

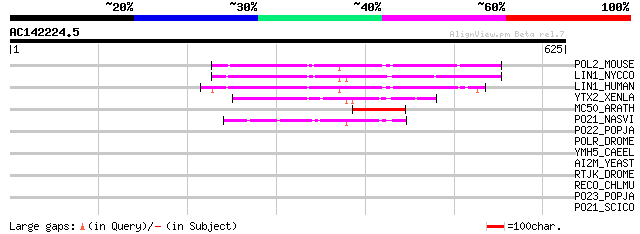
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142224.5 - phase: 0 /pseudo
(625 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 89 3e-17
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 89 3e-17
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 80 2e-14
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 65 4e-10
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 60 2e-08
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 59 3e-08
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 41 0.011
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 36 0.26
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 34 1.0
AI2M_YEAST (P03876) Putative COX1/OXI3 intron 2 protein 33 1.7
RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile elem... 33 2.2
RECO_CHLMU (Q9PJS3) DNA repair protein recO (Recombination prote... 32 6.5
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 32 6.5
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 31 8.5
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 89.4 bits (220), Expect = 3e-17
Identities = 92/345 (26%), Positives = 155/345 (44%), Gaps = 28/345 (8%)
Query: 228 YNKPFKLLLTKALIKLL*KPIVSEGSFPPNLIDTNIALIAKIDR-PKSMKHLCPVSLCNV 286
+ + FK L L KL K I EG+ P + + I LI K + P +++ P+SL N+
Sbjct: 506 FYQTFKEDLIPILHKLFHK-IEVEGTLPNSFYEATITLIPKPQKDPTKIENFRPISLMNI 564
Query: 287 IYKILSKVLANRLKQILHKCISDSQAAFVPSRFILENALTYFEVLHYMKCKTKGKEGNIA 346
KIL+K+LANR+++ + I Q F+P N V+HY+ K K K ++
Sbjct: 565 DAKILNKILANRIQEHIKAIIHPDQVGFIPGMQGWFNIRKSINVIHYIN-KLKDK-NHMI 622
Query: 347 LKQDVSKAFDRIKWSYLQAVMEKK-------------WDSQMFGL-TGSCNV*PIDPHCG 392
+ D KAFD+I+ ++ V+E+ + + + + I G
Sbjct: 623 ISLDAEKAFDKIQHPFMIKVLERSGIQGPYLNMIKAIYSKPVANIKVNGEKLEAIPLKSG 682
Query: 393 IRQGGPFSPYLYIICSEGLNSYIKHHENTGLIHGTRICRGSPSITQLLFADDSFLFYKAY 452
RQG P SPYL+ I E L I+ + I G +I G + L ADD ++
Sbjct: 683 TRQGCPLSPYLFNIVLEVLARAIRQQKE---IKGIQI--GKEEVKISLLADDMIVYISDP 737
Query: 453 VSEDTILKNILYTYEAASGQAINYRKS-SIAFSRNTDSIGHHNITHLLGVVESMWHGKYL 511
+ L N++ ++ G IN KS + +++N + T +V + + KYL
Sbjct: 738 KNSTRELLNLINSFGEVVGYKINSNKSMAFLYTKNKQAEKEIRETTPFSIVTN--NIKYL 795
Query: 512 G--LPSMVERDKKSIFSFIKERIWKNIQNWSAWSLSRAGKEVLLK 554
G L V+ F +K+ I ++++ W S G+ ++K
Sbjct: 796 GVTLTKEVKDLYDKNFKSLKKEIKEDLRRWKDLPCSWIGRINIVK 840
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 89.4 bits (220), Expect = 3e-17
