

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142498.7 + phase: 0
(1705 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
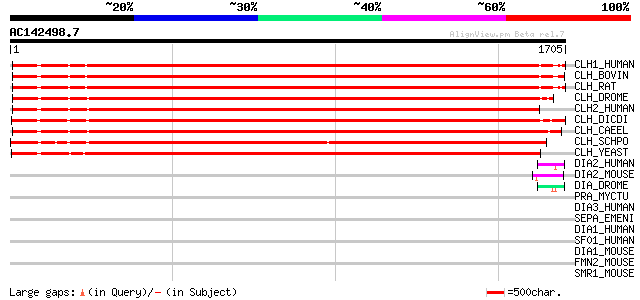
Score E
Sequences producing significant alignments: (bits) Value
CLH1_HUMAN (Q00610) Clathrin heavy chain 1 (CLH-17) 1891 0.0
CLH_BOVIN (P49951) Clathrin heavy chain 1889 0.0
CLH_RAT (P11442) Clathrin heavy chain 1887 0.0
CLH_DROME (P29742) Clathrin heavy chain 1881 0.0
CLH2_HUMAN (P53675) Clathrin heavy chain 2 (CLH-22) 1825 0.0
CLH_DICDI (P25870) Clathrin heavy chain 1809 0.0
CLH_CAEEL (P34574) Probable clathrin heavy chain 1801 0.0
CLH_SCHPO (Q10161) Probable clathrin heavy chain 1618 0.0
CLH_YEAST (P22137) Clathrin heavy chain 1453 0.0
DIA2_HUMAN (O60879) Diaphanous protein homolog 2 (Diaphanous-rel... 49 9e-05
DIA2_MOUSE (O70566) Diaphanous protein homolog 2 (Diaphanous-rel... 49 2e-04
DIA_DROME (P48608) Diaphanous protein 46 8e-04
PRA_MYCTU (O53426) Proline-rich antigen homolog 46 0.001
DIA3_HUMAN (Q9NSV4) Diaphanous protein homolog 3 (Diaphanous-rel... 45 0.002
SEPA_EMENI (P78621) Cytokinesis protein sepA (FH1/2 protein) (Fo... 44 0.003
DIA1_HUMAN (O60610) Diaphanous protein homolog 1 (Diaphanous-rel... 44 0.004
SF01_HUMAN (Q15637) Splicing factor 1 (Zinc finger protein 162) ... 43 0.007
DIA1_MOUSE (O08808) Diaphanous protein homolog 1 (Diaphanous-rel... 43 0.007
FMN2_MOUSE (Q9JL04) Formin 2 42 0.011
SMR1_MOUSE (Q61900) Submaxillary gland androgen regulated protei... 41 0.025
>CLH1_HUMAN (Q00610) Clathrin heavy chain 1 (CLH-17)
Length = 1674
Score = 1891 bits (4899), Expect = 0.0
Identities = 949/1700 (55%), Positives = 1255/1700 (73%), Gaps = 36/1700 (2%)
Query: 8 PILMREALTLPSIGINPQHITFTHVTMESDKYICVRE-TAPQNSVVIVDMNMPNQPLRRP 66
PI +E L L ++GINP +I F+ +TMESDK+IC+RE Q VVI+DMN P+ P+RRP
Sbjct: 5 PIRFQEHLQLQNLGINPANIGFSTLTMESDKFICIREKVGEQAQVVIIDMNDPSNPIRRP 64
Query: 67 ITADSALMNPNSRILALKAQLQGTTQDHLQIFNIELKAKMKSYQMPEQVVFWKWISPKLL 126
I+ADSA+MNP S+++ALKA G T LQIFNIE+K+KMK++ M + V FWKWIS +
Sbjct: 65 ISADSAIMNPASKVIALKA---GKT---LQIFNIEMKSKMKAHTMTDDVTFWKWISLNTV 118
Query: 127 GLVTQTSVYHWSIEGDSEPVKMFERTANLANNQIINYRCDPTEKWLVLIGIAPGSPERPQ 186
LVT +VYHWS+EG+S+PVKMF+R ++LA QIINYR D +KWL+L GI+ +
Sbjct: 119 ALVTDNAVYHWSMEGESQPVKMFDRHSSLAGCQIINYRTDAKQKWLLLTGISA----QQN 174
Query: 187 LVKGNMQLFSVEQQRSQALEAHAASFAQFKVPGNENPSTLISFATKTVNAGQVISKLHVI 246
V G MQL+SV+++ SQ +E HAASFAQFK+ GN STL FA + GQ KLH+I
Sbjct: 175 RVVGAMQLYSVDRKVSQPIEGHAASFAQFKMEGNAEESTLFCFAVR----GQAGGKLHII 230
Query: 247 ELGAQP-GKPSFTKKQADLFFPPDFADDFPVSMQISHKYSLIYVITKLGLLFVYDLETAT 305
E+G P G F KK D+FFPP+ +DFPV+MQIS K+ ++++ITK G + +YDLET T
Sbjct: 231 EVGTPPTGNQPFPKKAVDVFFPPEAQNDFPVAMQISEKHDVVFLITKYGYIHLYDLETGT 290
Query: 306 AVYRNRISPDPIFLTSEATSVGGFYAINRRGQVLLATVNEQTIVNFVSGQLNNLELAVNL 365
+Y NRIS + IF+T+ + G +NR+GQVL V E+ I+ +++ L N +LA+ +
Sbjct: 291 CIYMNRISGETIFVTAPHEATAGIIGVNRKGQVLSVCVEEENIIPYITNVLQNPDLALRM 350
Query: 366 AKRGNLPGAEKLVVERFHELFAQTKYKEAAELAAESPQGILRTPDTVAKFQSVPVQAGQT 425
A R NL GAE+L +F+ LFAQ Y EAA++AA +P+GILRTPDT+ +FQSVP Q GQT
Sbjct: 351 AVRNNLAGAEELFARKFNALFAQGNYSEAAKVAANAPKGILRTPDTIRRFQSVPAQPGQT 410
Query: 426 PPLLQYFGTLLTRGKLNAFESLELSRLVVNQNKKNLLENWLAEDKLECSEELGDLVKTVD 485
PLLQYFG LL +G+LN +ESLEL R V+ Q +K LLE WL EDKLECSEELGDLVK+VD
Sbjct: 411 SPLLQYFGILLDQGQLNKYESLELCRPVLQQGRKQLLEKWLKEDKLECSEELGDLVKSVD 470
Query: 486 NDLALKIYIKARATPKVVAAFAEKREFDKILIYSKQVGYTPDYLFLLQSILRTDPQGAVN 545
LAL +Y++A KV+ FAE + KI++Y+K+VGYTPD++FLL++++R P
Sbjct: 471 PTLALSVYLRANVPNKVIQCFAETGQVQKIVLYAKKVGYTPDWIFLLRNVMRISPDQGQQ 530
Query: 546 FALMMSQMEGGCPVDYNTITDLFLQRNLIREATAFLLDVLKPNLPEHGFLQTKVLEINLV 605
FA M+ Q E D I D+F++ NLI++ TAFLLD LK N P G LQT++LE+NL+
Sbjct: 531 FAQMLVQDEEPL-ADITQIVDVFMEYNLIQQCTAFLLDALKNNRPSEGPLQTRLLEMNLM 589
Query: 606 TFPNVADAILANGMFSHYDRPRIAQLCEKAGLYVRALQHYTELPDIKRVIVNTHAIEPQA 665
P VADAIL N MF+HYDR IAQLCEKAGL RAL+H+T+L DIKR +V+TH + P+
Sbjct: 590 HAPQVADAILGNQMFTHYDRAHIAQLCEKAGLLQRALEHFTDLYDIKRAVVHTHLLNPEW 649
Query: 666 LVEFFGTLSKEWALECMKDLLLVNLRGNLQIIVQVAKEYCEQLGVDACIKIFEQFRSYEG 725
LV +FG+LS E +LEC++ +L N+R NLQI VQVA +Y EQL + I++FE F+S+EG
Sbjct: 650 LVNYFGSLSVEDSLECLRAMLSANIRQNLQICVQVASKYHEQLSTQSLIELFESFKSFEG 709
Query: 726 LYFFLGSYLSSSEDPDIHFKYIEAAAKTGQIKEVERVTRESSFYDPEKTKNFLMEAKLPD 785
L++FLGS ++ S+DPD+HFKYI+AA KTGQIKEVER+ RES+ YDPE+ KNFL EAKL D
Sbjct: 710 LFYFLGSIVNFSQDPDVHFKYIQAACKTGQIKEVERICRESNCYDPERVKNFLKEAKLTD 769
Query: 786 ARPLINVCDRFGFVPDLTHYLYTSNMLRYIEGYVQKVNPGNAPLVVGQLLDDECPEDFIK 845
PLI VCDRF FV DL YLY +N+ +YIE YVQKVNP P+V+G LLD +C ED IK
Sbjct: 770 QLPLIIVCDRFDFVHDLVLYLYRNNLQKYIEIYVQKVNPSRLPVVIGGLLDVDCSEDVIK 829
Query: 846 GLILSVRSLLPVEPLVAECEKRNRLRLLTQFLEHLVSEGSQDVHVHNALGKIIIDSNNNP 905
LIL VR + LVAE EKRNRL+LL +LE + EG ++ HNAL KI IDSNNNP
Sbjct: 830 NLILVVRGQFSTDELVAEVEKRNRLKLLLPWLEARIHEGCEEPATHNALAKIYIDSNNNP 889
Query: 906 EHFLTTNPYYDSRVVGKYCEKRDPTLAVVAYRRGVCDDELINVTNKNSLFKLQARYVVER 965
E FL NPYYDSRVVGKYCEKRDP LA VAY RG CD ELINV N+NSLFK +RY+V R
Sbjct: 890 ERFLRENPYYDSRVVGKYCEKRDPHLACVAYERGQCDLELINVCNENSLFKSLSRYLVRR 949
Query: 966 MDADLWEKVLNPDNTYRRQLIDQVVSTALPESKSPEQVSASVKAFMTADLPHELIELLEK 1025
D +LW VL N YRR LIDQVV TAL E++ PE+VS +VKAFMTADLP+ELIELLEK
Sbjct: 950 KDPELWGSVLLESNPYRRPLIDQVVQTALSETQDPEEVSVTVKAFMTADLPNELIELLEK 1009
Query: 1026 IVLQNSAFSGNFNLQNLLILTAIKADSSRVMDYVNRLDNFDGPQVGEVAVEAELYEEAFA 1085
IVL NS FS + NLQNLLILTAIKAD +RVM+Y+NRLDN+D P + +A+ EL+EEAFA
Sbjct: 1010 IVLDNSVFSEHRNLQNLLILTAIKADRTRVMEYINRLDNYDAPDIANIAISNELFEEAFA 1069
Query: 1086 IFKKFNLNVQAVNVLLDNIHSIDRAVEFAFRVEEDAVWSQVAKAQLREGLVSDAIESFIR 1145
IF+KF++N AV VL+++I ++DRA EFA R E AVWSQ+AKAQL++G+V +AI+S+I+
Sbjct: 1070 IFRKFDVNTSAVQVLIEHIGNLDRAYEFAERCNEPAVWSQLAKAQLQKGMVKEAIDSYIK 1129
Query: 1146 ADDATQFLDVIRAAQDGNAYQDLVRYLLMVRQKTKEPKVDSELIYAYAKIDRLSDIEEFI 1205
ADD + +++V++AA +++LV+YL M R+K +E V++ELI+A AK +RL+++EEFI
Sbjct: 1130 ADDPSSYMEVVQAANTSGNWEELVKYLQMARKKARESYVETELIFALAKTNRLAELEEFI 1189
Query: 1206 LMPNVANLQNVGDRLYDEELYEAAKIIFAFISNWAKLAVTLVKLKQFQGAVDAARKANSA 1265
PN A++Q VGDR YDE++Y+AAK+++ +SN+ +LA TLV L ++Q AVD ARKANS
Sbjct: 1190 NGPNNAHIQQVGDRCYDEKMYDAAKLLYNNVSNFGRLASTLVHLGEYQAAVDGARKANST 1249
Query: 1266 KTWKEVCFACVDEEEFRLAQICGLNIIIQVDDLEEVSEYYQNRGCFNELISLMESGLGLE 1325
+TWKEVCFACVD +EFRLAQ+CGL+I++ D+LEE+ YYQ+RG F ELI+++E+ LGLE
Sbjct: 1250 RTWKEVCFACVDGKEFRLAQMCGLHIVVHADELEELINYYQDRGYFEELITMLEAALGLE 1309
Query: 1326 RAHMGIFTELGVLYARYRPEKLMEHIKLFATRLNIPKLIRACDEQQHWKELTYLYIQYDE 1385
RAHMG+FTEL +LY++++P+K+ EH++LF +R+NIPK++RA ++ W EL +LY +Y+E
Sbjct: 1310 RAHMGMFTELAILYSKFKPQKMREHLELFWSRVNIPKVLRAAEQAHLWAELVFLYDKYEE 1369
Query: 1386 FDNAATTIMNHSPEAWDHMQFKDVIAKVANVELYYKAVHFYLQEHPDLINDVLNVLALRV 1445
+DNA T+MNH +AW QFKD+I KVANVELYY+A+ FYL+ P L+ND+L VL+ R+
Sbjct: 1370 YDNAIITMMNHPTDAWKEGQFKDIITKVANVELYYRAIQFYLEFKPLLLNDLLMVLSPRL 1429
Query: 1446 DHARVVDIMRKAGHLRLVKPYMVAVQSNNVSAVNEALNEIYVEEEDYDRLRESIDLHDNF 1505
DH R V+ K L LVKPY+ +VQ++N +VNE+LN +++ EEDY LR SID +DNF
Sbjct: 1430 DHTRAVNYFSKVKQLPLVKPYLRSVQNHNNKSVNESLNNLFITEEDYQALRTSIDAYDNF 1489
Query: 1506 DQIGLAQKIEKHELLEMRRVAAYIYKKAGRWKQSIALSKKDNLYKDAMETASQSGERELA 1565
D I LAQ++EKHEL+E RR+AAY++K RWKQS+ L KKD+LYKDAM+ AS+S + ELA
Sbjct: 1490 DNISLAQRLEKHELIEFRRIAAYLFKGNNRWKQSVELCKKDSLYKDAMQYASESKDTELA 1549
Query: 1566 EELLVYFIDQGKKECFASCLFVCYDLIRVDVALELAWMHNMIDFAFPYLLQFIREYTGKV 1625
EELL +F+ + K+ECF +CLF CYDL+R DV LE AW HN++DFA PY +Q ++EY KV
Sbjct: 1550 EELLQWFLQEEKRECFGACLFTCYDLLRPDVVLETAWRHNIMDFAMPYFIQVMKEYLTKV 1609
Query: 1626 DELVKHKIESQNEVKAKEQEEKEVIAQQNMYAQLLPLALPAPPMPGMGGGMGGGYAPPPP 1685
D K+++ ++ +E++ E QL+ A P+ +P P P
Sbjct: 1610 D-----KLDASESLRKEEEQATETQPIVYGQPQLMLTAGPSVAVP-----------PQAP 1653
Query: 1686 MGGGMGMPPMPPFGMPMGGY 1705
G G PP+G P G+
Sbjct: 1654 FGYGY---TAPPYGQPQPGF 1670
>CLH_BOVIN (P49951) Clathrin heavy chain
Length = 1675
Score = 1889 bits (4894), Expect = 0.0
Identities = 950/1701 (55%), Positives = 1254/1701 (72%), Gaps = 37/1701 (2%)
Query: 8 PILMREALTLPSIGINPQHITFTHVTMESDKYICVRE-TAPQNSVVIVDMNMPNQPLRRP 66
PI +E L L ++GINP +I F+ +TMESDK+IC+RE Q VVI+DMN P+ P+RRP
Sbjct: 6 PIRFQEHLQLQNLGINPANIGFSTLTMESDKFICIREKVGEQAQVVIIDMNDPSNPIRRP 65
Query: 67 ITADSALMNPNSRILALKAQLQGTTQDHLQIFNIELKAKMKSYQMPEQVVFWKWISPKLL 126
I+ADSA+MNP S+++ALKA G T LQIFNIE+K+KMK++ M + V FWKWIS +
Sbjct: 66 ISADSAIMNPASKVIALKA---GKT---LQIFNIEMKSKMKAHTMTDDVTFWKWISLNTV 119
Query: 127 GLVTQTSVYHWSIEGDSEPVKMFERTANLANNQIINYRCDPTEKWLVLIGIAPGSPERPQ 186
LVT +VYHWS+EG+S+PVKMF+R ++LA QIINYR D +KWL+L GI+ +
Sbjct: 120 ALVTDNAVYHWSMEGESQPVKMFDRHSSLAGCQIINYRTDAKQKWLLLTGISA----QQN 175
Query: 187 LVKGNMQLFSVEQQRSQALEAHAASFAQFKVPGNENPSTLISFATKTVNAGQVISKLHVI 246
V G MQL+SV+++ SQ +E HAASFAQFK+ GN STL FA + GQ KLH+I
Sbjct: 176 RVVGAMQLYSVDRKVSQPIEGHAASFAQFKMEGNAEESTLFCFAVR----GQAGGKLHII 231
Query: 247 ELGAQP-GKPSFTKKQADLFFPPDFADDFPVSMQISHKYSLIYVITKLGLLFVYDLETAT 305
E+G P G F KK D+FFPP+ +DFPV+MQIS K+ ++++ITK G + +YDLET T
