

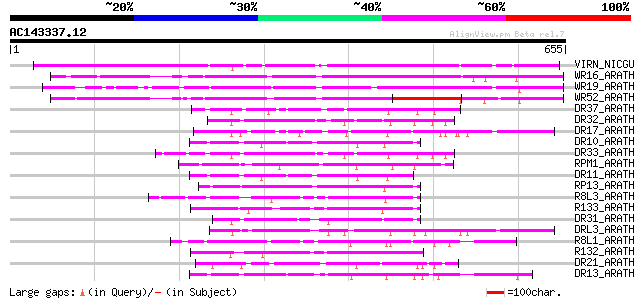
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC143337.12 - phase: 0
(655 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
VIRN_NICGU (Q40392) TMV resistance protein N 394 e-109
WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY ... 249 2e-65
WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY ... 241 4e-63
WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY ... 236 1e-61
DR37_ARATH (Q9LVT4) Probable disease resistance protein At5g47250 89 3e-17
DR32_ARATH (Q9FG91) Probable disease resistance protein At5g43730 79 5e-14
DR17_ARATH (O64790) Putative disease resistance protein At1g61300 77 1e-13
DR10_ARATH (Q8W3J8) Probable disease resistance protein RDL5/RF45 76 3e-13
DR33_ARATH (Q9FG90) Probable disease resistance protein At5g43740 75 5e-13
RPM1_ARATH (Q39214) Disease resistance protein RPM1 (Resistance ... 75 7e-13
DR11_ARATH (Q94HW3) Probable disease resistance protein RDL6/RF9 74 1e-12
RP13_ARATH (Q9M667) Disease resistance protein RPP13 (Resistance... 74 2e-12
R8L3_ARATH (Q9FJB5) Probable disease resistance RPP8-like protein 3 73 3e-12
R133_ARATH (Q9STE7) Putative disease resistance RPP13-like prote... 72 4e-12
DR31_ARATH (Q9FLB4) Putative disease resistance protein At5g05400 71 1e-11
DRL3_ARATH (Q9LMP6) Probable disease resistance protein At1g15890 70 2e-11
R8L1_ARATH (O04093) Putative disease resistance protein RPP8-lik... 69 3e-11
R132_ARATH (Q9STE5) Putative disease resistance RPP13-like prote... 69 3e-11
DR21_ARATH (Q9LRR5) Putative disease resistance protein At3g14460 69 3e-11
DR13_ARATH (Q9XIF0) Putative disease resistance protein At1g59780 69 3e-11
>VIRN_NICGU (Q40392) TMV resistance protein N
Length = 1144
Score = 394 bits (1012), Expect = e-109
Identities = 235/631 (37%), Positives = 369/631 (58%), Gaps = 30/631 (4%)
Query: 29 SDSNSIQSYRYDVFISFRGADTRSTFVDHLHAHLTTKGIFAFKDDKRLEKGESLSPQLLQ 88
+ S+S + YDVF+SFRG DTR TF HL+ L KGI F+DDKRLE G ++ +L +
Sbjct: 2 ASSSSSSRWSYDVFLSFRGEDTRKTFTSHLYEVLNDKGIKTFQDDKRLEYGATIPGELCK 61
Query: 89 AIQSSRISIVVFSKNYAESTLCLEEMATIAEYHTELKQTVFPIFYDADPSHVRKQSGVYQ 148
AI+ S+ +IVVFS+NYA S CL E+ I E T KQTV PIFYD DPSHVR Q +
Sbjct: 62 AIEESQFAIVVFSENYATSRWCLNELVKIMECKTRFKQTVIPIFYDVDPSHVRNQKESFA 121
Query: 149 NAFVLLQNKFKHDPNKVMRWVGAMESLAKLVG-WDVRNKPEFREIKNIVQEVINTMGHKF 207
AF + K+K D + RW A+ A L G D R+K + I+ IV ++ + +
Sbjct: 122 KAFEEHETKYKDDVEGIQRWRIALNEAANLKGSCDNRDKTDADCIRQIVDQISSKLCKIS 181
Query: 208 LGFADDLIGIQPRVEELESLLKLDSKDYEFRAIGIWGMAGIRKTTLASVLYDRV------ 261
L + +++GI +E++ESLL++ R +GIWGM G+ KTT+A ++D +
Sbjct: 182 LSYLQNIVGIDTHLEKIESLLEIGING--VRIMGIWGMGGVGKTTIARAIFDTLLGRMDS 239
Query: 262 SYQFDASCFIENVSKIYKDGGATAVQKQILRQTIDEKNLETYSPSEI-SGIIRKRLCNKK 320
SYQFD +CF++++ + G ++Q +L + + EK Y+ E + RL +KK
Sbjct: 240 SYQFDGACFLKDIKE--NKRGMHSLQNALLSELLREK--ANYNNEEDGKHQMASRLRSKK 295
Query: 321 FLVVLDNADLLEQ-MEELAINPELLGKGSRIIITTRDMHILSLSVSHDTCVSYDVYEVPL 379
L+VLD+ D + +E LA + + G GSRIIITTRD H++ +D +YEV
Sbjct: 296 VLIVLDDIDNKDHYLEYLAGDLDWFGNGSRIIITTRDKHLIE---KNDI-----IYEVTA 347
Query: 380 LNSNDARKLFYRKAFKSEDPTSGCVKLTPEVLKYAQGLPLAVRVVGSFLCTRDANQWRDA 439
L +++ +LF + AF E P KL+ EV+ YA+GLPLA++V GS L +W+ A
Sbjct: 348 LPDHESIQLFKQHAFGKEVPNENFEKLSLEVVNYAKGLPLALKVWGSLLHNLRLTEWKSA 407
Query: 440 LYRLRNNPDNNVMDVLQVSFEGLHSEDREIFLHIACFFKGEKEDYVKRILDACGLHPHIG 499
+ ++NN + ++D L++S++GL + +E+FL IACF +GE++DY+ +IL++C + G
Sbjct: 408 IEHMKNNSYSGIIDKLKISYDGLEPKQQEMFLDIACFLRGEEKDYILQILESCHIGAEYG 467
Query: 500 IQSLIERSFITIRN-NEILMHEMLQELGKKIVRQQFPFQPGSWSRLWLYDDFYSVMMTET 558
++ LI++S + I N++ MH+++Q++GK IV F PG SRLWL + VM T
Sbjct: 468 LRILIDKSLVFISEYNQVQMHDLIQDMGKYIV--NFQKDPGERSRLWLAKEVEEVMSNNT 525
Query: 559 GTNNINAIILDQKEHISEYPQLRAEALSIMRGLKILILLFHKNFSGSLTFLSNSLQYLLW 618
GT + AI + S + +A+ M+ L++ + + ++ +L N+L+ +
Sbjct: 526 GTMAMEAIWVSS---YSSTLRFSNQAVKNMKRLRV-FNMGRSSTHYAIDYLPNNLRCFVC 581
Query: 619 YGYPFASLPLNFEPFCLVELNMPYSSIQRLW 649
YP+ S P FE LV L + ++S++ LW
Sbjct: 582 TNYPWESFPSTFELKMLVHLQLRHNSLRHLW 612
>WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY
DNA-binding protein 16)
Length = 1372
Score = 249 bits (636), Expect = 2e-65
Identities = 181/627 (28%), Positives = 301/627 (47%), Gaps = 74/627 (11%)
Query: 49 DTRSTFVDHLHAHLTTKGIFAFKDDKRLEKGESLSPQLLQAIQSSRISIVVFSKNYAEST 108
