

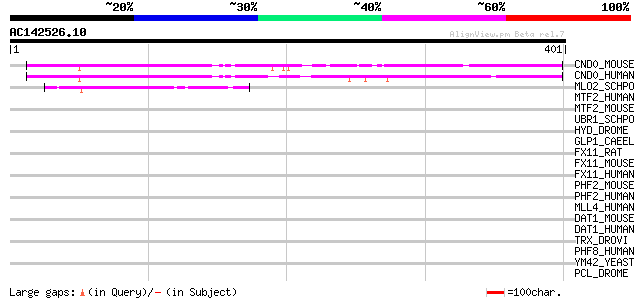
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142526.10 + phase: 0
(401 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CND0_MOUSE (Q8BU04) Protein C14orf130 homolog 252 1e-66
CND0_HUMAN (Q8N806) Protein C14orf130 247 4e-65
MLO2_SCHPO (Q09329) Protein mlo2 127 7e-29
MTF2_HUMAN (Q9Y483) Metal-response element-binding transcription... 41 0.006
MTF2_MOUSE (Q02395) Metal-response element-binding transcription... 40 0.011
UBR1_SCHPO (O60152) N-end-recognizing protein (Ubiquitin-protein... 39 0.018
HYD_DROME (P51592) Ubiquitin--protein ligase hyd (EC 6.3.2.-) (H... 39 0.024
GLP1_CAEEL (P13508) Glp-1 protein precursor 39 0.024
FX11_RAT (Q7TSL3) F-box only protein 11 38 0.053
FX11_MOUSE (Q7TPD1) F-box only protein 11 (Fragment) 38 0.053
FX11_HUMAN (Q86XK2) F-box only protein 11 (Vitiligo-associated p... 38 0.053
PHF2_MOUSE (Q9WTU0) PHD finger protein 2 (GRC5) 37 0.069
PHF2_HUMAN (O75151) PHD finger protein 2 (GRC5) 37 0.069
MLL4_HUMAN (Q9UMN6) Myeloid/lymphoid or mixed-lineage leukemia p... 37 0.069
DAT1_MOUSE (Q8C9B9) Death associated transcription factor 1 (Dea... 37 0.069
DAT1_HUMAN (Q9BTC0) Death associated transcription factor 1 37 0.090
TRX_DROVI (Q24742) Trithorax protein 37 0.12
PHF8_HUMAN (Q9UPP1) PHD finger protein 8 36 0.15
YM42_YEAST (Q03214) Hypothetical 162.7 kDa protein in SIP18-SPT2... 36 0.20
PCL_DROME (Q24459) Polycomb protein Pcl (Polycomblike protein) 35 0.26
>CND0_MOUSE (Q8BU04) Protein C14orf130 homolog
Length = 425
Score = 252 bits (644), Expect = 1e-66
Identities = 153/408 (37%), Positives = 216/408 (52%), Gaps = 46/408 (11%)
Query: 13 EADLVLGGDEGRECTYNKGYMKRQAIFSCITCTPDGN--AGVCTACSLSCHDGHQIVELW 70
EA VLGG + +C+Y++G + RQA+++C TCTP+G AG+C ACS CH H++ EL+
Sbjct: 33 EACAVLGGSDSEKCSYSQGSVGRQALYACSTCTPEGEEPAGICLACSYECHGSHKLFELY 92
Query: 71 TKRNFRCDCGNSKFGEFYCKIFPSKDIENVENSYNHNFKGLYCTCARPYPDPDAEEQIEM 130
TKRNFRCDCGNSKF CK+FP K N N YN NF GLYC C RPYPDP+ E EM
Sbjct: 93 TKRNFRCDCGNSKFKNLECKLFPDKSKVNSCNKYNDNFFGLYCVCKRPYPDPEDEVPDEM 152
Query: 131 IQCCLCEDWFHEEHLGLESSDEIPRDEEGEPLYEDFMCKACSEVCFFLKLYPEVILVA-- 188
IQC +CEDWFH HLG IP E G+ +++ +C+AC C FL Y + V
Sbjct: 153 IQCVVCEDWFHGRHLG-----AIP-PESGD--FQEMVCQACMRRCSFLWAYAAQLAVTRI 204
