

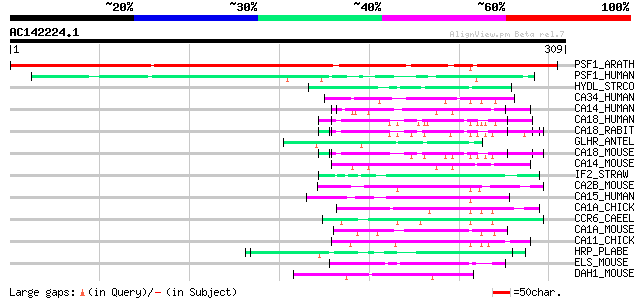
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142224.1 - phase: 0
(309 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
PSF1_ARATH (Q9M330) Putative proteasome inhibitor 317 3e-86
PSF1_HUMAN (Q92530) Proteasome inhibitor PI31 subunit (hPI31) 72 1e-12
HYDL_STRCO (P42534) Putative polyketide hydroxylase (EC 1.14.13.... 53 8e-07
CA34_HUMAN (Q01955) Collagen alpha 3(IV) chain precursor (Goodpa... 52 2e-06
CA14_HUMAN (P02462) Collagen alpha 1(IV) chain precursor 52 2e-06
CA18_HUMAN (P27658) Collagen alpha 1(VIII) chain precursor (Endo... 50 5e-06
CA18_RABIT (P14282) Collagen alpha 1(VIII) chain precursor (Endo... 50 9e-06
GLHR_ANTEL (P35409) Probable glycoprotein hormone G-protein coup... 49 2e-05
CA18_MOUSE (Q00780) Collagen alpha 1(VIII) chain precursor 49 2e-05
CA14_MOUSE (P02463) Collagen alpha 1(IV) chain precursor 49 2e-05
IF2_STRAW (Q82K53) Translation initiation factor IF-2 48 3e-05
CA2B_MOUSE (Q64739) Collagen alpha 2(XI) chain precursor 48 3e-05
CA15_HUMAN (P20908) Collagen alpha 1(V) chain precursor 48 3e-05
CA1A_CHICK (P08125) Collagen alpha 1(X) chain precursor 48 4e-05
CCR6_CAEEL (P20784) Cuticle collagen rol-6 47 5e-05
CA1A_MOUSE (Q05306) Collagen alpha 1(X) chain precursor 47 5e-05
CA11_CHICK (P02457) Collagen alpha 1(I) chain precursor 47 5e-05
HRP_PLABE (Q08168) 58 kDa phosphoprotein (Heat shock-related pro... 47 8e-05
ELS_MOUSE (P54320) Elastin precursor (Tropoelastin) 47 8e-05
DAH1_MOUSE (Q9QYB2) Dachshund homolog 1 (Dach1) 46 1e-04
>PSF1_ARATH (Q9M330) Putative proteasome inhibitor
Length = 302
Score = 317 bits (811), Expect = 3e-86
Identities = 169/311 (54%), Positives = 207/311 (66%), Gaps = 17/311 (5%)
Query: 1 MATDKSVLAVIRAARPTFRNQNDKIAFVVHSSFLASGYILTATGPQALSDTALSNPSNDE 60
MA ++V+A+IR ARP FRN +DK+AF +HSSF+ASGYILTATG A +D ALS+ S ++
Sbjct: 1 MANSQTVMAMIRLARPPFRNNHDKVAFAIHSSFVASGYILTATGRPAFADEALSSSSQND 60
Query: 61 VSVDHWNELNDEYAFVYANSEKEGLKKVLVKCLVMNGKLLVHALSEGFKEPLHLEINVAD 120
V ++ WNE EYAFVYAN K+G KK+LVKCL M+ KLLV A+++G EP HLEI V D
Sbjct: 61 VGIEGWNEFEGEYAFVYAN-PKKGSKKILVKCLAMDDKLLVDAIADGGAEPAHLEIKVGD 119
Query: 121 YAGEDGGSNFSQQFKNLDKLVKKIDADILSKLDGTAHASSSTKSSKTSDSTRPETSEPVP 180
YA E ++S QFKNLDKLV + ++I+ KLDG +S S + + P + P
Sbjct: 120 YAEESNEGDYSAQFKNLDKLVTDLQSEIIDKLDGKPKPVASRAQSSSETNEEPRYYDDTP 179
Query: 181 GFGEPAGPNDYHPGFIVPPVPAGSG-SDLFPGPGAGMYPSRGDHDFG-GSMLVGPNDPRW 238
P GP + G +VPP+P G SDLFPGPGAGMYP RG FG GSMLVGP DPR+
Sbjct: 180 ---NPLGPQIHPSGVVVPPIPGNGGYSDLFPGPGAGMYPGRG--GFGDGSMLVGPTDPRF 234
Query: 239 FSGGTGGGIGGGPGFIG----GLPGAPPGARFDPIGPPGVPGFEPNRFARNPRRPGFDAH 294
F G G PGF+G G+P PPGARFDP GPPGVPGFEP RF R P R H
Sbjct: 235 FPFGDG---SDRPGFMGPPHPGMP--PPGARFDPYGPPGVPGFEPGRFTRQPPRGPGGGH 289
Query: 295 PDLQHFRRDTD 305
PDL+HF +D
Sbjct: 290 PDLEHFPGGSD 300
>PSF1_HUMAN (Q92530) Proteasome inhibitor PI31 subunit (hPI31)
Length = 271
Score = 72.4 bits (176), Expect = 1e-12
Identities = 78/296 (26%), Positives = 117/296 (39%), Gaps = 54/296 (18%)
Query: 13 AARPTFRNQNDKIAFVVHSSFLASGYILTATGPQALSDTALSNPSNDEVSVDHWNELNDE 72
+A P + D + +H + GY G D N E+ WN D
Sbjct: 10 SAAPAITCRQDALVCFLHWEVVTHGYCGLGVG-----DQPGPNDKKSELLPAGWNNNKDL 64
Query: 73 YAFVYANSEKEGLKKVLVKCLVMNGKLLVHALSEGFKEPLHLEINVADYAGEDGGSNFSQ 132
Y Y K+G +K+LVK + + ++++ L G ++ L +N+ DY + +F +
Sbjct: 65 YVLRY--EYKDGSRKLLVKAITVESSMILNVLEYGSQQVADLTLNLDDYIDAEHLGDFHR 122
Query: 133 QFKNLDKLVKKIDADILSKLD--------GTAHASSSTKSSKTSDSTR-----PETSEPV 179
+KN ++L +I + I++ + + H +++ D R P TS
Sbjct: 123 TYKNSEELRSRIVSGIITPIHEQWEKANVSSPHREFPPATAREVDPLRIPPHHPHTSRQ- 181
Query: 180 PGFGEPAGPNDYHPGFIVPPVPAGSGSDLFP-GPGAGMYPSRGDHDFGGSMLVGPNDPRW 238
P + +P GP F+V G DL P GP G M+V P
Sbjct: 182 PPWCDPLGP------FVV------GGEDLDPFGPRR------------GGMIVDP----L 213
Query: 239 FSGGTGGGIGGGPGFIGGLP--GAPPGARFDPIGPPGVPGFEPNRFARNPRRPGFD 292
SG I G LP PPGARFDP GP G PN P PG+D
Sbjct: 214 RSGFPRALIDPSSGLPNRLPPGAVPPGARFDPFGPIGTSPPGPNPDHLPP--PGYD 267
>HYDL_STRCO (P42534) Putative polyketide hydroxylase (EC 1.14.13.-)
(WhiE ORF VIII)
Length = 627
Score = 53.1 bits (126), Expect = 8e-07
Identities = 42/113 (37%), Positives = 44/113 (38%), Gaps = 10/113 (8%)
Query: 167 TSDSTRPETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFG 226
T D+ R +E HPGF PPV G G PGAG P G
Sbjct: 361 TYDTERRPVAEATSARAAHRSVEHSHPGFAPPPVAGGGG------PGAGT-PGGAGRGTG 413
Query: 227 GSMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPGAPPGARFDPIGPPGVPGFEP 279
G GP P G G G GGPG GG PG P G R P G PG P
Sbjct: 414 GPG--GPGGPGGLGGPGGPGGTGGPGGPGG-PGGPDGPRGAGGAPGGGPGGGP 463
>CA34_HUMAN (Q01955) Collagen alpha 3(IV) chain precursor
(Goodpasture antigen)
Length = 1670
Score = 51.6 bits (122), Expect = 2e-06
Identities = 48/120 (40%), Positives = 54/120 (45%), Gaps = 23/120 (19%)
Query: 176 SEPVPGFGEPAGPNDYHPGFIVPPVPAGS----GSDLFPGPGAGMYPSRGDHDFGGSMLV 231
S PVPG P GP HPG PP GS G PGP G+ G
Sbjct: 651 STPVPGPPGPPGPPG-HPGPQGPPGIPGSLGKCGDPGLPGPD-------GEPGIPGIGFP 702
Query: 232 GPNDPRWFSG--GTGGGIGGGPGFIG--GLPGAP--PGARFDPI----GPPGVPGFEPNR 281
GP P+ G GT G +G PG +G GLPG P PGA+ +P G PG PGF R
Sbjct: 703 GPPGPKGDQGFPGTKGSLGC-PGKMGEPGLPGKPGLPGAKGEPAVAMPGGPGTPGFPGER 761
Score = 42.4 bits (98), Expect = 0.001
Identities = 40/105 (38%), Positives = 45/105 (42%), Gaps = 20/105 (19%)
Query: 205 GSDLFPGPGAGMYPSRGDHDFGGSM-LVGPNDPRWFSG-----GTGG--GIGGGPGFIG- 255