+ R +FV HL L KG+ +D ++ +SLS + ++ +R+S+++ N T
Sbjct: 15 EVRYSFVSHLSKALQRKGV----NDVFIDSDDSLSNESQSMVERARVSVMILPGN---RT 67
Query: 109 LCLEEMATIAEYHTELKQTVFPIFYDADPSHVRKQSGVYQNAFVLLQNKFKHDPNKVMRW 168
+ L+++ + + Q V P+ Y S S + F + + K + +
Sbjct: 68 VSLDKLVKVLDCQKNKDQVVVPVLYGVRSSETEWLSALDSKGFSSVHHSRKECSDSQL-- 125
Query: 169 VGAMESLAKLVGWDVRNKPEFREIKNIVQEVINTMGHKFLGFADDLIGIQPRVEELESLL 228
+K V++V + F + IGI ++ E+E ++
Sbjct: 126 -----------------------VKETVRDVYEKL------FYMERIGIYSKLLEIEKMI 156
Query: 229 KLDSKDYEFRAIGIWGMAGIRKTTLASVLYDRVSYQFDASCFIENVSKIYKDGGATAV-Q 287
+ + + R +GIWGM GI KTTLA ++D++S +FDA CFIE+ +K ++ G + +
Sbjct: 157 --NKQPLDIRCVGIWGMPGIGKTTLAKAVFDQMSGEFDAHCFIEDYTKAIQEKGVYCLLE 214
Query: 288 KQILRQTIDEKNLETYSPSEISGIIRKRLCNKKFLVVLDNADLLEQMEELAINPELLGKG 347
+Q L++ T ++R RL NK+ LVVLD+ +E + G
Sbjct: 215 EQFLKENAGASGTVTKL-----SLLRDRLNNKRVLVVLDDVRSPLVVESFLGGFDWFGPK 269
Query: 348 SRIIITTRDMHILSLSVSHDTCVSYDVYEVPLLNSNDARKLFYRKAFKSEDPTSGCVKLT 407
S IIIT++D + L C +YEV LN +A +LF A + +++
Sbjct: 270 SLIIITSKDKSVFRL------CRVNQIYEVQGLNEKEALQLFSLCASIDDMAEQNLHEVS 323
Query: 408 PEVLKYAQGLPLAVRVVGSFLCTRDAN-QWRDALYRLRNNPDNNVMDVLQVSFEGLHSED 466
+V+KYA G PLA+ + G L + + A +L+ P +D ++ S++ L+ +
Sbjct: 324 MKVIKYANGHPLALNLYGRELMGKKRPPEMEIAFLKLKECPPAIFVDAIKSSYDTLNDRE 383
Query: 467 REIFLHIACFFKGEKEDYVKRILDACGLHPHIGIQSLIERSFITIRNNEILMHEMLQELG 526
+ IFL IACFF+GE DYV ++L+ CG PH+GI L+E+S +TI N + MH ++Q++G
Sbjct: 384 KNIFLDIACFFQGENVDYVMQLLEGCGFFPHVGIDVLVEKSLVTISENRVRMHNLIQDVG 443
Query: 527 KKIVRQQFPFQPGSWSRLW-------LYDDFYSVMMTETGT--------NNINAIILDQK 571
++I+ ++ Q SRLW L +D E T I + LD
Sbjct: 444 RQIINRE-TRQTKRRSRLWEPCSIKYLLEDKEQNENEEQKTTFERAQVPEEIEGMFLDTS 502
Query: 572 EHISEYPQLRAEALSIMRGLKILIL---LFHKN--FSGSLTFLSNSLQYLLWYGYPFASL 626
+ + + + +R KI + H N GSL+ L N L+ L W YP L
Sbjct: 503 NLSFDIKHVAFDNMLNLRLFKIYSSNPEVHHVNNFLKGSLSSLPNVLRLLHWENYPLQFL 562
Query: 627 PLNFEPFCLVELNMPYSSIQRLWDGHK 653
P NF+P LVE+NMPYS +++LW G K
Sbjct: 563 PQNFDPIHLVEINMPYSQLKKLWGGTK 589
Score = 43.5 bits (101), Expect = 0.002
Identities = 27/87 (31%), Positives = 49/87 (56%), Gaps = 2/87 (2%)
Query: 449 NNVMDVLQVSFEGLHSEDREIFLHIACFFKGEKEDYVKRIL-DACGLHPHIGIQSLIERS 507
N +VL+V + GL + +FL+IA F E V ++ + + G++ L RS
Sbjct: 1045 NEDEEVLRVRYAGLQEIYKALFLYIAGLFNDEDVGLVAPLIANIIDMDVSYGLKVLAYRS 1104
Query: 508 FITIRNN-EILMHEMLQELGKKIVRQQ 533
I + +N EI+MH +L+++GK+I+ +
Sbjct: 1105 LIRVSSNGEIVMHYLLRQMGKEILHTE 1131
>WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY
DNA-binding protein 19)
Length = 1895
Score = 241 bits (615), Expect = 4e-63
Identities = 185/627 (29%), Positives = 306/627 (48%), Gaps = 59/627 (9%)
Query: 39 YDVFISFRGAD-TRSTFVDHLHAHLTTKGIFAFKDDKRLEKGESLSPQLLQAIQSSRISI 97
YDV I + AD + F+ HL A L +GI ++ ++ A+ R+ I
Sbjct: 668 YDVVIRYGRADISNEDFISHLRASLCRRGISVYEKFNEVD-----------ALPKCRVLI 716
Query: 98 VVFSKNYAESTLCLEEMATIAEYHTELKQTVFPIFYDADPSHVRKQSGVYQNAFVLLQNK 157
+V + Y S L + + HTE + V+PIFY P S Y+ ++
Sbjct: 717 IVLTSTYVPSNL----LNILEHQHTE-DRVVYPIFYRLSPYDFVCNSKNYERFYL----- 766
Query: 158 FKHDPNKVMRWVGAMESLAKLVGWDVRNKPEFREIKNIVQEVINTMGHKFLGFAD--DLI 215
+ +P K W A++ + ++ G+ + +K E I IV++ + K L AD ++I
Sbjct: 767 -QDEPKK---WQAALKEITQMPGYTLTDKSESELIDEIVRDAL-----KVLCSADKVNMI 817
Query: 216 GIQPRVEELESLLKLDSKDYEFRAIGIWGMAGIRKTTLASVLYDRVSYQFDASCFIENVS 275
G+ +VEE+ SLL ++S D R+IGIWG GI KTT+A ++ ++S Q++ ++++
Sbjct: 818 GMDMQVEEILSLLCIESLDV--RSIGIWGTVGIGKTTIAEEIFRKISVQYETCVVLKDLH 875
Query: 276 KIYKDGGATAVQKQILRQTID-EKNLETYSPSEISGIIRKRLCNKKFLVVLDNADLLEQM 334
K + G AV++ L + ++ E ++ S + S +R RL K+ LV+LD+ + +
Sbjct: 876 KEVEVKGHDAVRENFLSEVLEVEPHVIRISDIKTS-FLRSRLQRKRILVILDDVNDYRDV 934
Query: 335 EELAINPELLGKGSRIIITTRDMHILSLSVSHDTCVSYDVYEVPLLNSNDARKLFYRKAF 394
+ G GSRII+T+R+ + L C VYEV L+ + L R
Sbjct: 935 DTFLGTLNYFGPGSRIIMTSRNRRVFVL------CKIDHVYEVKPLDIPKSLLLLDRGTC 988
Query: 395 KSEDPTSGCVKLTPEVLKYAQGLPLAVRVVGSFLCTRDANQWRDALYRLRNNPDNNVMDV 454
+ L+ E++K++ G P ++ + S +W ++ + +
Sbjct: 989 QIVLSPEVYKTLSLELVKFSNGNPQVLQFLSSI-----DREWNKLSQEVKTTSPIYIPGI 1043
Query: 455 LQVSFEGLHSEDREIFLHIACFFKGEKEDYVKRILDACGLHPHIGIQSLIERSFITI-RN 513
+ S GL +R IFL IACFF +D V +LD CG H+G + L+++S +TI ++
Sbjct: 1044 FEKSCCGLDDNERGIFLDIACFFNRIDKDNVAMLLDGCGFSAHVGFRGLVDKSLLTISQH 1103
Query: 514 NEILMHEMLQELGKKIVRQQFPFQPGSWSRLWLYDDFYSVMMTETGTNNINAIILDQKEH 573
