

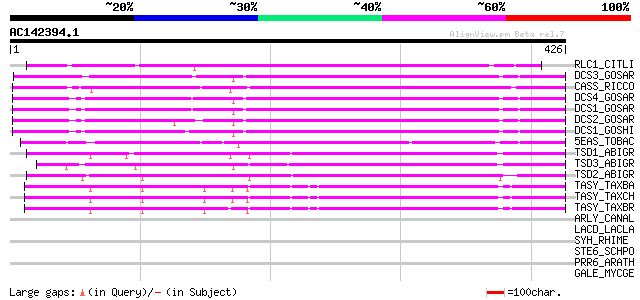
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142394.1 - phase: 0 /pseudo
(426 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RLC1_CITLI (Q8L5K3) (+)-limonene synthase 1 (EC 4.2.3.20) ((R)-l... 213 1e-54
DCS3_GOSAR (Q43714) (+)-delta-cadinene synthase isozyme A (EC 4.... 203 8e-52
CASS_RICCO (P59287) Casbene synthase, chloroplast precursor (EC ... 191 4e-48
DCS4_GOSAR (O49853) (+)-delta-cadinene synthase isozyme C2 (EC 4... 183 6e-46
DCS1_GOSAR (Q39761) (+)-delta-cadinene synthase isozyme XC1 (EC ... 182 1e-45
DCS2_GOSAR (Q39760) (+)-delta-cadinene synthase isozyme XC14 (EC... 181 3e-45
DCS1_GOSHI (P93665) (+)-delta-cadinene synthase (EC 4.2.3.13) (D... 179 2e-44
5EAS_TOBAC (Q40577) Aristolchene synthase (EC 4.2.3.9) (5-epi-ar... 172 1e-42
TSD1_ABIGR (O24475) Pinene synthase, chloroplast precursor (EC 4... 165 2e-40
TSD3_ABIGR (O22340) (4S)-limonene synthase, chloroplast precurso... 159 1e-38
TSD2_ABIGR (O24474) Myrcene synthase, chloroplast precursor (EC ... 159 1e-38
TASY_TAXBA (Q93YA3) Taxadiene synthase (EC 4.2.3.17) (Taxa-4(5),... 135 2e-31
TASY_TAXCH (Q9FT37) Taxadiene synthase (EC 4.2.3.17) (Taxa-4(5),... 134 6e-31
TASY_TAXBR (Q41594) Taxadiene synthase (EC 4.2.3.17) (Taxa-4(5),... 132 2e-30
ARLY_CANAL (P43061) Argininosuccinate lyase (EC 4.3.2.1) (Argino... 35 0.48
LACD_LACLA (P26593) Tagatose 1,6-diphosphate aldolase (EC 4.1.2.... 32 2.4
SYH_RHIME (Q92RR7) Histidyl-tRNA synthetase (EC 6.1.1.21) (Histi... 32 4.1
STE6_SCHPO (P26674) Ste6 protein 31 5.3
PRR6_ARATH (Q9C9F6) Putative two-component response regulator-li... 31 5.3
GALE_MYCGE (P47364) UDP-glucose 4-epimerase (EC 5.1.3.2) (Galact... 31 5.3
>RLC1_CITLI (Q8L5K3) (+)-limonene synthase 1 (EC 4.2.3.20)
((R)-limonene synthase 1)
Length = 606
Score = 213 bits (541), Expect = 1e-54
Identities = 138/400 (34%), Positives = 222/400 (55%), Gaps = 15/400 (3%)
Query: 14 DPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALAFRLLR 73
+P + LID +QRLG+ H F EI+ L I ++N LY +L FRLLR
Sbjct: 100 EPLDQLELIDNLQRLGLAHRFETEIRNILNN---IYNNNKDYNWRKENLYATSLEFRLLR 156
Query: 74 QGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLG-YLSCEL 132
Q GY V+ E+F+ K + +D KG+++L+EAS S+EGE + + + S L
Sbjct: 157 QHGYPVSQEVFNGFKDDQGGFIC---DDFKGILSLHEASYYSLEGESIMEEAWQFTSKHL 213
Query: 133 LQAWLPRH--QDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWICLDELAKMN 190
+ + ++ +D A L+ P+H+ + + + ++K L ELAKM
Sbjct: 214 KEVMISKNMEEDVFVAEQAKRALELPLHWKVPMLEARWFIHIYERREDKNHLLLELAKME 273
Query: 191 SSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELTK 250
+ ++ + + E E+ WW+D GL +++ FA + ++W M +P F+ R LT
Sbjct: 274 FNTLQAIYQEELKEISGWWKDTGLGEKLSFARNRLVASFLWSMGIAFEPQFAYCRRVLTI 333
Query: 251 PISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGA-ENLPNFMKVSLSSLYKVTNNFAE 309
I+L+ +IDDI+DV+GTLD+L +FT+AV RW+++ A ++LP +MK+ +LY N FA
Sbjct: 334 SIALITVIDDIYDVYGTLDELEIFTDAVERWDINYALKHLPGYMKMCFLALYNFVNEFAY 393
Query: 310 MVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVS-TGVHVVLIH 368
V K+ F+ + ++K +W+ L+ A+L EA W +S P EEYL NG+VS TG ++ I
Sbjct: 394 YVLKQQDFDLLLSIKNAWLGLIQAYLVEAKWYHSKYTPKLEEYLENGLVSITGPLIITIS 453
Query: 369 AFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDL 408
+ I K+ L L E P+I++ +KI RL DDL
Sbjct: 454 ---YLSGTNPIIKKELEFL-ESNPDIVHWSSKIFRLQDDL 489
>DCS3_GOSAR (Q43714) (+)-delta-cadinene synthase isozyme A (EC
4.2.3.13) (D-cadinene synthase)
Length = 555
Score = 203 bits (516), Expect = 8e-52
Identities = 129/427 (30%), Positives = 225/427 (52%), Gaps = 15/427 (3%)
Query: 4 QVHKKLVTS-EDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHEL 62