Query: 189 -----GKQPNATA----QVSK-------DKGILEDTPSTCGFEKPLGDTSYNSPKIDVAQ 232
G PNAT VSK D G+ ED P G S N K + Q
Sbjct: 205 SAEDDGLLPNATGMGDEDVSKPENGAPQDNGLKEDAPEH-------GRDSVNEVKAE--Q 255
Query: 233 ASVGSESISNRAGLPPGGSCNSSTCTEGASLHVNCLLGVNIVAASPVIHGKAMFLSKNWR 292
+ S S+ + L + TE S +C L + A V A + NWR
Sbjct: 256 KNEPCSSSSSESDLQTVFK-KENIKTEPQS---SCRL-QELQAKQFVKKDAATYWPLNWR 310
Query: 293 DALCKCKNCLEYYHQKRIAFLLDKEDSIVEYEKMAKQKREEKLQQQEGAELSLFNKLGHV 352
LC C++C++ Y + + FL D+ D+++ YE K ++ + + + + V
Sbjct: 311 SKLCTCQDCMKMYGELDVLFLTDECDTVLAYENKGK---NDQATDRRDPLMDTLSSMNRV 367
Query: 353 EKVEILKGIEDMKDGLRTFLES-ADSSKPISAADIHQFFDDIKNKRRR 399
++VE++ D+K L+ +L+ AD + DI QFF++ ++K+RR
Sbjct: 368 QQVELICEYNDLKTELKDYLKRFADEGTVVKREDIQQFFEEFQSKKRR 415
>CND0_HUMAN (Q8N806) Protein C14orf130
Length = 425
Score = 247 bits (630), Expect = 4e-65
Identities = 146/408 (35%), Positives = 211/408 (50%), Gaps = 46/408 (11%)
Query: 13 EADLVLGGDEGRECTYNKGYMKRQAIFSCITCTPDGN--AGVCTACSLSCHDGHQIVELW 70
EA VLGG + +C+Y++G +KRQA+++C TCTP+G AG+C ACS CH H++ EL+
Sbjct: 33 EACAVLGGSDSEKCSYSQGSVKRQALYACSTCTPEGEEPAGICLACSYECHGSHKLFELY 92
Query: 71 TKRNFRCDCGNSKFGEFYCKIFPSKDIENVENSYNHNFKGLYCTCARPYPDPDAEEQIEM 130
TKRNFRCDCGNSKF CK+ P K N N YN NF GLYC C RPYPDP+ E EM
Sbjct: 93 TKRNFRCDCGNSKFKNLECKLLPDKAKVNSGNKYNDNFFGLYCICKRPYPDPEDEIPDEM 152
Query: 131 IQCCLCEDWFHEEHLGLESSDEIPRDEEGEPLYEDFMCKACSEVCFFLKLYPEVILVAGK 190
IQC +CEDWFH HLG IP E G+ +++ +C+AC + C FL Y + V
Sbjct: 153 IQCVVCEDWFHGRHLG-----AIP-PESGD--FQEMVCQACMKRCSFLWAYAAQLAV--- 201
Query: 191 QPNATAQVSKDKGILEDTPSTCGFEKPLGDTSYNSPKIDVAQASVGSESISNRA------ 244
T ++D G++ + +GD P+ Q S E + +
Sbjct: 202 ----TKISTEDDGLVRNIDG-------IGDQEVIKPENGEHQDSTLKEDVPEQGKDDVRE 250
Query: 245 ------GLPPGGSCNSS---TCTEGASLHVNCLLGV---NIVAASPVIHGKAMFLSKNWR 292
P GS + S T + SL+ G + A + A + NWR
Sbjct: 251 VKVEQNSEPCAGSSSESDLQTVFKNESLNAESKSGCKLQELKAKQLIKKDTATYWPLNWR 310
Query: 293 DALCKCKNCLEYYHQKRIAFLLDKEDSIVEYEKMAKQKREEKLQQQEGAELSLFNKLGHV 352
LC C++C++ Y + FL D+ D+++ YE K + LS N+ V
Sbjct: 311 SKLCTCQDCMKMYGDLDVLFLTDEYDTVLAYENKGKIAQATDRSDPLMDTLSSMNR---V 367
Query: 353 EKVEILKGIEDMKDGLRTFLES-ADSSKPISAADIHQFFDDIKNKRRR 399
++VE++ D+K L+ +L+ AD + DI QFF++ ++K+RR
Sbjct: 368 QQVELICEYNDLKTELKDYLKRFADEGTVVKREDIQQFFEEFQSKKRR 415