N + M +Q G++IVRQ+ +PG SRLW D V + +TGT+ I I LD
Sbjct: 1104 NLVDMLSFIQATGREIVRQESADRPGDRSRLWNADYIRHVFINDTGTSAIEGIFLDM--- 1160
Query: 574 ISEYPQLRAEALSIMRGLKILILLFHK-------NFSGSLTFLSNSLQYLLWYGYPFASL 626
++ M L++L L K +F L +L + L+ L W YP +SL
Sbjct: 1161 LNLKFDANPNVFEKMCNLRLLKLYCSKAEEKHGVSFPQGLEYLPSKLRLLHWEYYPLSSL 1220
Query: 627 PLNFEPFCLVELNMPYSSIQRLWDGHK 653
P +F P LVELN+P S ++LW G K
Sbjct: 1221 PKSFNPENLVELNLPSSCAKKLWKGKK 1247
>WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY
DNA-binding protein 52) (Disease resistance protein
RRS1) (Resistance to Ralstonia solanacearum 1 protein)
Length = 1378
Score = 236 bits (603), Expect = 1e-61
Identities = 187/630 (29%), Positives = 301/630 (47%), Gaps = 76/630 (12%)
Query: 49 DTRSTFVDHLHAHLTTKGIFAFKDDKRLEKGESLSPQLLQAIQSSRISIVVFSKNYAEST 108
+ R +FV HL L KGI D ++ + L + I+ + +S++V N S
Sbjct: 18 EVRYSFVSHLSEALRRKGINNVVVDVDID--DLLFKESQAKIEKAGVSVMVLPGNCDPSE 75
Query: 109 LCLEEMATIAEYHTELK-QTVFPIFYDADPSHVRKQSGVYQNAFVLLQNKFKHDPNKVMR 167
+ L++ A + E K Q V + Y + S + + K + ++
Sbjct: 76 VWLDKFAKVLECQRNNKDQAVVSVLYGDSLLRDQWLSELDFRGLSRIHQSRKECSDSIL- 134
Query: 168 WVGAMESLAKLVGWDVRNKPEFREIKNIVQEVINTMGHKFLGFADDLIGIQPRVEELESL 227
++ IV++V T H ++G IGI ++ E+E++
Sbjct: 135 ------------------------VEEIVRDVYET--HFYVG----RIGIYSKLLEIENM 164
Query: 228 LKLDSKDYEFRAIGIWGMAGIRKTTLASVLYDRVSYQFDASCFIENVSKIYKDGGATAVQ 287
+ + + R +GIWGM GI KTTLA ++D++S FDASCFIE+ K + G +
Sbjct: 165 V--NKQPIGIRCVGIWGMPGIGKTTLAKAVFDQMSSAFDASCFIEDYDKSIHEKGLYCLL 222
Query: 288 KQILRQTIDEKNLETYSPSEISGIIRKRLCNKKFLVVLDNADLLEQMEELAINPELLGKG 347
++ L D ++ S +R RL +K+ LVVLD+ E + LG G
Sbjct: 223 EEQLLPGNDATIMKLSS-------LRDRLNSKRVLVVLDDVCNALVAESFLEGFDWLGPG 275
Query: 348 SRIIITTRDMHILSLSVSHDTCVSYDVYEVPLLNSNDARKLFYRKAFKSEDP-TSGCVKL 406
S IIIT+RD + L C +YEV LN +AR+LF A ED +L
Sbjct: 276 SLIIITSRDKQVFRL------CGINQIYEVQGLNEKEARQLFLLSASIMEDMGEQNLHEL 329
Query: 407 TPEVLKYAQGLPLAVRVVGSFLC-TRDANQWRDALYRLRNNPDNNVMDVLQVSFEGLHSE 465
+ V+ YA G PLA+ V G L + ++ A +L+ P ++D + S++ L
Sbjct: 330 SVRVISYANGNPLAISVYGRELKGKKKLSEMETAFLKLKRRPPFKIVDAFKSSYDTLSDN 389
Query: 466 DREIFLHIACFFKGEKEDYVKRILDACGLHPHIGIQSLIERSFITIRNNEILMHEMLQEL 525
++ IFL IACFF+GE +YV ++L+ CG PH+ I L+++ +TI N + +H++ Q++
Sbjct: 390 EKNIFLDIACFFQGENVNYVIQLLEGCGFFPHVEIDVLVDKCLVTISENRVWLHKLTQDI 449
Query: 526 GKKIV--------RQQFPFQPGSWSRLWLYDDFYSVMMTET------GTNNINAIILDQK 571
G++I+ R++ ++P S L Y++ + +T G+ I + LD
Sbjct: 450 GREIINGETVQIERRRRLWEPWSIKYLLEYNEHKANGEPKTTFKRAQGSEEIEGLFLDTS 509
Query: 572 EHISEYPQLRAEALSIMRGLKILILLFHK-------NF-SGSLTFLSNSLQYLLWYGYPF 623
+ L+ A M L++L + NF +GSL L N L+ L W YP
Sbjct: 510 NLRFD---LQPSAFKNMLNLRLLKIYCSNPEVHPVINFPTGSLHSLPNELRLLHWENYPL 566
Query: 624 ASLPLNFEPFCLVELNMPYSSIQRLWDGHK 653
SLP NF+P LVE+NMPYS +Q+LW G K
Sbjct: 567 KSLPQNFDPRHLVEINMPYSQLQKLWGGTK 596
Score = 54.7 bits (130), Expect = 8e-07
Identities = 28/82 (34%), Positives = 51/82 (62%), Gaps = 1/82 (1%)
Query: 453 DVLQVSFEGLHSEDREIFLHIACFFKGEKEDYVKRILDACGLHPHIGIQSLIERSFITIR 512
+VL+VS++ L D+ +FL+IA F E D+V ++ L G++ L + S I++
Sbjct: 1087 EVLRVSYDDLQEMDKVLFLYIASLFNDEDVDFVAPLIAGIDLDVSSGLKVLADVSLISVS 1146
Query: 513 NN-EILMHEMLQELGKKIVRQQ 533
+N EI+MH + +++GK+I+ Q
Sbjct: 1147 SNGEIVMHSLQRQMGKEILHGQ 1168
>DR37_ARATH (Q9LVT4) Probable disease resistance protein At5g47250
Length = 843
Score = 89.4 bits (220), Expect = 3e-17
Identities = 90/347 (25%), Positives = 166/347 (46%), Gaps = 49/347 (14%)
Query: 215 IGIQPRVEELESLLKLDSKDYEFRAIGIWGMAGIRKTTLASVLYDR---VSYQFDASCFI 271
+G+ +E+ L+ D E R +GI+GM G+ KTTL +++ ++ VS +D ++
Sbjct: 158 VGLDTTLEKTWESLRKD----ENRMLGIFGMGGVGKTTLLTLINNKFVEVSDDYDVVIWV 213
Query: 272 ENVSKIYKDGGATAVQKQI-LRQTIDEKNLETYS----PSEISGIIRKRLCNKKFLVVLD 326
E+ KD +Q I R I + N TYS SEIS ++R +F+++LD
Sbjct: 214 ESS----KDADVGKIQDAIGERLHICDNNWSTYSRGKKASEISRVLRDM--KPRFVLLLD 267
Query: 327 NADLLEQMEELAINPELLGKGSRIIITTRDMHILSLSVSHDTCVSYDVYEVPLLNSNDAR 386
DL E + AI +LGK +++ TTR + S+ +++ EV L+ NDA
Sbjct: 268 --DLWEDVSLTAIGIPVLGKKYKVVFTTRSKDVCSVMRANED------IEVQCLSENDAW 319
Query: 387 KLFYRKAFKSEDPTSGCVKLTPEVLKYAQGLPLAVRVVGSFLCTRD-ANQWRDALYRLRN 445
LF K D + + +++ GLPLA+ V+ + ++ QWR AL L +
Sbjct: 320 DLFDMKVHC--DGLNEISDIAKKIVAKCCGLPLALEVIRKTMASKSTVIQWRRALDTLES 377
Query: 446 ------NPDNNVMDVLQVSFEGLHSEDREIFLHIACFFKG---EKEDYVKRILDACGLHP 496
+ + VL++S++ L +++ + FL+ A F K ++++ V+ + +