QV K ++ +D + ID +QRLG+ ++F +EI+ LE + ++N D +L
Sbjct: 57 QVRKMIMEPVDDSNQKLPFIDAVQRLGVSYHFEKEIEDELENIYRDTNNNDADT----DL 112
Query: 63 YEVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGL 122
Y AL FRLLR+ G+ ++ + F+ K + + DV+GL+ LYEAS + + GED L
Sbjct: 113 YTTALRFRLLREHGFDISCDAFNKFKDEAGNFKASLTSDVQGLLELYEASYMRVHGEDIL 172
Query: 123 ND-LGYLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIF--INDLKAKNK 179
++ + + + +L A H H + V + L+ I GL R ++ DL++ NK
Sbjct: 173 DEAISFTTAQLTLALPTLH--HPLSEQVGHALKQSIRRGLPRVEARNFISIYQDLESHNK 230
Query: 180 WICLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDP 239
L + AK++ ++++ +++ E E+ +WW+DL +++ FA ++ Y W M + +P
Sbjct: 231 --SLLQFAKIDFNLLQLLHRKELSEICRWWKDLDFTRKLPFARDRVVEGYFWIMGVYFEP 288
Query: 240 CFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSS 299
+S R LTK I++ I+DD +D + T D+L +T A+ RW++ LPN+MK+S +
Sbjct: 289 QYSLGRKMLTKVIAMASIVDDTYDSYATYDELIPYTNAIERWDIKCMNQLPNYMKISYKA 348
Query: 300 LYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVS 359
L V +++ + ++ K + +RL+ A+L EA W + PT EE+ +N + +
Sbjct: 349 LLNVYEEMEQLLANQGRQYRVEYAKKAMIRLVQAYLLEAKWTHQNYKPTFEEFRDNALPT 408
Query: 360 TGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGL 419
+G ++ I AF M IT ET + P II + I R DD+ K +
Sbjct: 409 SGYAMLAITAFVGMGEV--ITPETFKWAASD-PKIIKASTIICRFMDDIAEHKFNHRRED 465
Query: 420 DGSYLNC 426
D S + C
Sbjct: 466 DCSAIEC 472
>CASS_RICCO (P59287) Casbene synthase, chloroplast precursor (EC
4.2.3.8)
Length = 601
Score = 191 bits (484), Expect = 4e-48
Identities = 120/430 (27%), Positives = 228/430 (52%), Gaps = 16/430 (3%)
Query: 3 KQVHKKLVTS-EDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHE 61
K+V L++S D E LID++ RLG+ ++F +I+ L K I +S P D V E
Sbjct: 100 KKVKDILISSTSDSVETVILIDLLCRLGVSYHFENDIEELLSK---IFNSQP-DLVDEKE 155
Query: 62 --LYEVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGE 119
LY A+ FR+ RQ G+ +++++F K S + D KG+++L+EAS LS+ GE
Sbjct: 156 CDLYTAAIVFRVFRQHGFKMSSDVFSKFKDSDGKFKESLRGDAKGMLSLFEASHLSVHGE 215
Query: 120 DGLNDLGYLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKS---IFINDLKA 176
D L + + + LQ+ +++ +++N L+ P H G+ R + ++ D++
Sbjct: 216 DILEEAFAFTKDYLQSSAVELFPNLKR-HITNALEQPFHSGVPRLEARKFIDLYEADIEC 274
Query: 177 KNKWICLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACF 236
+N+ + E AK++ + V+ +++ E + KWW+DL LA ++ +A + + W +A +
Sbjct: 275 RNETLL--EFAKLDYNRVQLLHQQELCQFSKWWKDLNLASDIPYARDRMAEIFFWAVAMY 332
Query: 237 TDPCFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVS 296
+P +++ R+ + K + L+ +IDD D + T+++ + EAV RW+M E LP++MKV
Sbjct: 333 FEPDYAHTRMIIAKVVLLISLIDDTIDAYATMEETHILAEAVARWDMSCLEKLPDYMKVI 392
Query: 297 LSSLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNG 356
L + F + + + + + ++ L+ + EA W + G +P+ ++YL NG
Sbjct: 393 YKLLLNTFSEFEKELTAEGKSYSVKYGREAFQELVRGYYLEAVWRDEGKIPSFDDYLYNG 452
Query: 357 IVSTGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQ 416
++TG+ +V +F + G+ + E P + Y+ +RL +DL + Q
Sbjct: 453 SMTTGLPLVSTASFMGVQEITGLNEFQWL---ETNPKLSYASGAFIRLVNDLTSHVTEQQ 509
Query: 417 NGLDGSYLNC 426
G S ++C
Sbjct: 510 RGHVASCIDC 519
>DCS4_GOSAR (O49853) (+)-delta-cadinene synthase isozyme C2 (EC
4.2.3.13) (D-cadinene synthase)
Length = 554
Score = 183 bits (465), Expect = 6e-46
Identities = 124/427 (29%), Positives = 220/427 (51%), Gaps = 15/427 (3%)
Query: 3 KQVHKKLVTS-EDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHE 61
++V K +V + T+ ID +QRLG+ ++F +EI+ LE + N D + ++
Sbjct: 57 EEVRKMIVAPMANSTQKLAFIDSVQRLGVSYHFTKEIEDELENIY----HNNND--AEND 110
Query: 62 LYEVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDG 121
LY +L FRLLR+ GY V+ ++F+ K + + + DV+GL+ LY+AS L + GED