G FPGP S G F G L GP P+ F G G+ G GI G PGF G
Sbjct: 49 GEKGFPGPPG----SPGQKGFTGPEGLPGPQGPKGFPGLPGLTGSKGVRGISGLPGFSGS 104
Query: 256 -GLPGAPPGARFDPIGPPGVPGFEPNR----FARNPRRPGFDAHP 295
GLPG P P G GVPG ++ F P PG+ P
Sbjct: 105 PGLPGTP--GNTGPYGLVGVPGCSGSKGEQGFPGLPGTPGYPGIP 147
Score = 41.6 bits (96), Expect = 0.003
Identities = 46/124 (37%), Positives = 55/124 (44%), Gaps = 20/124 (16%)
Query: 179 VPGF----GEPAGPNDYHP-GFIVPPVPAGS-GSDLFPG-PGAGMYP----SRGDHDFGG 227
+PGF G P P + P G + P +GS G FPG PG YP + G G
Sbjct: 98 LPGFSGSPGLPGTPGNTGPYGLVGVPGCSGSKGEQGFPGLPGTPGYPGIPGAAGLKGQKG 157
Query: 228 SMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPGAP--PGARFDPIGPPGVPGFEPNRFARN 285
+ G + G G + G PG GLPG P PG P+GPPG PGF A
Sbjct: 158 APAKGEDIELDAKGDPG--LPGAPG-PQGLPGPPGFPG----PVGPPGPPGFFGFPGAMG 210
Query: 286 PRRP 289
PR P
Sbjct: 211 PRGP 214
Score = 36.6 bits (83), Expect = 0.081
Identities = 36/103 (34%), Positives = 40/103 (37%), Gaps = 20/103 (19%)
Query: 183 GEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGG 242
GEP PG PP P G PG M G +GP P G
Sbjct: 1054 GEPGYSEGTRPG---PPGPTGD-----PGLPGDMGKKGEMGQPGPPGHLGPAGPEGAPGS 1105
Query: 243 TGG-GIGGGPGFIG-----GLPG--APPGARFDPIGPPGVPGF 277
G G+ G PG G G+ G PPG R GPPG+PGF
Sbjct: 1106 PGSPGLPGKPGPHGDLGFKGIKGLLGPPGIR----GPPGLPGF 1144
Score = 35.0 bits (79), Expect = 0.24
Identities = 38/123 (30%), Positives = 47/123 (37%), Gaps = 21/123 (17%)
Query: 179 VPGF--GEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAG------MYPSRGDHDFGGSML 230
VPGF E N PG + G D+ P G Y +GD
Sbjct: 304 VPGFPGSEGVKGNRGFPGLMGEDGIKGQKGDIGPPGFRGPTEYYDTYQEKGDEG-----T 358
Query: 231 VGPNDPRWFSGGTGGGIGGGPGFIGGLPGAPPGARFDPIGPPGVPGFEPNRFARNPRRPG 290
GP PR G G P G+PG+P +R G PG PG + ++ R RPG
Sbjct: 359 PGPPGPRGARGPQG------PSGPPGVPGSPGSSRPGLRGAPGWPGLKGSKGERG--RPG 410
Query: 291 FDA 293
DA
Sbjct: 411 KDA 413
Score = 33.1 bits (74), Expect = 0.90
Identities = 34/123 (27%), Positives = 46/123 (36%), Gaps = 22/123 (17%)
Query: 180 PGFGEPAGPNDYH-----PGFIVPPVPAGSGSDLFPGPGA--GMYPSRGDHDFGGSMLVG 232
PGF G + H PG +P +P G++ GP G G+ G + G
Sbjct: 755 PGFPGERGNSGEHGEIGLPG--LPGLPGTPGNEGLDGPRGDPGQPGPPGEQGPPGRCIEG 812
Query: 233 PNDPRWFSGGTGGGIGGGPGFIGGLPGAPPG-ARFDPIGPPGVPGFEPNRFARNPRRPGF 291
P G G PG + GL G + P G PG+PG + + F PG
Sbjct: 813 PR-----------GAQGLPG-LNGLKGQQGRRGKTGPKGDPGIPGLDRSGFPGETGSPGI 860
Query: 292 DAH 294
H
Sbjct: 861 PGH 863
Score = 33.1 bits (74), Expect = 0.90
Identities = 36/90 (40%), Positives = 39/90 (43%), Gaps = 8/90 (8%)
Query: 193 PGFIVPPVPAGS-GSDLFPG-PGA-GMYPSRGDHDFGGSMLVGPNDPRWFSGGTGG-GIG 248
PG PP P G GS PG PG G+ S G+ GS G F G G G+
Sbjct: 987 PGPAGPPGPRGDLGSTGNPGEPGLRGIPGSMGNMGMPGSK--GKRGTLGFPGRAGRPGLP 1044
Query: 249 GGPGFIG--GLPGAPPGARFDPIGPPGVPG 276
G G G G PG G R P GP G PG
Sbjct: 1045 GIHGLQGDKGEPGYSEGTRPGPPGPTGDPG 1074
Score = 33.1 bits (74), Expect = 0.90
Identities = 44/152 (28%), Positives = 50/152 (31%), Gaps = 48/152 (31%)
Query: 173 PETSEPVPGFGEPAGPNDYH-------------PGFIVPPVPAGS----------GSDLF 209
P S PG P GP H PG PP AG G+
Sbjct: 1243 PPGSRGSPGAPGPPGPPGSHVIGIKGDKGSMGHPGPKGPPGTAGDMGPPGRLGAPGTPGL 1302
Query: 210 PGPGA--------GMYPSRGDHDFGGSM----LVGPNDPRWFSGGTGG-------GIGGG 250
PGP G+ +G+ F GS+ +GP P G G G G
Sbjct: 1303 PGPRGDPGFQGFPGVKGEKGNPGFLGSIGPPGPIGPKGPPGVRGDPGTLKIISLPGSPGP 1362
Query: 251 PGFIG-----GLPGAP-PGARFDPIGPPGVPG 276
PG G G PG P P P GP G PG
Sbjct: 1363 PGTPGEPGMQGEPGPPGPPGNLGPCGPRGKPG 1394
Score = 33.1 bits (74), Expect = 0.90
Identities = 37/115 (32%), Positives = 45/115 (38%), Gaps = 14/115 (12%)
Query: 198 PPVPAGSGSDLFPG-PGA----GMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPG 252
P P +G FPG PGA G+ +G+ + P DP G+ G PG
Sbjct: 513 PGSPGNTGLPGFPGFPGAQGDPGLKGEKGETLQPEGQVGVPGDPGLRGQPGRKGLDGIPG 572
Query: 253 FIG--GLPG-----APPGARFD--PIGPPGVPGFEPNRFARNPRRPGFDAHPDLQ 298
+G GLPG A G + D P G PG PG P G P LQ
Sbjct: 573 TLGVKGLPGPKGELALSGEKGDQGPPGDPGSPGSPGPAGPAGPPGYGPQGEPGLQ 627
Score = 30.8 bits (68), Expect = 4.5
Identities = 37/118 (31%), Positives = 41/118 (34%), Gaps = 39/118 (33%)
Query: 183 GEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGG 242
G+P P GF P +GS PG M GP R + G
Sbjct: 839 GDPGIPGLDRSGF-----PGETGSPGIPGHQGEM---------------GPLGQRGYPGN 878
Query: 243 TGG-GIGGGPGFIG--GLPGAPPGARFDPIGPPGVPGFEPNRFARNPRRPGFDAHPDL 297
G G G G IG G PGA IGPPG PG NP PG P +
Sbjct: 879 PGILGPPGEDGVIGMMGFPGA--------IGPPGPPG--------NPGTPGQRGSPGI 920
Score = 30.0 bits (66), Expect = 7.6
Identities = 40/130 (30%), Positives = 43/130 (32%), Gaps = 30/130 (23%)
Query: 193 PGFIVPPVPAG-SGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGP 251
PG P P G G FPGP P G F G+M GP P+ G G G
Sbjct: 173 PGLPGAPGPQGLPGPPGFPGPVGPPGPP-GFFGFPGAM--GPRGPKGHMGERVIGHKGER 229
Query: 252 GFIG--GLPGAP---------PGARFD---------------PIGPPGVPGFEPNRFARN 285
G G G PG P P R D P GP G+PG
Sbjct: 230 GVKGLTGPPGPPGTVIVTLTGPDNRTDLKGEKGDKGAMGEPGPPGPSGLPGESYGSEKGA 289
Query: 286 PRRPGFDAHP 295
P PG P
Sbjct: 290 PGDPGLQGKP 299
>CA14_HUMAN (P02462) Collagen alpha 1(IV) chain precursor
Length = 1669
Score = 51.6 bits (122), Expect = 2e-06
Identities = 52/133 (39%), Positives = 56/133 (42%), Gaps = 24/133 (18%)
Query: 180 PGF-GEPAGPN-------DYHPGFIVP---PVPAGSGSDLFPGPGAGMYPSRGDHDFGGS 228
PGF G+P P D HPG P P G + P G G SRGD G
Sbjct: 546 PGFPGQPGMPGRAGSPGRDGHPGLPGPKGSPGSVGLKGERGPPGGVGFPGSRGDTGPPGP 605