Sbjct: 232 EVGTPPTGNQPFPKKAVDVFFPPEAQNDFPVAMQISEKHDVVFLITKYGYIHLYDLETGT 291
Query: 306 AVYRNRISPDPIFLTSEATSVGGFYAINRRGQVLLATVNEQTIVNFVSGQLNNLELAVNL 365
+Y NRIS + IF+T+ + G +NR+GQVL V E+ I+ +++ L N +LA+ +
Sbjct: 292 CIYMNRISGETIFVTAPHEATAGIIGVNRKGQVLSVCVEEENIIPYITNVLQNPDLALRM 351
Query: 366 AKRGNLPGAEKLVVERFHELFAQTKYKEAAELAAESPQGILRTPDTVAKFQSVPVQAGQT 425
A R NL GAE+L +F+ LFAQ Y EAA++AA +P+GILRTPDT+ +FQSVP Q GQT
Sbjct: 352 AVRNNLAGAEELFARKFNALFAQGNYSEAAKVAANAPKGILRTPDTIRRFQSVPAQPGQT 411
Query: 426 PPLLQYFGTLLTRGKLNAFESLELSRLVVNQNKKNLLENWLAEDKLECSEELGDLVKTVD 485
PLLQYFG LL +G+LN +ESLEL R V+ Q +K LLE WL EDKLECSEELGDLVK+VD
Sbjct: 412 SPLLQYFGILLDQGQLNKYESLELCRPVLQQGRKQLLEKWLKEDKLECSEELGDLVKSVD 471
Query: 486 NDLALKIYIKARATPKVVAAFAEKREFDKILIYSKQVGYTPDYLFLLQSILRTDPQGAVN 545
LAL +Y++A KV+ FAE + KI++Y+K+VGYTPD++FLL++++R P
Sbjct: 472 PTLALSVYLRANVPNKVIQCFAETGQVQKIVLYAKKVGYTPDWIFLLRNVMRISPDQGQQ 531
Query: 546 FALMMSQMEGGCPVDYNTITDLFLQRNLIREATAFLLDVLKPNLPEHGFLQTKVLEINLV 605
FA M+ Q E D I D+F++ NLI++ TAFLLD LK N P G LQT++LE+NL+
Sbjct: 532 FAQMLVQDEEPL-ADITQIVDVFMEYNLIQQCTAFLLDALKNNRPSEGPLQTRLLEMNLM 590
Query: 606 TFPNVADAILANGMFSHYDRPRIAQLCEKAGLYVRALQHYTELPDIKRVIVNTHAIEPQA 665
P VADAIL N MF+HYDR IAQLCEKAGL RAL+H+T+L DIKR +V+TH + P+
Sbjct: 591 HAPQVADAILGNQMFTHYDRAHIAQLCEKAGLLQRALEHFTDLYDIKRAVVHTHLLNPEW 650
Query: 666 LVEFFGTLSKEWALECMKDLLLVNLRGNLQIIVQVAKEYCEQLGVDACIKIFEQFRSYEG 725
LV +FG+LS E +LEC++ +L N+R NLQI VQVA +Y EQL + I++FE F+S+EG
Sbjct: 651 LVNYFGSLSVEDSLECLRAMLSANIRQNLQICVQVASKYHEQLSTQSLIELFESFKSFEG 710
Query: 726 LYFFLGSYLSSSEDPDIHFKYIEAAAKTGQIKEVERVTRESSFYDPEKTKNFLMEAKLPD 785
L++FLGS ++ S+DPD+HFKYI+AA KTGQIKEVER+ RES+ YDPE+ KNFL EAKL D
Sbjct: 711 LFYFLGSIVNFSQDPDVHFKYIQAACKTGQIKEVERICRESNCYDPERVKNFLKEAKLTD 770
Query: 786 ARPLINVCDRFGFVPDLTHYLYTSNMLRYIEGYVQKVNPGNAPLVVGQLLDDECPEDFIK 845
PLI VCDRF FV DL YLY +N+ +YIE YVQKVNP P+V+G LLD +C ED IK
Sbjct: 771 QLPLIIVCDRFDFVHDLVLYLYRNNLQKYIEIYVQKVNPSRLPVVIGGLLDVDCSEDVIK 830
Query: 846 GLILSVRSLLPVEPLVAECEKRNRLRLLTQFLEHLVSEGSQDVHVHNALGKIIIDSNNNP 905
LIL VR + LVAE EKRNRL+LL +LE + EG ++ HNAL KI IDSNNNP
Sbjct: 831 NLILVVRGQFSTDELVAEVEKRNRLKLLLPWLEARIHEGCEEPATHNALAKIYIDSNNNP 890
Query: 906 EHFLTTNPYYDSRVVGKYCEKRDPTLAVVAYRRGVCDDELINVTNKNSLFKLQARYVVER 965
E FL NPYYDSRVVGKYCEKRDP LA VAY RG CD ELINV N+NSLFK +RY+V R
Sbjct: 891 ERFLRENPYYDSRVVGKYCEKRDPHLACVAYERGQCDLELINVCNENSLFKSLSRYLVRR 950
Query: 966 MDADLWEKVLNPDNTYRRQLIDQVVSTALPESKSPEQVSASVKAFMTADLPHELIELLEK 1025
D +LW VL N YRR LIDQVV TAL E++ PE+VS +VKAFMTADLP+ELIELLEK
Sbjct: 951 KDPELWGSVLLESNPYRRPLIDQVVQTALSETQDPEEVSVTVKAFMTADLPNELIELLEK 1010
Query: 1026 IVLQNSAFSGNFNLQNLLILTAIKADSSRVMDYVNRLDNFDGPQVGEVAVEAELYEEAFA 1085
IVL NS FS + NLQNLLILTAIKAD +RVM+Y+NRLDN+D P + +A+ EL+EEAFA
Sbjct: 1011 IVLDNSVFSEHRNLQNLLILTAIKADRTRVMEYINRLDNYDAPDIANIAISNELFEEAFA 1070
Query: 1086 IFKKFNLNVQAVNVLLDNIHSIDRAVEFAFRVEEDAVWSQVAKAQLREGLVSDAIESFIR 1145
IF+KF++N AV VL+++I ++DRA EFA R E AVWSQ+AKAQL++G+V +AI+S+I+
Sbjct: 1071 IFRKFDVNTSAVQVLIEHIGNLDRAYEFAERCNEPAVWSQLAKAQLQKGMVKEAIDSYIK 1130
Query: 1146 ADDATQFLDVIRAAQDGNAYQDLVRYLLMVRQKTKEPKVDSELIYAYAKIDRLSDIEEFI 1205
ADD + +++V++AA +++LV+YL M R+K +E V++ELI+A AK +RL+++EEFI
Sbjct: 1131 ADDPSSYMEVVQAANTSGNWEELVKYLQMARKKARESYVETELIFALAKTNRLAELEEFI 1190
Query: 1206 LMPNVANLQNVGDRLYDEELYEAAKIIFAFISNWAKLAVTLVKLKQFQGAVDAARKANSA 1265
PN A++Q VGDR YDE++Y+AAK+++ +SN+ +LA TLV L ++Q AVD ARKANS
Sbjct: 1191 NGPNNAHIQQVGDRCYDEKMYDAAKLLYNNVSNFGRLASTLVHLGEYQAAVDGARKANST 1250
Query: 1266 KTWKEVCFACVDEEEFRLAQICGLNIIIQVDDLEEVSEYYQNRGCFNELISLMESGLGLE 1325
+TWKEVCFACVD +EFRLAQ+CGL+I++ D+LEE+ YYQ+RG F ELI+++E+ LGLE
Sbjct: 1251 RTWKEVCFACVDGKEFRLAQMCGLHIVVHADELEELINYYQDRGYFEELITMLEAALGLE 1310
Query: 1326 RAHMGIFTELGVLYARYRPEKLMEHIKLFATRLNIPKLIRACDEQQHWKELTYLYIQYDE 1385
RAHMG+FTEL +LY++++P+K+ EH++LF +R+NIPK++RA ++ W EL +LY +Y+E
Sbjct: 1311 RAHMGMFTELAILYSKFKPQKMREHLELFWSRVNIPKVLRAAEQAHLWAELVFLYDKYEE 1370
Query: 1386 FDNAATTIMNHSPEAWDHMQFKDVIAKVANVELYYKAVHFYLQEHPDLINDVLNVLALRV 1445
+DNA T+MNH +AW QFKD+I KVANVELYY+A+ FYL+ P L+ND+L VL+ R+
Sbjct: 1371 YDNAIITMMNHPTDAWKEGQFKDIITKVANVELYYRAIQFYLEFKPLLLNDLLMVLSPRL 1430
Query: 1446 DHARVVDIMRKAGHLRLVKPYMVAVQSNNVSAVNEALNEIYVEEEDYDRLRESIDLHDNF 1505
DH R V+ K L LVKPY+ +VQ++N +VNE+LN +++ EEDY LR SID +DNF
Sbjct: 1431 DHTRAVNYFSKVKQLPLVKPYLRSVQNHNNKSVNESLNNLFITEEDYQALRTSIDAYDNF 1490
Query: 1506 DQIGLAQKIEKHELLEMRRVAAYIYKKAGRWKQSIALSKKDNLYKDAMETASQSGERELA 1565
D I LAQ++EKHEL+E RR+AAY++K RWKQS+ L KKD+LYKDAM+ AS+S + ELA
Sbjct: 1491 DNISLAQRLEKHELIEFRRIAAYLFKGNNRWKQSVELCKKDSLYKDAMQYASESKDTELA 1550
Query: 1566 EELLVYFIDQGKKECFASCLFVCYDLIRVDVALELAWMHNMIDFAFPYLLQFIREYTGKV 1625
EELL +F+ + K+ECF +CLF CYDL+R DV LE AW HN++DFA PY +Q ++EY KV
Sbjct: 1551 EELLQWFLQEEKRECFGACLFTCYDLLRPDVVLETAWRHNIMDFAMPYFIQVMKEYLTKV 1610
Query: 1626 DELVKHKIESQNEVKAKEQEEKEVIAQQNMYAQLLPLALPAPPMPGMGGGMGGGYAPPPP 1685
D K+++ ++ +E++ E QL+ A P+ +P P P
Sbjct: 1611 D-----KLDASESLRKEEEQATETQPIVYGQPQLMLTAGPSVAVP-----------PQAP 1654
Query: 1686 MGGGMGMP----PMPPFGMPM 1702
G G P P P FG M
Sbjct: 1655 FGYGYTAPAYGQPQPGFGYSM 1675
>CLH_RAT (P11442) Clathrin heavy chain
Length = 1675
Score = 1887 bits (4887), Expect = 0.0
Identities = 948/1700 (55%), Positives = 1254/1700 (73%), Gaps = 36/1700 (2%)
Query: 8 PILMREALTLPSIGINPQHITFTHVTMESDKYICVRE-TAPQNSVVIVDMNMPNQPLRRP 66
PI +E L L ++GINP +I F+ +TMESDK+IC+RE Q VVI+DMN P+ P+RRP
Sbjct: 6 PIRFQEHLQLQNLGINPANIGFSTLTMESDKFICIREKVGEQAQVVIIDMNDPSNPIRRP 65
Query: 67 ITADSALMNPNSRILALKAQLQGTTQDHLQIFNIELKAKMKSYQMPEQVVFWKWISPKLL 126
I+ADSA+MNP S+++ALKA G T LQIFNIE+K+KMK++ M + V FWKWIS +
Sbjct: 66 ISADSAIMNPASKVIALKA---GKT---LQIFNIEMKSKMKAHTMTDDVTFWKWISLNTV 119
Query: 127 GLVTQTSVYHWSIEGDSEPVKMFERTANLANNQIINYRCDPTEKWLVLIGIAPGSPERPQ 186
LVT +VYHWS+EG+S+PVKMF+R ++LA QIINYR D +KWL+L GI+ +
Sbjct: 120 ALVTDNAVYHWSMEGESQPVKMFDRHSSLAGCQIINYRTDAKQKWLLLTGISA----QQN 175
Query: 187 LVKGNMQLFSVEQQRSQALEAHAASFAQFKVPGNENPSTLISFATKTVNAGQVISKLHVI 246
V G MQL+SV+++ SQ +E HAASFAQFK+ GN STL FA + GQ KLH+I
Sbjct: 176 RVVGAMQLYSVDRKVSQPIEGHAASFAQFKMEGNAEESTLFCFAVR----GQAGGKLHII 231
Query: 247 ELGAQP-GKPSFTKKQADLFFPPDFADDFPVSMQISHKYSLIYVITKLGLLFVYDLETAT 305
E+G P G F KK D+FFPP+ +DFPV+MQIS K+ ++++ITK G + +YDLET T
Sbjct: 232 EVGTPPTGNQPFPKKAVDVFFPPEAQNDFPVAMQISEKHDVVFLITKYGYIHLYDLETGT 291
Query: 306 AVYRNRISPDPIFLTSEATSVGGFYAINRRGQVLLATVNEQTIVNFVSGQLNNLELAVNL 365
+Y NRIS + IF+T+ + G +NR+GQVL V E+ I+ +++ L N +LA+ +
Sbjct: 292 CIYMNRISGETIFVTAPHEATAGIIGVNRKGQVLSVCVEEENIIPYITNVLQNPDLALRM 351
Query: 366 AKRGNLPGAEKLVVERFHELFAQTKYKEAAELAAESPQGILRTPDTVAKFQSVPVQAGQT 425
A R NL GAE+L +F+ LFAQ Y EAA++AA +P+GILRTPDT+ +FQSVP Q GQT
Sbjct: 352 AVRNNLAGAEELFARKFNALFAQGNYSEAAKVAANAPKGILRTPDTIRRFQSVPAQPGQT 411
Query: 426 PPLLQYFGTLLTRGKLNAFESLELSRLVVNQNKKNLLENWLAEDKLECSEELGDLVKTVD 485
PLLQYFG LL +G+LN +ESLEL R V+ Q +K LLE WL EDKLECSEELGDLVK+VD
Sbjct: 412 SPLLQYFGILLDQGQLNKYESLELCRPVLQQGRKQLLEKWLKEDKLECSEELGDLVKSVD 471
Query: 486 NDLALKIYIKARATPKVVAAFAEKREFDKILIYSKQVGYTPDYLFLLQSILRTDPQGAVN 545
LAL +Y++A KV+ FAE + KI++Y+K+VGYTPD++FLL++++R P
Sbjct: 472 PTLALSVYLRANVPNKVIQCFAETGQVQKIVLYAKKVGYTPDWIFLLRNVMRISPDQGQQ 531
Query: 546 FALMMSQMEGGCPVDYNTITDLFLQRNLIREATAFLLDVLKPNLPEHGFLQTKVLEINLV 605
FA M+ Q E D I D+F++ NLI++ TAFLLD LK N P G LQT++LE+NL+
Sbjct: 532 FAQMLVQDEEPL-ADITQIVDVFMEYNLIQQCTAFLLDALKNNRPSEGPLQTRLLEMNLM 590
Query: 606 TFPNVADAILANGMFSHYDRPRIAQLCEKAGLYVRALQHYTELPDIKRVIVNTHAIEPQA 665
P VADAIL N MF+HYDR IAQLCEKAGL RAL+H+T+L DIKR +V+TH + P+
Sbjct: 591 HAPQVADAILGNQMFTHYDRAHIAQLCEKAGLLQRALEHFTDLYDIKRAVVHTHLLNPEW 650
Query: 666 LVEFFGTLSKEWALECMKDLLLVNLRGNLQIIVQVAKEYCEQLGVDACIKIFEQFRSYEG 725
LV +FG+LS E +LEC++ +L N+R NLQI VQVA +Y EQL + I++FE F+S+EG
Sbjct: 651 LVNYFGSLSVEDSLECLRAMLSANIRQNLQIWVQVASKYHEQLSTQSLIELFESFKSFEG 710
Query: 726 LYFFLGSYLSSSEDPDIHFKYIEAAAKTGQIKEVERVTRESSFYDPEKTKNFLMEAKLPD 785
L++FLGS ++ S+DPD+HFKYI+AA KTGQIKEVER+ RES+ YDPE+ KNFL EAKL D
Sbjct: 711 LFYFLGSIVNFSQDPDVHFKYIQAACKTGQIKEVERICRESNCYDPERVKNFLKEAKLTD 770
Query: 786 ARPLINVCDRFGFVPDLTHYLYTSNMLRYIEGYVQKVNPGNAPLVVGQLLDDECPEDFIK 845
PLI VCDRF FV DL YLY +++ +YIE YVQKVNP P+V+G LLD +C ED IK
Sbjct: 771 QLPLIIVCDRFDFVHDLVLYLYRNSLQKYIEIYVQKVNPSRLPVVIGGLLDVDCSEDVIK 830
Query: 846 GLILSVRSLLPVEPLVAECEKRNRLRLLTQFLEHLVSEGSQDVHVHNALGKIIIDSNNNP 905
LIL VR + LVAE EKRNRL+LL +LE + EG ++ HNAL KI IDSNNNP
Sbjct: 831 NLILVVRGQFSTDELVAEVEKRNRLKLLLPWLEARIHEGCEEPATHNALAKIYIDSNNNP 890
Query: 906 EHFLTTNPYYDSRVVGKYCEKRDPTLAVVAYRRGVCDDELINVTNKNSLFKLQARYVVER 965
E FL NPYYDSRVVGKYCEKRDP LA VAY RG CD ELINV N+NSLFK +RY+V R
Sbjct: 891 ERFLRENPYYDSRVVGKYCEKRDPHLACVAYERGQCDLELINVCNENSLFKSLSRYLVRR 950
Query: 966 MDADLWEKVLNPDNTYRRQLIDQVVSTALPESKSPEQVSASVKAFMTADLPHELIELLEK 1025
D +LW VL N YRR LIDQVV TAL E++ PE+VS +VKAFMTADLP+ELIELLEK
Sbjct: 951 KDPELWGSVLLESNPYRRPLIDQVVQTALSETQDPEEVSVTVKAFMTADLPNELIELLEK 1010
Query: 1026 IVLQNSAFSGNFNLQNLLILTAIKADSSRVMDYVNRLDNFDGPQVGEVAVEAELYEEAFA 1085