Identities = 91/344 (26%), Positives = 146/344 (41%), Gaps = 26/344 (7%)
Query: 228 YNKPFKLLLTKALIKLL*KPIVSEGSFPPNLIDTNIALIAKIDR-PKSMKHLCPVSLCNV 286
+ + FK L L+ L + I EG P + NI LI K + P ++ P+SL N+
Sbjct: 479 FYQTFKEELVPILLNLF-QNIEKEGILPNTFYEANITLIPKPGKDPTRKENYRPISLMNI 537
Query: 287 IYKILSKVLANRLKQILHKCISDSQAAFVPSRFILENALTYFEVLHYMKCKTKGKEGNIA 346
KIL+K+L NR++Q + K I Q F+P N V+ ++ K K K+ ++
Sbjct: 538 DAKILNKILTNRIQQHIKKIIHHDQVGFIPGSQGWFNIRKSINVIQHIN-KLKNKD-HMI 595
Query: 347 LKQDVSKAFDRIKWSYLQAVMEK--------KWDSQMFG------LTGSCNV*PIDPHCG 392
L D KAFD I+ ++ ++K K ++ + + G
Sbjct: 596 LSIDAEKAFDNIQHPFMIRTLKKIGIEGTFLKLIEAIYSKPTANIILNGVKLKSFPLRSG 655
Query: 393 IRQGGPFSPYLYIICSEGLNSYIKHHENTGLIHGTRICRGSPSITQLLFADDSFLFYKAY 452
RQG P SP L+ I E L I+ + IH GS I LFADD ++ +
Sbjct: 656 TRQGCPLSPLLFNIVMEVLAIAIREEKAIKGIH-----IGSEEIKLSLFADDMIVYLENT 710
Query: 453 VSEDTILKNILYTYEAASGQAINYRKSSIAFSRNTDSIGHHNITHLLGVVESMWHGKYLG 512
T L ++ Y SG IN K S+AF ++ + + KYLG
Sbjct: 711 RDSTTKLLEVIKEYSNVSGYKINTHK-SVAFIYTNNNQAEKTVKDSIPFTVVPKKMKYLG 769
Query: 513 --LPSMVERDKKSIFSFIKERIWKNIQNWSAWSLSRAGKEVLLK 554
L V+ K + +++ I +++ W S G+ ++K
Sbjct: 770 VYLTKDVKDLYKENYETLRKEIAEDVNKWKNIPCSWLGRINIVK 813
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 80.1 bits (196), Expect = 2e-14
Identities = 93/353 (26%), Positives = 151/353 (42%), Gaps = 43/353 (12%)
Query: 216 MLQQQPNVKLP-------LYNKPFKLLLTKALIKLL*KPIVSEGSFPPNLIDTNIALIAK 268
++ PN K P + + +K L L+KL + I EG P + + +I LI K
Sbjct: 460 IINSLPNKKSPGPEGFTAEFYQRYKEELVPFLLKLF-QSIEKEGILPNSFYEASIILIPK 518
Query: 269 IDRPKSMK-HLCPVSLCNVIYKILSKVLANRLKQILHKCISDSQAAFVPSRFILENALTY 327
R + K + P+SL N+ KIL+K+LAN+++Q + K I Q F+P+ N
Sbjct: 519 PGRDTTKKENFRPISLMNIDAKILNKILANQIQQHIKKLIHHDQVGFIPAMQGWFNIRKS 578
Query: 328 FEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVMEKK-------------WDSQ 374
++ ++ +TK ++ + D KAFD+I+ ++ + K +D
Sbjct: 579 INIIQHIN-RTK-DTNHMIISIDAEKAFDKIQQPFMLKPLNKLGIDGTYLKIIRAIYDKP 636
Query: 375 MFGLTGSCNV*PIDP-HCGIRQGGPFSPYLYIICSEGLNSYIKHHENTGLIHGTRICRGS 433
+ + P G RQG P SP L I E L I+ + I G ++ G
Sbjct: 637 TANIILNGQKLEAPPLKTGTRQGCPLSPLLPNIVLEVLARAIRQEKE---IKGIQL--GK 691
Query: 434 PSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAINYRKSSIAFSRNTDSIGHH 493
+ LFADD ++ + + L ++ + SG IN +KS N
Sbjct: 692 EEVKLSLFADDMIVYLENPIVSAQNLLKLISNFSKVSGYKINVQKSQAFLYTNNRQTESQ 751