Query: 229 MLVGPNDP------RWFSGGTGG----GIGGGPGFIGGLPGAPPGARFDPIGPPGVPGFE 278
GP P F GG G G G PG I LPG PPGA P G PG PG +
Sbjct: 606 PGYGPAGPIGDKGQAGFPGGPGSPGLPGPKGEPGKIVPLPG-PPGAEGLP-GSPGFPGPQ 663
Query: 279 PNR-FARNPRRPG 290
+R F P RPG
Sbjct: 664 GDRGFPGTPGRPG 676
Score = 50.1 bits (118), Expect = 7e-06
Identities = 37/100 (37%), Positives = 46/100 (46%), Gaps = 12/100 (12%)
Query: 183 GEPAGPNDY-----HPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPR 237
GEP GP + PG PVP +G+ FPG +GD F G+ L GP+
Sbjct: 356 GEP-GPKGFPGLPGQPGPPGLPVPGQAGAPGFPGERG----EKGDRGFPGTSLPGPSGRD 410
Query: 238 WFSGGTGG-GIGGGPGFIGGLPGAPPGARFDPIGPPGVPG 276
G G G G PG+ G+ PG D GPPG+PG
Sbjct: 411 GLPGPPGSPGPPGQPGYTNGIVECQPGPPGDQ-GPPGIPG 449
Score = 39.7 bits (91), Expect = 0.010
Identities = 44/129 (34%), Positives = 50/129 (38%), Gaps = 36/129 (27%)
Query: 180 PGF-GEPAGPNDY-HPGFIVPPVPAGSGSDLFPG-PGAGMYPSRGDHDFGGSM----LVG 232
PG G P P GF+ PP P G PG PG +GD G L G
Sbjct: 1194 PGLAGSPGIPGSKGEQGFMGPPGPQGQPG--LPGSPGHATEGPKGDRGPQGQPGLPGLPG 1251
Query: 233 PNDPRWFSGGTG-GGIGGGPGFIGGLPGAP--PGARFDP--------------------I 269
P P G G G G PG+ PGAP PG + DP +
Sbjct: 1252 PMGPPGLPGIDGVKGDKGNPGW----PGAPGVPGPKGDPGFQGMPGIGGSPGITGSKGDM 1307
Query: 270 GPPGVPGFE 278
GPPGVPGF+
Sbjct: 1308 GPPGVPGFQ 1316
Score = 39.7 bits (91), Expect = 0.010
Identities = 44/117 (37%), Positives = 50/117 (42%), Gaps = 24/117 (20%)
Query: 176 SEPVPGFGEPA--GPNDYHPGFIVPPVPAG----SGSDLFPG-PGAGMYPSRGDHDFGGS 228
S VPG G P GP PG PP +G G FPG PG M +GD G
Sbjct: 795 SPGVPGIGPPGARGP----PGGQGPPGLSGPPGIKGEKGFPGFPGLDMPGPKGDKGAQGL 850
Query: 229 M-LVGPNDPRWFSGGTGG-GIGGGPGFIG-----GLPGAP--PGARFDPIGPPGVPG 276
+ G + G G GI G PG G G PG P PG P+G PG+PG
Sbjct: 851 PGITGQSGLPGLPGQQGAPGIPGFPGSKGEMGVMGTPGQPGSPG----PVGAPGLPG 903
Score = 38.5 bits (88), Expect = 0.021
Identities = 50/143 (34%), Positives = 59/143 (40%), Gaps = 32/143 (22%)
Query: 177 EPVPGFGEPAGPN--DYHPGFIVPP-VPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVG- 232
+P GF P GP D PG + PP P G + PG G+ +G+ G L G
Sbjct: 686 QPGIGFPGPPGPKGVDGLPGDMGPPGTPGRPGFNGLPG-NPGVQGQKGEPGVGLPGLKGL 744
Query: 233 ---PNDPRWFSGGTGGGIG-----------GGPGFIG--GLPGAP--PGARFDPIGPPGV 274
P P + G G IG G PG G G PG P PG+ +G PGV
Sbjct: 745 PGLPGIPG--TPGEKGSIGVPGVPGEHGAIGPPGLQGIRGEPGPPGLPGS----VGSPGV 798
Query: 275 PGFEPNRFARNPRRPGFDAHPDL 297
PG P AR P PG P L
Sbjct: 799 PGIGPPG-ARGP--PGGQGPPGL 818
Score = 36.2 bits (82), Expect = 0.11
Identities = 34/90 (37%), Positives = 37/90 (40%), Gaps = 15/90 (16%)
Query: 211 GPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTG-GGIGGGPGFIG--GLPGAP--PGAR 265
GP P D G L G R G +G G G PG G G PG P PG
Sbjct: 67 GPEGPQGPPGQKGDTGEPGLPGTKGTRGPPGASGYPGNPGLPGIPGQDGPPGPPGIPGCN 126
Query: 266 FD-----PIGPPGVPGFEPNRFARNPRRPG 290
P+GPPG+PG FA NP PG
Sbjct: 127 GTKGERGPLGPPGLPG-----FAGNPGPPG 151
Score = 34.3 bits (77), Expect = 0.40
Identities = 36/122 (29%), Positives = 46/122 (37%), Gaps = 19/122 (15%)
Query: 180 PGF-GEPAGPNDYHPGFIVPPVPAGSGSDLFPG----PGAGMYPSRGDHDFGGSMLVGPN 234
PG G+P + + + G FPG PG P R H G
Sbjct: 518 PGLIGQPGAKGEPGEFYFDLRLKGDKGDPGFPGQPGMPGRAGSPGRDGHP-------GLP 570
Query: 235 DPRWFSGGTG-GGIGGGPGFIGGLPGAPPGARFDPIGPPGVPGFEPNRFARNPRRPGFDA 293
P+ G G G G PG +G PG+R D GPPG PG+ P + + GF
Sbjct: 571 GPKGSPGSVGLKGERGPPGGVGF-----PGSRGDT-GPPGPPGYGPAGPIGDKGQAGFPG 624
Query: 294 HP 295
P
Sbjct: 625 GP 626
Score = 33.9 bits (76), Expect = 0.53
Identities = 32/109 (29%), Positives = 38/109 (34%), Gaps = 14/109 (12%)
Query: 180 PGF-GEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRW 238
PGF GEP +PG I P G + P G+ G +G R
Sbjct: 301 PGFPGEPG-----YPGLIGRQGPQGEKGEAGPPGPPGIVIGTGP--------LGEKGERG 347
Query: 239 FSGGTGGGIGGGPGFIGGLPGAPPGARFDPIGPPGVPGFEPNRFARNPR 287
+ G G GP GLPG P G G PGF R + R
Sbjct: 348 YPGTPGPRGEPGPKGFPGLPGQPGPPGLPVPGQAGAPGFPGERGEKGDR 396
Score = 33.9 bits (76), Expect = 0.53
Identities = 34/104 (32%), Positives = 39/104 (36%), Gaps = 18/104 (17%)
Query: 179 VPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPG----PGAGMYPSRGDHDFGGSMLVGPN 234
VPGF P G PG + + G PG PG P D G L GP
Sbjct: 1312 VPGFQGPKGL----PG--LQGIKGDQGDQGVPGAKGLPGPPGPPGPYDIIKGEPGLPGPE 1365
Query: 235 DPRWFSGGTGGGIGGGPGFIGGLPGAPPGARFDPIGPPGVPGFE 278
P G G G + GL G P GPPG+PGF+
Sbjct: 1366 GPPGLKGLQGLPGPKGQQGVTGLVGIP--------GPPGIPGFD 1401
Score = 32.3 bits (72), Expect = 1.5
Identities = 45/140 (32%), Positives = 52/140 (37%), Gaps = 30/140 (21%)
Query: 185 PAGPNDYHPGFIVPPVPAG-SGSDLFPGPGAGMYPS---RGDHDFGGSM----------L 230
P GP PGF P P G G PG G P +G+ F G L
Sbjct: 134 PLGPPGL-PGFAGNPGPPGLPGMKGDPGEILGHVPGMLLKGERGFPGIPGTPGPPGLPGL 192
Query: 231 VGPNDPRWFSGGTGG-GIGGGPGFIGGLPGAPPGARFDP-----IGPPGVPG----FEPN 280
GP P F+G G G G PG G + + G + D GPPGVPG E
Sbjct: 193 QGPVGPPGFTGPPGPPGPPGPPGEKGQMGLSFQGPKGDKGDQGVSGPPGVPGQAQVQEKG 252
Query: 281 RFARNPRR-----PGFDAHP 295
FA + PGF P
Sbjct: 253 DFATKGEKGQKGEPGFQGMP 272
Score = 32.0 bits (71), Expect = 2.0
Identities = 36/105 (34%), Positives = 40/105 (37%), Gaps = 21/105 (20%)
Query: 179 VPGFGEPAGPNDY---HPGFIVPPVPAG-SGSDLFPGPGAGMYPSRGDHDFGGSMLVGPN 234
+PG P GP D PG P P G G PGP +G G LVG
Sbjct: 1342 LPGPPGPPGPYDIIKGEPGLPGPEGPPGLKGLQGLPGP-------KGQQGVTG--LVGIP 1392
Query: 235 DPRWFSGGTGGGIGGGPGFIGGL-PGAPPGARF--DPIGPPGVPG 276
P G G PG G + P P G R P GP G+PG