IVL NS FS + NLQNLLILTAIKAD +RVM+Y+NRLDN+D P + +A+ EL+EEAFA
Sbjct: 1011 IVLDNSVFSEHRNLQNLLILTAIKADRTRVMEYINRLDNYDAPDIANIAISNELFEEAFA 1070
Query: 1086 IFKKFNLNVQAVNVLLDNIHSIDRAVEFAFRVEEDAVWSQVAKAQLREGLVSDAIESFIR 1145
IF+KF++N AV VL+++I ++DRA EFA R E AVWSQ+AKAQL++G+V +AI+S+I+
Sbjct: 1071 IFRKFDVNTSAVQVLIEHIGNLDRAYEFAERCNEPAVWSQLAKAQLQKGMVKEAIDSYIK 1130
Query: 1146 ADDATQFLDVIRAAQDGNAYQDLVRYLLMVRQKTKEPKVDSELIYAYAKIDRLSDIEEFI 1205
ADD + +++V++AA +++LV+YL M R+K +E V++ELI+A AK +RL+++EEFI
Sbjct: 1131 ADDPSSYMEVVQAANTSGNWEELVKYLQMARKKARESYVETELIFALAKTNRLAELEEFI 1190
Query: 1206 LMPNVANLQNVGDRLYDEELYEAAKIIFAFISNWAKLAVTLVKLKQFQGAVDAARKANSA 1265
PN A++Q VGDR YDE++Y+AAK+++ +SN+ +LA TLV L ++Q AVD ARKANS
Sbjct: 1191 NGPNNAHIQQVGDRCYDEKMYDAAKLLYNNVSNFGRLASTLVHLGEYQAAVDGARKANST 1250
Query: 1266 KTWKEVCFACVDEEEFRLAQICGLNIIIQVDDLEEVSEYYQNRGCFNELISLMESGLGLE 1325
+TWKEVCFACVD +EFRLAQ+CGL+I++ D+LEE+ YYQ+RG F ELI+++E+ LGLE
Sbjct: 1251 RTWKEVCFACVDGKEFRLAQMCGLHIVVHADELEELINYYQDRGYFEELITMLEAALGLE 1310
Query: 1326 RAHMGIFTELGVLYARYRPEKLMEHIKLFATRLNIPKLIRACDEQQHWKELTYLYIQYDE 1385
RAHMG+FTEL +LY++++P+K+ EH++LF +R+NIPK++RA ++ W EL +LY +Y+E
Sbjct: 1311 RAHMGMFTELAILYSKFKPQKMREHLELFWSRVNIPKVLRAAEQAHLWAELVFLYDKYEE 1370
Query: 1386 FDNAATTIMNHSPEAWDHMQFKDVIAKVANVELYYKAVHFYLQEHPDLINDVLNVLALRV 1445
+DNA T+MNH +AW QFKD+I KVANVELYYKA+ FYL+ P L+ND+L VL+ R+
Sbjct: 1371 YDNAIITMMNHPTDAWKEGQFKDIITKVANVELYYKAIQFYLEFKPLLLNDLLMVLSPRL 1430
Query: 1446 DHARVVDIMRKAGHLRLVKPYMVAVQSNNVSAVNEALNEIYVEEEDYDRLRESIDLHDNF 1505
H R V+ K L LVKPY+ +VQ++N +VNE+LN +++ EEDY LR SID +DNF
Sbjct: 1431 AHTRAVNYFSKVKQLPLVKPYLRSVQNHNNKSVNESLNNLFITEEDYQALRTSIDAYDNF 1490
Query: 1506 DQIGLAQKIEKHELLEMRRVAAYIYKKAGRWKQSIALSKKDNLYKDAMETASQSGERELA 1565
D I LAQ++EKHEL+E RR+AAY++K RWKQS+ L KKD+LYKDAM+ AS+S + ELA
Sbjct: 1491 DNISLAQRLEKHELIEFRRIAAYLFKGNNRWKQSVELCKKDSLYKDAMQYASESKDTELA 1550
Query: 1566 EELLVYFIDQGKKECFASCLFVCYDLIRVDVALELAWMHNMIDFAFPYLLQFIREYTGKV 1625
EELL +F+ + K+ECF +CLF CYDL+R DV LE AW HN++DFA PY +Q ++EY KV
Sbjct: 1551 EELLQWFLQEEKRECFGACLFTCYDLLRPDVVLETAWRHNIMDFAMPYFIQVMKEYLTKV 1610
Query: 1626 DELVKHKIESQNEVKAKEQEEKEVIAQQNMYAQLLPLALPAPPMPGMGGGMGGGYAPPPP 1685
D K+++ ++ +E++ E QL+ A P+ +P P P
Sbjct: 1611 D-----KLDASESLRKEEEQATETQPIVYGQPQLMLTAGPSVAVP-----------PQAP 1654
Query: 1686 MGGGMGMPPMPPFGMPMGGY 1705
G G PP+G P G+
Sbjct: 1655 FGYGY---TAPPYGQPQPGF 1671
>CLH_DROME (P29742) Clathrin heavy chain
Length = 1678
Score = 1881 bits (4873), Expect = 0.0
Identities = 949/1666 (56%), Positives = 1238/1666 (73%), Gaps = 23/1666 (1%)
Query: 8 PILMREALTLPSIGINPQHITFTHVTMESDKYICVRETAPQNS-VVIVDMNMPNQPLRRP 66
PI +E L L ++GIN +F+ +TMESDK+ICVRE + VVI+DMN P RRP
Sbjct: 6 PIRFQEHLQLTNVGINANSFSFSTLTMESDKFICVREKVNDTAQVVIIDMNDATNPTRRP 65
Query: 67 ITADSALMNPNSRILALKAQLQGTTQDHLQIFNIELKAKMKSYQMPEQVVFWKWISPKLL 126
I+ADSA+MNP S+++ALKAQ LQIFNIE+K+KMK++ M E VVFWKWIS L
Sbjct: 66 ISADSAIMNPASKVIALKAQKT------LQIFNIEMKSKMKAHTMNEDVVFWKWISLNTL 119
Query: 127 GLVTQTSVYHWSIEGDSEPVKMFERTANLANNQIINYRCDPTEKWLVLIGIAPGSPERPQ 186
LVT+TSV+HWS+EGDS P KMF+R ++L QIINYRC+ +++WL+L+GI+ P
Sbjct: 120 ALVTETSVFHWSMEGDSMPQKMFDRHSSLNGCQIINYRCNASQQWLLLVGISA----LPS 175
Query: 187 LVKGNMQLFSVEQQRSQALEAHAASFAQFKVPGNENPSTLISFATKTVNAGQVISKLHVI 246
V G MQL+SVE++ SQA+E HAASFA FK+ N+ P+TL FA +T G KLH+I
Sbjct: 176 RVAGAMQLYSVERKVSQAIEGHAASFATFKIDANKEPTTLFCFAVRTATGG----KLHII 231
Query: 247 ELGAQP-GKPSFTKKQADLFFPPDFADDFPVSMQISHKYSLIYVITKLGLLFVYDLETAT 305
E+GA P G F KK D+FFPP+ +DFPV+MQ+S KY IY+ITK G + +YD+ETAT
Sbjct: 232 EVGAPPNGNQPFAKKAVDVFFPPEAQNDFPVAMQVSAKYDTIYLITKYGYIHLYDMETAT 291
Query: 306 AVYRNRISPDPIFLTSEATSVGGFYAINRRGQVLLATVNEQTIVNFVSGQLNNLELAVNL 365
+Y NRIS D IF+T+ + GG +NR+GQVL TV+E+ I+ +++ L N +LA+ +
Sbjct: 292 CIYMNRISADTIFVTAPHEASGGIIGVNRKGQVLSVTVDEEQIIPYINTVLQNPDLALRM 351
Query: 366 AKRGNLPGAEKLVVERFHELFAQTKYKEAAELAAESPQGILRTPDTVAKFQSVPVQAGQT 425
A R NL GAE L V +F++LF +Y EAA++AA +P+ ILRTP T+ +FQ V AG T
Sbjct: 352 AVRNNLAGAEDLFVRKFNKLFTAGQYAEAAKVAALAPKAILRTPQTIQRFQQVQTPAGST 411
Query: 426 -PPLLQYFGTLLTRGKLNAFESLELSRLVVNQNKKNLLENWLAEDKLECSEELGDLVKTV 484
PPLLQYFG LL +GKLN FESLEL R V+ Q KK L E WL E+KLECSEELGDLVK
Sbjct: 412 TPPLLQYFGILLDQGKLNKFESLELCRPVLLQGKKQLCEKWLKEEKLECSEELGDLVKAS 471
Query: 485 DNDLALKIYIKARATPKVVAAFAEKREFDKILIYSKQVGYTPDYLFLLQSILRTDPQGAV 544
D LAL IY++A KV+ FAE +F KI++Y+K+V YTPDY+FLL+S++R++P+
Sbjct: 472 DLTLALSIYLRANVPNKVIQCFAETGQFQKIVLYAKKVNYTPDYVFLLRSVMRSNPEQGA 531
Query: 545 NFALMMSQMEGGCPVDYNTITDLFLQRNLIREATAFLLDVLKPNLPEHGFLQTKVLEINL 604
FA M+ E D N I D+F++ +++++ TAFLLD LK N P G LQT++LE+NL
Sbjct: 532 GFASMLVAEEEPL-ADINQIVDIFMEHSMVQQCTAFLLDALKHNRPAEGALQTRLLEMNL 590
Query: 605 VTFPNVADAILANGMFSHYDRPRIAQLCEKAGLYVRALQHYTELPDIKRVIVNTHAIEPQ 664
++ P VADAIL N MF+HYDR IAQLCEKAGL RAL+HYT+L DIKR +V+TH + +
Sbjct: 591 MSAPQVADAILGNAMFTHYDRAHIAQLCEKAGLLQRALEHYTDLYDIKRAVVHTHMLNAE 650
Query: 665 ALVEFFGTLSKEWALECMKDLLLVNLRGNLQIIVQVAKEYCEQLGVDACIKIFEQFRSYE 724
LV FFGTLS E +LEC+K +L NLR NLQI VQ+A +Y EQL A I +FE F+SY+
Sbjct: 651 WLVSFFGTLSVEDSLECLKAMLTANLRQNLQICVQIATKYHEQLTNKALIDLFEGFKSYD 710
Query: 725 GLYFFLGSYLSSSEDPDIHFKYIEAAAKTGQIKEVERVTRESSFYDPEKTKNFLMEAKLP 784
GL++FL S ++ S+DP++HFKYI+AA KT QIKEVER+ RES+ Y+PE+ KNFL EAKL
Sbjct: 711 GLFYFLSSIVNFSQDPEVHFKYIQAACKTNQIKEVERICRESNCYNPERVKNFLKEAKLT 770
Query: 785 DARPLINVCDRFGFVPDLTHYLYTSNMLRYIEGYVQKVNPGNAPLVVGQLLDDECPEDFI 844
D PLI VCDRF FV DL YLY +N+ +YIE YVQKVNP P+VVG LLD +C ED I
Sbjct: 771 DQLPLIIVCDRFDFVHDLVLYLYRNNLQKYIEIYVQKVNPSRLPVVVGGLLDVDCSEDII 830
Query: 845 KGLILSVRSLLPVEPLVAECEKRNRLRLLTQFLEHLVSEGSQDVHVHNALGKIIIDSNNN 904
K LIL V+ + LV E EKRNRL+LL +LE V EG + HNAL KI IDSNNN
Sbjct: 831 KNLILVVKGQFSTDELVEEVEKRNRLKLLLPWLESRVHEGCVEPATHNALAKIYIDSNNN 890
Query: 905 PEHFLTTNPYYDSRVVGKYCEKRDPTLAVVAYRRGVCDDELINVTNKNSLFKLQARYVVE 964
PE +L N YYDSRVVG+YCEKRDP LA VAY RG+CD ELI V N+NSLFK +ARY+V
Sbjct: 891 PERYLKENQYYDSRVVGRYCEKRDPHLACVAYERGLCDRELIAVCNENSLFKSEARYLVG 950
Query: 965 RMDADLWEKVLNPDNTYRRQLIDQVVSTALPESKSPEQVSASVKAFMTADLPHELIELLE 1024
R DA+LW +VL+ N Y+RQLIDQVV TAL E++ P+ +S +VKAFMTADLP+ELIELLE
Sbjct: 951 RRDAELWAEVLSESNPYKRQLIDQVVQTALSETQDPDDISVTVKAFMTADLPNELIELLE 1010
Query: 1025 KIVLQNSAFSGNFNLQNLLILTAIKADSSRVMDYVNRLDNFDGPQVGEVAVEAELYEEAF 1084
KI+L +S FS + NLQNLLILTAIKAD +RVMDY+NRL+N+D P + +A+ +LYEEAF
Sbjct: 1011 KIILDSSVFSDHRNLQNLLILTAIKADRTRVMDYINRLENYDAPDIANIAISNQLYEEAF 1070
Query: 1085 AIFKKFNLNVQAVNVLLDNIHSIDRAVEFAFRVEEDAVWSQVAKAQLREGLVSDAIESFI 1144
AIFKKF++N A+ VL+D +++++RA EFA R E AVWSQ+AKAQL++GLV +AI+S+I
Sbjct: 1071 AIFKKFDVNTSAIQVLIDQVNNLERANEFAERCNEPAVWSQLAKAQLQQGLVKEAIDSYI 1130
Query: 1145 RADDATQFLDVIRAAQDGNAYQDLVRYLLMVRQKTKEPKVDSELIYAYAKIDRLSDIEEF 1204
+ADD + ++DV+ A ++ DLVRYL M R+K +E ++SELIYAYA+ RL+D+EEF
Sbjct: 1131 KADDPSAYVDVVDVASKVESWDDLVRYLQMARKKARESYIESELIYAYARTGRLADLEEF 1190
Query: 1205 ILMPNVANLQNVGDRLYDEELYEAAKIIFAFISNWAKLAVTLVKLKQFQGAVDAARKANS 1264
I PN A++Q +G+R + + +Y+AAK+++ +SN+A+LA+TLV LK+FQGAVD+ARKANS
Sbjct: 1191 ISGPNHADIQKIGNRCFSDGMYDAAKLLYNNVSNFARLAITLVYLKEFQGAVDSARKANS 1250
Query: 1265 AKTWKEVCFACVDEEEFRLAQICGLNIIIQVDDLEEVSEYYQNRGCFNELISLMESGLGL 1324
+TWKEVCFACVD EEFRLAQ+CGL+I++ D+LE++ YYQNRG F+ELI+L+ES LGL
Sbjct: 1251 TRTWKEVCFACVDAEEFRLAQMCGLHIVVHADELEDLINYYQNRGYFDELIALLESALGL 1310
Query: 1325 ERAHMGIFTELGVLYARYRPEKLMEHIKLFATRLNIPKLIRACDEQQHWKELTYLYIQYD 1384
ERAHMG+FTEL +LY++++P K+ EH++LF +R+NIPK++RA + W EL +LY +Y+
Sbjct: 1311 ERAHMGMFTELAILYSKFKPSKMREHLELFWSRVNIPKVLRAAESAHLWSELVFLYDKYE 1370
Query: 1385 EFDNAATTIMNHSPEAWDHMQFKDVIAKVANVELYYKAVHFYLQEHPDLINDVLNVLALR 1444
E+DNA +M H EAW FKD+I KVAN+ELYYKA+ FYL P L+ND+L VLA R
Sbjct: 1371 EYDNAVLAMMAHPTEAWREGHFKDIITKVANIELYYKAIEFYLDFKPLLLNDMLLVLAPR 1430
Query: 1445 VDHARVVDIMRKAGHLRLVKPYMVAVQSNNVSAVNEALNEIYVEEEDYDRLRESIDLHDN 1504
+DH R V K G+L LVKPY+ +VQS N A+NEALN + ++EEDY LR SID DN
Sbjct: 1431 MDHTRAVSYFSKTGYLPLVKPYLRSVQSLNNKAINEALNGLLIDEEDYQGLRNSIDGFDN 1490
Query: 1505 FDQIGLAQKIEKHELLEMRRVAAYIYKKAGRWKQSIALSKKDNLYKDAMETASQSGEREL 1564
FD I LAQK+EKHEL E RR+AAY+YK RWKQS+ L KKD LYKDAME A++S ++++
Sbjct: 1491 FDNIALAQKLEKHELTEFRRIAAYLYKGNNRWKQSVELCKKDKLYKDAMEYAAESCKQDI 1550
Query: 1565 AEELLVYFIDQGKKECFASCLFVCYDLIRVDVALELAWMHNMIDFAFPYLLQFIREYTGK 1624
AEELL +F+++ +CFA+CL+ CYDL+R DV LELAW H ++DFA PYL+Q +REYT K
Sbjct: 1551 AEELLGWFLERDAYDCFAACLYQCYDLLRPDVILELAWKHKIVDFAMPYLIQVLREYTTK 1610
Query: 1625 VDELVKHKIESQNEVKAKEQEEKEVIAQQNMYAQLLPLALPAPPMP 1670
VD+L + E+Q E + E K +I M QL+ A PA +P
Sbjct: 1611 VDKLELN--EAQREKEDDSTEHKNII---QMEPQLMITAGPAMGIP 1651
>CLH2_HUMAN (P53675) Clathrin heavy chain 2 (CLH-22)
Length = 1640
Score = 1825 bits (4728), Expect = 0.0
Identities = 909/1624 (55%), Positives = 1211/1624 (73%), Gaps = 19/1624 (1%)
Query: 8 PILMREALTLPSIGINPQHITFTHVTMESDKYICVRE-TAPQNSVVIVDMNMPNQPLRRP 66
P+ +E L ++GINP +I F+ +TMESDK+IC+RE Q V I+DM+ P P+RRP
Sbjct: 6 PVRFQEHFQLQNLGINPANIGFSTLTMESDKFICIREKVGEQAQVTIIDMSDPMAPIRRP 65