Query: 494 NITHLLGVVESMWHGKYLGLPSMVERDKKSIF--------SFIKE--RIWKNI 536
++ L + S KYLG+ + RD K +F + IKE WKNI
Sbjct: 752 IMSELPFTIASK-RIKYLGI--QLTRDVKDLFKENYKPLLNEIKEDTNKWKNI 801
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 65.5 bits (158), Expect = 4e-10
Identities = 67/244 (27%), Positives = 110/244 (44%), Gaps = 23/244 (9%)
Query: 251 EGSFPPNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISDS 310
+G P + ++L+ K + +K+ PVSL + YKI++K ++ RLK +L + I
Sbjct: 498 KGELPLSCRRAVLSLLPKKGDLRLIKNWRPVSLLSTDYKIVAKAISLRLKSVLAEVIHPD 557
Query: 311 QAAFVPSRFILENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYLQAVMEK- 369
Q+ VP R I +N ++LH+ + +T ++L Q+ KAFDR+ YL ++
Sbjct: 558 QSYTVPGRTIFDNVFLIRDLLHFAR-RTGLSLAFLSLDQE--KAFDRVDHQYLIGTLQAY 614
Query: 370 KWDSQMFG------LTGSCNV-------*PIDPHCGIRQGGPFSPYLYIICSEGLNSYIK 416
+ Q G + C V P+ G+RQG P S LY + E ++
Sbjct: 615 SFGPQFVGYLKTMYASAECLVKINWSLTAPLAFGRGVRQGCPLSGQLYSLAIEPFLCLLR 674
Query: 417 HHENTGLIHGTRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAINY 476
TGL+ + + +ADD L + V + + Y AAS IN+
Sbjct: 675 -KRLTGLV----LKEPDMRVVLSAYADDVILVAQDLVDLER-AQECQEVYAAASSARINW 728
Query: 477 RKSS 480
KSS
Sbjct: 729 SKSS 732
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 59.7 bits (143), Expect = 2e-08
Identities = 26/59 (44%), Positives = 37/59 (62%)
Query: 387 IDPHCGIRQGGPFSPYLYIICSEGLNSYIKHHENTGLIHGTRICRGSPSITQLLFADDS 445
+ P G+RQG P SPYL+I+C+E L+ + + G + G R+ SP I LLFADD+
Sbjct: 22 VTPSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSNNSPRINHLLFADDT 80
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 59.3 bits (142), Expect = 3e-08
Identities = 65/223 (29%), Positives = 97/223 (43%), Gaps = 31/223 (13%)
Query: 241 IKLL*KPIVSEGSFPPNLIDTNIALIAKIDRPKSMKHLC--PVSLCNVIYKILSKVLANR 298
IK L I+ G P +D+ LI K P +M C P+S+ +V + ++LANR
Sbjct: 352 IKSLFNMIMYHGQCPRRYLDSRTVLIPK--EPGTMDPACFRPLSIASVALRHFHRILANR 409
Query: 299 LKQILHKCISDSQAAFVPSRFILENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRI 358
+ + H + Q AF+ + + EN ++ + K KG IA+ DV KAFD +
Sbjct: 410 IGE--HGLLDTRQRAFIVADGVAENTSLLSAMIKEARMKIKGL--YIAI-LDVKKAFDSV 464
Query: 359 KW-SYLQAVMEKKWDSQMFGL-------------TGSCNV*PIDPHCGIRQGGPFSPYLY 404
+ S L A+ KK +M I P G+RQG P SP L+
Sbjct: 465 EHRSILDALRRKKLPLEMRNYIMWVYRNSKTRLEVVKTKGRWIRPARGVRQGDPLSPLLF 524
Query: 405 IICSEGLNSYIKHHENTGLIHGTRICRGSPSITQLLFADDSFL 447
+ + + ENTG + G+ I L+FADD L