Sbjct: 1393 GPPGIPG-----FDGAPGQKGEMGPAGPTGPRGFPGPPGPDGLPG 1432
Score = 32.0 bits (71), Expect = 2.0
Identities = 37/107 (34%), Positives = 43/107 (39%), Gaps = 14/107 (13%)
Query: 173 PETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVG 232
P T G P P ++ G I PP G + PGP G+ S G G +G
Sbjct: 751 PGTPGEKGSIGVPGVPGEH--GAIGPPGLQGIRGE--PGP-PGLPGSVGSPGVPG---IG 802
Query: 233 PNDPRWFSGGTGG-GIGGGPGFIG--GLPGAPPGARFDPIGPPGVPG 276
P R GG G G+ G PG G G PG P D GP G G
Sbjct: 803 PPGARGPPGGQGPPGLSGPPGIKGEKGFPGFPG---LDMPGPKGDKG 846
>CA18_HUMAN (P27658) Collagen alpha 1(VIII) chain precursor
(Endothelial collagen)
Length = 744
Score = 50.4 bits (119), Expect = 5e-06
Identities = 42/100 (42%), Positives = 46/100 (46%), Gaps = 19/100 (19%)
Query: 180 PGFGEPAGPNDYH-PGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRW 238
PGF P G + PG PP G G FPGP +GD GG + G PR
Sbjct: 327 PGF--PGGKGEQGLPGLPGPPGLPGIGKPGFPGP-------KGDRGMGG--VPGALGPRG 375
Query: 239 FSGGTGG-GIGGGPGFIGGLPGAPPGARFDPIGPPGVPGF 277
G G GIGG PG GLPG P P+GPPG GF
Sbjct: 376 EKGPIGAPGIGGPPGE-PGLPGIP-----GPMGPPGAIGF 409
Score = 48.1 bits (113), Expect = 3e-05
Identities = 50/139 (35%), Positives = 59/139 (41%), Gaps = 27/139 (19%)
Query: 173 PETSEPVPGFGEPAGPNDYHPGFIVPPVPAGS-GSDLFPGPGAGMYPSRGDHDFG--GSM 229
P +PG G+P G PG P P G G PGP G+ +GD FG G+
Sbjct: 198 PPGPHGLPGIGKPGG-----PGLPGQPGPKGDRGPKGLPGP-QGLRGPKGDKGFGMPGAP 251
Query: 230 LV-------GPNDPRWFSGGTGGGIGGGPGFIG--GLPGAP--PGARFDPI------GPP 272
V GP P G G+ G PG G G PGAP PG + PI GPP
Sbjct: 252 GVKGPPGMHGPPGPVGLPGVGKPGVTGFPGPQGPLGKPGAPGEPGPQ-GPIGVPGVQGPP 310
Query: 273 GVPGFEPNRFARNPRRPGF 291
G+PG P +PGF
Sbjct: 311 GIPGIGKPGQDGIPGQPGF 329
Score = 47.4 bits (111), Expect = 5e-05
Identities = 50/148 (33%), Positives = 60/148 (39%), Gaps = 35/148 (23%)
Query: 173 PETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFP----GPGA--GMYPSRGDHDFG 226
P +PG P GP PG I P P G G + P GP G+ G F
Sbjct: 388 PPGEPGLPGIPGPMGP----PGAIGFPGPKGEGGIVGPQGPPGPKGEPGLQGFPGKPGFL 443
Query: 227 GSM-------LVGPNDPRWFSGGTG-GGIGGGPGFIG-----GLPG-----APPG----- 263
G + L GP P+ +G G G+ G PG +G G+PG PPG
Sbjct: 444 GEVGPPGMRGLPGPIGPKGEAGQKGVPGLPGVPGLLGPKGEPGIPGDQGLQGPPGIPGIG 503
Query: 264 ARFDPIGPPGVPGFEPNRFARNPRRPGF 291
PIGPPG+PG P P PGF
Sbjct: 504 GPSGPIGPPGIPG--PKGEPGLPGPPGF 529
Score = 42.4 bits (98), Expect = 0.001
Identities = 40/115 (34%), Positives = 46/115 (39%), Gaps = 16/115 (13%)
Query: 179 VPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRW 238
+PG P GP+ PG P P G PGP +GD G L GP R
Sbjct: 192 IPGPQGPPGPHGL-PGIGKPGGPGLPGQ---PGP-------KGDR--GPKGLPGPQGLRG 238
Query: 239 FSGGTGGGIGGGPGFIG--GLPGAPPGARFDPIGPPGVPGFE-PNRFARNPRRPG 290
G G G+ G PG G G+ G P +G PGV GF P P PG
Sbjct: 239 PKGDKGFGMPGAPGVKGPPGMHGPPGPVGLPGVGKPGVTGFPGPQGPLGKPGAPG 293
Score = 40.8 bits (94), Expect = 0.004
Identities = 44/119 (36%), Positives = 49/119 (40%), Gaps = 18/119 (15%)
Query: 181 GFGEPAGPN-DYHPGFIVPPVPAG------SGSDLFPGPGA--GMYPSRGDHDFGGSMLV 231
GFG P P PG PP P G G FPGP G + G+ G + V
Sbjct: 244 GFGMPGAPGVKGPPGMHGPPGPVGLPGVGKPGVTGFPGPQGPLGKPGAPGEPGPQGPIGV 303
Query: 232 -GPNDPRWFSG-GTGG--GIGGGPGFIGG-----LPGAPPGARFDPIGPPGVPGFEPNR 281
G P G G G GI G PGF GG LPG P IG PG PG + +R
Sbjct: 304 PGVQGPPGIPGIGKPGQDGIPGQPGFPGGKGEQGLPGLPGPPGLPGIGKPGFPGPKGDR 362
Score = 35.0 bits (79), Expect = 0.24
Identities = 44/108 (40%), Positives = 48/108 (43%), Gaps = 19/108 (17%)
Query: 180 PGFGEPAGPNDYHPGFIVPPVPAG----SGSDLFPG-PGA-GMYPSRGDHDFGGSM-LVG 232
PGF GP PG P P G +G PG PG G+ +G+ G L G
Sbjct: 440 PGFLGEVGP----PGMRGLPGPIGPKGEAGQKGVPGLPGVPGLLGPKGEPGIPGDQGLQG 495
Query: 233 PNDPRWFSGGTGGGIG--GGPGFIG--GLPGAPPGARFDPIGPPGVPG 276
P GG G IG G PG G GLPG PPG F IG PGV G
Sbjct: 496 PPGIPGI-GGPSGPIGPPGIPGPKGEPGLPG-PPG--FPGIGKPGVAG 539
Score = 33.1 bits (74), Expect = 0.90
Identities = 39/121 (32%), Positives = 42/121 (34%), Gaps = 14/121 (11%)
Query: 178 PVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPG----PGAGMYPSRGDHDFGGSMLVGP 233
P+ GP PG PP P G PG PG YP G G P
Sbjct: 112 PLASLRGEQGPRG-EPGPRGPPGPPGLPGHGIPGIKGKPGPQGYPGVGKPGMPGM----P 166
Query: 234 NDPRWFSGGTGGGIGGGPGFIG--GLPG--APPGARFDP-IGPPGVPGFEPNRFARNPRR 288
P G G G IG G+PG PPG P IG PG PG + R
Sbjct: 167 GKPGAMGMPGAKGEIGQKGEIGPMGIPGPQGPPGPHGLPGIGKPGGPGLPGQPGPKGDRG 226
Query: 289 P 289
P
Sbjct: 227 P 227
Score = 29.6 bits (65), Expect = 9.9
Identities = 26/100 (26%), Positives = 36/100 (36%), Gaps = 19/100 (19%)
Query: 173 PETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVG 232
P+ +PG P GP P ++PP P G + P G G+ + H +G
Sbjct: 552 PQGQPGLPGPPGPPGPPG--PPAVMPPTPPPQG-EYLPDMGLGIDGVKPPHAYGAKK--- 605
Query: 233 PNDPRWFSGGTGGGIGGGPGFIGGLPGAPPGARFDPIGPP 272
G GGP + A A F P+G P
Sbjct: 606 -------------GKNGGPAYEMPAFTAELTAPFPPVGAP 632
>CA18_RABIT (P14282) Collagen alpha 1(VIII) chain precursor
(Endothelial collagen)
Length = 744
Score = 49.7 bits (117), Expect = 9e-06
Identities = 41/100 (41%), Positives = 46/100 (46%), Gaps = 19/100 (19%)
Query: 180 PGFGEPAGPNDYH-PGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRW 238
PGF P G + PG PP G G FPGP +GD GG + G PR
Sbjct: 327 PGF--PGGKGEQGLPGLPGPPGLPGVGKPGFPGP-------KGDRGIGG--VPGALGPRG 375
Query: 239 FSGGTGG-GIGGGPGFIGGLPGAPPGARFDPIGPPGVPGF 277
G G G+GG PG GLPG P P+GPPG GF
Sbjct: 376 EKGPVGAPGMGGPPGE-PGLPGIP-----GPMGPPGAIGF 409
Score = 47.8 bits (112), Expect = 4e-05