Sbjct: 378 YRSEMKGTEKGIFQVLKLSYDYLKTKNAKCFLYCALFPKAYYIKQDELVEYWIGEGFIDE 437
Query: 497 HIG-----------IQSLIERSFITIRNNEILMHEMLQELGKKIVRQ 532
G I +L+ + N ++ MH+M++++ IV +
Sbjct: 438 KDGRERAKDRGYEIIDNLVGAGLLLESNKKVYMHDMIRDMALWIVSE 484
>DR32_ARATH (Q9FG91) Probable disease resistance protein At5g43730
Length = 848
Score = 78.6 bits (192), Expect = 5e-14
Identities = 77/324 (23%), Positives = 152/324 (46%), Gaps = 46/324 (14%)
Query: 234 DYEFRAIGIWGMAGIRKTTLASVLYDR---VSYQFDASCFIENVSKIYKDGGATAVQKQI 290
D E R +G++GM GI KTTL L ++ + +FD ++ + KD +Q QI
Sbjct: 169 DDEIRTLGLYGMGGIGKTTLLESLNNKFVELESEFDVVIWVV----VSKDFQLEGIQDQI 224
Query: 291 LRQTIDEKNLETYSPSEISGIIRKRLCNKKFLVVLDNADLLEQMEELAINPELLGKGSRI 350
L + +K E + S+ + +I L KKF+++LD+ + ++ + P GS+I
Sbjct: 225 LGRLRPDKEWERETESKKASLINNNLKRKKFVLLLDDLWSEVDLIKIGVPPPSRENGSKI 284
Query: 351 IITTRDMHILSLSVSHDTCVSYDVYEVPLLNSNDARKLFYRKA----FKSEDPTSGCVKL 406
+ TTR + + D + D L+ ++A +LF +S ++
Sbjct: 285 VFTTRSKEVCK-HMKADKQIKVD-----CLSPDEAWELFRLTVGDIILRSHQDIPALARI 338
Query: 407 TPEVLKYAQGLPLAVRVVG-SFLCTRDANQWRDALYRLRNNP-------DNNVMDVLQVS 458
V GLPLA+ V+G + +C +WR A+ + N+P + ++ +L+ S
Sbjct: 339 ---VAAKCHGLPLALNVIGKAMVCKETVQEWRHAI-NVLNSPGHKFPGMEERILPILKFS 394
Query: 459 FEGL-HSEDREIFLHIACF---FKGEKEDYVKRILDACGLHPH----------IGIQSLI 504
++ L + E + FL+ + F F+ EK+ ++ + ++P+ I L+
Sbjct: 395 YDSLKNGEIKLCFLYCSLFPEDFEIEKDKLIEYWICEGYINPNRYEDGGTNQGYDIIGLL 454
Query: 505 ERSFITIR---NNEILMHEMLQEL 525
R+ + I +++ MH++++E+
Sbjct: 455 VRAHLLIECELTDKVKMHDVIREM 478
>DR17_ARATH (O64790) Putative disease resistance protein At1g61300
Length = 766
Score = 77.0 bits (188), Expect = 1e-13
Identities = 114/488 (23%), Positives = 218/488 (44%), Gaps = 86/488 (17%)
Query: 218 QPRVEELESLLKLDSKDYEFRA--IGIWGMAGIRKTTLASVLYDR---VSYQFDASCFI- 271
QP + + E L K ++ E R +G+ GM G+ KTTL ++++ +S +FD +I
Sbjct: 40 QPTIGQEEMLEKAWNRLMEDRVGIMGLHGMGGVGKTTLFKKIHNKFAKMSSRFDIVIWIV 99
Query: 272 ----ENVSKIYKDGGATAVQKQILRQTIDEKNLETYSPSEISGIIRKRLCNKKFLVVLDN 327
+SK+ +D +K L + + E+ ++I +++ K+F+++LD
Sbjct: 100 VSKGAKLSKLQED----IAEKLHLCDDLWKNKNESDKATDIHRVLK----GKRFVLMLD- 150
Query: 328 ADLLEQMEELAIN---PELLGKGSRIIITTRDMHILSLSVSHDTCVSYDVYEVPLLNSND 384
D+ E+++ AI P + K ++ TTRD + H +V L D
Sbjct: 151 -DIWEKVDLEAIGVPYPSEVNK-CKVAFTTRDQKVCGEMGDHKPM------QVKCLEPED 202
Query: 385 ARKLFYRKAFKS---EDPTSGCVKLTPEVLKYAQGLPLAVRVVGSFLCTRD-ANQWRDAL 440
A +LF K + DP V+L EV + +GLPLA+ V+G + ++ +W A+
Sbjct: 203 AWELFKNKVGDNTLRSDPV--IVELAREVAQKCRGLPLALSVIGETMASKTMVQEWEHAI 260
Query: 441 YRLR------NNPDNNVMDVLQVSFEGLHSED-REIFLHIACFFKGEKEDYVKRILDACG 493
L +N N ++ +L+ S++ L E + FL+ A F + E Y ++++D
Sbjct: 261 DVLTRSAAEFSNMGNKILPILKYSYDSLGDEHIKSCFLYCA-LFPEDDEIYNEKLIDYWI 319
Query: 494 LHPHIGIQSLIERS---------FITIRN-------NEILMHEMLQEL--------GKK- 528
IG +I+R+ +T+ N ++MH++++E+ GK+
Sbjct: 320 CEGFIGEDQVIKRARNKGYEMLGTLTLANLLTKVGTEHVVMHDVVREMALWIASDFGKQK 379
Query: 529 ---IVRQQFPFQP-------GSWSRLWLYDDFYSVMMTETGTNNINAIILDQKEHISEYP 578
+VR + G+ R+ L D+ + E+ + + + L ++
Sbjct: 380 ENFVVRARVGLHERPEAKDWGAVRRMSLMDNHIEEITCESKCSELTTLFLQS----NQLK 435
Query: 579 QLRAEALSIMRGLKILILLFHKNFSGSLTFLSN--SLQYLLWYGYPFASLPLNFEPF-CL 635
L E + M+ L +L L ++++F+ +S SLQ+L LP+ + L
Sbjct: 436 NLSGEFIRYMQKLVVLDLSYNRDFNKLPEQISGLVSLQFLDLSNTSIKQLPVGLKKLKKL 495
Query: 636 VELNMPYS 643
LN+ Y+
Sbjct: 496 TFLNLAYT 503
>DR10_ARATH (Q8W3J8) Probable disease resistance protein RDL5/RF45
Length = 855
Score = 75.9 bits (185), Expect = 3e-13
Identities = 83/295 (28%), Positives = 139/295 (46%), Gaps = 41/295 (13%)
Query: 213 DLIGIQPRVEELESLLKLDSKDYEFRAIGIWGMAGIRKTTLASVLYDR--VSYQFDASCF 270
D +G++ V++L L ++ + + I GM G+ KTTLA +++ V +QFD +
Sbjct: 162 DFVGLEANVKKLVGYLVDEAN---VQVVSITGMGGLGKTTLAKQVFNHEDVKHQFDGLSW 218
Query: 271 IENVSKIYKDGGATAVQKQILRQTI---DEKNLETYSPSEISGIIRKRLCNKKFLVVLDN 327
+ + +D V ++ILR +EK + + + G + + L K L+VLD
Sbjct: 219 V----CVSQDFTRMNVWQKILRDLKPKEEEKKIMEMTQDTLQGELIRLLETSKSLIVLD- 273
Query: 328 ADLLEQMEELAINPELLG-KGSRIIITTRDMHILSLSVSHDTCVSYDVYEVPLLNSNDAR 386
D+ E+ + I P KG ++++T+R+ SV+ SY ++ L + D+
Sbjct: 274 -DIWEKEDWELIKPIFPPTKGWKVLLTSRNE-----SVAMRRNTSYINFKPECLTTEDSW 327
Query: 387 KLFYRKAFKSEDPTSGCVKLTPE-----VLKYAQGLPLAVRVVGSFLCTR-DANQWRDAL 440
LF R A +D + E ++K+ GLPLA+RV+G L + ++ WR
Sbjct: 328 TLFQRIALPMKDAAEFKIDEEKEELGKLMIKHCGGLPLAIRVLGGMLAEKYTSHDWRRLS 387
