 457 NCKGEGHRSAECPEPPRGCFNCGEQGHRSNECP----NPAK-PREGAEGEGPKATYVPVE 511
Query: 264 EEKGDVSHL 272
+ +V ++
Sbjct: 512 DNMEEVFNM 520
>GAG_MMTVC (P11284) Gag polyprotein [Contains: Protein p10;
Phosphorylated protein pp21; Protein p3; Protein p8;
Major core protein p27; Nucleic acid binding protein
p14]
Length = 591
Score = 35.0 bits (79), Expect = 0.20
Identities = 15/37 (40%), Positives = 21/37 (56%), Gaps = 4/37 (10%)
Query: 205 GGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPKINGN 241
GGG+G + G CF+C K GH K+DC + G+
Sbjct: 514 GGGKGGQGSKGPV----CFSCGKTGHIKRDCKEEKGS 546
>RAP1_RAT (O35550) Rab GTPase binding effector protein 1
(Rabaptin-5) (Rabaptin-5alpha)
Length = 862
Score = 34.7 bits (78), Expect = 0.26
Identities = 30/117 (25%), Positives = 54/117 (45%), Gaps = 5/117 (4%)
Query: 24 EAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVDKARSAVVLCLGDKVLREVAKEATAASM 83
E ++ QK +L+G+ SL V++ AE + D + L L K + TAA
Sbjct: 640 EELVRLQKDNDSLQGKHSLHVSLQLAEDFILPDTVQVLRELVL--KYRENIVHVRTAADH 697
Query: 84 WA---KLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQLTEFNKILDDLENIEVQ 137
K E L++ + + Q LK+ L +E + E++ + + +LE I+V+
Sbjct: 698 MEEKLKAEILFLKEQIQAEQCLKENLEETLQLEIENCKEEIASISSLKAELERIKVE 754
>GRP2_NICSY (P27484) Glycine-rich protein 2
Length = 214
Score = 34.7 bits (78), Expect = 0.26
Identities = 15/38 (39%), Positives = 18/38 (46%), Gaps = 3/38 (7%)
Query: 203 GNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPKING 240
G GGG G G CF C + GHF +DC + G
Sbjct: 143 GGGGGYGGGGSGGGSG---CFKCGESGHFARDCSQSGG 177
>GLH1_CAEEL (P34689) ATP-dependent RNA helicase glh-1 (EC 3.6.1.-)
(Germline helicase-1)
Length = 763
Score = 34.7 bits (78), Expect = 0.26
Identities = 15/43 (34%), Positives = 18/43 (40%)
Query: 195 GEGLSVTRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPK 237
G G G G G G + CFNC + GH DCP+
Sbjct: 133 GSGGGSFGGGNSGFGEGGHGGGERNNNCFNCQQPGHRSSDCPE 175
Score = 32.3 bits (72), Expect = 1.3
Identities = 21/69 (30%), Positives = 31/69 (44%), Gaps = 5/69 (7%)
Query: 204 NGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPKINGNSAQIVSEGYEDAGALMVWCCLE 263
N G G+R + CFNC + GH +CP N A+ EG E G + +E
Sbjct: 246 NCKGEGHRSAECPEPPRGCFNCGEQGHRSNECP----NPAK-PREGVEGEGPKATYVPVE 300
Query: 264 EEKGDVSHL 272
+ DV ++
Sbjct: 301 DNMEDVFNM 309
Score = 32.3 bits (72), Expect = 1.3
Identities = 16/45 (35%), Positives = 21/45 (46%), Gaps = 2/45 (4%)
Query: 195 GEGLSVTRGNGGGRGNRRKSGNKSR--FECFNCHKMGHFKKDCPK 237
G G GN G G+ G + R +CFNC GH +CP+
Sbjct: 215 GAGFGNNGGNDGFGGDGGFGGGEERGPMKCFNCKGEGHRSAECPE 259
>RAP1_MOUSE (O35551) Rab GTPase binding effector protein 1
(Rabaptin-5) (Rabaptin-5alpha)
Length = 862
Score = 34.3 bits (77), Expect = 0.34
Identities = 30/117 (25%), Positives = 53/117 (44%), Gaps = 5/117 (4%)
Query: 24 EAVLIQQKCEKALKGEGSLPVTMSRAEKTKMVDKARSAVVLCLGDKVLREVAKEATAASM 83
E ++ QK +L+G+ SL V++ AE + D L L K + TAA
Sbjct: 640 EELVRLQKDNDSLQGKHSLHVSLQLAEDFILPDTVEVLRELVL--KYRENIVHVRTAADH 697