>MLO2_SCHPO (Q09329) Protein mlo2
Length = 329
Score = 127 bits (318), Expect = 7e-29
Identities = 64/150 (42%), Positives = 88/150 (58%), Gaps = 8/150 (5%)
Query: 26 CTYNKGYMKRQAIFSCITCTPDGNA--GVCTACSLSCHDGHQIVELWTKRNFRCDCGNSK 83
CTY+ GY+K Q +++C+TC + VC +CS+SCH H +V+L+ KR+FRCDCG ++
Sbjct: 35 CTYSMGYLK-QPLYACLTCQKASGSLNAVCYSCSISCHADHDLVDLFNKRHFRCDCGTTR 93
Query: 84 FGEFYCKIFPSKDIENVENSYNHNFKGLYCTCARPYPDPDAEEQIEMIQCCLCEDWFHEE 143
C + S D EN YNHNF+G +C C Y +P+ EE M QC LCEDWFHE+
Sbjct: 94 THSIPCNLRKSVDECGSENDYNHNFEGRFCICDTVY-NPETEEG-TMFQCILCEDWFHEK 151
Query: 144 HLGLESSDEIPRDEEGEPLYEDFMCKACSE 173
L + D E +E +C CSE
Sbjct: 152 CLQKTNKGIAIPDAE---TFEWLVCSECSE 178
Score = 35.8 bits (81), Expect = 0.20
Identities = 28/119 (23%), Positives = 52/119 (43%), Gaps = 13/119 (10%)
Query: 285 MFLSKNWRDALCKCKNCLEYYHQKRIAFLL---------DKEDSIVEYEKMAKQKR---E 332
+FLS+N+R+ LC C++C+ + + + D ED I E + + E
Sbjct: 200 LFLSENFRENLCPCESCISLRNLEMPMLVAEEPIYEPPEDSEDGISEMNEDPSESGEMIE 259
Query: 333 EKLQQQEGAELSLFNKLGHVEKVEILKGIEDMKDGLRTFLES-ADSSKPISAADIHQFF 390
+ + L + ++L V+ E + +K L FL A ++ ++ DI FF
Sbjct: 260 QVISSTMNDVLRILDRLPRVQANESVYAYNRLKSELTDFLTPFARENRVVTKEDISNFF 318
>MTF2_HUMAN (Q9Y483) Metal-response element-binding transcription
factor 2 (Metal-response element DNA-binding protein
M96)
Length = 593
Score = 40.8 bits (94), Expect = 0.006
Identities = 22/76 (28%), Positives = 33/76 (42%), Gaps = 10/76 (13%)
Query: 107 NFKGLYCTCARPYPDPDAEEQIEMIQCCLCEDWFHEEHLGLESSDEIPRDEEGEPLYEDF 166
N + YC C P + ++M+QCC C+ WFHE + + D + F
Sbjct: 198 NVQQCYCYCGGP-----GDWYLKMLQCCKCKQWFHEACVQCLQKPMLFGDR-----FYTF 247
Query: 167 MCKACSEVCFFLKLYP 182
+C CS +LK P
Sbjct: 248 ICSVCSSGPEYLKRLP 263
>MTF2_MOUSE (Q02395) Metal-response element-binding transcription
factor 2 (Zinc-regulated factor 1) (ZiRF1)
(Metal-response element DNA-binding protein M96)
(Fragment)
Length = 373
Score = 40.0 bits (92), Expect = 0.011
Identities = 22/76 (28%), Positives = 32/76 (41%), Gaps = 10/76 (13%)
Query: 107 NFKGLYCTCARPYPDPDAEEQIEMIQCCLCEDWFHEEHLGLESSDEIPRDEEGEPLYEDF 166
N YC C P + ++M+QCC C+ WFHE + + D + F
Sbjct: 96 NVPECYCYCGGP-----GDWYLKMLQCCKCKQWFHEACVQCLQKPMLFGDR-----FYTF 145
Query: 167 MCKACSEVCFFLKLYP 182
+C CS +LK P
Sbjct: 146 ICSVCSSGPEYLKRLP 161