Identities = 45/129 (34%), Positives = 57/129 (43%), Gaps = 21/129 (16%)
Query: 179 VPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGA-GMYPSRGD----HDFGGSMLVGP 233
+PG GP Y PG P +P G PGA GM ++G+ + G + GP
Sbjct: 142 IPGIKGKPGPQGY-PGVGKPGMPGMPGK-----PGAMGMPGAKGEIGPKGEIGPMGIPGP 195
Query: 234 NDPRWFSGGTGGGIGGGPGFIGGLPGAPPGARFD--PIGPPGVPGFEPNRFARN---PRR 288
P G G G GGPG LPG P GA+ D P GPPG PG + + + P
Sbjct: 196 QGPPGPHGLPGIGKPGGPG----LPGQP-GAKGDRGPKGPPGPPGLQGPKGEKGFGMPGL 250
Query: 289 PGFDAHPDL 297
PG P +
Sbjct: 251 PGLKGPPGM 259
Score = 47.4 bits (111), Expect = 5e-05
Identities = 50/148 (33%), Positives = 60/148 (39%), Gaps = 35/148 (23%)
Query: 173 PETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFP----GPGA--GMYPSRGDHDFG 226
P +PG P GP PG I P P G G + P GP G+ G F
Sbjct: 388 PPGEPGLPGIPGPMGP----PGAIGFPGPKGEGGIVGPQGPPGPKGEPGLQGFPGKPGFL 443
Query: 227 GSM-------LVGPNDPRWFSGGTG-GGIGGGPGFIG-----GLPG-----APPG----- 263
G + L GP P+ +G G G+ G PG +G G+PG PPG
Sbjct: 444 GEVGPPGIRGLPGPIGPKGEAGHKGLPGLPGVPGLLGPKGEPGIPGDQGLQGPPGIPGIT 503
Query: 264 ARFDPIGPPGVPGFEPNRFARNPRRPGF 291
PIGPPG+PG P P PGF
Sbjct: 504 GPSGPIGPPGIPG--PKGEPGLPGPPGF 529
Score = 43.5 bits (101), Expect = 7e-04
Identities = 45/134 (33%), Positives = 53/134 (38%), Gaps = 18/134 (13%)
Query: 173 PETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVG 232
P +PG G+P GP PG P G PGP G+ +G+ FG L G
Sbjct: 198 PPGPHGLPGIGKPGGPG--LPG--QPGAKGDRGPKGPPGP-PGLQGPKGEKGFGMPGLPG 252
Query: 233 PNDPRWFSGGTG---------GGIGGGPGFIG--GLPGAPPGARFDPIGPPGVPGFEPNR 281
P G G G+ G PG G G PG PPG P GP GVPG +
Sbjct: 253 LKGPPGMHGPPGPVGLPGVGKPGVTGFPGPQGPLGKPG-PPGEP-GPQGPIGVPGVQGPP 310
Query: 282 FARNPRRPGFDAHP 295
+PG D P
Sbjct: 311 GLPGVGKPGQDGIP 324
Score = 38.1 bits (87), Expect = 0.028
Identities = 43/119 (36%), Positives = 48/119 (40%), Gaps = 18/119 (15%)
Query: 181 GFGEPAGPN-DYHPGFIVPPVPAG------SGSDLFPGPGA--GMYPSRGDHDFGGSMLV 231
GFG P P PG PP P G G FPGP G G+ G + V
Sbjct: 244 GFGMPGLPGLKGPPGMHGPPGPVGLPGVGKPGVTGFPGPQGPLGKPGPPGEPGPQGPIGV 303
Query: 232 -GPNDPRWFSG-GTGG--GIGGGPGFIGG-----LPGAPPGARFDPIGPPGVPGFEPNR 281
G P G G G GI G PGF GG LPG P +G PG PG + +R
Sbjct: 304 PGVQGPPGLPGVGKPGQDGIPGQPGFPGGKGEQGLPGLPGPPGLPGVGKPGFPGPKGDR 362
Score = 36.2 bits (82), Expect = 0.11
Identities = 44/131 (33%), Positives = 47/131 (35%), Gaps = 15/131 (11%)
Query: 178 PVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPG----PGAGMYPSRGDHDFGGSMLVGP 233
P+ GP PG PP P G PG PG YP G G P
Sbjct: 112 PLASLRGEQGPRG-EPGPRGPPGPPGLPGQGIPGIKGKPGPQGYPGVGKPGMPGM----P 166
Query: 234 NDPRWFSGGTGGGIGGGPGFIG--GLPG--APPGARFDP-IGPPGVPGFEPNRFARNPRR 288
P G G G IG G+PG PPG P IG PG PG A+ R
Sbjct: 167 GKPGAMGMPGAKGEIGPKGEIGPMGIPGPQGPPGPHGLPGIGKPGGPGLPGQPGAKGDRG 226
Query: 289 P-GFDAHPDLQ 298
P G P LQ
Sbjct: 227 PKGPPGPPGLQ 237
Score = 35.0 bits (79), Expect = 0.24
Identities = 41/109 (37%), Positives = 46/109 (41%), Gaps = 21/109 (19%)
Query: 180 PGFGEPAGPNDYHPGFIVPPVPAG----SGSDLFPG-PGA-GMYPSRGDHDFGGSMLVGP 233
PGF GP PG P P G +G PG PG G+ +G+ G G
Sbjct: 440 PGFLGEVGP----PGIRGLPGPIGPKGEAGHKGLPGLPGVPGLLGPKGEPGIPGDQ--GL 493
Query: 234 NDPRWFSGGTG-GGIGGGPGFIG-----GLPGAPPGARFDPIGPPGVPG 276
P G TG G G PG G GLPG PPG F +G PGV G
Sbjct: 494 QGPPGIPGITGPSGPIGPPGIPGPKGEPGLPG-PPG--FPGVGKPGVAG 539
>GLHR_ANTEL (P35409) Probable glycoprotein hormone G-protein coupled
receptor precursor
Length = 925
Score = 48.9 bits (115), Expect = 2e-05
Identities = 46/129 (35%), Positives = 52/129 (39%), Gaps = 23/129 (17%)
Query: 153 DGTAHASSSTKSSKTSD-------STRPETSEPVPGFGEPAGPNDYHPG----------F 195
+G ASS TK TS ST+P T+ G G P G + G
Sbjct: 309 NGQTTASSPTKEPATSGLGGGTHLSTQPHTTSGFGGGGFPGGGGGFPGGGGFPAGGSKTS 368
Query: 196 IVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGI-GGGPGFI 254
P +G G FPG G G +P G GGS P SG GGG GGG GF
Sbjct: 369 TQPHTTSGFGGGGFPGGGGG-FPGGGGFPAGGSKT--STQPHTTSGFGGGGFPGGGGGFP 425
Query: 255 GGLPGAPPG 263
GG G PG
Sbjct: 426 GG--GGFPG 432
Score = 40.8 bits (94), Expect = 0.004
Identities = 32/81 (39%), Positives = 34/81 (41%), Gaps = 5/81 (6%)
Query: 198 PPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGI-GGGPGFIGG 256
P +G G FPG G G +P G GGS P SG GGG GGG GF GG
Sbjct: 336 PHTTSGFGGGGFPGGGGG-FPGGGGFPAGGSKT--STQPHTTSGFGGGGFPGGGGGFPGG 392
Query: 257 LPGAPPGARFDPIGPPGVPGF 277
G P G P GF
Sbjct: 393 -GGFPAGGSKTSTQPHTTSGF 412
Score = 38.1 bits (87), Expect = 0.028
Identities = 35/100 (35%), Positives = 42/100 (42%), Gaps = 12/100 (12%)
Query: 165 SKTSDSTRPETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFP----GPGAGMYPSR 220
SKTS T+P T+ G G P G + G P GS + P G G G +P
Sbjct: 365 SKTS--TQPHTTSGFGGGGFPGGGGGFPGGGGFPA--GGSKTSTQPHTTSGFGGGGFPGG 420
Query: 221 GDHDFGGSMLVGPND----PRWFSGGTGGGIGGGPGFIGG 256
G GG G ++ P S GGG GG GF GG
Sbjct: 421 GGGFPGGGGFPGGSNTSTQPHTTSNSGGGGFPGGGGFPGG 460
>CA18_MOUSE (Q00780) Collagen alpha 1(VIII) chain precursor
Length = 743
Score = 48.9 bits (115), Expect = 2e-05
Identities = 41/100 (41%), Positives = 46/100 (46%), Gaps = 19/100 (19%)
Query: 180 PGFGEPAGPNDYH-PGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRW 238
PGF P G + PG PP G G FPGP +GD GG + G PR
Sbjct: 327 PGF--PGGKGEQGLPGLPGPPGLPGVGKPGFPGP-------KGDRGIGG--VPGVLGPRG 375
Query: 239 FSGGTGG-GIGGGPGFIGGLPGAPPGARFDPIGPPGVPGF 277
G G G+GG PG GLPG P P+GPPG GF
Sbjct: 376 EKGPIGAPGMGGPPGE-PGLPGIP-----GPMGPPGAIGF 409