Query: 67 ITADSALMNPNSRILALKAQLQGTTQDHLQIFNIELKAKMKSYQMPEQVVFWKWISPKLL 126
I+A+SA+MNP S+++ALKA G T LQIFNIE+K+KMK++ M E+V+FWKW+S +
Sbjct: 66 ISAESAIMNPASKVIALKA---GKT---LQIFNIEMKSKMKAHTMAEEVIFWKWVSVNTV 119
Query: 127 GLVTQTSVYHWSIEGDSEPVKMFERTANLANNQIINYRCDPTEKWLVLIGIAPGSPERPQ 186
LVT+T+VYHWS+EGDS+P+KMF+R +L Q+I+YR D +KWL+L+GI+ +
Sbjct: 120 ALVTETAVYHWSMEGDSQPMKMFDRHTSLVGCQVIHYRTDEYQKWLLLVGISA----QQN 175
Query: 187 LVKGNMQLFSVEQQRSQALEAHAASFAQFKVPGNENPSTLISFATKTVNAGQVISKLHVI 246
V G MQL+SV+++ SQ +E HAA+FA+FK+ GN P+TL FA + G KLH+I
Sbjct: 176 RVVGAMQLYSVDRKVSQPIEGHAAAFAEFKMEGNAKPATLFCFAVRNPTGG----KLHII 231
Query: 247 ELGAQP--GKPSFTKKQADLFFPPDFADDFPVSMQISHKYSLIYVITKLGLLFVYDLETA 304
E+G QP G F KK D+FFPP+ +DFPV+MQI K+ +IY+ITK G L +YDLE+
Sbjct: 232 EVG-QPAAGNQPFVKKAVDVFFPPEAQNDFPVAMQIGAKHGVIYLITKYGYLHLYDLESG 290
Query: 305 TAVYRNRISPDPIFLTSEATSVGGFYAINRRGQVLLATVNEQTIVNFVSGQLNNLELAVN 364
+ NRIS D IF+T+ G +N++GQVL V E IVN+ + L N +L +
Sbjct: 291 VCICMNRISADTIFVTAPHKPTSGIIGVNKKGQVLSVCVEEDNIVNYATNVLQNPDLGLR 350
Query: 365 LAKRGNLPGAEKLVVERFHELFAQTKYKEAAELAAESPQGILRTPDTVAKFQSVPVQAGQ 424
LA R NL GAEKL V +F+ LFAQ Y EAA++AA +P+GILRT +TV KFQS+P Q+GQ
Sbjct: 351 LAVRSNLAGAEKLFVRKFNTLFAQGSYAEAAKVAASAPKGILRTRETVQKFQSIPAQSGQ 410
Query: 425 TPPLLQYFGTLLTRGKLNAFESLELSRLVVNQNKKNLLENWLAEDKLECSEELGDLVKTV 484
PLLQYFG LL +G+LN ESLEL LV+ Q +K LLE WL EDKLECSEELGDLVKT
Sbjct: 411 ASPLLQYFGILLDQGQLNKLESLELCHLVLQQGRKQLLEKWLKEDKLECSEELGDLVKTT 470
Query: 485 DNDLALKIYIKARATPKVVAAFAEKREFDKILIYSKQVGYTPDYLFLLQSILRTDPQGAV 544
D LAL +Y++A KV+ FAE +F KI++Y+K+VGYTPD++FLL+ +++ P+ +
Sbjct: 471 DPMLALSVYLRANVPSKVIQCFAETGQFQKIVLYAKKVGYTPDWIFLLRGVMKISPEQGL 530
Query: 545 NFALMMSQMEGGCPVDYNTITDLFLQRNLIREATAFLLDVLKPNLPEHGFLQTKVLEINL 604
F+ M+ Q E + + I D+F++ +LI++ T+FLLD LK N P G LQT +LE+NL
Sbjct: 531 QFSRMLVQDEEPL-ANISQIVDIFMENSLIQQCTSFLLDALKNNRPAEGLLQTWLLEMNL 589
Query: 605 VTFPNVADAILANGMFSHYDRPRIAQLCEKAGLYVRALQHYTELPDIKRVIVNTHAIEPQ 664
V P VADAIL N MF+HYDR IAQLCEKAGL +AL+HYT+L DIKR +V+TH + P+
Sbjct: 590 VHAPQVADAILGNKMFTHYDRAHIAQLCEKAGLLQQALEHYTDLYDIKRAVVHTHLLNPE 649
Query: 665 ALVEFFGTLSKEWALECMKDLLLVNLRGNLQIIVQVAKEYCEQLGVDACIKIFEQFRSYE 724
LV FFG+LS E ++EC+ +L N+R NLQ+ VQVA +Y EQLG A +++FE F+SY+
Sbjct: 650 WLVNFFGSLSVEDSVECLHAMLSANIRQNLQLCVQVASKYHEQLGTQALVELFESFKSYK 709
Query: 725 GLYFFLGSYLSSSEDPDIHFKYIEAAAKTGQIKEVERVTRESSFYDPEKTKNFLMEAKLP 784
GL++FLGS ++ S+DPD+H KYI+AA KTGQIKEVER+ RESS Y+PE+ KNFL EAKL
Sbjct: 710 GLFYFLGSIVNFSQDPDVHLKYIQAACKTGQIKEVERICRESSCYNPERVKNFLKEAKLT 769
Query: 785 DARPLINVCDRFGFVPDLTHYLYTSNMLRYIEGYVQKVNPGNAPLVVGQLLDDECPEDFI 844
D PLI VCDRFGFV DL YLY +N+ RYIE YVQKVNP P V+G LLD +C E+ I
Sbjct: 770 DQLPLIIVCDRFGFVHDLVLYLYRNNLQRYIEIYVQKVNPSRTPAVIGGLLDVDCSEEVI 829
Query: 845 KGLILSVRSLLPVEPLVAECEKRNRLRLLTQFLEHLVSEGSQDVHVHNALGKIIIDSNNN 904
K LI++VR + LVAE EKRNRL+LL +LE + EG ++ HNAL KI IDSNN+
Sbjct: 830 KHLIMAVRGQFSTDELVAEVEKRNRLKLLLPWLESQIQEGCEEPATHNALAKIYIDSNNS 889
Query: 905 PEHFLTTNPYYDSRVVGKYCEKRDPTLAVVAYRRGVCDDELINVTNKNSLFKLQARYVVE 964
PE FL N YYDS VVG+YCEKRDP LA VAY RG CD ELI V N+NSLFK +ARY+V
Sbjct: 890 PECFLRENAYYDSSVVGRYCEKRDPHLACVAYERGQCDLELIKVCNENSLFKSEARYLVC 949
Query: 965 RMDADLWEKVLNPDNTYRRQLIDQVVSTALPESKSPEQVSASVKAFMTADLPHELIELLE 1024
R D +LW VL N RRQLIDQVV TAL E++ PE++S +VKAFMTADLP+ELIELLE
Sbjct: 950 RKDPELWAHVLEETNPSRRQLIDQVVQTALSETRDPEEISVTVKAFMTADLPNELIELLE 1009
Query: 1025 KIVLQNSAFSGNFNLQNLLILTAIKADSSRVMDYVNRLDNFDGPQVGEVAVEAELYEEAF 1084
KIVL NS FS + NLQNLLILTAIKAD +RVM+Y++RLDN+D + +AV + LYEEAF
Sbjct: 1010 KIVLDNSVFSEHRNLQNLLILTAIKADRTRVMEYISRLDNYDALDIASIAVSSALYEEAF 1069
Query: 1085 AIFKKFNLNVQAVNVLLDNIHSIDRAVEFAFRVEEDAVWSQVAKAQLREGLVSDAIESFI 1144
+F KF++N A+ VL+++I ++DRA EFA R E AVWSQ+A+AQL++ LV +AI S+I
Sbjct: 1070 TVFHKFDMNASAIQVLIEHIGNLDRAYEFAERCNEPAVWSQLAQAQLQKDLVKEAINSYI 1129
Query: 1145 RADDATQFLDVIRAAQDGNAYQDLVRYLLMVRQKTKEPKVDSELIYAYAKIDRLSDIEEF 1204
R DD + +L+V+++A N ++DLV++L M R+K +E +++ELI+A AK R+S++E+F
Sbjct: 1130 RGDDPSSYLEVVQSASRSNNWEDLVKFLQMARKKGRESYIETELIFALAKTSRVSELEDF 1189
Query: 1205 ILMPNVANLQNVGDRLYDEELYEAAKIIFAFISNWAKLAVTLVKLKQFQGAVDAARKANS 1264
I PN A++Q VGDR Y+E +YEAAK++++ +SN+A+LA TLV L ++Q AVD +RKA+S
Sbjct: 1190 INGPNNAHIQQVGDRCYEEGMYEAAKLLYSNVSNFARLASTLVHLGEYQAAVDNSRKASS 1249
Query: 1265 AKTWKEVCFACVDEEEFRLAQICGLNIIIQVDDLEEVSEYYQNRGCFNELISLMESGLGL 1324
+TWKEVCFAC+D +EFR AQ+CGL+I+I D+LEE+ YYQ+RG F ELI L+E+ LGL
Sbjct: 1250 TRTWKEVCFACMDGQEFRFAQLCGLHIVIHADELEELMCYYQDRGYFEELILLLEAALGL 1309
Query: 1325 ERAHMGIFTELGVLYARYRPEKLMEHIKLFATRLNIPKLIRACDEQQHWKELTYLYIQYD 1384
ERAHMG+FTEL +LY++++P+K++EH++LF +R+NIPK++RA ++ W EL +LY +Y+
Sbjct: 1310 ERAHMGMFTELAILYSKFKPQKMLEHLELFWSRVNIPKVLRAAEQAHLWAELVFLYDKYE 1369
Query: 1385 EFDNAATTIMNHSPEAWDHMQFKDVIAKVANVELYYKAVHFYLQEHPDLINDVLNVLALR 1444
E+DNA T+M+H EAW QFKD+I KVANVEL Y+A+ FYL P LIND+L VL+ R
Sbjct: 1370 EYDNAVLTMMSHPTEAWKEGQFKDIITKVANVELCYRALQFYLDYKPLLINDLLLVLSPR 1429
Query: 1445 VDHARVVDIMRKAGHLRLVKPYMVAVQSNNVSAVNEALNEIYVEEEDYDRLRESIDLHDN 1504
+DH V KAG L LVKPY+ +VQS+N +VNEALN + EEEDY LR SID +DN
Sbjct: 1430 LDHTWTVSFFSKAGQLPLVKPYLRSVQSHNNKSVNEALNHLLTEEEDYQGLRASIDAYDN 1489
Query: 1505 FDQIGLAQKIEKHELLEMRRVAAYIYKKAGRWKQSIALSKKDNLYKDAMETASQSGEREL 1564
FD I LAQ++EKH+L+E R +AAY+YK W QS+ L KKD+LYKDAM+ A++S + EL
Sbjct: 1490 FDNISLAQQLEKHQLMEFRCIAAYLYKGNNWWAQSVELCKKDHLYKDAMQHAAESRDAEL 1549
Query: 1565 AEELLVYFIDQGKKECFASCLFVCYDLIRVDVALELAWMHNMIDFAFPYLLQFIREYTGK 1624
A++LL +F+++GK+ECFA+CLF CYDL+R D+ LELAW HN++D A PY +Q +REY K
Sbjct: 1550 AQKLLQWFLEEGKRECFAACLFTCYDLLRPDMVLELAWRHNLVDLAMPYFIQVMREYLSK 1609
Query: 1625 VDEL 1628
VD+L
Sbjct: 1610 VDKL 1613
>CLH_DICDI (P25870) Clathrin heavy chain
Length = 1694
Score = 1809 bits (4686), Expect = 0.0
Identities = 915/1709 (53%), Positives = 1236/1709 (71%), Gaps = 36/1709 (2%)
Query: 6 NAPILMREALTLPSIGINPQHITFTHVTMESDKYICVRETAP--QNSVVIVDMNMPNQPL 63
N PI +E L L ++GI I F+ +TMES+KYIC+RET P +N+VVI+D + P+Q L
Sbjct: 3 NLPIRFQEVLQLTNLGIGSNSIGFSTLTMESEKYICIRETTPDDKNNVVIIDTDNPSQIL 62
Query: 64 RRPITADSALMNPNSRILALKAQLQGTTQDHLQIFNIELKAKMKSYQMPEQVVFWKWISP 123
R+ + D+A+MNP ILALK LQ+ +IE K ++KS QM E + FWKWISP
Sbjct: 63 RKQMKTDAAIMNPKEPILALKIG------QVLQLISIEQKMQLKSCQMQEPLEFWKWISP 116
Query: 124 KLLGLVTQTSVYHWSIEGDSEPVKMFERTANLANNQIINYRCDPTEKWLVLIGIAPGSPE 183
L LVT TSV+HW+ EG+S+PVK+F+R +L N +IINYR D T+ WLVL+ I +
Sbjct: 117 NTLALVTATSVFHWTKEGNSDPVKVFDRHPDLQNTEIINYRSDSTQNWLVLVAIH----Q 172
Query: 184 RPQLVKGNMQLFSVEQQRSQALEAHAASFAQFKVPGNENPSTLISFATKTVNAGQVISKL 243
R V G +QL+SVE+Q SQ++E HAA FA + VPG PSTL + +++T NA SK+
Sbjct: 173 RDGRVVGRIQLYSVEKQISQSIEGHAACFANYIVPGATRPSTLFAISSRTQNA----SKI 228
Query: 244 HVIELGAQPGKPSFTKKQADLFFPPDF-ADDFPVSMQISHKYSLIYVITKLGLLFVYDLE 302
V+E+ G P+F K+ +D+F+PP+ A DFPV+MQ+S KY +IY++TKLG + ++DL
Sbjct: 229 LVLEVSKGDG-PNFQKRASDVFYPPEIGASDFPVAMQVSEKYEVIYMVTKLGYIHLFDLG 287
Query: 303 TATAVYRNRISPDPIFLTSEATSVGGFYAINRRGQVLLATVNEQTIVNFVSGQLNNLELA 362
TA +YRNRIS + IF+T+ S G A+NR+GQVL +++++ I+ ++ LNNLELA
Sbjct: 288 TANLIYRNRISNENIFVTAFEESTNGIIAVNRKGQVLSVSIDDKNIIPYICNTLNNLELA 347
Query: 363 VNLAKRGNLPGAEKLVVERFHELFAQTKYKEAAELAAESPQGILRTPDTVAKFQSVPVQA 422
+++A + NLPGAE L+ +F F Q +YKEAA++AA+SP ILR T+ KFQS+P
Sbjct: 348 ISMACKNNLPGAEGLLTTQFERYFQQGQYKEAAKVAADSPGSILRNLQTIQKFQSIPPIP 407
Query: 423 GQTPPLLQYFGTLLTRGKLNAFESLELSRLVVNQNKKNLLENWLAEDKLECSEELGDLVK 482
Q LLQYFG LL +GKLN ESLEL R V+ Q KK +LE WL EDKLECSE+LGD V+
Sbjct: 408 DQPSALLQYFGMLLEKGKLNKVESLELVRPVLAQGKKPILEKWLTEDKLECSEQLGDEVR 467
Query: 483 TVDNDLALKIYIKARATPKVVAAFAEKREFDKILIYSKQVGYTPDYLFLLQSILRTDPQG 542
D LAL IY +A A+ KV+ FAE EFDKI+ Y K+ Y PD++FLLQ + +P G
Sbjct: 468 PHDRKLALSIYYRANASDKVITLFAETGEFDKIIAYCKKFNYKPDFMFLLQRMANANPMG 527
Query: 543 AVNFALMMSQMEGGCPVDYNTITDLFLQRNLIREATAFLLDVLKPNLPEHGFLQTKVLEI 602
A +FA+ + + EGG +D N + +LF RN+I+E + FL +L + P+ LQTK+LEI
Sbjct: 528 AADFAVKLVKEEGGPYIDANQVVELFSARNMIQETSNFLFAILDGDRPQDANLQTKLLEI 587
Query: 603 NLVTFPNVADAILANGMFSHYDRPRIAQLCEKAGLYVRALQHYTELPDIKRVIVNT-HAI 661
NL+ P ADAI+ F+HY+R RI LCEKAGLY RAL+HYT+L DIKRV+ + H +
Sbjct: 588 NLLHAPQNADAIMGGQKFTHYNRLRIGGLCEKAGLYQRALEHYTDLADIKRVLSHAGHMV 647
Query: 662 EPQALVEFFGTLSKEWALECMKDLLLVNLRGNLQIIVQVAKEYCEQLGVDACIKIFEQFR 721
+ LV +FG+L+ E +ECM+D L N R NLQ++V +A Y +Q+ +A I +FE FR
Sbjct: 648 NQEFLVSYFGSLNPEDRMECMRDFLRTNPRQNLQLVVAIAVSYSDQITPEAIIAMFESFR 707
Query: 722 SYEGLYFFLGSYLSSSEDPDIHFKYIEAAAKTGQIKEVERVTRESSFYDPEKTKNFLMEA 781
YEGLY +L + +S P++HFKYIEAAAK Q KEVER+ R+S++YDPEKT++FL EA
Sbjct: 708 LYEGLYLYLTQVVVTSTSPEVHFKYIEAAAKINQFKEVERMCRDSNYYDPEKTRDFLKEA 767
Query: 782 KLPDARPLINVCDRFGFVPDLTHYLYTSNMLRYIEGYVQKVNPGNAPLVVGQLLDDECPE 841
KLPD PLI VCDR+ F+ DLT+YLY +N+ +YIE YVQK+NP N PLVVG LLD +C E
Sbjct: 768 KLPDQLPLIIVCDRYEFISDLTNYLYKNNLSKYIEAYVQKINPVNTPLVVGALLDLDCQE 827
Query: 842 DFIKGLILSVRSLLPVEPLVAECEKRNRLRLLTQFLEHLVSEGSQDVHVHNALGKIIIDS 901
D+++ LI+SVR++ P + LV + EKRNRL+LL +LE V+EG+ + +HNAL KI IDS
Sbjct: 828 DYLRNLIMSVRNMCPADSLVEQVEKRNRLKLLLPWLEARVAEGNIEPAIHNALAKIYIDS 887