Sbjct: 111 LYTTSLRFRLLREHGYNVSCDVFNKFKDEQGNFKSSVTSDVQGLLELYQASYLRVHGEDI 170
Query: 122 LNDLGYLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIF--INDLKAKNK 179
L++ + L + DH + VS+ L+ I GL R + D+++ NK
Sbjct: 171 LDEAISFTTNHLSLAVSS-LDHPLSEEVSHALKQSIRRGLPRVEARHYLSVYQDIESHNK 229
Query: 180 WICLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDP 239
L E AK++ ++++F+++ E E+ +WW+DL +++ +A ++ Y W + +P
Sbjct: 230 --ALLEFAKIDFNMLQFLHRKELSEICRWWKDLDFQRKLPYARDRVVEGYFWISGVYFEP 287
Query: 240 CFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSS 299
+S R LTK I++ I+DD +D + T ++L +T A+ RW++ + LP +MK S +
Sbjct: 288 QYSLGRKMLTKVIAMASIVDDTYDSYATYEELIPYTNAIERWDIKCIDELPEYMKPSYKA 347
Query: 300 LYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVS 359
L V ++V + ++ K + +RL ++L EA W P+ EE+ N + +
Sbjct: 348 LLDVYKEMEQLVAEHGRQYRVEYAKNAMIRLAQSYLVEARWTLQNYKPSFEEFKANALPT 407
Query: 360 TGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGL 419
G ++ I +F M +T ET + P II + I R DD+ K +
Sbjct: 408 CGYAMLAITSFVGMGDI--VTPETFKWAAND-PKIIQASTIICRFMDDVAEHKFKHRRED 464
Query: 420 DGSYLNC 426
D S + C
Sbjct: 465 DCSAIEC 471
>DCS1_GOSAR (Q39761) (+)-delta-cadinene synthase isozyme XC1 (EC
4.2.3.13) (D-cadinene synthase)
Length = 554
Score = 182 bits (462), Expect = 1e-45
Identities = 122/427 (28%), Positives = 220/427 (50%), Gaps = 15/427 (3%)
Query: 3 KQVHKKLVTS-EDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHE 61
++V K +V + T+ ID +QRLG+ ++F +EI+ LE + N D + ++
Sbjct: 57 EEVRKMIVAPMANSTQKLAFIDSVQRLGVSYHFTKEIEDELENIY----HNNND--AEND 110
Query: 62 LYEVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDG 121
LY ++ FRLLR+ GY V+ ++F+ K + + + DV+GL+ LY+AS L + GED
Sbjct: 111 LYTTSIRFRLLREHGYNVSCDVFNKFKDEQGNFKSSVTSDVRGLLELYQASYLRVHGEDI 170
Query: 122 LNDLGYLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIF--INDLKAKNK 179
L++ + L + DH + VS+ L+ I GL R + D+++ NK
Sbjct: 171 LDEAISFTTHHLSLAVAS-LDHPLSEEVSHALKQSIRRGLPRVEARHYLSVYQDIESHNK 229
Query: 180 WICLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDP 239
L E AK++ ++++F+++ E E+ +WW+DL +++ +A ++ Y W + +P
Sbjct: 230 --ALLEFAKIDFNMLQFLHRKELSEICRWWKDLDFQRKLPYARDRVVEGYFWISGVYFEP 287
Query: 240 CFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSS 299
+S R LTK I++ I+DD +D + T ++L +T A+ RW++ + +P +MK S +
Sbjct: 288 QYSLGRKMLTKVIAMASIVDDTYDSYATYEELIPYTNAIERWDIKCIDEIPEYMKPSYKA 347
Query: 300 LYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVS 359
L V ++V + ++ K + +RL ++L EA W P+ EE+ N + +
Sbjct: 348 LLDVYEEMVQLVAEHGRQYRVEYAKNAMIRLAQSYLVEAKWTLQNYKPSFEEFKANALPT 407
Query: 360 TGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGL 419
G ++ I +F M +T ET + P II + I R DD+ K +
Sbjct: 408 CGYAMLAITSFVGMGDI--VTPETFKWAASD-PKIIQASTIICRFMDDVAEHKFKHRRED 464
Query: 420 DGSYLNC 426
D S + C
Sbjct: 465 DCSAIEC 471
>DCS2_GOSAR (Q39760) (+)-delta-cadinene synthase isozyme XC14 (EC
4.2.3.13) (D-cadinene synthase)
Length = 554
Score = 181 bits (459), Expect = 3e-45
Identities = 124/432 (28%), Positives = 221/432 (50%), Gaps = 25/432 (5%)
Query: 3 KQVHKKLVTS-EDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHE 61
++V K +V + T+ ID +QRLG+ ++F +EI+ LE + N D + ++
Sbjct: 57 EEVRKMIVAPMANSTQKLAFIDSVQRLGVSYHFTKEIEDELENIY----HNNND--AEND 110
Query: 62 LYEVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDG 121
LY +L FRLLR+ G+ V+ ++F+ K + + + DV+GL+ LY+AS L + GED
Sbjct: 111 LYTTSLRFRLLREHGFNVSCDVFNKFKDEQGNFKSSVTSDVRGLLELYQASYLRVHGEDI 170
Query: 122 LNDL-----GYLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIF--INDL 174
L++ +LS + P ++ VS+ L+ I GL R + D+
Sbjct: 171 LDEAISFTTNHLSLAVASLDYPLSEE------VSHALKQSIRRGLPRVEARHYLSVYQDI 224