Sbjct: 525 NCVMDAVLRRLP--ENTGFL------MGAEKIGALVFADDLVL 559
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 40.8 bits (94), Expect = 0.011
Identities = 60/257 (23%), Positives = 109/257 (42%), Gaps = 38/257 (14%)
Query: 280 PVSLCNVIYKILSKVLANRLKQILHKCISDSQAAFVPSRFILENALTYFEVLHYMKCKTK 339
P+++ + + ++L ++LA RL+ + + +Q + L N+L + Y+ + +
Sbjct: 78 PITIASALQRLLHRILAKRLEAAVE--LHPAQKGYARIDGTLVNSLL---LDTYISSRRE 132
Query: 340 GKEGNIALKQDVSKAFDRIKWSYLQAVMEKKW--DSQMFGLTGSCN-------V*P---- 386
++ + DV KAFD + S + +++ + +TGS + V P
Sbjct: 133 QRKTYNVVSLDVRKAFDTVSHSSICRALQRLGIDEGTSNYITGSLSDSTTTIRVGPGSQT 192
Query: 387 --IDPHCGIRQGGPFSPYLYIICSEGLNSYIKHHENTGLIHGTRICRGSPSITQLLFADD 444
I G++QG P SP+L+ + L+ + ++T I GT G I L FADD
Sbjct: 193 RKICIRRGVKQGDPLSPFLF---NAVLDELLCSLQSTPGIGGT---IGEEKIPVLAFADD 246
Query: 445 SFLFYKAYVSEDTILKNILYTYEAASGQAINYRKS---------SIAFSRNTDSIGHHNI 495
L V T L + + G ++N +KS + R + N+
Sbjct: 247 LLLLEDNDVLLPTTLATVA-NFFRLRGMSLNAKKSVSISVAASGGVCIPRTKPFLRVDNV 305
Query: 496 THLLGVVESMWHGKYLG 512
LL V + M +YLG
Sbjct: 306 --LLPVTDRMQTFRYLG 320
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 36.2 bits (82), Expect = 0.26
Identities = 54/242 (22%), Positives = 87/242 (35%), Gaps = 59/242 (24%)
Query: 252 GSFPPNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISDSQ 311
G+ P ++ I K K + P+S+ +V+ + L+ +LA RL ++ Q
Sbjct: 389 GNLPHSIRLARTVFIPKTVTAKRPQDFRPISVPSVLVRQLNAILATRLNSSINW--DPRQ 446
Query: 312 AAFVPSRFILENALTYFEVL-----HYMKCKTKGKEGNIALKQDVSKAFDRIKWS----- 361
F+P+ +NA VL H+ C DVSKAFD + +
Sbjct: 447 RGFLPTDGCADNATIVDLVLRHSHKHFRSC--------YIANLDVSKAFDSLSHASIYDT 498
Query: 362 ------------YLQAVME--------KKWDSQMFGLTGSCNV*PIDPHCGIRQGGPFSP 401
Y+Q E W S+ F P G++QG P SP
Sbjct: 499 LRAYGAPKGFVDYVQNTYEGGGTSLNGDGWSSEEFV-----------PARGVKQGDPLSP 547
Query: 402 YLYIICSEGLNSYIKHHENTGLIHGTRICRGSPSITQLLFADDSFLFYKAYVSEDTILKN 461
L+ + + L + G ++ G+ FADD LF + + +L
Sbjct: 548 ILFNLVMDRLLRTLPSE------IGAKV--GNAITNAAAFADDLVLFAETRMGLQVLLDK 599
Query: 462 IL 463
L
Sbjct: 600 TL 601
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 34.3 bits (77), Expect = 1.0
Identities = 37/171 (21%), Positives = 69/171 (39%), Gaps = 16/171 (9%)
Query: 250 SEGSFPPNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISD 309
S+ + P I I K P S + P+SL + +I+ +++ +R++ +S
Sbjct: 657 SDSAIPNRWKHAVIIPIPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSHLLSP 716