Query: 902 NNNPEHFLTTNPYYDSRVVGKYCEKRDPTLAVVAYRRGVCDDELINVTNKNSLFKLQARY 961
N NPE FL N +YDS+VVGKYCEKRDP L+ VAY+RG+CD ELI VTNKN+LFK QARY
Sbjct: 888 NKNPEAFLLHNQFYDSKVVGKYCEKRDPYLSFVAYKRGLCDYELIEVTNKNTLFKNQARY 947
Query: 962 VVERMDADLWEKVLNPDNTYRRQLIDQVVSTALPESKSPEQVSASVKAFMTADLPHELIE 1021
+VER D DLW VL+ N Y+R LIDQVV TALPES + +VSA+VKAFM A+LP+ELIE
Sbjct: 948 LVERQDPDLWAYVLSDQNEYKRSLIDQVVQTALPESTNATEVSATVKAFMDANLPNELIE 1007
Query: 1022 LLEKIVLQNSAFSGNFNLQNLLILTAIKADSSRVMDYVNRLDNFDGPQVGEVAVEAELYE 1081
LLEKIVL+ F LQNLLILTAI+AD SRV DY+NRLDNFDG ++ +A+E++LYE
Sbjct: 1008 LLEKIVLEGKEFKTAKELQNLLILTAIRADKSRVTDYINRLDNFDGSKLAPIAIESQLYE 1067
Query: 1082 EAFAIFKKFNLNVQAVNVLLDNIHSIDRAVEFAFRVEEDAVWSQVAKAQLREGLVSDAIE 1141
EAF ++KKF NV+A++VL+ +I SI+RA +FA R + V+S++ AQL+ +V + IE
Sbjct: 1068 EAFFMYKKFQFNVEAIDVLITHIGSIERAHDFAERCNQTEVYSKLGVAQLKAEMVKECIE 1127
Query: 1142 SFIRADDATQFLDVIRAAQDGNAYQDLVRYLLMVRQKTKEPKVDSELIYAYAKIDRLSDI 1201
SFI+A+D + +V+ AA+ + Y+DLV++L M R+K KEP ++SELI+AYAK+++L+++
Sbjct: 1128 SFIKANDTEHYQEVVAAAERKDEYEDLVKFLQMCRKKIKEPAIESELIFAYAKVNKLAEM 1187
Query: 1202 EEFILMPNVANLQNVGDRLYDEELYEAAKIIFAFISNWAKLAVTLVKLKQFQGAVDAARK 1261
E+FI PN A++Q VGDR ++ LYEAAK+++ ISN+++L LVKL Q+Q AVDAARK
Sbjct: 1188 EDFINSPNSAHIQVVGDRCFENGLYEAAKVLYTNISNFSRLTSCLVKLGQYQAAVDAARK 1247
Query: 1262 ANSAKTWKEVCFACVDEEEFRLAQICGLNIIIQVDDLEEVSEYYQNRGCFNELISLMESG 1321
ANS KTWKEV AC+D +EFRLAQ+CG+NII+ D+LEE+ Y++RG FNELISL+ESG
Sbjct: 1248 ANSTKTWKEVSAACIDAKEFRLAQVCGINIIVHGDELEELIRQYEDRGYFNELISLLESG 1307
Query: 1322 LGLERAHMGIFTELGVLYARYRPEKLMEHIKLFATRLNIPKLIRACDEQQHWKELTYLYI 1381
L ERAH+G+FTEL +LY++Y+ EKLMEH+KLF +RLN+PK+I+AC Q W +LTYLYI
Sbjct: 1308 LASERAHVGMFTELAILYSKYKEEKLMEHLKLFYSRLNVPKVIKACQANQQWPQLTYLYI 1367
Query: 1382 QYDEFDNAATTIMNHSPEAWDHMQFKDVIAKVANVELYYKAVHFYLQEHPDLINDVLNVL 1441
YDE DNA T++NHS EAWDH+ FK+ I KVA ++LYY A+ FYL+E P LIND+L+VL
Sbjct: 1368 HYDEHDNAIQTMINHSIEAWDHVLFKETIPKVAKLDLYYSAISFYLEEQPLLINDLLSVL 1427
Query: 1442 ALRVDHARVVDIMRKAGHLRLVKPYMVAVQSNNVSAVNEALNEIYVEEEDYDRLRESIDL 1501
+ R+DH R V ++R GHL LVKPY+V+ Q NV+A+NEALNE+YVEEEDY+ LR SID
Sbjct: 1428 SPRIDHTRAVTLIRSLGHLPLVKPYLVSAQDQNVAALNEALNELYVEEEDYESLRSSIDA 1487
Query: 1502 HDNFDQIGLAQKIEKHELLEMRRVAAYIYKKAGRWKQSIALSKKDNLYKDAMETASQSGE 1561
+ NF I LAQK+EKHELLE RR+AAY+YKK RW QS+ LSKKD LYKDA+++AS S
Sbjct: 1488 NSNFGTIALAQKLEKHELLEFRRIAAYLYKKNNRWAQSVELSKKDKLYKDAIQSASDSKN 1547
Query: 1562 RELAEELLVYFIDQGKKECFASCLFVCYDLIRVDVALELAWMHNMIDFAFPYLLQFIREY 1621
+ EELL YF+DQ FA+CL+ CYD ++ D +ELAW +N+++++FPYL+Q+++EY
Sbjct: 1548 PAIGEELLQYFVDQQNNSAFAACLYTCYDFLKPDAVIELAWRNNILNYSFPYLIQYVKEY 1607
Query: 1622 TGKVDELVKHKIESQNEVKAKEQEEKEVIAQQNMYAQLLPLALPAPPMPGMGGG---MGG 1678
T KVD+LV ++ KA++++ +E QQN+ + P + + G GG
Sbjct: 1608 TTKVDQLV-------DDFKARQKKTEEEKEQQNIESSQY-----QPDLTNLSYGYAATGG 1655
Query: 1679 GYAPPPPMGGGMGMPPMPPF--GMPMGGY 1705
A PP +G P + MGG+
Sbjct: 1656 MLALPPAVGYQQQQQPQQMYNPNQMMGGF 1684
>CLH_CAEEL (P34574) Probable clathrin heavy chain
Length = 1681
Score = 1801 bits (4666), Expect = 0.0
Identities = 914/1693 (53%), Positives = 1231/1693 (71%), Gaps = 25/1693 (1%)
Query: 8 PILMREALTLPSIGINPQHITFTHVTMESDKYICVRET-APQNSVVIVDMNMPNQPLRRP 66
PI E L LP+ GI +ITF++VTMESDK I VRE Q VVI+D+ P RRP
Sbjct: 4 PIKFHEHLQLPNAGIRVPNITFSNVTMESDKNIVVREMIGDQQQVVIIDLADTANPTRRP 63
Query: 67 ITADSALMNPNSRILALKAQLQGTTQDHLQIFNIELKAKMKSYQMPEQVVFWKWISPKLL 126
I+ADS +M+P ++ILALK+ G T LQIFNIELKAK+K++Q E VV+WKWIS K +
Sbjct: 64 ISADSVIMHPTAKILALKS---GKT---LQIFNIELKAKVKAHQNVEDVVYWKWISEKTI 117
Query: 127 GLVTQTSVYHWSIEGDSEPVKMFERTANLANNQIINYRCDPTEKWLVLIGIAPGSPERPQ 186
LV+ T+VYHWSIEGD+ PVKMF+R +LA QIINYR D KWLVLIGI+ +
Sbjct: 118 ALVSDTAVYHWSIEGDAAPVKMFDRHQSLAGTQIINYRADAENKWLVLIGISA----KDS 173
Query: 187 LVKGNMQLFSVEQQRSQALEAHAASFAQFKVPGNENPSTLISFATKTVNAGQVISKLHVI 246
V G+MQL+S E++ SQ +E HAA F +FKV GN+NPS L F+ KT N G KLHVI
Sbjct: 174 RVVGSMQLYSTERKVSQPIEGHAACFVRFKVDGNQNPSNLFCFSVKTDNGG----KLHVI 229
Query: 247 ELGAQP-GKPSFTKKQADLFFPPDFADDFPVSMQISHKYSLIYVITKLGLLFVYDLETAT 305
E+G G F KK D+ + D A DFPVSMQ+S K +IY++TK G + +YD+E+ T
Sbjct: 230 EVGTPAAGNTPFQKKNVDVPYTADTAGDFPVSMQVSAKQGIIYLVTKQGYVHLYDVESGT 289
Query: 306 AVYRNRISPDPIFLTSEATSVGGFYAINRRGQVLLATVNEQTIVNFVSGQLNNLELAVNL 365
+Y NRIS D +F+T E T+ GG INR+GQVL +++E +V FV+ QL N +LA+ L
Sbjct: 290 RIYSNRISTDTVFVTCEYTATGGIMGINRKGQVLSVSIDEANLVPFVTNQLQNPDLALKL 349
Query: 366 AKRGNLPGAEKLVVERFHELFAQTKYKEAAELAAESPQGILRTPDTVAKFQSVPVQAGQT 425
A R +LPGAE+L V +F+ LF+ ++ E+A++AA +PQGILRTP T+ KFQ P
Sbjct: 350 AVRCDLPGAEELFVRKFNLLFSNGQFGESAKVAASAPQGILRTPATIQKFQQCPSTGPGP 409
Query: 426 PPLLQYFGTLLTRGKLNAFESLELSRLVVNQNKKNLLENWLAEDKLECSEELGDLVKTVD 485
PLLQYFG LL +GKLN +E+LEL R V+ Q +K L+ WL + KLEC EELGDL+K D
Sbjct: 410 SPLLQYFGILLDQGKLNKYETLELCRPVLAQGRKELITKWLNDQKLECCEELGDLIKPHD 469
Query: 486 NDLALKIYIKARATPKVVAAFAEKREFDKILIYSKQVGYTPDYLFLLQSILR-TDPQGAV 544
+ AL +Y++ KVV +FAE +FDKI++Y+K+VG+ PDYLF L+ ILR ++P
Sbjct: 470 VNTALSVYLRGNVPHKVVQSFAETGQFDKIVMYAKRVGFQPDYLFQLRQILRNSNPDHGA 529
Query: 545 NFA-LMMSQMEGGCPV-DYNTITDLFLQRNLIREATAFLLDVLKPNLPEHGFLQTKVLEI 602
FA L++S+ E G P+ D + I D F++ ++ T+FLL+VLK + PE G LQT++LE+
Sbjct: 530 KFAQLLVSESENGEPLADLSQIIDCFMEVQAVQPCTSFLLEVLKGDKPEEGHLQTRLLEM 589
Query: 603 NLVTFPNVADAILANGMFSHYDRPRIAQLCEKAGLYVRALQHYTELPDIKRVIVNTHAIE 662
NL+ P VADAILAN MFSHYDR I QLCEKAGL RAL+H+T+L DIKR +V+TH ++
Sbjct: 590 NLLAAPAVADAILANKMFSHYDRAAIGQLCEKAGLLQRALEHFTDLYDIKRTVVHTHLLK 649
Query: 663 PQALVEFFGTLSKEWALECMKDLLLVNLRGNLQIIVQVAKEYCEQLGVDACIKIFEQFRS 722
P LV +FG+LS E ++EC+K +L N+R NLQ++VQ+A +Y EQLG D I++FE +S
Sbjct: 650 PDWLVGYFGSLSVEDSVECLKAMLTQNIRQNLQVVVQIASKYHEQLGADKLIEMFENHKS 709
Query: 723 YEGLYFFLGSYLSSSEDPDIHFKYIEAAAKTGQIKEVERVTRESSFYDPEKTKNFLMEAK 782
YEGL++FLGS ++ S+DP++HFKYI+AA +TGQIKEVER+ RES YD E+ KNFL EAK
Sbjct: 710 YEGLFYFLGSIVNFSQDPEVHFKYIQAATRTGQIKEVERICRESQCYDAERVKNFLKEAK 769
Query: 783 LPDARPLINVCDRFGFVPDLTHYLYTSNMLRYIEGYVQKVNPGNAPLVVGQLLDDECPED 842
L D PLI VCDR V DL YLY + + +YIE +VQKVN P+VVG LLD +C ED
Sbjct: 770 LNDQLPLIIVCDRHNMVHDLVLYLYRNQLQKYIEVFVQKVNAARLPIVVGALLDVDCSED 829
Query: 843 FIKGLILSVRSLLPVEPLVAECEKRNRLRLLTQFLEHLVSEGSQDVHVHNALGKIIIDSN 902
IK LI++ R ++ LV E EKRNRL+LL +LE + EG+ D HNA+ KI IDSN
Sbjct: 830 AIKQLIINTRGKFDIDELVEEVEKRNRLKLLNHWLESKIQEGATDAATHNAMAKIYIDSN 889
Query: 903 NNPEHFLTTNPYYDSRVVGKYCEKRDPTLAVVAYRRGVCDDELINVTNKNSLFKLQARYV 962
NNPE FL NPYYDS+VVGKYCEKRDP A ++Y RG CD ELINV N+NSLFK ARY+
Sbjct: 890 NNPERFLKENPYYDSKVVGKYCEKRDPHYAFLSYERGQCDAELINVCNENSLFKNLARYL 949
Query: 963 VERMDADLWEKVLNPDNTYRRQLIDQVVSTALPESKSPEQVSASVKAFMTADLPHELIEL 1022
V+R D LWE+VLN +N +RRQLIDQVV TAL E++ PE +S +VKAFM ADLP+ELIEL
Sbjct: 950 VKRRDFTLWEQVLNEENVHRRQLIDQVVQTALSETQDPEDISVTVKAFMAADLPNELIEL 1009
Query: 1023 LEKIVLQNSAFSGNFNLQNLLILTAIKADSSRVMDYVNRLDNFDGPQVGEVAVEAELYEE 1082
LEKIVL NSAFS + NLQNLLILTA++AD +RVM+Y+ +LDN+D P + +A+ +ELYEE
Sbjct: 1010 LEKIVLDNSAFSEHRNLQNLLILTAMRADRTRVMEYIQKLDNYDAPDIANIAITSELYEE 1069
Query: 1083 AFAIFKKFNLNVQAVNVLLDNIHSIDRAVEFAFRVEEDAVWSQVAKAQLREGLVSDAIES 1142
AFAIFKKF++N A+NVL++N++++DRA EFA + + VW+ +AKAQL++ LV +A++S
Sbjct: 1070 AFAIFKKFDVNSSAINVLIENVNNLDRAYEFAEKCNQSDVWASLAKAQLQQNLVKEAVDS 1129
Query: 1143 FIRADDATQFLDVIRAAQDGNAYQDLVRYLLMVRQKTKEPKVDSELIYAYAKIDRLSDIE 1202
FI+ADD +++V+ ++DLVRYL M R+K++E +++EL++A AK RL+++E
Sbjct: 1130 FIKADDPGAYMEVVNKCSQTEHWEDLVRYLQMARKKSRESYIETELVFALAKTGRLTELE 1189
Query: 1203 EFILMPNVANLQNVGDRLYDEELYEAAKIIFAFISNWAKLAVTLVKLKQFQGAVDAARKA 1262
EFI PN A + +GDR +D ++++AKI+F +SN+AKL+VTLV+L ++QGAVDAARKA
Sbjct: 1190 EFIAGPNHAQIGQIGDRCFDNGMFDSAKILFNNVSNFAKLSVTLVRLGEYQGAVDAARKA 1249
Query: 1263 NSAKTWKEVCFACVDEEEFRLAQICGLNIIIQVDDLEEVSEYYQNRGCFNELISLMESGL 1322
NS KTWK+VCF+CV+ EFRLAQ+CGL+I++ D+LEE+ +YQ+RG F ELI+L+E+ L
Sbjct: 1250 NSTKTWKQVCFSCVENGEFRLAQMCGLHIVVHADELEELINFYQDRGHFEELIALLEAAL 1309
Query: 1323 GLERAHMGIFTELGVLYARYRPEKLMEHIKLFATRLNIPKLIRACDEQQHWKELTYLYIQ 1382
GLERAHMG+FTEL +LY++Y+PEK+ EH++LF +R+NIPK++RA ++ W EL +LY +
Sbjct: 1310 GLERAHMGMFTELAILYSKYKPEKMREHLELFWSRVNIPKVLRAAEQAHLWSELVFLYDK 1369
Query: 1383 YDEFDNAATTIMNHSPEAWDHMQFKDVIAKVANVELYYKAVHFYLQEHPDLINDVLNVLA 1442
Y+E+DNAA T+M H E+W FK+VIAKVANVELYYKA+ FYL P L+ND+L VL+
Sbjct: 1370 YEEYDNAALTMMQHPTESWREQHFKEVIAKVANVELYYKAMQFYLDYKPLLLNDLLTVLS 1429
Query: 1443 LRVDHARVVDIMRKAGHLRLVKPYMVAVQSNNVSAVNEALNEIYVEEEDYDRLRESIDLH 1502
R+DH+R V K + LVKPY+ VQ+ N A+NEALN++ ++EED+ LR SI+
Sbjct: 1430 PRLDHSRTVLFFNKLKQIPLVKPYLRQVQNLNNKAINEALNQLLIDEEDHAGLRSSIEAQ 1489
Query: 1503 DNFDQIGLAQKIEKHELLEMRRVAAYIYKKAGRWKQSIALSKKDNLYKDAMETASQSGER 1562
DNFD I LAQ++EKH L+E RR++AY++K RWKQSI L KKD LYKDAME A++S
Sbjct: 1490 DNFDNITLAQQLEKHPLVEFRRISAYLFKGNNRWKQSIELCKKDKLYKDAMEYAAESRNG 1549
Query: 1563 ELAEELLVYFIDQGKKECFASCLFVCYDLIRVDVALELAWMHNMIDFAFPYLLQFIREYT 1622
ELAEELL +F+D+ +CFA+ L+ CYDL+ DV +ELAW H ++D+A PY++Q +R+Y
Sbjct: 1550 ELAEELLSFFLDEKLYDCFAASLYHCYDLLHPDVIMELAWKHKIMDYAMPYMIQVMRDYQ 1609