Query: 441 Y---------RLRNNPDNN--VMDVLQVSFEGLHSEDREIFLHIACFFKGEKEDY 484
R N DNN +VL +SFE L S + FL++A F EDY
Sbjct: 388 ENIGSHLVGGRTNFNDDNNNTCNNVLSLSFEELPSYLKHCFLYLAHF----PEDY 438
>DR33_ARATH (Q9FG90) Probable disease resistance protein At5g43740
Length = 862
Score = 75.1 bits (183), Expect = 5e-13
Identities = 85/384 (22%), Positives = 174/384 (45%), Gaps = 52/384 (13%)
Query: 173 ESLAKLVGWDVRNKPEFREIKNIVQEVINTMGHKFLGFADDLIGIQPRVEELESLLKLDS 232
E ++K++ +V+ ++ + + QE+I+ + K + +G+ VE S L D
Sbjct: 115 EKVSKMLE-EVKELLSKKDFRMVAQEIIHKVEKKLI---QTTVGLDKLVEMAWSSLMND- 169
Query: 233 KDYEFRAIGIWGMAGIRKTTLASVLYDR---VSYQFDASCFIENVSKIYKDGGATAVQKQ 289
E +G++GM G+ KTTL L ++ + +FD ++ + KD +Q Q
Sbjct: 170 ---EIGTLGLYGMGGVGKTTLLESLNNKFVELESEFDVVIWVV----VSKDFQFEGIQDQ 222
Query: 290 ILRQTIDEKNLETYSPSEISGIIRKRLCNKKFLVVLDNADLLEQMEELAINPELLGKGSR 349
IL + +K E + S+ + +I L KKF+++LD+ M ++ + P GS+
Sbjct: 223 ILGRLRSDKEWERETESKKASLIYNNLERKKFVLLLDDLWSEVDMTKIGVPPPTRENGSK 282
Query: 350 IIITTRDMHILSLSVSHDTCVSYDVYEVPLLNSNDARKLFYRKA----FKSEDPTSGCVK 405
I+ TTR + + D + +V L+ ++A +LF +S +
Sbjct: 283 IVFTTRSTEVCK-HMKADKQI-----KVACLSPDEAWELFRLTVGDIILRSHQDIPALAR 336
Query: 406 LTPEVLKYAQGLPLAVRVVGSFL-CTRDANQWRDALYRLRN------NPDNNVMDVLQVS 458
+ V GLPLA+ V+G + C +W A+ L + + ++ +L+ S
Sbjct: 337 I---VAAKCHGLPLALNVIGKAMSCKETIQEWSHAINVLNSAGHEFPGMEERILPILKFS 393
Query: 459 FEGL-HSEDREIFLHIACFFKGE---KEDYVKRILDACGLHPH----------IGIQSLI 504
++ L + E + FL+ + F + KE +++ + ++P+ I L+
Sbjct: 394 YDSLKNGEIKLCFLYCSLFPEDSEIPKEKWIEYWICEGFINPNRYEDGGTNHGYDIIGLL 453
Query: 505 ERSFITIR---NNEILMHEMLQEL 525
R+ + I + + MH++++E+
Sbjct: 454 VRAHLLIECELTDNVKMHDVIREM 477
>RPM1_ARATH (Q39214) Disease resistance protein RPM1 (Resistance to
Pseudomonas syringae protein 3)
Length = 926
Score = 74.7 bits (182), Expect = 7e-13
Identities = 85/350 (24%), Positives = 168/350 (47%), Gaps = 44/350 (12%)
Query: 200 INTMGHKFLGFADD-LIGIQPRVEELESLLKLDSKDYEFRAIGIWGMAGIRKTTLASVLY 258
+N + L F+++ L+GI +L + +L S + + + + GM G KTTL++ ++
Sbjct: 157 VNNISESSLFFSENSLVGIDAPKGKL--IGRLLSPEPQRIVVAVVGMGGSGKTTLSANIF 214
Query: 259 DRVSYQ--FDASCFIENVSKIYKDGGATAVQKQILRQTIDE--KNLETYSPSEISGIIRK 314
S + F++ ++ +SK Y V + + R I E K +T P+E+ + +
Sbjct: 215 KSQSVRRHFESYAWV-TISKSY-------VIEDVFRTMIKEFYKEADTQIPAELYSLGYR 266
Query: 315 RLC--------NKKFLVVLDNADLLEQMEELAINPELLGKGSRIIITTRDMHILSLSVSH 366
L +K+++VVLD+ E++I GSR+++TTRDM++ S
Sbjct: 267 ELVEKLVEYLQSKRYIVVLDDVWTTGLWREISIALPDGIYGSRVMMTTRDMNVASFPYG- 325
Query: 367 DTCVSYDVYEVPLLNSNDARKLFYRKAFKSEDPTSGCVKLTP---EVLKYAQGLPLAVRV 423
+ +E+ LL ++A LF KAF + L P ++++ QGLPLA+
Sbjct: 326 ---IGSTKHEIELLKEDEAWVLFSNKAFPASLEQCRTQNLEPIARKLVERCQGLPLAIAS 382
Query: 424 VGSFLCTRD-ANQWRDALYRLRNNPDNN-----VMDVLQVSFEGLHSEDREIFLHIACF- 476
+GS + T+ ++W+ L +NN V ++ +SF L + FL+ + F
Sbjct: 383 LGSMMSTKKFESEWKKVYSTLNWELNNNHELKIVRSIMFLSFNDLPYPLKRCFLYCSLFP 442
Query: 477 --FKGEKEDYVKRILDACGLHPHIGIQS-LIERSFITIRNNEILMHEMLQ 523
++ +++ ++ + + P G+++ + S++ NE++ MLQ
Sbjct: 443 VNYRMKRKRLIRMWMAQRFVEPIRGVKAEEVADSYL----NELVYRNMLQ 488
>DR11_ARATH (Q94HW3) Probable disease resistance protein RDL6/RF9
Length = 1049
Score = 73.9 bits (180), Expect = 1e-12
Identities = 78/287 (27%), Positives = 137/287 (47%), Gaps = 37/287 (12%)
Query: 213 DLIGIQPRVEELESLLKLDSKDYEFRAIGIWGMAGIRKTTLASVLYDR--VSYQFDASCF 270
D +G++ V++L L ++ + + I GM G+ KTTLA +++ V +QFD +
Sbjct: 162 DFVGLEANVKKLVGYLVDEAN---VQVVSITGMGGLGKTTLAKQVFNHEDVKHQFDGLSW 218
Query: 271 IENVSKIYKDGGATAVQKQILRQTI---DEKNLETYSPSEISGIIRKRLCNKKFLVVLDN 327
+ + +D V ++ILR +EK + + + G + + L K L+VLD
Sbjct: 219 V----CVSQDFTRMNVWQKILRDLKPKEEEKKIMEMTQDTLQGELIRLLETSKSLIVLD- 273
Query: 328 ADLLEQMEELAINPELLG-KGSRIIITTRDMHILSLSVSHDTCVSYDVYEVPLLNSNDAR 386
D+ E+ + I P KG ++++T+R+ SV+ SY ++ L + D+
Sbjct: 274 -DIWEKEDWELIKPIFPPTKGWKVLLTSRNE-----SVAMRRNTSYINFKPECLTTEDSW 327
Query: 387 KLFYRKAFKSEDPTSGCVKLTPE-----VLKYAQGLPLAVRVVGSFLCTR-DANQWRDAL 440
LF R A +D + E ++K+ GLPLA+RV+G L + ++ WR
Sbjct: 328 TLFQRIALPMKDAAEFKIDEEKEELGKLMIKHCGGLPLAIRVLGGMLAEKYTSHDWRRLS 387
Query: 441 YRLR----------NNPDNNVMD-VLQVSFEGLHSEDREIFLHIACF 476
+ N+ +NN + VL +SFE L S + FL++A F
Sbjct: 388 ENIGSHLVGGRTNFNDDNNNTCNYVLSLSFEELPSYLKHCFLYLAHF 434
>RP13_ARATH (Q9M667) Disease resistance protein RPP13 (Resistance to
Peronospora parasitica protein 13)