Query: 175 KAKNKWICLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMA 234
++ NK L E AK++ ++V+ +++ E E+ +WW+DL +++ +A ++ Y W
Sbjct: 225 ESHNK--VLLEFAKIDFNMVQLLHRKELSEISRWWKDLDFQRKLPYARDRVVEGYFWISG 282
Query: 235 CFTDPCFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMK 294
+ +P +S R LTK I++ I+DD +D + T ++L +T+A+ RW++ + LP +MK
Sbjct: 283 VYFEPQYSLGRKMLTKVIAMASIVDDTYDSYATYEELIPYTKAIERWDIKCIDELPEYMK 342
Query: 295 VSLSSLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLN 354
S +L V ++V K ++ K + +RL ++L EA W P+ EE+
Sbjct: 343 PSYKALLDVYEEMEQLVAKHGRQYRVEYAKNAMIRLAQSYLVEARWTLQNYKPSFEEFKA 402
Query: 355 NGIVSTGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSG 414
N + + G ++ I +F M +T ET + P II + I R DD+ K
Sbjct: 403 NALPTCGYAMLAITSFVGMGDI--VTPETFKWAAND-PKIIQASTIICRFMDDVAEHKFK 459
Query: 415 DQNGLDGSYLNC 426
+ D S + C
Sbjct: 460 HRREDDCSAIEC 471
>DCS1_GOSHI (P93665) (+)-delta-cadinene synthase (EC 4.2.3.13)
(D-cadinene synthase)
Length = 554
Score = 179 bits (453), Expect = 2e-44
Identities = 124/428 (28%), Positives = 220/428 (50%), Gaps = 17/428 (3%)
Query: 3 KQVHKKLVTS-EDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHE 61
++V K +V + T ID +Q LG+ ++F +EI+ LE + N D + ++
Sbjct: 57 EEVRKMIVAPMANSTLKLAFIDSVQGLGVSYHFTKEIEDELENIY----HNNND--AEND 110
Query: 62 LYEVALAFRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDG 121
LY +L FRLLR+ G++V+ ++F+ K + + + DV+GL+ LY+AS L + GED
Sbjct: 111 LYTTSLRFRLLREHGFHVSCDVFNKFKDEQGNFKSSVTSDVRGLLELYQASYLRVHGEDI 170
Query: 122 LND-LGYLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIF--INDLKAKN 178
L++ + + S L A DH + VS+ L+ I GL R + D+++ N
Sbjct: 171 LDEAISFTSNHLSLA--VASLDHPLSEEVSHALKQSIRRGLPRVEARHYLSVYQDIESHN 228
Query: 179 KWICLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTD 238
K L E AK++ ++V+ +++ E E+ +WW+DL +++ +A ++ Y W + +
Sbjct: 229 K--VLLEFAKIDFNMVQLLHRKELSEISRWWKDLDFQRKLPYARDRVVEGYFWISGVYFE 286
Query: 239 PCFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLS 298
P +S R LTK I++ I+DD +D + T ++L +T A+ RW++ + LP +MK S
Sbjct: 287 PQYSLGRKMLTKVIAMASIVDDTYDSYATYEELIPYTNAIERWDIKCIDELPEYMKPSYK 346
Query: 299 SLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIV 358
+L V ++V + ++ K + +RL ++L EA W P+ EE+ N +
Sbjct: 347 ALLDVYEEMEQLVAEHGRQYRVEYAKNAMIRLAQSYLVEARWTLQNYKPSFEEFKANALP 406
Query: 359 STGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNG 418
+ G ++ I +F M +T ET + P II + I R DD+ K +
Sbjct: 407 TCGYAMLAITSFVGMGDI--VTPETFKWAAND-PKIIQASTIICRFMDDVTEHKFKHRRE 463
Query: 419 LDGSYLNC 426
D S + C
Sbjct: 464 DDCSAIEC 471
>5EAS_TOBAC (Q40577) Aristolchene synthase (EC 4.2.3.9)
(5-epi-aristolochene synthase) (EAS)
Length = 548
Score = 172 bits (436), Expect = 1e-42
Identities = 117/421 (27%), Positives = 214/421 (50%), Gaps = 17/421 (4%)
Query: 9 LVTSEDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSHELYEVALA 68
L T + +LID I+RLGI ++F +EI L+ Q +SN D +S AL
Sbjct: 58 LATGMKLADTLNLIDTIERLGISYHFEKEIDDILD-QIYNQNSNCNDLCTS------ALQ 110
Query: 69 FRLLRQGGYYVNAELFDCLKCSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLNDLGYL 128
FRLLRQ G+ ++ E+F + + DV GL+ LYEAS + +D L D
Sbjct: 111 FRLLRQHGFNISPEIFSKFQDENGKFKESLASDVLGLLNLYEASHVRTHADDILEDALAF 170
Query: 129 SCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDL--KAKNKWICLDEL 186
S L++ P + ++ V++ L+ +H G+ R ++ FI+ + K ++K L
Sbjct: 171 STIHLESAAPHLKSPLRE-QVTHALEQCLHKGVPR-VETRFFISSIYDKEQSKNNVLLRF 228
Query: 187 AKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERV 246
AK++ ++++ ++K E +V +WW+DL + +A ++ Y W + + +P +S RV
Sbjct: 229 AKLDFNLLQMLHKQELAQVSRWWKDLDFVTTLPYARDRVVECYFWALGVYFEPQYSQARV 288
Query: 247 ELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSLYKVTNN 306