>UBR1_SCHPO (O60152) N-end-recognizing protein (Ubiquitin-protein
ligase E3 component) (N-recognin)
Length = 1958
Score = 39.3 bits (90), Expect = 0.018
Identities = 29/102 (28%), Positives = 45/102 (43%), Gaps = 8/102 (7%)
Query: 9 FNLQEADLVLGGDEGRE--CTYNKGYM--KRQAIFSCITCTPDGNAGVCTAC-SLSCHDG 63
F L+ A GDE R C G++ K + + C TC+ D N+ +C C + H
Sbjct: 73 FLLRRAQGHSEGDEYRHGTCESKCGHIFRKGEVFYRCKTCSVDSNSALCVKCFRATSHKD 132
Query: 64 HQI-VELWTKRNFRCDCGNSK--FGEFYCKIFPSKDIENVEN 102
H+ + CDCGN+ G+ CKI ++ + N
Sbjct: 133 HETSFTVSAGSGGCCDCGNAAAWIGDVSCKIHSHEEDATISN 174
>HYD_DROME (P51592) Ubiquitin--protein ligase hyd (EC 6.3.2.-)
(Hyperplastic discs protein)
Length = 2895
Score = 38.9 bits (89), Expect = 0.024
Identities = 18/44 (40%), Positives = 20/44 (44%)
Query: 36 QAIFSCITCTPDGNAGVCTACSLSCHDGHQIVELWTKRNFRCDC 79
Q IF C TC G+ CT C+ CH GH T CDC
Sbjct: 1224 QNIFECKTCGLTGSLCCCTECARVCHKGHDCKLKRTAPTAYCDC 1267
>GLP1_CAEEL (P13508) Glp-1 protein precursor
Length = 1295
Score = 38.9 bits (89), Expect = 0.024
Identities = 31/91 (34%), Positives = 41/91 (44%), Gaps = 6/91 (6%)
Query: 54 TACSLS---CHDGHQIVELWTKRNFRCDCGNSKFGEFYCKIFPSKDIEN--VENSYNHNF 108
T C+L C+ G I +NFRC C + GEF K IE V NS N
Sbjct: 192 TECALMGNICNHGRCIPNRDEDKNFRCVCDSGYEGEFCNKDKNECLIEETCVNNSTCFNL 251
Query: 109 KGLY-CTCARPYPDPDAEEQIEMIQCCLCED 138
G + CTC Y EE I+M + +C++
Sbjct: 252 HGDFTCTCKPGYAGKYCEEAIDMCKDYVCQN 282
>FX11_RAT (Q7TSL3) F-box only protein 11
Length = 843
Score = 37.7 bits (86), Expect = 0.053
Identities = 16/42 (38%), Positives = 18/42 (42%), Gaps = 1/42 (2%)
Query: 39 FSCITCTPDGNAGVCTACSLSCHDGHQIVELWTKRNFRCDCG 80
+ C TC +C C CH GH VE F CDCG
Sbjct: 767 YRCHTCNTTDRNAICVNCIKKCHQGHD-VEFIRHDRFFCDCG 807
>FX11_MOUSE (Q7TPD1) F-box only protein 11 (Fragment)
Length = 521
Score = 37.7 bits (86), Expect = 0.053
Identities = 16/42 (38%), Positives = 18/42 (42%), Gaps = 1/42 (2%)
Query: 39 FSCITCTPDGNAGVCTACSLSCHDGHQIVELWTKRNFRCDCG 80
+ C TC +C C CH GH VE F CDCG
Sbjct: 445 YRCHTCNTTDRNAICVNCIKKCHQGHD-VEFIRHDRFFCDCG 485
>FX11_HUMAN (Q86XK2) F-box only protein 11 (Vitiligo-associated
protein VIT-1)
Length = 843
Score = 37.7 bits (86), Expect = 0.053
Identities = 16/42 (38%), Positives = 18/42 (42%), Gaps = 1/42 (2%)
Query: 39 FSCITCTPDGNAGVCTACSLSCHDGHQIVELWTKRNFRCDCG 80