Query: 84 WA---KLESLYMTKSLAHRQFLKQQLYSFRMVESKAIMEQLTEFNKILDDLENIEVQ 137
K E L++ + + Q LK+ L +E + E++ + + +LE I+V+
Sbjct: 698 MEEKLKAEILFLKEQIQAEQCLKENLEETLQLEIENCKEEIASISSLKAELERIKVE 754
>GAG_SIVA1 (P27972) Gag polyprotein [Contains: Core protein p17;
Core protein p24; Core protein p15]
Length = 520
Score = 34.3 bits (77), Expect = 0.34
Identities = 14/33 (42%), Positives = 19/33 (57%), Gaps = 5/33 (15%)
Query: 205 GGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPK 237
GGGRG R +C+NC K GH ++ CP+
Sbjct: 388 GGGRGRPRPPP-----KCYNCGKFGHMQRQCPE 415
>GK12_HUMAN (P63130) HERV-K_1q22 provirus ancestral Gag polyprotein
(Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III)
Gag protein) [Contains: Matrix protein; Capsid protein;
Nucleocapsid protein]
Length = 665
Score = 33.9 bits (76), Expect = 0.44
Identities = 11/25 (44%), Positives = 18/25 (72%)
Query: 221 ECFNCHKMGHFKKDCPKINGNSAQI 245
+C+NC ++GH KK+CP +N + I
Sbjct: 544 KCYNCGQIGHLKKNCPVLNKQNITI 568
>GK11_HUMAN (P63145) HERV-K_22q11.21 provirus ancestral Gag
polyprotein (Gag polyprotein) (HERV-K101 Gag protein)
[Contains: Matrix protein; Capsid protein; Nucleocapsid
protein]
Length = 665
Score = 33.9 bits (76), Expect = 0.44
Identities = 11/25 (44%), Positives = 18/25 (72%)
Query: 221 ECFNCHKMGHFKKDCPKINGNSAQI 245
+C+NC ++GH KK+CP +N + I
Sbjct: 544 KCYNCGQIGHLKKNCPVLNKQNITI 568
>GK10_HUMAN (P87889) HERV-K_5q33.3 provirus ancestral Gag
polyprotein (Gag polyprotein) (HERV-K10 Gag protein)
(HERV-K107 Gag protein) [Contains: Matrix protein;
Capsid protein; Nucleocapsid protein]
Length = 665
Score = 33.9 bits (76), Expect = 0.44
Identities = 11/25 (44%), Positives = 18/25 (72%)
Query: 221 ECFNCHKMGHFKKDCPKINGNSAQI 245
+C+NC ++GH KK+CP +N + I
Sbjct: 544 KCYNCGQIGHLKKNCPVLNKQNITI 568
>GAK6_HUMAN (P62685) HERV-K_8p23.1 provirus ancestral Gag
polyprotein (Gag polyprotein) (HERV-K115 Gag protein)
[Contains: Matrix protein; Capsid protein; Nucleocapsid
protein]
Length = 646
Score = 33.9 bits (76), Expect = 0.44
Identities = 11/25 (44%), Positives = 18/25 (72%)
Query: 221 ECFNCHKMGHFKKDCPKINGNSAQI 245
+C+NC ++GH KK+CP +N + I
Sbjct: 544 KCYNCGQIGHLKKNCPVLNKQNITI 568
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.132 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,673,452
Number of Sequences: 164201
Number of extensions: 1283836
Number of successful extensions: 4953
Number of sequences better than 10.0: 99
Number of HSP's better than 10.0 without gapping: 66
Number of HSP's successfully gapped in prelim test: 35
Number of HSP's that attempted gapping in prelim test: 4798
Number of HSP's gapped (non-prelim): 184
length of query: 276
length of database: 59,974,054
effective HSP length: 109
effective length of query: 167
effective length of database: 42,076,145
effective search space: 7026716215
effective search space used: 7026716215
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 65 (29.6 bits)
Medicago: description of AC143337.6