L K IS++ I+DD FD +GT+ +L +T+A+ RW+++ + LP++MK+S ++ + +
Sbjct: 289 MLVKTISMISIVDDTFDAYGTVKELEAYTDAIQRWDINEIDRLPDYMKISYKAILDLYKD 348
Query: 307 FAEMVYKKHGFNPIDTLKISWVR-LLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVV 365
+ E G + I I ++ ++ + E+ W G P EYL+N + +T + +
Sbjct: 349 Y-EKELSSAGRSHIVCHAIERMKEVVRNYNVESTWFIEGYTPPVSEYLSNALATTTYYYL 407
Query: 366 LIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDLEGVKSGDQNGLDGSYLN 425
++ M K T++ L + P I+ + I R+ DD + G + +
Sbjct: 408 ATTSYLGM---KSATEQDFEWLSKN-PKILEASVIICRVIDDTATYEVEKSRGQIATGIE 463
Query: 426 C 426
C
Sbjct: 464 C 464
>TSD1_ABIGR (O24475) Pinene synthase, chloroplast precursor (EC
4.2.3.14) (Beta-geraniolene synthase)
((-)-(1S,5S)-pinene synthase)
Length = 628
Score = 165 bits (418), Expect = 2e-40
Identities = 113/430 (26%), Positives = 208/430 (48%), Gaps = 25/430 (5%)
Query: 14 DPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSH---ELYEVALAFR 70
D + ++D ++RLGI +F +EIK AL+ + N I +L AL R
Sbjct: 122 DLIQRLWIVDSLERLGIHRHFKDEIKSALDYVYSYWGENGIGCGRESVVTDLNSTALGLR 181
Query: 71 LLRQGGYYVNAELFDCLK-------CSKKSLRVKYGEDVKGLIALYEASQLSIEGEDGLN 123
LR GY V++++F K CS+ ++ E+++G++ L+ AS ++ GE ++
Sbjct: 182 TLRLHGYPVSSDVFKAFKGQNGQFSCSEN---IQTDEEIRGVLNLFRASLIAFPGEKIMD 238
Query: 124 DLGYLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKS---IFINDLKAKNKW 180
+ S + L+ L + + + + L+Y H L R ++ +F D + +
Sbjct: 239 EAEIFSTKYLKEALQKIPVSSLSREIGDVLEYGWHTYLPRLEARNYIQVFGQDTENTKSY 298
Query: 181 IC---LDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFT 237
+ L ELAK+ +I + + K E + +WW++ G EM F + +++Y
Sbjct: 299 VKSKKLLELAKLEFNIFQSLQKRELESLVRWWKESGFP-EMTFCRHRHVEYYTLASCIAF 357
Query: 238 DPCFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSL 297
+P S R+ K L+ ++DD++D GT+D+L LFT + RW+ + LP +MK
Sbjct: 358 EPQHSGFRLGFAKTCHLITVLDDMYDTFGTVDELELFTATMKRWDPSSIDCLPEYMKGVY 417
Query: 298 SSLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGI 357
++Y N A + G + + + +W ++++++EA W+ +G LP+ +EY NG
Sbjct: 418 IAVYDTVNEMAREAEEAQGRDTLTYAREAWEAYIDSYMQEARWIATGYLPSFDEYYENGK 477
Query: 358 VSTGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVA-KILRLSDDLEGVKSGDQ 416
VS G + + MD I + + +FP+ + +A ILRL D K+
Sbjct: 478 VSCGHRISALQPILTMD----IPFPDHILKEVDFPSKLNDLACAILRLRGDTRCYKADRA 533
Query: 417 NGLDGSYLNC 426
G + S ++C
Sbjct: 534 RGEEASSISC 543
>TSD3_ABIGR (O22340) (4S)-limonene synthase, chloroplast precursor
(EC 4.2.3.16) ((-)-(4S)-limonene synthase)
Length = 637
Score = 159 bits (403), Expect = 1e-38
Identities = 114/429 (26%), Positives = 203/429 (46%), Gaps = 33/429 (7%)
Query: 21 LIDIIQRLGIEHYFVEEIKVAL--------EKQFLILSSNPIDFVSSHELYEVALAFRLL 72
++D I+RLGI+ +F EI+VAL EK+ + + + +L ALA R L
Sbjct: 134 MVDSIERLGIDRHFQNEIRVALDYVYSYWKEKEGIGCGRDS----TFPDLNSTALALRTL 189
Query: 73 RQGGYYVNAELFDCLKCSKKSLR----VKYGEDVKGLIALYEASQLSIEGEDGLNDLGYL 128
R GY V++++ + K K + G+ + ++ LY AS ++ GE + +
Sbjct: 190 RLHGYNVSSDVLEYFKDEKGHFACPAILTEGQITRSVLNLYRASLVAFPGEKVMEEAEIF 249
Query: 129 SCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIF----------INDLKAKN 178
S L+ L + + + L+Y H L R ++ +N++ N
Sbjct: 250 SASYLKKVLQKIPVSNLSGEIEYVLEYGWHTNLPRLEARNYIEVYEQSGYESLNEMPYMN 309
Query: 179 KWICLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTD 238
L +LAK+ +I + E + +WW++ G + ++ F + +++Y
Sbjct: 310 MKKLL-QLAKLEFNIFHSLQLRELQSISRWWKESG-SSQLTFTRHRHVEYYTMASCISML 367
Query: 239 PCFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLS 298
P S R+E K LV ++DDI+D GT+++L LFT+A+ RW++ LP +MK
Sbjct: 368 PKHSAFRMEFVKVCHLVTVLDDIYDTFGTMNELQLFTDAIKRWDLSTTRWLPEYMKGVYM 427
Query: 299 SLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIV 358