+ C TC +C C CH GH VE F CDCG
Sbjct: 767 YRCHTCNTTDRNAICVNCIKKCHQGHD-VEFIRHDRFFCDCG 807
>PHF2_MOUSE (Q9WTU0) PHD finger protein 2 (GRC5)
Length = 1096
Score = 37.4 bits (85), Expect = 0.069
Identities = 20/63 (31%), Positives = 29/63 (45%), Gaps = 13/63 (20%)
Query: 111 LYCTCARPYPDPDAEEQIEMIQCCLCEDWFHEEHLGLESSDEIPRDEEGEPLYEDFMCKA 170
+YC C PY + MI+C C+DWFH +G+E EE P + + C
Sbjct: 6 VYCVCRLPY-----DVTRFMIECDACKDWFHGSCVGVE--------EEEAPDIDIYHCPN 52
Query: 171 CSE 173
C +
Sbjct: 53 CEK 55
>PHF2_HUMAN (O75151) PHD finger protein 2 (GRC5)
Length = 1101
Score = 37.4 bits (85), Expect = 0.069
Identities = 20/63 (31%), Positives = 29/63 (45%), Gaps = 13/63 (20%)
Query: 111 LYCTCARPYPDPDAEEQIEMIQCCLCEDWFHEEHLGLESSDEIPRDEEGEPLYEDFMCKA 170
+YC C PY + MI+C C+DWFH +G+E EE P + + C
Sbjct: 6 VYCVCRLPY-----DVTRFMIECDACKDWFHGSCVGVE--------EEEAPDIDIYHCPN 52
Query: 171 CSE 173
C +
Sbjct: 53 CEK 55
>MLL4_HUMAN (Q9UMN6) Myeloid/lymphoid or mixed-lineage leukemia
protein 4 (Trithorax homolog 2)
Length = 2715
Score = 37.4 bits (85), Expect = 0.069
Identities = 19/44 (43%), Positives = 23/44 (52%), Gaps = 3/44 (6%)
Query: 109 KGLYCT-CARPYPDPDAEEQIEMIQCCLCEDWFHEEHLGLESSD 151
KG YC C R Y D D E + M+QC C+ W H + GL D
Sbjct: 1334 KGNYCPICTRCYEDNDYESK--MMQCAQCDHWVHAKCEGLSDED 1375
>DAT1_MOUSE (Q8C9B9) Death associated transcription factor 1 (Death
inducer-obliterator-1) (DIO-1)
Length = 614
Score = 37.4 bits (85), Expect = 0.069
Identities = 25/93 (26%), Positives = 40/93 (42%), Gaps = 22/93 (23%)
Query: 111 LYCTCARPYPDPDAEEQIEMIQCCLCEDWFHEEHLGLESSDEIPRDEEGEPLYEDFMCKA 170
LYC C +P+ + MI C CE+WFH + +G+ + + G ED++C
Sbjct: 266 LYCICRQPHNNRF------MICCDRCEEWFHGDCVGISEARGRLLERNG----EDYICPN 315
Query: 171 CSEVCFFLKLYPEVILVAGKQPNATAQVSKDKG 203
C+ IL + N +A +D G
Sbjct: 316 CT------------ILQVQDETNGSATDEQDSG 336
>DAT1_HUMAN (Q9BTC0) Death associated transcription factor 1
Length = 1225
Score = 37.0 bits (84), Expect = 0.090
Identities = 19/62 (30%), Positives = 31/62 (49%), Gaps = 10/62 (16%)
Query: 111 LYCTCARPYPDPDAEEQIEMIQCCLCEDWFHEEHLGLESSDEIPRDEEGEPLYEDFMCKA 170
LYC C +P+ + MI C CE+WFH + +G+ + + G ED++C
Sbjct: 269 LYCICRQPHNNRF------MICCDRCEEWFHGDCVGISEARGRLLERNG----EDYICPN 318
Query: 171 CS 172
C+
Sbjct: 319 CT 320
>TRX_DROVI (Q24742) Trithorax protein