Score = 47.0 bits (110), Expect = 6e-05
Identities = 50/148 (33%), Positives = 56/148 (37%), Gaps = 35/148 (23%)
Query: 173 PETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSM--- 229
P +PG P GP PG I P P G G + P G G F G
Sbjct: 388 PPGEPGLPGIPGPMGP----PGAIGFPGPKGEGGVVGPQGPPGPKGEPGLQGFPGKPGFL 443
Query: 230 -LVGPNDPRWFSG-----GTGG-----GIGGGPGFIG-----GLPG-----APPG----- 263
VGP R G G GG G+ G PG +G G+PG PPG
Sbjct: 444 GEVGPPGMRGLPGPIGPKGEGGHKGLPGLPGVPGLLGPKGEPGIPGDQGLQGPPGIPGIV 503
Query: 264 ARFDPIGPPGVPGFEPNRFARNPRRPGF 291
PIGPPG+PG P P PGF
Sbjct: 504 GPSGPIGPPGIPG--PKGEPGLPGPPGF 529
Score = 44.3 bits (103), Expect = 4e-04
Identities = 43/129 (33%), Positives = 56/129 (43%), Gaps = 21/129 (16%)
Query: 179 VPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGA-GMYPSRGD----HDFGGSMLVGP 233
+PG GP Y PG P +P G PGA GM ++G+ + G + GP
Sbjct: 142 MPGIKGKPGPQGY-PGIGKPGMPGMPGK-----PGAMGMPGAKGEIGPKGEIGPMGIPGP 195
Query: 234 NDPRWFSGGTGGGIGGGPGFIGGLPGAPPGARFD-----PIGPPGVPGFEPNRFARNPRR 288
P G G G GGPG LPG P GA+ + P GPPG+ G + + P
Sbjct: 196 QGPPGPHGLPGIGKPGGPG----LPGQP-GAKGERGPKGPPGPPGLQGPKGEKGFGMPGL 250
Query: 289 PGFDAHPDL 297
PG P +
Sbjct: 251 PGLKGPPGM 259
Score = 40.8 bits (94), Expect = 0.004
Identities = 38/106 (35%), Positives = 44/106 (40%), Gaps = 9/106 (8%)
Query: 173 PETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVG 232
P +PG G+P GP PG P G PGP G+ +G+ FG L G
Sbjct: 198 PPGPHGLPGIGKPGGPG--LPG--QPGAKGERGPKGPPGP-PGLQGPKGEKGFGMPGLPG 252
Query: 233 PNDPRWFSGGTG--GGIGGGPGFIGGLPGAPPGARFDPIGPPGVPG 276
P G G G G G + G PG P G P GPPG PG
Sbjct: 253 LKGPPGMHGPPGPVGLPGVGKPGVTGFPG-PQGPLGKP-GPPGEPG 296
Score = 40.0 bits (92), Expect = 0.007
Identities = 39/119 (32%), Positives = 46/119 (37%), Gaps = 18/119 (15%)
Query: 181 GFGEPAGPN-DYHPGFIVPPVPAG------SGSDLFPGPGAGMYPSRGDHDFGGSMLVGP 233
GFG P P PG PP P G G FPGP + + G L+G
Sbjct: 244 GFGMPGLPGLKGPPGMHGPPGPVGLPGVGKPGVTGFPGPQGPLGKPGPPGEPGPQGLIGV 303
Query: 234 NDPRWFSGGTG------GGIGGGPGFIGG-----LPGAPPGARFDPIGPPGVPGFEPNR 281
+ G G GI G PGF GG LPG P +G PG PG + +R
Sbjct: 304 PGVQGPPGMPGVGKPGQDGIPGQPGFPGGKGEQGLPGLPGPPGLPGVGKPGFPGPKGDR 362
Score = 35.8 bits (81), Expect = 0.14
Identities = 44/131 (33%), Positives = 47/131 (35%), Gaps = 15/131 (11%)
Query: 178 PVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPG----PGAGMYPSRGDHDFGGSMLVGP 233
P+ GP PG PP P G PG PG YP G G P
Sbjct: 112 PLASLRGEQGPRG-EPGPRGPPGPPGLPGHGMPGIKGKPGPQGYPGIGKPGMPGM----P 166
Query: 234 NDPRWFSGGTGGGIGGGPGFIG--GLPG--APPGARFDP-IGPPGVPGFEPNRFARNPRR 288
P G G G IG G+PG PPG P IG PG PG A+ R
Sbjct: 167 GKPGAMGMPGAKGEIGPKGEIGPMGIPGPQGPPGPHGLPGIGKPGGPGLPGQPGAKGERG 226
Query: 289 P-GFDAHPDLQ 298
P G P LQ
Sbjct: 227 PKGPPGPPGLQ 237
Score = 33.1 bits (74), Expect = 0.90
Identities = 42/109 (38%), Positives = 46/109 (41%), Gaps = 21/109 (19%)
Query: 180 PGFGEPAGPNDYHPGFIVPPVPAG----SGSDLFPG-PGA-GMYPSRGDHDFGGSMLVGP 233
PGF GP PG P P G G PG PG G+ +G+ G G
Sbjct: 440 PGFLGEVGP----PGMRGLPGPIGPKGEGGHKGLPGLPGVPGLLGPKGEPGIPGDQ--GL 493
Query: 234 NDPRWFSG--GTGGGIG--GGPGFIG--GLPGAPPGARFDPIGPPGVPG 276
P G G G IG G PG G GLPG PPG F +G PGV G
Sbjct: 494 QGPPGIPGIVGPSGPIGPPGIPGPKGEPGLPG-PPG--FPGVGKPGVAG 539
>CA14_MOUSE (P02463) Collagen alpha 1(IV) chain precursor
Length = 1669
Score = 48.5 bits (114), Expect = 2e-05
Identities = 51/133 (38%), Positives = 56/133 (41%), Gaps = 24/133 (18%)
Query: 180 PGF-GEPAGPN-------DYHPGFIVP---PVPAGSGSDLFPGPGAGMYPSRGDHDFGGS 228
PGF G+P P D HPG P P G + P G G SRGD G
Sbjct: 546 PGFPGQPGMPGRAGTPGRDGHPGLPGPKGSPGSIGLKGERGPPGGVGFPGSRGDIGPPGP 605
Query: 229 MLVGPNDP------RWFSGGTGG----GIGGGPGFIGGLPGAPPGARFDPIGPPGVPGFE 278
VGP P F GG G G G G + LPG PPGA P G PG PG +
Sbjct: 606 PGVGPIGPVGEKGQAGFPGGPGSPGLPGPKGEAGKVVPLPG-PPGAAGLP-GSPGFPGPQ 663
Query: 279 PNR-FARNPRRPG 290
+R F P RPG
Sbjct: 664 GDRGFPGTPGRPG 676
Score = 42.0 bits (97), Expect = 0.002
Identities = 41/115 (35%), Positives = 51/115 (43%), Gaps = 16/115 (13%)
Query: 173 PETSEPVPG-FGEPAGPNDYHPGFI-VPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSML 230
P+ + +PG G P P PGF +P P G PG G+ +G G +
Sbjct: 697 PKGVDGLPGEIGRPGSPG--RPGFNGLPGNPGPQGQK--GEPGIGLPGLKGQPGLPG--I 750
Query: 231 VGPNDPRWFSGGTG----GGIGGGPGFIG--GLPGAPPGARFDPIGPPGVPGFEP 279
G + GG G G+ G PG G G PG PPG + P GPPGVPG P
Sbjct: 751 PGTPGEKGSIGGPGVPGEQGLTGPPGLQGIRGDPG-PPGVQ-GPAGPPGVPGIGP 803
Score = 41.6 bits (96), Expect = 0.003
Identities = 42/121 (34%), Positives = 46/121 (37%), Gaps = 34/121 (28%)
Query: 177 EPVP-GF-GEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPN 234
EP P GF G P P PGF P P +G+ FPG +GD F G L GP+
Sbjct: 357 EPGPKGFPGTPGQPGP--PGF---PTPGQAGAPGFPGERG----EKGDQGFPGVSLPGPS 407
Query: 235 DPRWFSGGTGGGIGGGPGFIGGLPGAPPGARFDPIGPPGVPGFEPNRFARNPRRPGFDAH 294
G G PG PPG P GPPG PG P PG
Sbjct: 408 -----------GRDGAPG--------PPG----PPGPPGQPGHTNGIVECQPGPPGDQGP 444
Query: 295 P 295
P
Sbjct: 445 P 445
Score = 37.0 bits (84), Expect = 0.062
Identities = 42/133 (31%), Positives = 51/133 (37%), Gaps = 20/133 (15%)
Query: 176 SEPVPGF----GEPAGPNDYHPGFIVPPVPAGSGSDL-FPGPGA--GMYPSRGDHDFGGS 228
S+ +PG GE P PGF GS ++ FPG G+ +G+ F G
Sbjct: 1155 SDGIPGSAGEKGEQGVPGRGFPGFPGSKGDKGSKGEVGFPGLAGSPGIPGVKGEQGFMGP 1214
Query: 229 MLVGPNDPRWFSGGTGGGIGGGPGFIG-----GLPGAPPGARFDPIGPPGVPGFE-PNRF 282
GP G G + G G G GLPG P P+GPPG PG P