Query: 1623 GKVDELVKHKIESQNEVKAKEQEEKEVIAQQNMYAQLLPLALPAPPM--PGMGGGMGGGY 1680
++++L + + E + E KA++Q+ + + + +L PAP M PG GG GG
Sbjct: 1610 TRLEKLERSEHERKEE-KAEQQQNNGMTMEPQL---MLTYGAPAPQMTYPGTTGGYGGQP 1665
Query: 1681 APPPPMGGGMGMP 1693
A P G P
Sbjct: 1666 AYGQPGQPGYNAP 1678
>CLH_SCHPO (Q10161) Probable clathrin heavy chain
Length = 1666
Score = 1618 bits (4190), Expect = 0.0
Identities = 808/1647 (49%), Positives = 1150/1647 (69%), Gaps = 21/1647 (1%)
Query: 4 AANAPILMREALTLPSIGINPQHITFTHVTMESDKYICVRETAPQ-NSVVIVDMNMPNQP 62
A PI E L L S+GI P F +VT+ESDKY+CVR+ N VVIVD+ P+
Sbjct: 2 AQQLPIRFSEVLQLASVGIQPSSFGFANVTLESDKYVCVRDNPNGVNQVVIVDLEDPSNV 61
Query: 63 LRRPITADSALMNPNSRILALKAQLQGTTQDHLQIFNIELKAKMKSYQMPEQVVFWKWIS 122
LRRPI+ADS +++P +I+ALKAQ Q LQ+F++E KAK+ SY M + VV+W WIS
Sbjct: 62 LRRPISADSVILHPKKKIIALKAQRQ------LQVFDLEAKAKINSYVMNQDVVYWTWIS 115
Query: 123 PKLLGLVTQTSVYHWSIEGDSEPVKMFERTANLANNQIINYRCDPTEKWLVLIGIAPGSP 182
++G+VT TSV+HW++ G S+PVKMF+R ++L QII+Y+ + E+W LIGI+
Sbjct: 116 DSVIGMVTDTSVFHWTVSG-SDPVKMFDRHSSLNGTQIISYKSNYNEEWFTLIGIS---- 170
Query: 183 ERPQLVKGNMQLFSVEQQRSQALEAHAASFAQFKVPGNENPSTLISFATKTVNAGQVISK 242
R + GN+QL+S +++ SQ LE+HA++FA + G ++ +++ A++ SK
Sbjct: 171 SRDNRIAGNLQLYSKKRKVSQPLESHASAFAVIQPEGVDHEVQVLALASRLPTG----SK 226
Query: 243 LHVIELGAQPGKPSFTKKQADLFFPPDFADDFPVSMQISHKYSLIYVITKLGLLFVYDLE 302
L ++E+ P P+F K DLFFPP+ +DFP++++I Y++ YV+TK G + VYDLE
Sbjct: 227 LSIVEVDRNPNNPAFATKTVDLFFPPEAVNDFPIAIEIGSTYNVAYVVTKYGFIHVYDLE 286
Query: 303 TATAVYRNRISPDPIFLTSEATSVGGFYAINRRGQVLLATVNEQTIVNFVSGQLNNLELA 362
TA +Y NR+S + IF+T+ SV G AINR+GQVL ++N +TI+ ++ LN+ LA
Sbjct: 287 TAKCIYMNRVSGESIFVTTAHKSVNGLMAINRKGQVLSVSINPETIIPYILSNLNDPGLA 346
Query: 363 VNLAKRGNLPGAEKLVVERFHELFAQTKYKEAAELAAESPQGILRTPDTVAKFQSVPVQA 422
V +A NLPGA+ L +++F +L AQ Y EAA++AA SP+GILRT + +F+ +
Sbjct: 347 VRMASHANLPGADNLYMQQFQQLMAQGNYSEAAKVAASSPRGILRTSQVIDQFKLIQAAP 406
Query: 423 GQTPPLLQYFGTLLTRGKLNAFESLELSRLVVNQNKKNLLENWLAEDKLECSEELGDLVK 482
GQ P+LQYFGTLL +G LN E++EL+R V+ QN+ LLE W E+KL C+E LGDLVK
Sbjct: 407 GQIAPILQYFGTLLDKGPLNEHETIELARPVLAQNRIQLLEKWYGENKLACTEALGDLVK 466
Query: 483 TVDNDLALKIYIKARATPKVVAAFAEKREFDKILIYSKQVGYTPDYLFLLQSILRTDPQG 542
+ ALKIY A KVV +E +F K+ Y+ Q TPDY+ LLQ+++R +P
Sbjct: 467 PYNTPFALKIYETANVPNKVVMCLSELGDFGKLATYTSQQNITPDYVSLLQNLVRVNPDQ 526
Query: 543 AVNFALMMSQMEGGCPVDYNTITDLFLQRNLIREATAFLLDVLKPNLPEHGFLQTKVLEI 602
A FA M ++ I D+F+ +NL+++ATAFLLD LK + PEH LQT++LEI
Sbjct: 527 AAEFATQM--FNSNPSINLEKIVDIFMSQNLVQQATAFLLDALKDDNPEHSHLQTRLLEI 584
Query: 603 NLVTFPNVADAILANGMFSHYDRPRIAQLCEKAGLYVRALQHYTELPDIKRVIVNTHAIE 662
NL+ P VADAIL N MF+H+DR IA LCE+AGL RAL+ Y + DIKRVIV+++ +
Sbjct: 585 NLINAPQVADAILGNQMFTHFDRAVIASLCERAGLVQRALELYDKPADIKRVIVHSNLLN 644
Query: 663 PQALVEFFGTLSKEWALECMKDLLLVNLRGNLQIIVQVAKEYCEQLGVDACIKIFEQFRS 722
P+ L+ +F S + + ++++L NLR NLQI+VQ+A Y + +G I++FE+F++
Sbjct: 645 PEWLMNYFSRFSPDEVYDYLREMLRSNLRQNLQIVVQIATRYSDLVGAQRIIEMFEKFKT 704
Query: 723 YEGLYFFLGSYLSSSEDPDIHFKYIEAAAKTGQIKEVERVTRESSFYDPEKTKNFLMEAK 782
+EGLY++LGS ++ +EDP++ +KYI+AA Q EVER+ R+++ Y+PEK KN L EAK
Sbjct: 705 FEGLYYYLGSIVNITEDPEVVYKYIQAACLMNQFTEVERICRDNNVYNPEKVKNLLKEAK 764
Query: 783 LPDARPLINVCDRFGFVPDLTHYLYTSNMLRYIEGYVQKVNPGNAPLVVGQLLDDECPED 842
L D PLI VCDR+ FV DL YL+ +NM ++IE YVQ++NP P VVG LLD +C E+
Sbjct: 765 LADQLPLILVCDRYDFVNDLVFYLFRNNMFQFIEIYVQRINPSKTPQVVGALLDIDCDEE 824
Query: 843 FIKGLILSVRSLLPVEPLVAECEKRNRLRLLTQFLEHLVSEGSQDVHVHNALGKIIIDSN 902
++ L++SV +PV+ LV E E+RNRL+LL +LE L+ GSQD +++AL KI IDSN
Sbjct: 825 LVQNLLMSVVGQVPVDELVEEVERRNRLKLLLPYLESLLQSGSQDRAIYDALAKIYIDSN 884
Query: 903 NNPEHFLTTNPYYDSRVVGKYCEKRDPTLAVVAYRRGVCDDELINVTNKNSLFKLQARYV 962
NNPE FL N +YD+ VGKYCEKRDP LA +AY +G D E+IN+ N+NS+FK ARY+
Sbjct: 885 NNPEVFLKENNFYDTLTVGKYCEKRDPYLAFIAYEKGGNDTEIINLCNENSMFKQLARYL 944
Query: 963 VERMDADLWEKVLNPDNTYRRQLIDQVVSTALPESKSPEQVSASVKAFMTADLPHELIEL 1022
++R D++LW +VL D+ YRR L+DQV++TA+PES PE VS VKA M DLP +LIEL
Sbjct: 945 LKRSDSNLWSEVLQ-DSAYRRPLLDQVIATAVPESSDPEAVSIVVKALMEVDLPSQLIEL 1003
Query: 1023 LEKIVLQNSAFSGNFNLQNLLILTAIKADSSRVMDYVNRLDNFDGPQVGEVAVEAELYEE 1082
LEKIVLQ S+FS N NLQNLL LTAIKAD SRVM+Y+++LD +D ++ E+A+E LYEE
Sbjct: 1004 LEKIVLQPSSFSENANLQNLLFLTAIKADKSRVMEYIDKLDKYDVDEIAEIAIENGLYEE 1063
Query: 1083 AFAIFKKFNLNVQAVNVLLDNIHSIDRAVEFAFRVEEDAVWSQVAKAQLREGLVSDAIES 1142
AF I+K N + QA+ VL+++I S+DRA ++A VE+ VWS++AKAQL + DAIES
Sbjct: 1064 AFRIYKIHNKHEQAMKVLVEDIVSLDRAQDYAETVEQPEVWSRLAKAQLDGIRIPDAIES 1123
Query: 1143 FIRADDATQFLDVIRAAQDGNAYQDLVRYLLMVRQKTKEPKVDSELIYAYAKIDRLSDIE 1202
+++ADD + + +VI A Y++L++YLLM R K EP VDS L+ AYAK ++L+++E
Sbjct: 1124 YLKADDPSNYSEVIELASRAGKYEELIKYLLMARSKMHEPDVDSALLIAYAKTNQLTEME 1183
Query: 1203 EFILMPNVANLQNVGDRLYDEELYEAAKIIFAFISNWAKLAVTLVKLKQFQGAVDAARKA 1262
F++ NVA+++ VGD ++ + YEAAK++++ ISNW+ LA TLV L ++QGAVD ARKA
Sbjct: 1184 TFLIGSNVADVKAVGDECFESKNYEAAKLMYSSISNWSMLATTLVYLGEYQGAVDCARKA 1243
Query: 1263 NSAKTWKEVCFACVDEEEFRLAQICGLNIIIQVDDLEEVSEYYQNRGCFNELISLMESGL 1322
NS K WK+V AC+D+ EFRLAQICGLN+I+ ++L + Y+ RG F E+ISLME+GL
Sbjct: 1244 NSIKVWKQVGTACIDKREFRLAQICGLNLIVHAEELPGLIRLYEERGYFEEVISLMEAGL 1303
Query: 1323 GLERAHMGIFTELGVLYARYRPEKLMEHIKLFATRLNIPKLIRACDEQQHWKELTYLYIQ 1382
GLERAHM +TEL +LYA+Y+PE++MEH+KLF RLN+ K+IRACD+ W E +LY+
Sbjct: 1304 GLERAHMAFYTELAILYAKYKPERMMEHLKLFWGRLNMAKVIRACDQMHLWNEAVFLYVH 1363
Query: 1383 YDEFDNAATTIMNHSPEAWDHMQFKDVIAKVANVELYYKAVHFYLQEHPDLINDVLNVLA 1442
+DNAA +M PEA+DH FKD+I VAN+ELYY+A++FYL++HP L+ D+L L
Sbjct: 1364 DQSYDNAA-AVMMEQPEAFDHQSFKDIIVHVANLELYYRALNFYLEQHPMLLTDLLAALT 1422
Query: 1443 LRVDHARVVDIMRKAGHLRLVKPYMVAVQSNNVSAVNEALNEIYVEEEDYDRLRESIDLH 1502
R+DH RV+ I K+ + L+ +MVA+Q N+ AVN A N++ +E EDY L++SI+ +
Sbjct: 1423 PRIDHPRVIRIFEKSENTPLILNFMVAIQHLNIQAVNHAYNDLLIEMEDYQSLQDSIENY 1482
Query: 1503 DNFDQIGLAQKIEKHELLEMRRVAAYIYKKAGRWKQSIALSKKDNLYKDAMETASQSGER 1562
D+FD I LA+++EKH LLE RR+AAYIY+K RW QSI LSK+D YKDA+ TA S +
Sbjct: 1483 DHFDAIALARRLEKHSLLEFRRIAAYIYRKNKRWTQSIELSKQDRFYKDAIITARDSDQT 1542
Query: 1563 ELAEELLVYFIDQGKKECFASCLFVCYDLIRVDVALELAWMHNMIDFAFPYLLQFIREYT 1622
+AE+L+ YF++ G ECFA+ L+ CY L+R D+ +E++W + D+A+PY + F E
Sbjct: 1543 TIAEDLMKYFVEIGNYECFAAILYTCYHLLRNDLVMEISWRKGLQDYAYPYFINFQCEMF 1602
Query: 1623 GKVDELVKHKIESQNEVKAKEQEEKEV 1649
KV L K ++ + VK++E+ +
Sbjct: 1603 SKVLNLEK-DLKDRQAVKSEEESASTI 1628
>CLH_YEAST (P22137) Clathrin heavy chain
Length = 1653
Score = 1453 bits (3761), Expect = 0.0
Identities = 725/1632 (44%), Positives = 1103/1632 (67%), Gaps = 18/1632 (1%)
Query: 5 ANAPILMREALTLPSIGINPQHITFTHVTMESDKYICVRETAP-QNSVVIVDMNMPNQPL 63
++ PI E + L S+GI+PQ + F T ESD ++ VRET NSV IVD+ N+
Sbjct: 2 SDLPIEFTELVDLMSLGISPQFLDFRSTTFESDHFVTVRETKDGTNSVAIVDLAKGNEVT 61
Query: 64 RRPITADSALMNPNSRILALKAQLQGTTQDHLQIFNIELKAKMKSYQMPEQVVFWKWISP 123
R+ + DSA+M+P+ +++++A GT +QIFN+E K+K+KS+ + E V+FW+W+S
Sbjct: 62 RKNMGGDSAIMHPSQMVISVRAN--GTI---VQIFNLETKSKLKSFTLDEPVIFWRWLSE 116
Query: 124 KLLGLVTQTSVYHWSI---EGDSEPVKMFERTANLANNQIINYRCDPTEKWLVLIGIAPG 180
LG VT S+ ++ +++P + R ANL N QIIN+ + W ++GI
Sbjct: 117 TTLGFVTARSILTSNVFDGNVNAKPQLLTLRHANLNNTQIINFVANKNLDWFAVVGILQE 176
Query: 181 SPERPQLVKGNMQLFSVEQQRSQALEAHAASFAQFKVPGNENPSTLISFATKTVNAGQVI 240
+ + G +QLFS ++ SQA++ H A F + GN + + F T NA
Sbjct: 177 NGR----IAGRIQLFSKQRNISQAIDGHVAIFTNILLEGNGSTPVQV-FVTGNRNATTGA 231
Query: 241 SKLHVIELGAQPGKPS-FTKKQADLFFPPDFADDFPVSMQISHKYSLIYVITKLGLLFVY 299
+L +IE+ PS + K+ D+FFPPD +DFP+++Q+S KY +IY++TK G + +Y
Sbjct: 232 GELRIIEIDHDASLPSQYQKETTDIFFPPDATNDFPIAVQVSEKYGIIYLLTKYGFIHLY 291
Query: 300 DLETATAVYRNRISPDPIFLTSEATSVGGFYAINRRGQVLLATVNEQTIVNFVSGQLNNL 359
+LET T ++ NRI+ + +F + G IN++GQVL ++ IV ++ +L+N+
Sbjct: 292 ELETGTNLFVNRITAESVFTAAPYNHENGIACINKKGQVLAVEISTSQIVPYILNKLSNV 351
Query: 360 ELAVNLAKRGNLPGAEKLVVERFHELFAQTKYKEAAELAAESPQGILRTPDTVAKFQSVP 419
LA+ +A RG LPGA+ L ++F L Q Y+ AA++AA S LR +T+ + +++
Sbjct: 352 ALALIVATRGGLPGADDLFQKQFESLLLQNDYQNAAKVAASSTS--LRNQNTINRLKNIQ 409
Query: 420 VQAGQTPPLLQYFGTLLTRGKLNAFESLELSRLVVNQNKKNLLENWLAEDKLECSEELGD 479
G P+L YF TLL +GKLN E++EL+R V+ Q++K L E WL EDKLECSEELGD
Sbjct: 410 APPGAISPILLYFSTLLDKGKLNKEETIELARPVLQQDRKQLFEKWLKEDKLECSEELGD 469
Query: 480 LVKTVDNDLALKIYIKARATPKVVAAFAEKREFDKILIYSKQVGYTPDYLFLLQSILRTD 539
+VK D LAL Y++A A KV++ AE ++F+KI+ Y ++VGY P++L L+ S++R+
Sbjct: 470 IVKPFDTTLALACYLRAGAHAKVISCLAELQQFEKIIPYCQKVGYQPNFLVLISSLIRSS 529
Query: 540 PQGAVNFALMMSQM-EGGCPVDYNTITDLFLQRNLIREATAFLLDVLKPNLPEHGFLQTK 598
P A FA+ + Q E +D I DLF +N I++ T+ LLD LK + P+ G LQT+
Sbjct: 530 PDRASEFAVSLLQNPETASQIDIEKIADLFFSQNHIQQGTSLLLDALKGDTPDQGHLQTR 589
Query: 599 VLEINLVTFPNVADAILANGMFSHYDRPRIAQLCEKAGLYVRALQHYTELPDIKRVIVNT 658
VLE+NL+ P VADAIL N +FSHYD+P IA L EKAGLY RAL++YT++ DIKR +V+T
Sbjct: 590 VLEVNLLHAPQVADAILGNNIFSHYDKPTIASLSEKAGLYQRALENYTDIKDIKRCVVHT 649
Query: 659 HAIEPQALVEFFGTLSKEWALECMKDLLLVNLRGNLQIIVQVAKEYCEQLGVDACIKIFE 718
+A+ LV +FG L+ E +L C+K L+ N++ N+Q +VQVA ++ + +G IK+FE
Sbjct: 650 NALPIDWLVGYFGKLNVEQSLACLKALMDNNIQANIQTVVQVATKFSDLIGPSTLIKLFE 709