Length = 3828
Score = 36.6 bits (83), Expect = 0.12
Identities = 23/68 (33%), Positives = 34/68 (49%), Gaps = 5/68 (7%)
Query: 109 KGLYCT-CARPYPDPDAEEQIEMIQCCLCEDWFHEEHLGLESSDEIPRDEEGEPLYEDFM 167
KG +C C + Y D D + ++M++C C W H + GL SDE P +F+
Sbjct: 1407 KGNFCPICQKCYDDNDFD--LKMMECGDCNQWVHSKCEGL--SDEQYNLLSTLPESIEFI 1462
Query: 168 CKACSEVC 175
CK C+ C
Sbjct: 1463 CKKCARRC 1470
>PHF8_HUMAN (Q9UPP1) PHD finger protein 8
Length = 1024
Score = 36.2 bits (82), Expect = 0.15
Identities = 15/38 (39%), Positives = 22/38 (57%), Gaps = 5/38 (13%)
Query: 111 LYCTCARPYPDPDAEEQIEMIQCCLCEDWFHEEHLGLE 148
+YC C PY + MI+C +C+DWFH +G+E
Sbjct: 6 VYCLCRLPY-----DVTRFMIECDMCQDWFHGSCVGVE 38
>YM42_YEAST (Q03214) Hypothetical 162.7 kDa protein in SIP18-SPT21
intergenic region
Length = 1411
Score = 35.8 bits (81), Expect = 0.20
Identities = 25/99 (25%), Positives = 45/99 (45%), Gaps = 19/99 (19%)
Query: 82 SKFGEFYCKIFPSKDIEN-----VENSYNHNFKGLYCTCARPYPDPDAEEQIEMIQCCLC 136
+KF E IF D+++ +E+ K YC C R EE M++C +C
Sbjct: 1207 NKFIEILPSIFRCLDLKSDKYIPLESCSKRQTK--YCFCRR------VEEGTAMVECEIC 1258
Query: 137 EDWFHEEHLGLESSDEIPRDEEGEPLYEDFMCKACSEVC 175
++W+H + + + + +P D+ F+C C+ C
Sbjct: 1259 KEWYHVD--CISNGELVPPDDPNVL----FVCSICTPPC 1291
>PCL_DROME (Q24459) Polycomb protein Pcl (Polycomblike protein)
Length = 1043
Score = 35.4 bits (80), Expect = 0.26
Identities = 13/43 (30%), Positives = 22/43 (50%), Gaps = 5/43 (11%)
Query: 101 ENSYNHNFKGLYCTCARPYPDPDAEEQIEMIQCCLCEDWFHEE 143
+ + N + +YC C +P + M+QCC C +WFH +
Sbjct: 503 DEKHRVNEEQIYCYCGKP-----GKFDHNMLQCCKCRNWFHTQ 540
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.137 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 51,541,901
Number of Sequences: 164201
Number of extensions: 2338584
Number of successful extensions: 6453
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 43
Number of HSP's that attempted gapping in prelim test: 6403
Number of HSP's gapped (non-prelim): 77
length of query: 401
length of database: 59,974,054
effective HSP length: 112
effective length of query: 289
effective length of database: 41,583,542
effective search space: 12017643638
effective search space used: 12017643638
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC142526.10