Sbjct: 1215 P--GPQGQPGLPGTPGHPVEGPKGDRGPQGQPGLPGHP-----GPMGPPGFPGINGPKGD 1267
Query: 283 ARNPRRPGFDAHP 295
N PG P
Sbjct: 1268 KGNQGWPGAPGVP 1280
Score = 35.0 bits (79), Expect = 0.24
Identities = 40/126 (31%), Positives = 47/126 (36%), Gaps = 27/126 (21%)
Query: 180 PGF-GEPAGPNDYHPGFIVPPVPAGSGSDLFPG----PGAGMYPSRGDHDFGGSMLVGPN 234
PG G+P + F + G FPG PG P R H L GP
Sbjct: 518 PGLIGQPGAKGEPGEIFFDMRLKGDKGDPGFPGQPGMPGRAGTPGRDGHPG----LPGPK 573
Query: 235 DPRWFSGGTGGGIG-----GGPGFIGGLPGAPPGARFDPIGPPGVPGFEPNRFARNPRRP 289
G+ G IG G PG +G PG+R D IGPPG PG P +
Sbjct: 574 -------GSPGSIGLKGERGPPGGVGF-----PGSRGD-IGPPGPPGVGPIGPVGEKGQA 620
Query: 290 GFDAHP 295
GF P
Sbjct: 621 GFPGGP 626
Score = 35.0 bits (79), Expect = 0.24
Identities = 34/121 (28%), Positives = 41/121 (33%), Gaps = 19/121 (15%)
Query: 179 VPGFGEPAGPNDYHPGFIV-----------PPVPAGSGSDLFPGPGAGMYPSRGDHDFGG 227
VPG+GE P P P +P SG PG G +G+ G
Sbjct: 271 VPGYGEKGEPGKQGPRGKPGKDGEKGERGSPGIPGDSGYPGLPGR-QGPQGEKGEAGLPG 329
Query: 228 -------SMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPGAPPGARFDPIGPPGVPGFEPN 280
+M +G R + G G GP G PG P F G G PGF
Sbjct: 330 PPGTVIGTMPLGEKGDRGYPGAPGLRGEPGPKGFPGTPGQPGPPGFPTPGQAGAPGFPGE 389
Query: 281 R 281
R
Sbjct: 390 R 390
Score = 35.0 bits (79), Expect = 0.24
Identities = 38/116 (32%), Positives = 44/116 (37%), Gaps = 21/116 (18%)
Query: 179 VPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSM-LVGPNDPR 237
+PG G G +P VP G PG G+ +GD G L GP P
Sbjct: 1291 MPGIGGSPGITGSKGDMGLPGVPGFQGQKGLPGL-QGVKGDQGDQGVPGPKGLQGPPGPP 1349
Query: 238 WFSGGTGGGIGGGPGFIG--GLPG-----APPGARFDP--------IGPPGVPGFE 278
G I G PG G G PG PPG + GPPGVPGF+
Sbjct: 1350 ----GPYDVIKGEPGLPGPEGPPGLKGLQGPPGPKGQQGVTGSVGLPGPPGVPGFD 1401
Score = 34.7 bits (78), Expect = 0.31
Identities = 34/95 (35%), Positives = 36/95 (37%), Gaps = 12/95 (12%)
Query: 211 GPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTG-GGIGGGPGFIG--GLPGAP--PGAR 265
GP P D G L G R G G G G PG G G PG P PG
Sbjct: 67 GPEGPHGPPGQKGDAGEPGLPGTKGTRGPPGAAGYPGNPGLPGIPGQDGPPGPPGIPGCN 126
Query: 266 FD-----PIGPPGVPGFEPNRFARNPRRPGFDAHP 295
P+GPPG+PGF N P PG P
Sbjct: 127 GTKGERGPLGPPGLPGFSGN--PGPPGLPGMKGDP 159
Score = 34.7 bits (78), Expect = 0.31
Identities = 45/140 (32%), Positives = 51/140 (36%), Gaps = 30/140 (21%)
Query: 179 VPGFGEPAGPNDY-----HPGFIVPPVPAG--SGSDLFPG-PGAGMYPSRGDHDFGGSML 230
+PGF GP PG I+ VP G FPG PG P G L
Sbjct: 140 LPGFSGNPGPPGLPGMKGDPGEILGHVPGTLLKGERGFPGIPGMPGSP-------GLPGL 192
Query: 231 VGPNDPRWFSGGTGG-GIGGGPGFIGGLPGAPPGARFDP-----IGPPGVPG-------- 276
GP P F+G G G G PG G + + G + D GPPGVPG
Sbjct: 193 QGPVGPPGFTGPPGPPGPPGPPGEKGQMGSSFQGPKGDKGEQGVSGPPGVPGQAQVKEKG 252
Query: 277 -FEPNRFARNPRRPGFDAHP 295
F P PGF P
Sbjct: 253 DFAPTGEKGQKGEPGFPGVP 272
Score = 32.7 bits (73), Expect = 1.2
Identities = 30/89 (33%), Positives = 35/89 (38%), Gaps = 13/89 (14%)
Query: 200 VPAGSGSDLFPGPGAGMYPSRGDHDFGGSM-------LVGPNDPRWFSGGTGG-GIGGGP 251
VP G PGP +G+ G L GP P+ G TG G+ G P
Sbjct: 1336 VPGPKGLQGPPGPPGPYDVIKGEPGLPGPEGPPGLKGLQGPPGPKGQQGVTGSVGLPGPP 1395
Query: 252 GFIGGLPGAP----PGARFDPIGPPGVPG 276
G + G GAP F P GP G PG
Sbjct: 1396 G-VPGFDGAPGQKGETGPFGPPGPRGFPG 1423
Score = 32.3 bits (72), Expect = 1.5
Identities = 34/105 (32%), Positives = 38/105 (35%), Gaps = 8/105 (7%)
Query: 180 PGF-GEPAGPNDY----HPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPN 234
PGF G P P PG +P + G PG G G L GP
Sbjct: 716 PGFNGLPGNPGPQGQKGEPGIGLPGLKGQPGLPGIPGTPGEKGSIGGPGVPGEQGLTGPP 775
Query: 235 DPRWFSGGTGGGIGGGPGFIGGLPG-APPGARFDP--IGPPGVPG 276
+ G G GP G+PG PPGA P GPPG G
Sbjct: 776 GLQGIRGDPGPPGVQGPAGPPGVPGIGPPGAMGPPGGEGPPGSSG 820
Score = 32.3 bits (72), Expect = 1.5
Identities = 37/94 (39%), Positives = 39/94 (41%), Gaps = 17/94 (18%)
Query: 185 PAGPNDYHPGFIVPPVPAG-SGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGT 243
P GP PGF P P G G PG G P G++L G R F G
Sbjct: 134 PLGPPGL-PGFSGNPGPPGLPGMKGDPGEILGHVP--------GTLLKGE---RGFPGIP 181
Query: 244 G-GGIGGGPGFIGGLPGAPPGARFDPIGPPGVPG 276
G G G PG G P PPG P GPPG PG
Sbjct: 182 GMPGSPGLPGLQG--PVGPPGFTGPP-GPPGPPG 212
Score = 30.8 bits (68), Expect = 4.5
Identities = 39/117 (33%), Positives = 48/117 (40%), Gaps = 17/117 (14%)
Query: 173 PETSEPVPGFGEPAGPNDYHPGFI-VPPVPAGSGSDLFPG-PGA-------GMYPSRGDH 223
P+ + PG G P PG +P P GS PG PG G+ RGD
Sbjct: 727 PQGQKGEPGIGLPGLKGQ--PGLPGIPGTPGEKGSIGGPGVPGEQGLTGPPGLQGIRGDP 784
Query: 224 DFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPGA--PPGARFDPIGPPGVPGFE 278
G + GP P G G G PG G PG+ PPG + + G PG PG +
Sbjct: 785 --GPPGVQGPAGPPGVPGIGPPGAMGPPGGEGP-PGSSGPPGIKGEK-GFPGFPGLD 837
>IF2_STRAW (Q82K53) Translation initiation factor IF-2
Length = 1046
Score = 48.1 bits (113), Expect = 3e-05
Identities = 44/123 (35%), Positives = 47/123 (37%), Gaps = 14/123 (11%)
Query: 173 PETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVG 232
P P PG G P G N +PG ++P PA P PG G RG G G
Sbjct: 289 PRPQAPRPG-GAPGG-NRPNPG-MMPQRPAAG-----PRPGGGGPGGRGPGGGGRPGGPG 340
Query: 233 PNDPRWFSGGTGGGIGGGPGFIGGLPGAPPGARFDPIGPPGVPGFEPNRFARNPRRPGFD 292
R GG G GGG G G PG P G G PG PG RPGF
Sbjct: 341 GGGGRPGGGGFAGRPGGGGGGFAGRPGGPGGGGGGFAGRPGGPG------GGGGGRPGFG 394
Query: 293 AHP 295
P
Sbjct: 395 GRP 397
Score = 42.4 bits (98), Expect = 0.001
Identities = 37/99 (37%), Positives = 38/99 (38%), Gaps = 11/99 (11%)