Length = 835
Score = 73.6 bits (179), Expect = 2e-12
Identities = 73/270 (27%), Positives = 129/270 (47%), Gaps = 21/270 (7%)
Query: 224 LESLLKLDSKDYEFRAIGIWGMAGIRKTTLASVLYDR--VSYQFDASCFIENVSKIYKDG 281
LE LL + K+ I I+GM G+ KT LA LY+ V +F+ + VS+ YK G
Sbjct: 174 LEKLLDYEEKNRFI--ISIFGMGGLGKTALARKLYNSRDVKERFEYRAWTY-VSQEYKTG 230
Query: 282 GATAVQKQILRQTIDEK--NLETYSPSEISGIIRKRLCNKKFLVVLDNADLLEQMEELAI 339
+ L T E+ + ++ E+ + L KK+LVV+D+ E + L
Sbjct: 231 DILMRIIRSLGMTSGEELEKIRKFAEEELEVYLYGLLEGKKYLVVVDDIWEREAWDSLKR 290
Query: 340 NPELLGKGSRIIITTRDMHILSLSVSHDTCVSYDVYEVPLLNSNDARKLFYRKAFKS-ED 398
+GSR+IITTR +V+ + +++ L ++ +LF ++AF++ +
Sbjct: 291 ALPCNHEGSRVIITTRIK-----AVAEGVDGRFYAHKLRFLTFEESWELFEQRAFRNIQR 345
Query: 399 PTSGCVKLTPEVLKYAQGLPLAVRVVGSFLCTRDANQWRDAL----YRLRNNPDNNVMDV 454
+K E+++ +GLPL + V+ L + ++W D RL+++ + V
Sbjct: 346 KDEDLLKTGKEMVQKCRGLPLCIVVLAGLLSRKTPSEWNDVCNSLWRRLKDDSIHVAPIV 405
Query: 455 LQVSFEGLHSEDREIFLHIACFFKGEKEDY 484
+SF+ L E + FL+++ F EDY
Sbjct: 406 FDLSFKELRHESKLCFLYLSIF----PEDY 431
>R8L3_ARATH (Q9FJB5) Probable disease resistance RPP8-like protein 3
Length = 901
Score = 72.8 bits (177), Expect = 3e-12
Identities = 85/348 (24%), Positives = 164/348 (46%), Gaps = 61/348 (17%)
Query: 164 KVMRWVGAMESLAKLVGWDVRNKPEFREIKNIVQEVINTMGHKFLGFADDLIGIQPRVEE 223
++ + +G M+SL + + + +++I +E+ T + DL+G++ VEE
Sbjct: 118 RISKVIGEMQSLG--IQQQIIDGGRSLSLQDIQREIRQTFPNSS---ESDLVGVEQSVEE 172
Query: 224 LES-LLKLDSKDYEFRAIGIWGMAGIRKTTLASVLY--DRVSYQFDASCFIENVSKIYKD 280
L ++++D+ + + I GM GI KTTLA ++ D V FD ++
Sbjct: 173 LVGPMVEIDN----IQVVSISGMGGIGKTTLARQIFHHDLVRRHFDGFAWV--------- 219
Query: 281 GGATAVQKQILRQTIDEKNLETYSPSE----------ISGIIRKRLCNKKFLVVLDNADL 330
V +Q ++ + ++ L+ P + I G + + L ++LVVLD+
Sbjct: 220 ----CVSQQFTQKHVWQRILQELRPHDGEILQMDEYTIQGKLFQLLETGRYLVVLDDVWK 275
Query: 331 LEQMEELA-INPELLGKGSRIIITTRDMHILSLSVSHD-TCVSYDVYEVPLLNSNDARKL 388
E + + + P +G ++++T+R+ + + D TC+S+ +LN ++ KL
Sbjct: 276 EEDWDRIKEVFPRK--RGWKMLLTSRNEGV---GLHADPTCLSF---RARILNPKESWKL 327
Query: 389 FYRKA-FKSEDPTSGCVKLTPEVLKYAQGLPLAVRVVGSFLCTR-DANQWRDALYR---- 442
F R ++E + E++ Y GLPLAV+V+G L + A++W+
Sbjct: 328 FERIVPRRNETEYEEMEAIGKEMVTYCGGLPLAVKVLGGLLANKHTASEWKRVSENIGAQ 387
Query: 443 ------LRNNPDNNVMDVLQVSFEGLHSEDREIFLHIACFFKGEKEDY 484
L +N N+V +L +S+E L ++ + FL++A F EDY
Sbjct: 388 IVGKSCLDDNSLNSVYRILSLSYEDLPTDLKHCFLYLAHF----PEDY 431
>R133_ARATH (Q9STE7) Putative disease resistance RPP13-like protein
3
Length = 847
Score = 72.4 bits (176), Expect = 4e-12
Identities = 75/288 (26%), Positives = 137/288 (47%), Gaps = 35/288 (12%)
Query: 214 LIGIQPRVEELESLLKLDSKDYEFRAIGIWGMAGIRKTTLASVLYDR--VSYQFDASCFI 271
++G++ V+ L L D++ + I I+GM G+ KT LA LY+ V +FD +
Sbjct: 162 VVGLEDDVKILLVKLLSDNEKDKSYIISIFGMGGLGKTALARKLYNSGDVKRRFDCRAW- 220
Query: 272 ENVSKIYKD-----------GGATAVQKQILRQTIDEKNLETYSPSEISGIIRKRLCNKK 320
VS+ YK G +A + + ++ +++ LE Y + G K
Sbjct: 221 TYVSQEYKTRDILIRIIRSLGIVSAEEMEKIKMFEEDEELEVYLYGLLEG--------KN 272
Query: 321 FLVVLDNADLLEQMEELAINPELLGKGSRIIITTRDMHILSLSVSHDTCVSYDVYEVPLL 380
++VV+D+ + E L +GS++IITTR I +++ + V +++ L
Sbjct: 273 YMVVVDDVWDPDAWESLKRALPCDHRGSKVIITTR---IRAIAEGVEGTVY--AHKLRFL 327
Query: 381 NSNDARKLFYRKAFKSEDPTSGCVKLT-PEVLKYAQGLPLAVRVVGSFLCTRDANQWRD- 438
++ LF RKAF + + ++ T E++K GLPLA+ V+ L + N+W +
Sbjct: 328 TFEESWTLFERKAFSNIEKVDEDLQRTGKEMVKKCGGLPLAIVVLSGLLSRKRTNEWHEV 387
Query: 439 --ALYRLRNNPDNNVMDVLQVSFEGLHSEDREIFLHIACFFKGEKEDY 484
+L+R + ++ V +SF+ + E + FL+ + F EDY
Sbjct: 388 CASLWRRLKDNSIHISTVFDLSFKEMRHELKLCFLYFSVF----PEDY 431
>DR31_ARATH (Q9FLB4) Putative disease resistance protein At5g05400
Length = 874
Score = 70.9 bits (172), Expect = 1e-11
Identities = 67/254 (26%), Positives = 124/254 (48%), Gaps = 28/254 (11%)
Query: 240 IGIWGMAGIRKTTLASVLYDR---VSYQFDASCFIENVSKIYKDGGATAVQKQI-LRQTI 295
+GI+GM G+ KTTL S + ++ VS FD + ++ + K+ +Q+ I R +
Sbjct: 178 LGIYGMGGVGKTTLLSQINNKFRTVSNDFDIAIWVV----VSKNPTVKRIQEDIGKRLDL 233
Query: 296 DEKNLETYSPSEISGIIRKRLCNKKFLVVLDNADLLEQMEELAINPELLGKGSRIIITTR 355
+ E + +EI+ I++ L NKK++++LD+ + + I P GS+I T+R
Sbjct: 234 YNEGWEQKTENEIASTIKRSLENKKYMLLLDDMWTKVDLANIGI-PVPKRNGSKIAFTSR 292
Query: 356 DMHILSLSVSHDTCVSYDV---YEVPLLNSNDARKLFYRKAFKSEDPTSGCVKLTPEVLK 412
S++ C V EV L +DA LF R ++ + ++ + +
Sbjct: 293 ---------SNEVCGKMGVDKEIEVTCLMWDDAWDLFTRNMKETLESHPKIPEVAKSIAR 343