Query: 719 QFRSYEGLYFFLGSYLSSSEDPDIHFKYIEAAAKTGQIKEVERVTRESSFYDPEKTKNFL 778
+ + EGLY++L S ++ +ED D+ +KYIEAAAK Q +E+ER+ ++++ YDPE+ KNFL
Sbjct: 710 DYNATEGLYYYLASLVNLTEDKDVVYKYIEAAAKMKQYREIERIVKDNNVYDPERVKNFL 769
Query: 779 MEAKLPDARPLINVCDRFGFVPDLTHYLYTSNMLRYIEGYVQKVNPGNAPLVVGQLLDDE 838
+A L D PL+ VCDRF FV ++ YLY S L++IE YVQ+VNP VVG LLD +
Sbjct: 770 KDANLEDQLPLVIVCDRFDFVHEMILYLYKSQNLKFIETYVQQVNPSKTAQVVGALLDMD 829
Query: 839 CPEDFIKGLILSVRSLLPVEPLVAECEKRNRLRLLTQFLEHLVSEGSQDVHVHNALGKII 898
C E FI+ L+ SV +P+ L E EKRNRL++L FLE +S+G QD V+NAL KI
Sbjct: 830 CDEAFIQSLLQSVLGQVPINELTTEVEKRNRLKILLPFLEQSLSQGIQDQAVYNALAKIY 889
Query: 899 IDSNNNPEHFLTTNPYYDSRVVGKYCEKRDPTLAVVAYRRGVCDDELINVTNKNSLFKLQ 958
IDSNN+PE FL N YD+ VG YCEKRDP LA +AY +G DD+LI +TN+NS++K Q
Sbjct: 890 IDSNNSPEKFLKENDQYDTLDVGHYCEKRDPYLAYIAYEKGQNDDDLIRITNENSMYKYQ 949
Query: 959 ARYVVERMDADLWEKVLNPDNTYRRQLIDQVVSTALPESKSPEQVSASVKAFMTADLPHE 1018
ARY++ER D DLW KVLN +N +RRQLID V+S +PE PE VS +V+AFMT L E
Sbjct: 950 ARYLLERSDLDLWNKVLNQENIHRRQLIDSVISVGIPELTDPEPVSLTVQAFMTNGLKLE 1009
Query: 1019 LIELLEKIVLQNSAFSGNFNLQNLLILTAIKADSSRVMDYVNRLDNFDGPQVGEVAVEAE 1078
LIELLEKI+L+ S F+ N LQ LL+L+AIK + ++V Y+ +LDN+D ++ + +E +
Sbjct: 1010 LIELLEKIILEPSPFNENVALQGLLLLSAIKYEPTKVSSYIEKLDNYDADEIAPLCIEHD 1069
Query: 1079 LYEEAFAIFKKFNLNVQAVNVLLDNIHSIDRAVEFAFRVEEDAVWSQVAKAQLREGLVSD 1138
L EEAF I+ K + +A+ VL+++I S+DRA +A ++ +WSQ+ AQL + D
Sbjct: 1070 LKEEAFEIYDKHEMYGKALKVLIEDIMSLDRAASYADKINTPELWSQIGTAQLDGLRIPD 1129
Query: 1139 AIESFIRADDATQFLDVIRAAQDGNAYQDLVRYLLMVRQKTKEPKVDSELIYAYAKIDRL 1198
AIES+I+A+D + + +VI A+ Y++L+ +LLM R+ KEPK+D LI AYA+++++
Sbjct: 1130 AIESYIKAEDPSNYENVIDIAEQAGKYEELIPFLLMARKTLKEPKIDGALILAYAELNKI 1189
Query: 1199 SDIEEFILMPNVANLQNVGDRLYDEELYEAAKIIFAFISNWAKLAVTLVKLKQFQGAVDA 1258
+IE + NVANL +VGD+L++ + Y+AA++ ++ +SN++KLA TLV L +Q AVD
Sbjct: 1190 HEIENLLAGSNVANLDHVGDKLFENKEYKAARLCYSAVSNYSKLASTLVYLGDYQAAVDT 1249
Query: 1259 ARKANSAKTWKEVCFACVDEEEFRLAQICGLNIIIQVDDLEEVSEYYQNRGCFNELISLM 1318
ARKA++ K WK V AC++++EF+LAQICGLN+I+ ++L+E+ E Y++ G F ELISL
Sbjct: 1250 ARKASNIKVWKLVNDACIEKKEFKLAQICGLNLIVHAEELDELVERYESNGYFEELISLF 1309
Query: 1319 ESGLGLERAHMGIFTELGVLYARYRPEKLMEHIKLFATRLNIPKLIRACDEQQHWKELTY 1378
E+GLGLERAHMG+FTEL +LY++Y P+K EH+KLF +R+NIPK+IRA ++ W EL +
Sbjct: 1310 EAGLGLERAHMGMFTELAILYSKYEPDKTFEHLKLFWSRINIPKVIRAVEQAHLWSELVF 1369
Query: 1379 LYIQYDEFDNAATTIMNHSPEAWDHMQFKDVIAKVANVELYYKAVHFYLQEHPDLINDVL 1438
LY YDE+DNAA T++ S + DH FK+V+ KV+N+E+YYKA++FY++ HP L+ D+L
Sbjct: 1370 LYAHYDEWDNAALTLIEKSTKDLDHAYFKEVVVKVSNLEIYYKAINFYVKFHPSLLVDLL 1429
Query: 1439 NVLALRVDHARVVDIMRKAGHLRLVKPYMVAVQSNNVSAVNEALNEIYVEEEDYDRLRES 1498
L R+D R V I K+ +L L+KP+++ V N S VN+A +++ +EEEDY L+++
Sbjct: 1430 TSLTPRLDIPRTVKIFSKSDNLPLIKPFLINVLPKNNSVVNQAYHDLMIEEEDYKALQDA 1489
Query: 1499 IDLHDNFDQIGLAQKIEKHELLEMRRVAAYIYKKAGRWKQSIALSKKDNLYKDAMETASQ 1558
+D +D FDQ+GLA ++E H+L+ +++ A +Y++ +W +S+++ K++ L+KDA+ETA+
Sbjct: 1490 VDSYDKFDQLGLASRLESHKLIFFKKIGALLYRRNKKWAKSLSILKEEKLWKDAIETAAI 1549
Query: 1559 SGERELAEELLVYFIDQGKKECFASCLFVCYDLIRVDVALELAWMHNMIDFAFPYLLQFI 1618
S + ++ E LL YF++ G +E F + L+ Y+L+R++ LE++WM+++ D+ P+ +
Sbjct: 1550 SQDPKVVEALLTYFVETGNREGFVALLYAAYNLVRIEFVLEISWMNSLEDYIKPFEISIK 1609
Query: 1619 REYTGKVDELVK 1630
+E + ++ +
Sbjct: 1610 KEQNDSIKKITE 1621
>DIA2_HUMAN (O60879) Diaphanous protein homolog 2 (Diaphanous-related
formin 2) (DRF2)
Length = 1101
Score = 49.3 bits (116), Expect = 9e-05
Identities = 36/104 (34%), Positives = 54/104 (51%), Gaps = 19/104 (18%)
Query: 1620 EYTGKVDELVKHKIESQNEVKAKEQEEKEV---IAQQNMYAQLL------PLALPAPPMP 1670
E++ K DE + E+Q E++ ++++ KE+ I Q AQ+L P APP+P
Sbjct: 499 EFSKKFDEEFTARQEAQAELQKRDEKIKELEAEIQQLRTQAQVLSSSSGIPGPPAAPPLP 558
Query: 1671 GMGGG-------MGGGYA---PPPPMGGGMGMPPMPPFGMPMGG 1704
G+G + GG PPPP+ G MG+PP PP + GG
Sbjct: 559 GVGPPPPPPAPPLPGGAPLPPPPPPLPGMMGIPPPPPPPLLFGG 602
>DIA2_MOUSE (O70566) Diaphanous protein homolog 2 (Diaphanous-related
formin 2) (DRF2) (mDia3)
Length = 1098
Score = 48.5 bits (114), Expect = 2e-04
Identities = 37/115 (32%), Positives = 55/115 (47%), Gaps = 20/115 (17%)
Query: 1607 IDFAFPYLL-------------QFIREYTGKVDELVKHKIESQNEVKAKEQEEKEVIAQ- 1652
IDF F +LL Q E++ K DE + E+Q E++ ++++ KE+ +
Sbjct: 472 IDFDFTHLLDACVNKAKVEENEQKAMEFSKKFDEEFTARQEAQAELQKRDEKIKELETEI 531
Query: 1653 QNMYAQLLPLALPAPPMPGMGGGMGG------GYAPPPPMGGGMGMPPMPPFGMP 1701
Q + Q +P A+P PP P G G PPPP+ G + PP P GMP
Sbjct: 532 QQLRGQGVPSAIPGPPPPPPLPGAGPCPPPPPPPPPPPPLPGVVPPPPPPLPGMP 586
>DIA_DROME (P48608) Diaphanous protein
Length = 1091
Score = 46.2 bits (108), Expect = 8e-04
Identities = 40/119 (33%), Positives = 46/119 (38%), Gaps = 36/119 (30%)
Query: 1620 EYTGKVDELVKHKIESQNEVKAKEQEEKEVIAQQNMYAQLLPLALPA------------- 1666
EY K+++L K E+ E KA EEK + + N A P LP
Sbjct: 464 EYEKKIEQLESAKQEA--EAKAAHLEEKVKLMEANGVAAPSPNKLPKVNIPMPPPPPGGG 521
Query: 1667 -------PPMPGMGGG-------------MGGGYAPPPPMG-GGMGMPPMPPFGMPMGG 1704
PPMPG GG GG PPPP G GG PPMP P GG
Sbjct: 522 GAPPPPPPPMPGRAGGGPPPPPPPPMPGRAGGPPPPPPPPGMGGPPPPPMPGMMRPGGG 580
Score = 34.3 bits (77), Expect = 3.0
Identities = 20/41 (48%), Positives = 22/41 (52%), Gaps = 9/41 (21%)
Query: 1665 PAPPMPGMGG-------GM--GGGYAPPPPMGGGMGMPPMP 1696
P PP PGMGG GM GG PPPPM G +P +P
Sbjct: 556 PPPPPPGMGGPPPPPMPGMMRPGGGPPPPPMMMGPMVPVLP 596
>PRA_MYCTU (O53426) Proline-rich antigen homolog
Length = 240
Score = 45.8 bits (107), Expect = 0.001
Identities = 24/49 (48%), Positives = 24/49 (48%), Gaps = 12/49 (24%)
Query: 1665 PAPPMPGMGGGM---------GGGYAPPPPMGGGMGMPPMPPFGMPMGG 1704
P PP PG GG G GYAPPPP G G PP PP P GG
Sbjct: 13 PPPPPPGPSGGHEPPPAAPPGGSGYAPPPPPSSGSGYPPPPP---PPGG 58
Score = 37.7 bits (86), Expect = 0.27
Identities = 17/33 (51%), Positives = 17/33 (51%), Gaps = 5/33 (15%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
P PP PG GG Y PPPP GG PP P
Sbjct: 51 PPPPPPG-----GGAYPPPPPSAGGYAPPPPGP 78
>DIA3_HUMAN (Q9NSV4) Diaphanous protein homolog 3 (Diaphanous-related
formin 3) (DRF3) (Fragment)
Length = 853
Score = 44.7 bits (104), Expect = 0.002
Identities = 35/117 (29%), Positives = 53/117 (44%), Gaps = 14/117 (11%)
Query: 1594 VDVALELAWMHNMIDFAFPYLLQFIREYTGKVDELVKHKIESQNEVKAKEQEEKEVIAQQ 1653
VD+ ++ A + + A +F +E+T H+ E+Q E++ KE + E+ A+
Sbjct: 236 VDICIDQAKLEEFEEKASELYKKFEKEFTD-------HQ-ETQAELQKKEAKINELQAEL 287
Query: 1654 NMYAQ---LLPLALPAPPMPGMGGGMGGGYAPPPP---MGGGMGMPPMPPFGMPMGG 1704
+ LP P P GG G PPPP GGG+ PP PP P+ G
Sbjct: 288 QAFKSQFGALPADCNIPLPPSKEGGTGHSALPPPPPLPSGGGVPPPPPPPPPPPLPG 344
Score = 43.1 bits (100), Expect = 0.007
Identities = 20/40 (50%), Positives = 24/40 (60%), Gaps = 4/40 (10%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMG--GGMGMPPMP--PFGM 1700
P PP+PGM G PPPP+G GG PP+P PFG+
Sbjct: 338 PPPPLPGMRMPFSGPVPPPPPLGFLGGQNSPPLPILPFGL 377
>SEPA_EMENI (P78621) Cytokinesis protein sepA (FH1/2 protein) (Forced
expression inhibition of growth A)
Length = 1790
Score = 44.3 bits (103), Expect = 0.003
Identities = 20/41 (48%), Positives = 22/41 (52%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMGGY 1705
P PP PG G PPPP GG+G PP PP P GG+
Sbjct: 1040 PPPPPPGAGAAPPPPPPPPPPPPGGLGGPPPPPPPPPPGGF 1080
Score = 43.9 bits (102), Expect = 0.004
Identities = 22/43 (51%), Positives = 23/43 (53%), Gaps = 6/43 (13%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP------FGMP 1701
P PP P GG GG PPPP GG G PP PP FG+P
Sbjct: 1069 PPPPPPPPPGGFGGPPPPPPPPGGFGGPPPPPPPPPGGAFGVP 1111
Score = 42.4 bits (98), Expect = 0.011
Identities = 21/42 (50%), Positives = 24/42 (57%), Gaps = 1/42 (2%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGM-PPMPPFGMPMGGY 1705
P PP P GG G PPPP GG G+ PP PP G +GG+
Sbjct: 1083 PPPPPPPPGGFGGPPPPPPPPPGGAFGVPPPPPPPGTVIGGW 1124
Score = 42.0 bits (97), Expect = 0.015
Identities = 22/42 (52%), Positives = 24/42 (56%), Gaps = 3/42 (7%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPM-GGGMGMPPMPPFGMPMGGY 1705
P PP P GG+GG PPPP GG G PP PP P GG+
Sbjct: 1054 PPPPPPPPPGGLGGPPPPPPPPPPGGFGGPPPPP--PPPGGF 1093
Score = 36.6 bits (83), Expect = 0.61
Identities = 24/68 (35%), Positives = 27/68 (39%), Gaps = 3/68 (4%)
Query: 1640 KAKEQEEKEVIAQQNMYAQLLPLALPAPPMPGMGGGMGGGYAPPPPMGGGMGM---PPMP 1696
K EQ + +A A P P P PG+ G PPPP G G PP P
Sbjct: 997 KGDEQGVDDTVAVDKATAAPPPPPPPPPAHPGLSGAAPPPPPPPPPPPPGAGAAPPPPPP 1056
Query: 1697 PFGMPMGG 1704
P P GG
Sbjct: 1057 PPPPPPGG 1064
>DIA1_HUMAN (O60610) Diaphanous protein homolog 1 (Diaphanous-related
formin 1) (DRF1)
Length = 1248
Score = 43.9 bits (102), Expect = 0.004
Identities = 21/38 (55%), Positives = 23/38 (60%), Gaps = 6/38 (15%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP-FGMP 1701
P PP+PG G G PPPP GG G+PP PP GMP
Sbjct: 695 PPPPLPG-----GPGIPPPPPFPGGPGIPPPPPGMGMP 727
Score = 43.1 bits (100), Expect = 0.007
Identities = 20/43 (46%), Positives = 23/43 (52%), Gaps = 9/43 (20%)
Query: 1665 PAPPMPGMGG---------GMGGGYAPPPPMGGGMGMPPMPPF 1698
P PP+PG G G G PPPP+ GG G+PP PPF