LY+ N E K G + ++ ++ +W L + F++EA W++S LPT EEYL N V
Sbjct: 428 DLYQCINEMVEEAEKTQGRDMLNYIQNAWEALFDTFMQEAKWISSSYLPTFEEYLKNAKV 487
Query: 359 STGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVA-KILRLSDDLEGVKSGDQN 417
S+G + + +D + + + ++P+ +A ILRL D K+
Sbjct: 488 SSGSRIATLQPILTLD----VPLPDYILQEIDYPSRFNELASSILRLRGDTRCYKADRAR 543
Query: 418 GLDGSYLNC 426
G + S ++C
Sbjct: 544 GEEASAISC 552
>TSD2_ABIGR (O24474) Myrcene synthase, chloroplast precursor (EC
4.2.3.15)
Length = 627
Score = 159 bits (403), Expect = 1e-38
Identities = 115/433 (26%), Positives = 197/433 (44%), Gaps = 32/433 (7%)
Query: 14 DPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPI----DFVSSHELYEVALAF 69
D + ++D ++RLGI +F EI AL+ F N I D + + +L AL F
Sbjct: 122 DLMQRLWIVDSVERLGIARHFKNEITSALDYVFRYWEENGIGCGRDSIVT-DLNSTALGF 180
Query: 70 RLLRQGGYYVNAELFDCLKCSKKSLRVKYGE---DVKGLIALYEASQLSIEGEDGLNDLG 126
R LR GY V+ E+ + G+ +++ ++ LY AS ++ GE + +
Sbjct: 181 RTLRLHGYTVSPEVLKAFQDQNGQFVCSPGQTEGEIRSVLNLYRASLIAFPGEKVMEEAE 240
Query: 127 YLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWIC---- 182
S L+ L + + + ++Y H L R ++ K + W+
Sbjct: 241 IFSTRYLKEALQKIPVSALSQEIKFVMEYGWHTNLPRLEARNYIDTLEKDTSAWLNKNAG 300
Query: 183 --LDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPC 240
L ELAK+ +I + + E + +WW++ L K + FA + +++Y DP
Sbjct: 301 KKLLELAKLEFNIFNSLQQKELQYLLRWWKESDLPK-LTFARHRHVEFYTLASCIAIDPK 359
Query: 241 FSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSSL 300
S R+ K LV ++DDI+D GT+D+L LFT A+ RW E+LP +MK +
Sbjct: 360 HSAFRLGFAKMCHLVTVLDDIYDTFGTIDELELFTSAIKRWNSSEIEHLPEYMKCVYMVV 419
Query: 301 YKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVST 360
++ N K G N ++ ++ +W +++++EA W+++G LP EEY NG VS+
Sbjct: 420 FETVNELTREAEKTQGRNTLNYVRKAWEAYFDSYMEEAKWISNGYLPMFEEYHENGKVSS 479
Query: 361 GVHVVLIHAFFLMDH------AKGITKETLSILDEEFPNIIYSVA-KILRLSDDLEGVKS 413
V + ++ KGI +FP+ +A LRL D K+
Sbjct: 480 AYRVATLQPILTLNAWLPDYILKGI----------DFPSRFNDLASSFLRLRGDTRCYKA 529
Query: 414 GDQNGLDGSYLNC 426
G + S ++C
Sbjct: 530 DRDRGEEASCISC 542
>TASY_TAXBA (Q93YA3) Taxadiene synthase (EC 4.2.3.17)
(Taxa-4(5),11(12)-diene synthase)
Length = 862
Score = 135 bits (340), Expect = 2e-31
Identities = 111/437 (25%), Positives = 203/437 (46%), Gaps = 30/437 (6%)
Query: 12 SEDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSH---ELYEVALA 68
S D E L+D I+ LGI +F +EIKVAL+ + S I + +L AL
Sbjct: 349 SIDLLERLSLVDNIEHLGIGRHFKQEIKVALDYVYRHWSERGIGWGRDSLVPDLNTTALG 408
Query: 69 FRLLRQGGYYVNAELFDCLKCSKKSLRVKYGE---DVKGLIALYEASQLSIEGEDGLNDL 125
R LR GY V++++ + K G+ +++ ++ L+ AS L+ E ++D
Sbjct: 409 LRTLRTHGYDVSSDVLNNFKDENGRFFSSAGQTHVELRSVVNLFRASDLAFPDEGAMDDA 468
Query: 126 GYLSCELLQAWLPRHQDHIQAIY--VSNTLQYPIHYGLSRFMDKSI---FINDLKAKNKW 180
+ L+ L +Y + ++YP H + R +S + +D + K
Sbjct: 469 RKFAEPYLRDALATKISTNTKLYKEIEYVVEYPWHMSIPRLEARSYIDSYDDDYVWQRKT 528
Query: 181 I----------CLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYM 230
+ CL ELAK++ +IV+ +++ E + +WW++ G+A ++ F + + Y
Sbjct: 529 LYRMPSLSNSKCL-ELAKLDFNIVQSLHQEELKLLTRWWKESGMA-DINFTRHRVAEVY- 585
Query: 231 WPMACFTDPCFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLP 290
+ A F +P +S R+ TK L + DD+ D+ TLD+L FTE V RW+ +P
Sbjct: 586 FSSATF-EPEYSATRIAFTKIGCLQVLFDDMADIFATLDELKSFTEGVKRWDTSLLHEIP 644
Query: 291 NFMKVSLSSLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTE 350
M+ +K+ V K G + + ++ W N +++E WL +G +PT E
Sbjct: 645 ECMQTCFKVWFKLMEEVNNDVVKVQGRDMLAHIRKPWELYFNCYVQEREWLEAGYIPTFE 704
Query: 351 EYLNNGIVSTGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKI-LRLSDDLE 409
EYL +S G+ + LM + K+ + + +P+ ++ + + RL++D +
Sbjct: 705 EYLKTYAISVGLGPCTLQPILLMGE---LVKDDV-VEKVHYPSNMFELVSLSWRLTNDTK 760