Query: 413 YAQGLPLAVRVVGSFLCTRDA-NQWRDALYRLRNNPDNNVMDVLQVSFEGLHSE-DREIF 470
GLPLA+ V+G + + + +W DA+ + + + +++ +L+ S++ L E + F
Sbjct: 344 KCNGLPLALNVIGETMARKKSIEEWHDAV-GVFSGIEADILSILKFSYDDLKCEKTKSCF 402
Query: 471 LHIACFFKGEKEDY 484
L A F EDY
Sbjct: 403 LFSALF----PEDY 412
>DRL3_ARATH (Q9LMP6) Probable disease resistance protein At1g15890
Length = 921
Score = 69.7 bits (169), Expect = 2e-11
Identities = 102/466 (21%), Positives = 206/466 (43%), Gaps = 80/466 (17%)
Query: 236 EFRAIGIWGMAGIRKTTLASVLYDRV---SYQFDASCFIENVSKIYKDGGATAVQKQILR 292
E R +G++GM G+ KTTL + + ++ FD ++ VSK ++ G +Q+QIL
Sbjct: 243 ERRTLGLYGMGGVGKTTLLASINNKFLEGMNGFDLVIWVV-VSKDLQNEG---IQEQILG 298
Query: 293 QTIDEKNLETYSPSEISGIIRKRLCNKKFLVVLDNADLLEQMEELAINPELLGKGSRIII 352
+ + + + E + I L KKF+++LD+ +E++ + P GS+I+
Sbjct: 299 RLGLHRGWKQVTEKEKASYICNILNVKKFVLLLDDLWSEVDLEKIGVPPLTRENGSKIVF 358
Query: 353 TTRDMHILSLSVSHDTCVSYDV---YEVPLLNSNDARKLFYRKA------FKSEDPTSGC 403
TTR S D C +V +V L ++A +LF +K + PT
Sbjct: 359 TTR---------SKDVCRDMEVDGEMKVDCLPPDEAWELFQKKVGPIPLQSHEDIPT--- 406
Query: 404 VKLTPEVLKYAQGLPLAVRVVGSFLCTRD-ANQWRDALYRLRNNP------DNNVMDVLQ 456
L +V + GLPLA+ V+G + +R+ +W+ ++ L ++ + ++ VL+
Sbjct: 407 --LARKVAEKCCGLPLALSVIGKAMASRETVQEWQHVIHVLNSSSHEFPSMEEKILPVLK 464
Query: 457 VSFEGLHSEDREI-FLHIACF---FKGEKEDYVKRIL-----------DACGLHPHIGIQ 501
S++ L E ++ FL+ + F ++ KE+ ++ + D H I
Sbjct: 465 FSYDDLKDEKVKLCFLYCSLFPEDYEVRKEELIEYWMCEGFIDGNEDEDGANNKGHDIIG 524
Query: 502 SLIERSFIT--IRNNEILMHEMLQELGKKIV------RQQFPFQPG-------------S 540
SL+ + ++ MH++++E+ I ++ +PG S
Sbjct: 525 SLVRAHLLMDGELTTKVKMHDVIREMALWIASNFGKQKETLCVKPGVQLCHIPKDINWES 584
Query: 541 WSRLWLYDDFYSVMMTETGTNNINAIILDQKEHISEYPQLRAEALSIMRGLKILILLFHK 600
R+ L + + + + + + N++ ++L + + + + M L +L L +
Sbjct: 585 LRRMSLMCNQIANISSSSNSPNLSTLLLQNNKLV----HISCDFFRFMPALVVLDLSRNS 640
Query: 601 NFSGSLTFLS--NSLQYLLWYGYPFASLPLNFEPF-CLVELNMPYS 643
+ S +S SLQY+ LP++F+ L+ LN+ ++
Sbjct: 641 SLSSLPEAISKLGSLQYINLSTTGIKWLPVSFKELKKLIHLNLEFT 686
>R8L1_ARATH (O04093) Putative disease resistance protein RPP8-like
protein 1
Length = 821
Score = 69.3 bits (168), Expect = 3e-11
Identities = 103/447 (23%), Positives = 189/447 (42%), Gaps = 74/447 (16%)
Query: 190 REIKNIVQEVINTMGHKFLGFADDLIGIQPRVEELESLLKLDSKDYEFRAIGIWGMAGIR 249
RE K I Q N+ DL+G++ VE L L ++ + + I GM GI
Sbjct: 123 REQKEIRQTFANSS-------ESDLVGVEQSVEALAGHLV---ENDNIQVVSISGMGGIG 172
Query: 250 KTTLASVLY--DRVSYQFDASCFIENVSKIYKDGGATAVQKQILRQTIDEKNLETYSPSE 307
KTTLA ++ D V FD ++ + + + +++ Q D +++ +
Sbjct: 173 KTTLARQVFHHDMVQRHFDGFAWVFVSQQFTQKHVWQRIWQELQPQNGDISHMDEHI--- 229
Query: 308 ISGIIRKRLCNKKFLVVLDNADLLEQMEEL-AINPELLGKGSRIIITTRDMHILSLSVSH 366
+ G + K L ++LVVLD+ E + + A+ P +G ++++T+R+ + H
Sbjct: 230 LQGKLFKLLETGRYLVVLDDVWKEEDWDRIKAVFPRK--RGWKMLLTSRNEGV----GIH 283
Query: 367 DTCVSYDVYEVPLLNSNDARKLFYRKAFKSEDPT---SGCVKLTPEVLKYAQGLPLAVRV 423
S+ ++ +L ++ KL + F D T S + E++ GLPLAV+V
Sbjct: 284 ADPKSFG-FKTRILTPEESWKLCEKIVFHRRDETGTLSDMEAMGKEMVTCCGGLPLAVKV 342
Query: 424 VGSFLCTR-DANQWRDALYRL------RNNPD---NNVMDVLQVSFEGLHSEDREIFLHI 473
+G L T+ +W+ + R++ D N++ VL +S+E L + FL++
Sbjct: 343 LGGLLATKHTVPEWKRVYDNIGPHLAGRSSLDDNLNSIYRVLSLSYENLPMCLKHCFLYL 402
Query: 474 ACFFKGEKEDYVKRILDACGLHPHIG---------------IQSLIERSFITIRNNEIL- 517
A F E +VKR+ + I ++ L R+ ITI N +
Sbjct: 403 A-HFPEYYEIHVKRLFNYLAAEGIITSSDDGTTIQDKGEDYLEELARRNMITIDKNYMFL 461
Query: 518 ------MHEMLQELGKKIVRQQFPFQPGSWSRLWLYDDFYSVMMTETGTNNINAIILDQK 571
MH+M++E+ ++ ++F + T T+ INA L +
Sbjct: 462 RKKHCQMHDMMREVCLSKAKE---------------ENFLEIFKVSTATSAINARSLSKS 506
Query: 572 EHISEYPQLRAEALSIMRGLKILILLF 598
+S + +L K+ LL+
Sbjct: 507 RRLSVHGGNALPSLGQTINKKVRSLLY 533
>R132_ARATH (Q9STE5) Putative disease resistance RPP13-like protein
2
Length = 847
Score = 69.3 bits (168), Expect = 3e-11
Identities = 71/287 (24%), Positives = 129/287 (44%), Gaps = 27/287 (9%)
Query: 214 LIGIQPRVEELESLLKLDSKDYEFRAIGIWGMAGIRKTTLASVLYD----RVSYQFDASC 269
++G+ + L + L D D + I I+GM G+ KT+LA L++ + S+++
Sbjct: 161 VVGLTDDAKVLLTKLLDDDGDNKIYMISIFGMEGLGKTSLARKLFNSSDVKESFEY---- 216
Query: 270 FIENVSKIYKDGGATAVQKQILRQTID------EKNLETYSPSEISGIIRKRLCNKKFLV 323
+++ + + IL + I E LE + E+ + L K++LV
Sbjct: 217 ------RVWTNVSGECNTRDILMRIISSLEETSEGELEKMAQQELEVYLHDILQEKRYLV 270
Query: 324 VLDNADLLEQMEELAINPELLGKGSRIIITTRDMHILSLSVSHDTCVSYDVYEVPLLNSN 383