Query: 310 SQAAFV-----PSRFILENALTY--------FEVLHYMKCKTKGKEGN-IALKQDVSKAF 355
Q F+ PS + +L + ++L + K K + I LK+
Sbjct: 717 HQHGFLNFRSCPSSLVRSISLYHSILKNEKSLDILFFDFAKAFDKVSHPILLKKLALFGL 776
Query: 356 DRIKWSYLQAVMEKKWDSQMFGLTGSCNV*PIDPHCGIRQGGPFSPYLYII 406
D++ S+ + + + S S N PI G+ QG P L+I+
Sbjct: 777 DKLTCSWFKEFLHLRTFSVKINKFVSSNAYPIS--SGVPQGSVSGPLLFIL 825
>AI2M_YEAST (P03876) Putative COX1/OXI3 intron 2 protein
Length = 789
Score = 33.5 bits (75), Expect = 1.7
Identities = 57/253 (22%), Positives = 104/253 (40%), Gaps = 31/253 (12%)
Query: 259 IDTNI---ALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQILHKCISDSQAAFV 315
I+TN+ + + +++ PK+ P+S+ N KI+ + + L+ I + S F
Sbjct: 268 INTNMFKFSPVRRVEIPKTSGGFRPLSVGNPREKIVQESMRMMLEIIYNNSFSYYSHGFR 327
Query: 316 PSRFILENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWSYL-----QAVMEKK 370
P+ L + +CK + N +K D++K FD I + L + + +K
Sbjct: 328 PNLSCLTAII---------QCKNYMQYCNWFIKVDLNKCFDTIPHNMLINVLNERIKDKG 378
Query: 371 WDSQMFGLTGSCNV*PIDPH----CGIRQGGPFSPYLYIICSEGLNSYIKHHENTGLIHG 426
+ ++ L + V + + GI QG SP L I + L+ Y+++ G
Sbjct: 379 FMDLLYKLLRAGYVDKNNNYHNTTLGIPQGSVVSPILCNIFLDKLDKYLENKFENEFNTG 438
Query: 427 TRICRGSPSITQLLFADDSFLFYKAYVSEDTILKNILYTYEAASGQAINYRKSSIAFSRN 486
RG I L S ++ +SE L + Y+ G +++++ F R
Sbjct: 439 NMSNRGRNPIYNSL---SSKIYRCKLLSEKLKLIRLRDHYQRNMGSDKSFKRA--YFVRY 493
Query: 487 TDSI-----GHHN 494
D I G HN
Sbjct: 494 ADDIIIGVMGSHN 506
>RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 33.1 bits (74), Expect = 2.2
Identities = 38/147 (25%), Positives = 66/147 (44%), Gaps = 9/147 (6%)
Query: 217 LQQQPNVKLPLYNKPFKLLLTKAL--IKLL*KPIVSEGSFPPNLIDTNIALIAKIDR-PK 273
L++ P L L N+ +LL +AL + L+ ++ G FP +I +I K + P
Sbjct: 452 LKKAPGEDL-LDNRTIRLLPDQALQFLALIFNSVLDVGYFPKAWKSASIIMIHKTGKTPT 510
Query: 274 SMKHLCPVSLCNVIYKILSKVLANRLK--QILHKCISDSQAAFVPSRFILENALTYFEVL 331
+ P SL + KI+ +++ NRL + + K I Q F R V+
Sbjct: 511 DVDSYRPTSLLPSLGKIMERLILNRLLTCKDVTKAIPKFQFGF---RLQHGTPEQLHRVV 567
Query: 332 HYMKCKTKGKEGNIALKQDVSKAFDRI 358
++ + KE + D+ +AFDR+
Sbjct: 568 NFALEAMENKEYAVGAFLDIQQAFDRV 594
>RECO_CHLMU (Q9PJS3) DNA repair protein recO (Recombination protein
O)
Length = 234
Score = 31.6 bits (70), Expect = 6.5
Identities = 29/126 (23%), Positives = 51/126 (40%), Gaps = 25/126 (19%)
Query: 347 LKQDVSKAFDRIKWSY---------LQAVMEKKWDS----QMFGLTGSCNV*PIDPHCGI 393
L+ ++ F IK SY +QA+++ +W Q+F L