Query: 410 GVKSGDQNGLDGSYLNC 426
++ G S + C
Sbjct: 761 TYQAEKARGQQASGIAC 777
>TASY_TAXCH (Q9FT37) Taxadiene synthase (EC 4.2.3.17)
(Taxa-4(5),11(12)-diene synthase)
Length = 862
Score = 134 bits (336), Expect = 6e-31
Identities = 110/437 (25%), Positives = 204/437 (46%), Gaps = 30/437 (6%)
Query: 12 SEDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSH---ELYEVALA 68
S D E L+D I+ LGI +F +EIKVAL+ + S I + +L AL
Sbjct: 349 SIDLLERLSLVDNIEHLGIGRHFKQEIKVALDYVYRHWSERGIGWGRDSLVPDLNTTALG 408
Query: 69 FRLLRQGGYYVNAELFDCLKCSKKSLRVKYGE---DVKGLIALYEASQLSIEGEDGLNDL 125
R LR GY V++++ + K G+ +++ ++ L+ AS L+ E ++D
Sbjct: 409 LRTLRTHGYDVSSDVLNNFKDENGRFFSSAGQTHVELRSVVILFRASDLAFPDEGAMDDA 468
Query: 126 GYLSCELLQAWLPRHQDHIQAIY--VSNTLQYPIHYGLSRFMDKSI---FINDLKAKNKW 180
+ L+ L ++ + ++YP H + R +S + +D + K
Sbjct: 469 RKFAEPYLRDALATKISTNTKLFKEIEYVVEYPWHMSIPRSEARSYIDSYDDDYVWERKT 528
Query: 181 I----------CLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWYM 230
+ CL ELAK++ +IV+ +++ E + +WW++ G+A ++ F + + Y
Sbjct: 529 LYRMPSLSNSKCL-ELAKLDFNIVQSLHQEELKLLTRWWKESGMA-DINFTRHRVAEVY- 585
Query: 231 WPMACFTDPCFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENLP 290
+ A F +P +S R+ TK L + DD+ D+ TLD+L FTE V RW+ +P
Sbjct: 586 FSSATF-EPEYSATRIAFTKIGCLQVLFDDMADIFATLDELKSFTEGVKRWDTSLLHEIP 644
Query: 291 NFMKVSLSSLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTE 350
M+ +K+ V K G + + ++ W N +++E WL++G +PT E
Sbjct: 645 ECMQTCFKVWFKLIEEVNNDVVKVQGRDMLAHIRKPWELYFNCYVQEREWLDAGYIPTFE 704
Query: 351 EYLNNGIVSTGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKI-LRLSDDLE 409
EYL +S G+ + LM + K+ + + +P+ ++ + + RL++D +
Sbjct: 705 EYLKTYAISVGLGPCTLQPILLMGE---LVKDDV-VEKVHYPSNMFELVSLSWRLTNDTK 760
Query: 410 GVKSGDQNGLDGSYLNC 426
++ G S + C
Sbjct: 761 TYQAEKARGQQASGIAC 777
>TASY_TAXBR (Q41594) Taxadiene synthase (EC 4.2.3.17)
(Taxa-4(5),11(12)-diene synthase)
Length = 862
Score = 132 bits (331), Expect = 2e-30
Identities = 109/438 (24%), Positives = 200/438 (44%), Gaps = 32/438 (7%)
Query: 12 SEDPTENFHLIDIIQRLGIEHYFVEEIKVALEKQFLILSSNPIDFVSSH---ELYEVALA 68
S D E L+D I+ LGI +F +EIK AL+ + S I + +L AL
Sbjct: 349 SIDLLERLSLVDNIEHLGIGRHFKQEIKGALDYVYRHWSERGIGWGRDSLVPDLNTTALG 408
Query: 69 FRLLRQGGYYVNAELFDCLKCSKKSLRVKYGE---DVKGLIALYEASQLSIEGEDGLNDL 125
R LR GY V++++ + K G+ +++ ++ L+ AS L+ E ++D
Sbjct: 409 LRTLRMHGYNVSSDVLNNFKDENGRFFSSAGQTHVELRSVVNLFRASDLAFPDERAMDDA 468
Query: 126 GYLSCELLQAWLPRHQDHIQAIY--VSNTLQYPIHYGLSRFMDKSIFINDLKAKNKWI-- 181
+ L+ L ++ + ++YP H + R +S +I+ W
Sbjct: 469 RKFAEPYLREALATKISTNTKLFKEIEYVVEYPWHMSIPRLEARS-YIDSYDDNYVWQRK 527
Query: 182 ------------CLDELAKMNSSIVKFMNKNESVEVFKWWEDLGLAKEMKFAGYNPLKWY 229
CL ELAK++ +IV+ +++ E + +WW++ G+A ++ F + + Y
Sbjct: 528 TLYRMPSLSNSKCL-ELAKLDFNIVQSLHQEELKLLTRWWKESGMA-DINFTRHRVAEVY 585
Query: 230 MWPMACFTDPCFSNERVELTKPISLVYIIDDIFDVHGTLDQLTLFTEAVNRWEMDGAENL 289
+ A F +P +S R+ TK L + DD+ D+ TLD+L FTE V RW+ +
Sbjct: 586 -FSSATF-EPEYSATRIAFTKIGCLQVLFDDMADIFATLDELKSFTEGVKRWDTSLLHEI 643
Query: 290 PNFMKVSLSSLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTT 349
P M+ +K+ V K G + + ++ W N +++E WL +G +PT
Sbjct: 644 PECMQTCFKVWFKLMEEVNNDVVKVQGRDMLAHIRKPWELYFNCYVQEREWLEAGYIPTF 703
Query: 350 EEYLNNGIVSTGVHVVLIHAFFLMDHAKGITKETLSILDEEFPNIIYSVAKI-LRLSDDL 408
EEYL +S G+ + LM + K+ + + +P+ ++ + + RL++D
Sbjct: 704 EEYLKTYAISVGLGPCTLQPILLMGE---LVKDDV-VEKVHYPSNMFELVSLSWRLTNDT 759
Query: 409 EGVKSGDQNGLDGSYLNC 426
+ ++ G S + C
Sbjct: 760 KTYQAEKARGQQASGIAC 777
>ARLY_CANAL (P43061) Argininosuccinate lyase (EC 4.3.2.1)
(Arginosuccinase) (ASAL)
Length = 468
Score = 34.7 bits (78), Expect = 0.48
Identities = 28/101 (27%), Positives = 49/101 (47%), Gaps = 2/101 (1%)