Sbjct: 669 PPPPLPGSAGIPPPPPPLPGEAGMPPPPPPLPGGPGIPPPPPF 711
Score = 38.5 bits (88), Expect = 0.16
Identities = 24/52 (46%), Positives = 25/52 (47%), Gaps = 17/52 (32%)
Query: 1665 PAPPMPGMGGGM-------GGGYAPPPP--MGG--------GMGMPPMPPFG 1699
P PP+PG G GG PPPP GG GMGMPP PPFG
Sbjct: 682 PPPPLPGEAGMPPPPPPLPGGPGIPPPPPFPGGPGIPPPPPGMGMPPPPPFG 733
Score = 37.4 bits (85), Expect = 0.36
Identities = 24/80 (30%), Positives = 35/80 (43%), Gaps = 8/80 (10%)
Query: 1618 IREYTGKVDELVKHKIESQNEVKAKEQEEKEVIAQQNMYAQLLPLALPAPPMPGMGGGMG 1677
+ + TG+V +L K E++ ++E + A +P P PP P + G
Sbjct: 532 VSQLTGEVAKLTK-------ELEDAKKEMASLSAAAITVPPSVPSRAPVPPAPPLPGD-S 583
Query: 1678 GGYAPPPPMGGGMGMPPMPP 1697
G PPPP G PP PP
Sbjct: 584 GTIIPPPPAPGDSTTPPPPP 603
Score = 36.6 bits (83), Expect = 0.61
Identities = 19/41 (46%), Positives = 23/41 (55%), Gaps = 6/41 (14%)
Query: 1665 PAPPMPGMGGGM-----GGGYAPPPPMGGGMGMPPMPPFGM 1700
P PP PG G G+ G G PPPP G G+ P+ PFG+
Sbjct: 707 PPPPFPG-GPGIPPPPPGMGMPPPPPFGFGVPAAPVLPFGL 746
Score = 36.6 bits (83), Expect = 0.61
Identities = 16/39 (41%), Positives = 21/39 (53%), Gaps = 5/39 (12%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMG 1703
P PP+PG G +PPPP+ G +PP PP +G
Sbjct: 608 PPPPLPG-----GTAISPPPPLSGDATIPPPPPLPEGVG 641
Score = 33.1 bits (74), Expect = 6.7
Identities = 18/40 (45%), Positives = 23/40 (57%), Gaps = 7/40 (17%)
Query: 1665 PAPPMP-GMG----GGMGGGYA--PPPPMGGGMGMPPMPP 1697
P PP+P G+G + GG A PPPP+ G +PP PP
Sbjct: 632 PPPPLPEGVGIPSPSSLPGGTAIPPPPPLPGSARIPPPPP 671
>SF01_HUMAN (Q15637) Splicing factor 1 (Zinc finger protein 162)
(Transcription factor ZFM1) (Zinc finger gene in MEN1
locus) (Mammalian branch point binding protein mBBP)
(BBP)
Length = 639
Score = 43.1 bits (100), Expect = 0.007
Identities = 24/53 (45%), Positives = 25/53 (46%), Gaps = 11/53 (20%)
Query: 1660 LPLALPAPPMPGMGGGMGGGYAPPPPMGG-----------GMGMPPMPPFGMP 1701
LP P PP P G G YAPPPP GMG+ MPPFGMP
Sbjct: 577 LPPGAPPPPPPPPPGSAGMMYAPPPPPPPPMDPSNFVTMMGMGVAGMPPFGMP 629
>DIA1_MOUSE (O08808) Diaphanous protein homolog 1 (Diaphanous-related
formin 1) (DRF1) (mDIA1) (p140mDIA)
Length = 1255
Score = 43.1 bits (100), Expect = 0.007
Identities = 27/80 (33%), Positives = 44/80 (54%), Gaps = 7/80 (8%)
Query: 1618 IREYTGKVDELVKHKIESQNEVKAKEQEEKEVIAQQNMYAQLLPLALPAPPMPGMGGGMG 1677
+ + TG+V +L K +++NE+ + V+A + +P PAPP+PG G +
Sbjct: 532 VSKLTGEVAKLSKELEDAKNEMASLSAV---VVAPSVSSSAAVP---PAPPLPGDSGTVI 585
Query: 1678 GGYAPPPPMGGGMGMPPMPP 1697
PPPP+ GG+ +PP PP
Sbjct: 586 PPPPPPPPLPGGV-VPPSPP 604
Score = 40.4 bits (93), Expect = 0.042
Identities = 21/49 (42%), Positives = 24/49 (48%), Gaps = 11/49 (22%)
Query: 1660 LPLALPAPPMPGMGGGMG-----------GGYAPPPPMGGGMGMPPMPP 1697
LP A PP P + GG G G PPPP+ GG G+PP PP
Sbjct: 665 LPGATAIPPPPPLPGGTGIPPPPPPLPGSVGVPPPPPLPGGPGLPPPPP 713
Score = 39.3 bits (90), Expect = 0.094
Identities = 22/51 (43%), Positives = 26/51 (50%), Gaps = 14/51 (27%)
Query: 1660 LPLALPAPPMPGMGGGMG-----------GGYAPPPPMGGGMGMPPMPPFG 1699
LP ++ PP P + GG G G PPPP GMG+PP PPFG
Sbjct: 690 LPGSVGVPPPPPLPGGPGLPPPPPPFPGAPGIPPPPP---GMGVPPPPPFG 737
Score = 38.1 bits (87), Expect = 0.21
Identities = 18/38 (47%), Positives = 21/38 (54%), Gaps = 2/38 (5%)
Query: 1660 LPLALPAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
LP A PP P + G PPPP+ GG G+PP PP
Sbjct: 653 LPGATAIPPPPPLPGAT--AIPPPPPLPGGTGIPPPPP 688
Score = 37.0 bits (84), Expect = 0.47
Identities = 18/40 (45%), Positives = 21/40 (52%), Gaps = 4/40 (10%)
Query: 1665 PAPPMPGMGG----GMGGGYAPPPPMGGGMGMPPMPPFGM 1700
P PP PG G G G PPPP G G+ P+ PFG+
Sbjct: 711 PPPPFPGAPGIPPPPPGMGVPPPPPFGFGVPAAPVLPFGL 750
Score = 34.7 bits (78), Expect = 2.3
Identities = 14/33 (42%), Positives = 18/33 (54%), Gaps = 5/33 (15%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
P PP+PG+ PPPP+ G +PP PP
Sbjct: 637 PPPPLPGVAS-----IPPPPPLPGATAIPPPPP 664
Score = 33.9 bits (76), Expect = 4.0
Identities = 17/40 (42%), Positives = 21/40 (52%), Gaps = 6/40 (15%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGM-PPMPPFGMPMG 1703
P PP+PG PPPP+ GG G+ PP PP +G
Sbjct: 661 PPPPLPG-----ATAIPPPPPLPGGTGIPPPPPPLPGSVG 695
>FMN2_MOUSE (Q9JL04) Formin 2
Length = 1567
Score = 42.4 bits (98), Expect = 0.011
Identities = 23/45 (51%), Positives = 26/45 (57%), Gaps = 10/45 (22%)
Query: 1660 LPLALPAPPMPGMG-------GGMGGGYAPPPPMGGGMGMPPMPP 1697
+P PAPP+PGMG GMG PPPP GMG+PP PP
Sbjct: 907 VPPPPPAPPLPGMGIPPPPPLPGMG---IPPPPPLPGMGIPPPPP 948
Score = 39.3 bits (90), Expect = 0.094
Identities = 19/37 (51%), Positives = 21/37 (56%)
Query: 1661 PLALPAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
P LPAPP P G+G PP P GMG+PP PP
Sbjct: 890 PEMLPAPPQPPPLPGLGVPPPPPAPPLPGMGIPPPPP 926
Score = 38.5 bits (88), Expect = 0.16
Identities = 21/50 (42%), Positives = 26/50 (52%), Gaps = 13/50 (26%)
Query: 1658 QLLPLALPAPPMPGMG----------GGMGGGYAPPPPMGGGMGMPPMPP 1697
++LP PP+PG+G GMG PPPP GMG+PP PP
Sbjct: 891 EMLPAPPQPPPLPGLGVPPPPPAPPLPGMG---IPPPPPLPGMGIPPPPP 937
Score = 38.1 bits (87), Expect = 0.21
Identities = 17/33 (51%), Positives = 20/33 (60%), Gaps = 7/33 (21%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
P PP+PG+G PPPP GMG+PP PP
Sbjct: 1011 PPPPLPGVG-------IPPPPPLPGMGIPPPPP 1036
Score = 37.7 bits (86), Expect = 0.27
Identities = 18/37 (48%), Positives = 22/37 (58%), Gaps = 4/37 (10%)
Query: 1665 PAPPMPGMG----GGMGGGYAPPPPMGGGMGMPPMPP 1697
P PP+PGMG + G PPPP G+G+PP PP
Sbjct: 934 PPPPLPGMGIPPPPPLPGVGIPPPPPLPGVGIPPPPP 970
Score = 37.0 bits (84), Expect = 0.47
Identities = 17/37 (45%), Positives = 21/37 (55%), Gaps = 4/37 (10%)
Query: 1665 PAPPMPGMG----GGMGGGYAPPPPMGGGMGMPPMPP 1697
P PP+PGMG + G PPPP G+ +PP PP
Sbjct: 1022 PPPPLPGMGIPPPPPLPGSGIPPPPALPGVAIPPPPP 1058
Score = 36.6 bits (83), Expect = 0.61
Identities = 16/33 (48%), Positives = 20/33 (60%), Gaps = 7/33 (21%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
P PP+PG+G PPPP G+G+PP PP
Sbjct: 967 PPPPLPGVG-------IPPPPPLPGVGIPPPPP 992
Score = 36.6 bits (83), Expect = 0.61
Identities = 16/33 (48%), Positives = 20/33 (60%), Gaps = 7/33 (21%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
P PP+PG+G PPPP G+G+PP PP
Sbjct: 989 PPPPLPGVG-------IPPPPPLPGVGIPPPPP 1014
Score = 36.6 bits (83), Expect = 0.61
Identities = 16/33 (48%), Positives = 20/33 (60%), Gaps = 7/33 (21%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
P PP+PG+G PPPP G+G+PP PP
Sbjct: 956 PPPPLPGVG-------IPPPPPLPGVGIPPPPP 981
Score = 36.6 bits (83), Expect = 0.61
Identities = 16/33 (48%), Positives = 20/33 (60%), Gaps = 7/33 (21%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
P PP+PG+G PPPP G+G+PP PP
Sbjct: 1000 PPPPLPGVG-------IPPPPPLPGVGIPPPPP 1025
Score = 36.6 bits (83), Expect = 0.61
Identities = 16/33 (48%), Positives = 20/33 (60%), Gaps = 7/33 (21%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPP 1697
P PP+PG+G PPPP G+G+PP PP
Sbjct: 978 PPPPLPGVG-------IPPPPPLPGVGIPPPPP 1003
Score = 35.0 bits (79), Expect = 1.8
Identities = 17/37 (45%), Positives = 20/37 (53%), Gaps = 4/37 (10%)
Query: 1665 PAPPMPGMG----GGMGGGYAPPPPMGGGMGMPPMPP 1697
P PP+PG G + G PPPP GMG+PP P
Sbjct: 1033 PPPPLPGSGIPPPPALPGVAIPPPPPLPGMGVPPPAP 1069
Score = 32.7 bits (73), Expect = 8.8
Identities = 18/37 (48%), Positives = 21/37 (56%), Gaps = 4/37 (10%)
Query: 1665 PAPPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMP 1701
P PP+PGMG APPPP G G+ PP+ P P
Sbjct: 1055 PPPPLPGMGVPPP---APPPP-GAGIPPPPLLPGSGP 1087
>SMR1_MOUSE (Q61900) Submaxillary gland androgen regulated protein 1
precursor (Salivary protein MSG1)
Length = 147
Score = 41.2 bits (95), Expect = 0.025
Identities = 19/37 (51%), Positives = 23/37 (61%), Gaps = 2/37 (5%)
Query: 1667 PPMPGMGGGMGGGYAPPPPMGGGMGMPPMPPFGMPMG 1703
PP P G G+G + PPP G G+G PP PPFG +G
Sbjct: 37 PPPPPHGPGIGRPH--PPPFGPGIGRPPPPPFGPGIG 71
Score = 38.9 bits (89), Expect = 0.12
Identities = 21/44 (47%), Positives = 23/44 (51%), Gaps = 7/44 (15%)
Query: 1661 PLALPAPPM-PGMGG------GMGGGYAPPPPMGGGMGMPPMPP 1697
P P PP PG+G G G G PPPP G G+G PP PP
Sbjct: 34 PFPPPPPPHGPGIGRPHPPPFGPGIGRPPPPPFGPGIGRPPPPP 77
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.137 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 196,768,431
Number of Sequences: 164201
Number of extensions: 8574490
Number of successful extensions: 30364
Number of sequences better than 10.0: 143
Number of HSP's better than 10.0 without gapping: 42
Number of HSP's successfully gapped in prelim test: 106
Number of HSP's that attempted gapping in prelim test: 29087
Number of HSP's gapped (non-prelim): 738
length of query: 1705
length of database: 59,974,054
effective HSP length: 124
effective length of query: 1581
effective length of database: 39,613,130
effective search space: 62628358530
effective search space used: 62628358530
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 73 (32.7 bits)
Medicago: description of AC142498.7