Sbjct: 70 LQGELKNPFTTIKNSYRLLQSTGKMIQAILKTQWQEKPSPQLFSL-----------FLNF 118
Query: 394 RQGGPFSPYLYIICSEGLNSYIKHHENTGLIHGTRICRGSPSITQLLFADDSFLFYKAYV 453
Q P +P+ Y S L ++H + L H +C+ S + ++ + LF + +
Sbjct: 119 LQRIPETPHPYFFSSMFLLKLLQHEGSLDLSHSCTLCKSSLE-SSTVYRHEGSLFCEKHA 177
Query: 454 SEDTIL 459
E TIL
Sbjct: 178 HEKTIL 183
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 31.6 bits (70), Expect = 6.5
Identities = 36/131 (27%), Positives = 56/131 (42%), Gaps = 22/131 (16%)
Query: 332 HYMKCKT-KGKEGNIALKQDVSKAFDRIKW-SYLQA-------------VMEKKWDSQMF 376
HY+K + KGK N+ + D+ KAFD + + L+A +M D+
Sbjct: 11 HYIKLRRLKGKTYNV-VSLDIRKAFDTVSHPAILRAMRAFGIDDGMQDFIMSTITDAYTN 69
Query: 377 GLTGSCNV*PIDPHCGIRQGGPFSPYLYIICSEGLNSYIKHHENTGLIHGTRICRGSPSI 436
+ G I G++QG P SP L+ I + L + + + + T C+ I
Sbjct: 70 IVVGGRTTNKIYIRNGVKQGDPLSPVLFNIVLDELVTRLNDEQPGASM--TPACK----I 123
Query: 437 TQLLFADDSFL 447
L FADD L
Sbjct: 124 ASLAFADDLLL 134
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 31.2 bits (69), Expect = 8.5
Identities = 44/183 (24%), Positives = 75/183 (40%), Gaps = 22/183 (12%)
Query: 242 KLL*KPIVSEGSFPPNLIDTNIALIAKIDRPKSMKHLCPVSLCNVIYKILSKVLANRLKQ 301
+LL V G P + + KI+ P+S+C+VI + +K+LA R
Sbjct: 194 RLLYNIFVFYGRVPSPIKGSRTVFTPKIEGGPDPGVFRPLSICSVILREFNKILARRF-- 251
Query: 302 ILHKCISDSQAAFVPSRFILENALTYFEVLHYMKCKTKGKEGNIALKQDVSKAFDRIKWS 361
+ + Q A++P + N ++ + K KE +IA+ D+ KAF+ + S
Sbjct: 252 VSCYTYDERQTAYLPIDGVCINVSMLTAII--AEAKRLRKELHIAI-LDLVKAFNSVYHS 308
Query: 362 YL-QAVMEK-------KWDSQMFG-------LTGSCNV*PIDPHCGIRQGGPFSPYLYII 406
L A+ E + + M+ G C + I G+ QG P S L+ +
Sbjct: 309 ALIDAITEAGCPPGVVDYIADMYNNVITEMQFEGKCELASI--LAGVYQGDPLSGPLFTL 366
Query: 407 CSE 409
E
Sbjct: 367 AYE 369
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.342 0.149 0.502
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 66,607,145
Number of Sequences: 164201
Number of extensions: 2608621
Number of successful extensions: 8259
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 8238
Number of HSP's gapped (non-prelim): 22
length of query: 625
length of database: 59,974,054
effective HSP length: 116
effective length of query: 509
effective length of database: 40,926,738
effective search space: 20831709642
effective search space used: 20831709642
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 69 (31.2 bits)
Medicago: description of AC142224.5