Query: 286 AENLPNFMKVSLSSLYKVTNNFAEMVYKKHGFNPIDTLKISWVRLLNAF-LKEAHWLNSG 344
A ++ F++ SL +L K+ + F + ++ ID L + L A ++ AHWL+S
Sbjct: 125 ATDMRIFVRESLLNLSKILHQFITAILER-AHKEIDVLMPGYTHLQKAQPIRWAHWLSSY 183
Query: 345 ILPTTEEYLNNGIVSTGVHVVLIHAFFLMDHAKGITKETLS 385
TE+Y + T V+ + + L H GI +E L+
Sbjct: 184 ATYFTEDYKRLQEIITRVNQSPLGSGALAGHPYGIDREFLA 224
>LACD_LACLA (P26593) Tagatose 1,6-diphosphate aldolase (EC 4.1.2.40)
(Tagatose-bisphosphate aldolase)
(D-tagatose-1,6-bisphosphate aldolase)
Length = 326
Score = 32.3 bits (72), Expect = 2.4
Identities = 38/168 (22%), Positives = 73/168 (42%), Gaps = 21/168 (12%)
Query: 203 VEVFKWWEDLGLAKEMKFAGYNPLKWYMWPMACFTDPCFSNERVELTKPISLVYIIDDIF 262
+E+ + E++ A +++A P K + M F+DP F+ + +++ P+++ Y
Sbjct: 162 LEILAYDEEISDAGSVEYAKVKPRK-VIEAMKVFSDPRFNIDVLKVEVPVNVKY------ 214
Query: 263 DVHGTLDQLTLFTEAVNRWEMDGAENLPNFMKVSLSS-----LYKVTNNFAEMVYKKHGF 317
V G D ++++A E N + LS+ L++ T FA K F
Sbjct: 215 -VEGFADGEVVYSKAEAADFFKAQEEATNLPYIYLSAGVSAKLFQETLQFAHDSGAK--F 271
Query: 318 NPIDTLKISWVRLLNAFLKEA-----HWLNSGILPTTEEYLNNGIVST 360
N + + +W + ++KE WL + +E LN +V T
Sbjct: 272 NGVLCGRATWAGSVEPYIKEGEKAAREWLRTTGFENIDE-LNKVLVKT 318
>SYH_RHIME (Q92RR7) Histidyl-tRNA synthetase (EC 6.1.1.21)
(Histidine--tRNA ligase) (HisRS)
Length = 504
Score = 31.6 bits (70), Expect = 4.1
Identities = 12/41 (29%), Positives = 24/41 (58%)
Query: 311 VYKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEE 351
VY+++GF+P++T + L FL ++ N G+ T++
Sbjct: 40 VYERYGFDPVETPLFEYTDALGKFLPDSDRPNEGVFSLTDD 80
>STE6_SCHPO (P26674) Ste6 protein
Length = 911
Score = 31.2 bits (69), Expect = 5.3
Identities = 26/97 (26%), Positives = 40/97 (40%), Gaps = 1/97 (1%)
Query: 312 YKKHGFNPIDTLKISWVRLLNAFLKEAHWLNSGILPTTEEYLNNGIVSTGVHVVLIHAFF 371
++KHG I+++ S L + H L GIL TT ++ I G V++
Sbjct: 95 FEKHGVQAINSIPSSEEFLRKNLQNDIHHLVKGIL-TTAAAVSQSIKKEGTQVIVFGIET 153
Query: 372 LMDHAKGITKETLSILDEEFPNIIYSVAKILRLSDDL 408
+ LS LDE F + + V L L +L
Sbjct: 154 VRSMVLSFPLIILSTLDENFLSEVAQVFSSLNLLPEL 190
>PRR6_ARATH (Q9C9F6) Putative two-component response regulator-like
APRR6 (Pseudo-response regulator 6)
Length = 663
Score = 31.2 bits (69), Expect = 5.3
Identities = 22/124 (17%), Positives = 48/124 (37%), Gaps = 15/124 (12%)
Query: 109 YEASQLSIEGEDGLNDLGYLSCELLQAWLPRHQDHIQAIYVSNTLQYPIHYGLSRFMDKS 168
Y + + E N++G++SCE+ P+ I SN L + ++ +
Sbjct: 495 YSGGSIPPQEETNTNNVGFVSCEIHSEIPPKTNMAILETNYSNPLDWVFPEDITSLETNT 554
Query: 169 IFINDLKAKNKWICLDELAKMNSS---------------IVKFMNKNESVEVFKWWEDLG 213
I + + + + LD + + ++ ++ F ++F W ED G
Sbjct: 555 IQKSLVSCETSYDALDNMVPLETNMEEMNDALYDISIEDLISFDIDANEKDIFSWLEDNG 614
Query: 214 LAKE 217
++E
Sbjct: 615 FSEE 618
>GALE_MYCGE (P47364) UDP-glucose 4-epimerase (EC 5.1.3.2)
(Galactowaldenase) (UDP-galactose 4-epimerase)
Length = 340
Score = 31.2 bits (69), Expect = 5.3
Identities = 13/34 (38%), Positives = 23/34 (67%)
Query: 3 KQVHKKLVTSEDPTENFHLIDIIQRLGIEHYFVE 36
+Q K +VT D +N H+I +++++GIE YF +
Sbjct: 27 EQNDKLIVTVIDNNKNNHVIKLLKKIGIEFYFAD 60
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.139 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 51,047,007
Number of Sequences: 164201
Number of extensions: 2148021
Number of successful extensions: 5285
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 5231
Number of HSP's gapped (non-prelim): 24
length of query: 426
length of database: 59,974,054
effective HSP length: 113
effective length of query: 313
effective length of database: 41,419,341
effective search space: 12964253733
effective search space used: 12964253733
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Medicago: description of AC142394.1