V+D+ E +E L +GSR+IITT I ++ D V + + L
Sbjct: 271 VVDDIWESEALESLKRALPCSYQGSRVIITTS---IRVVAEGRDKRVY--THNIRFLTFK 325
Query: 384 DARKLFYRKAFKSE-DPTSGCVKLTPEVLKYAQGLPLAVRVVGSFLCTRDANQWRDALYR 442
++ LF +KAF+ K+ E+++ GLP V+ + + N+W D
Sbjct: 326 ESWNLFEKKAFRYILKVDQELQKIGKEMVQKCGGLPRTTVVLAGLMSRKKPNEWNDVWSS 385
Query: 443 LRNNPDN-NVMDVLQVSFEGLHSEDREIFLHIACFFKGEKEDYVKRI 488
LR DN +V + +SF+ + E + FL+++ F + + D K I
Sbjct: 386 LRVKDDNIHVSSLFDLSFKDMGHELKLCFLYLSVFPEDYEVDVEKLI 432
>DR21_ARATH (Q9LRR5) Putative disease resistance protein At3g14460
Length = 1424
Score = 69.3 bits (168), Expect = 3e-11
Identities = 83/348 (23%), Positives = 166/348 (46%), Gaps = 58/348 (16%)
Query: 220 RVEELESLLKLDSKDYEFR-----AIGIWGMAGIRKTTLASVLYD--RVSYQFDASCFIE 272
RVE+ +L+ L D E I + GM G+ KTTL ++++ RV+ F+ +I
Sbjct: 171 RVEDKLALVNLLLSDDEISIGKPAVISVVGMPGVGKTTLTEIVFNDYRVTEHFEVKMWIS 230
Query: 273 -----NVSKIYKDGGATAVQKQILRQTIDEKNLETYSPSEISGIIRKRLCNKKFLVVLDN 327
NV + K AV + I ++ ++L + ++K L K+FL+VLD+
Sbjct: 231 AGINFNVFTVTK-----AVLQDITSSAVNTEDLPSLQIQ-----LKKTLSGKRFLLVLDD 280
Query: 328 --ADLLEQMEELAINPELLGKGSRIIITTRDMHILSLSVSHDTCVSYDVYEVPLLNSNDA 385
++ + E + +GS+I++TTR + +++ + +Y++ L+ + +
Sbjct: 281 FWSESDSEWESFQVAFTDAEEGSKIVLTTRSEIVSTVAKAE------KIYQMKLMTNEEC 334
Query: 386 RKLFYRKAFKSEDPTSGCVKLT---PEVLKYAQGLPLAVRVVGSFLCTR-DANQWRDALY 441
+L R AF + S +L + + +GLPLA R + S L ++ + + W A+
Sbjct: 335 WELISRFAFGNISVGSINQELEGIGKRIAEQCKGLPLAARAIASHLRSKPNPDDWY-AVS 393
Query: 442 RLRNNPDNNVMDVLQVSFEGLHSEDREIFLHIACFFKG---EKEDYV------------- 485
+ ++ N+++ VL++S++ L + + F + F KG ++E+ V
Sbjct: 394 KNFSSYTNSILPVLKLSYDSLPPQLKRCFALCSIFPKGHVFDREELVLLWMAIDLLYQPR 453
Query: 486 -KRILDACGLHPHIG---IQSLIERSFITIRNNEILMHEMLQELGKKI 529
R L+ G + ++G QS +R IT+ +MH+++ +L K +
Sbjct: 454 SSRRLEDIG-NDYLGDLVAQSFFQRLDITM--TSFVMHDLMNDLAKAV 498
>DR13_ARATH (Q9XIF0) Putative disease resistance protein At1g59780
Length = 906
Score = 69.3 bits (168), Expect = 3e-11
Identities = 102/458 (22%), Positives = 192/458 (41%), Gaps = 91/458 (19%)
Query: 213 DLIGIQPRVEEL-ESLLKLDSKDYEFRAIGIWGMAGIRKTTLASVLYD--RVSYQFDASC 269
+L+G++ VE+L E L+ DS + I G+ G+ KTTLA ++D +V FD
Sbjct: 159 NLVGLEKNVEKLVEELVGNDSS----HGVSITGLGGLGKTTLARQIFDHDKVKSHFDGLA 214
Query: 270 FIENVSKIYKDGGATAVQKQILRQTIDEKNLETYSPSEISGIIRKRLCNKKFLVVLDNAD 329
++ + ++ V K IL + +I + + L KK L+V D D
Sbjct: 215 WV----CVSQEFTRKDVWKTILGNLSPKYKDSDLPEDDIQKKLFQLLETKKALIVFD--D 268
Query: 330 LLEQMEELAINPELLGK--GSRIIITTRDMHILSLSVSHDTCVSYDVYEVPLLNSNDARK 387
L ++ + I P + G ++++T+R+ I H CV++ + LL ++ K
Sbjct: 269 LWKREDWYRIAPMFPERKAGWKVLLTSRNDAI------HPHCVTF---KPELLTHDECWK 319
Query: 388 LFYRKAFKSEDPTSG------CVKLTPEVLKYAQGLPLAVRVVGSFLCTRDA-NQWRDAL 440
L R AF + +G VK+ E+ K+ + LPLAV+++G L + QW+
Sbjct: 320 LLQRIAFSKQKTITGYIIDKEMVKMAKEMTKHCKRLPLAVKLLGGLLDAKHTLRQWKLIS 379
Query: 441 YRL-----------RNNPDNNVMDVLQVSFEGLHSEDREIFLHIACF-----FKGEKEDY 484
+ N ++V VL +SFEGL + L++A + + E+ Y
Sbjct: 380 ENIISHIVVGGTSSNENDSSSVNHVLSLSFEGLPGYLKHCLLYLASYPEDHEIEIERLSY 439
Query: 485 V--------------KRILDACGLHPHIGIQSLI-------ERSFITIRNNEILMHEMLQ 523
V I D L+ I+ L+ ER +T R + +H++++
Sbjct: 440 VWAAEGITYPGNYEGATIRDVADLY----IEELVKRNMVISERDALTSRFEKCQLHDLMR 495
Query: 524 ELGKKIVRQQFPFQPGSWSRLWLYDDFYSVMMTETGTNNINAIILDQKEHISEYPQLRAE 583
E+ ++ ++F ++ T +++++++ + + Y
Sbjct: 496 EICLLKAKE---------------ENFLQIVTDPTSSSSVHSLASSRSRRLVVYNTSIFS 540
Query: 584 ALSIMRGLKILILLF----HKNFSGSLTFLSNSLQYLL 617
+ M+ K+ LLF + FS F+ L +L
Sbjct: 541 GENDMKNSKLRSLLFIPVGYSRFSMGSNFIELPLLRVL 578
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.138 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 75,585,977
Number of Sequences: 164201
Number of extensions: 3187748
Number of successful extensions: 7800
Number of sequences better than 10.0: 85
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 72
Number of HSP's that attempted gapping in prelim test: 7651
Number of HSP's gapped (non-prelim): 106
length of query: 655
length of database: 59,974,054
effective HSP length: 117
effective length of query: 538
effective length of database: 40,762,537
effective search space: 21930244906
effective search space used: 21930244906
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 69 (31.2 bits)
Medicago: description of AC143337.12