Query: 178 PVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPR 237
P PG G P G G P P G G PG G + R GG P P
Sbjct: 318 PRPGGGGPGGRGPGGGGR--PGGPGGGGGR----PGGGGFAGRPGGG-GGGFAGRPGGP- 369
Query: 238 WFSGGTGGGIGGGPGFIGGLPGAPPGARFDPIGPPGVPG 276
GG GGG G PG GG G PG P GP G G
Sbjct: 370 ---GGGGGGFAGRPGGPGGGGGGRPGFGGRPGGPGGRGG 405
Score = 32.0 bits (71), Expect = 2.0
Identities = 41/139 (29%), Positives = 46/139 (32%), Gaps = 26/139 (18%)
Query: 170 STRPETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSM 229
+ RP P PG PAGP + F G P PG P GG+
Sbjct: 195 AARPGGQAPRPG-ARPAGPRPGNNPF-TSGGSTGMARPQAPRPGGAPRP-------GGAG 245
Query: 230 LVGPNDPRWFSGGTGGGIG-----GGPGFIGGLPGAPPGARFDPIG--------PPGVPG 276
G P+ G GG GGPG G P GAR P G P G PG
Sbjct: 246 APGGPRPQ----GAGGQDRAPRPQGGPGGAPRPQGGPGGARPTPGGMPRPQAPRPGGAPG 301
Query: 277 FEPNRFARNPRRPGFDAHP 295
P+RP P
Sbjct: 302 GNRPNPGMMPQRPAAGPRP 320
>CA2B_MOUSE (Q64739) Collagen alpha 2(XI) chain precursor
Length = 1736
Score = 48.1 bits (113), Expect = 3e-05
Identities = 50/139 (35%), Positives = 59/139 (41%), Gaps = 29/139 (20%)
Query: 172 RPETSEP-VPGFGEPAGPNDYHPGFIVPPVPAGS-GSDLFPGPGA--GMYPSRGDHD-FG 226
R E EP PG P G N PP P G G PGP G ++GD G
Sbjct: 1293 RGEDGEPGQPGSPGPTGENG-------PPGPLGKRGPAGTPGPEGRQGEKGAKGDPGAVG 1345
Query: 227 GSMLVGPNDPRWFSGGTG-GGIGGGPGFIG--GLPGA-----PPGARFDPIGPPGVPGFE 278
GP P +G G G+ G PG +G G PGA PPG P+GPPG+PG
Sbjct: 1346 APGKTGPVGPAGLAGKPGPDGLRGLPGSVGQQGRPGATGQAGPPG----PVGPPGLPGLR 1401
Query: 279 PNRFARNPRRPGFDAHPDL 297
+ A+ G HP L
Sbjct: 1402 GDAGAK-----GEKGHPGL 1415
Score = 39.3 bits (90), Expect = 0.013
Identities = 41/117 (35%), Positives = 48/117 (40%), Gaps = 16/117 (13%)
Query: 181 GFGEPAGPNDYHPGFIVPPVPAGS-GSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWF 239
G G P GP PG P P G+ G PG + P+ D G L GP P
Sbjct: 1024 GSGGPIGP----PGRPGPQGPPGAAGEKGVPGEKGPIGPTGRDGVQGPVGLPGPAGPPGV 1079
Query: 240 SGGTGG-GIGGGPGFIG--------GLPGAPPGARFDPIGPPGVPGFEPNRFARNPR 287
+G G G G PG G G PG PPG P+G PG G + AR P+
Sbjct: 1080 AGEDGDKGEVGDPGQKGTKGNKGEHGPPG-PPGP-IGPVGQPGAAGADGEPGARGPQ 1134
Score = 36.2 bits (82), Expect = 0.11
Identities = 36/100 (36%), Positives = 42/100 (42%), Gaps = 17/100 (17%)
Query: 180 PGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWF 239
PG AGP PG + PP G D AG +G G L+GP +
Sbjct: 1380 PGATGQAGP----PGPVGPPGLPGLRGD------AGAKGEKGHPGLIG--LIGPTGEQGE 1427
Query: 240 SGGTG-GGIGGGPGFIG--GLPGAPPGARFDPIGPPGVPG 276
G G G G PG G G+PGA P GPPG+PG
Sbjct: 1428 KGDRGLPGPQGSPGQKGETGIPGA--SGPIGPGGPPGLPG 1465
Score = 35.8 bits (81), Expect = 0.14
Identities = 33/96 (34%), Positives = 40/96 (41%), Gaps = 11/96 (11%)
Query: 194 GFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTG-GGIGGGPG 252
GF PP P G PGP +G+ GG M GP P G G G G G
Sbjct: 1147 GFNGPPGPIGLQG--LPGPSG----EKGETGDGGPM--GPPGPPGPRGPAGPNGADGPQG 1198
Query: 253 FIGGLPG-APPGARFDPIGPPGVPGFEPNRFARNPR 287
GG+ PPG + +P G G PG + + PR
Sbjct: 1199 SPGGVGNLGPPGEKGEP-GESGSPGVQGEPGVKGPR 1233
Score = 35.4 bits (80), Expect = 0.18
Identities = 38/116 (32%), Positives = 45/116 (38%), Gaps = 21/116 (18%)
Query: 181 GFGEPAGP-----NDYHPGFIVPPVPAGS----GSDLFPGPGAGMYPSRGDHDFGGSMLV 231
G PAGP D G + P G+ G PGP + P G
Sbjct: 1069 GLPGPAGPPGVAGEDGDKGEVGDPGQKGTKGNKGEHGPPGPPGPIGPVGQPGAAGADGEP 1128
Query: 232 GPNDPRWFSGGTGG----GIGGGPGFIG--GLPGAPPGARFD-----PIGPPGVPG 276
G P+ G G G G PG IG GLPG P G + + P+GPPG PG
Sbjct: 1129 GARGPQGHFGAKGDEGTRGFNGPPGPIGLQGLPG-PSGEKGETGDGGPMGPPGPPG 1183
Score = 35.4 bits (80), Expect = 0.18
Identities = 44/124 (35%), Positives = 51/124 (40%), Gaps = 28/124 (22%)
Query: 185 PAGPNDYHPGFIVPPVPAG----SGSDLFPG-PGA-GMYPSRGDHDFGGSMLVGPNDPRW 238
P GP G PP AG +G+D G PG GM +GD F G L G +
Sbjct: 521 PRGPQ----GLTGPPGKAGRRGRAGADGARGMPGEPGM---KGDRGFDG--LPGLPGEKG 571
Query: 239 FSGGTGG-GIGGGPGFIG-----------GLPG-APPGARFDPIGPPGVPGFEPNRFARN 285
G TG G+ G PG G GLPG + P P GPPG+PG R
Sbjct: 572 QRGDTGAQGLPGPPGEDGERGDDGEIGPRGLPGESGPRGLLGPKGPPGIPGPPGVRGMDG 631
Query: 286 PRRP 289
P P
Sbjct: 632 PHGP 635
Score = 35.0 bits (79), Expect = 0.24
Identities = 34/93 (36%), Positives = 38/93 (40%), Gaps = 17/93 (18%)
Query: 198 PPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSM-LVGPNDPRWFSGGTG-GGIGGGPGFIG 255
PP P G PG +G ++GD G+ GP R F G G G GGPG G
Sbjct: 947 PPGPPGEQG--LPGT-SGKEGTKGDPGPPGAPGKDGPAGLRGFPGERGLPGTAGGPGLKG 1003
Query: 256 GL-PGAPPGARFDP-----------IGPPGVPG 276
P PPG P IGPPG PG
Sbjct: 1004 NEGPAGPPGPAGSPGERGAAGSGGPIGPPGRPG 1036
Score = 33.1 bits (74), Expect = 0.90
Identities = 28/75 (37%), Positives = 32/75 (42%), Gaps = 3/75 (4%)
Query: 204 SGSDLFPGPGAGMYPSRGDH-DFGGSMLVGPNDPRWFSGGTGG-GIGGGPGFIGGLPGAP 261
+G+ PGP G RGD + G L G + PR G G GI G PG G
Sbjct: 576 TGAQGLPGP-PGEDGERGDDGEIGPRGLPGESGPRGLLGPKGPPGIPGPPGVRGMDGPHG 634
Query: 262 PGARFDPIGPPGVPG 276
P P G PG PG
Sbjct: 635 PKGSLGPQGEPGPPG 649
Score = 30.8 bits (68), Expect = 4.5
Identities = 24/55 (43%), Positives = 27/55 (48%), Gaps = 6/55 (10%)
Query: 227 GSMLVGPNDPRWFSGGTGG-GIGGGPGFIGGLPG--APPGARFDP--IGPPGVPG 276
G + GP P +G G GI G PG +G PG PPG P GPPG PG
Sbjct: 394 GMFVEGPPGPEGPAGLAGPPGIQGNPGPVGD-PGERGPPGRAGLPGSDGPPGPPG 447
Score = 30.8 bits (68), Expect = 4.5
Identities = 39/148 (26%), Positives = 51/148 (34%), Gaps = 19/148 (12%)
Query: 140 LVKKIDADILSKLDGTAHASSSTKSSKTSDSTRPETSEPVPGFGEPAGPNDY--HPGFIV 197
L K D L L G+ ++ + P +PG AG HPG I
Sbjct: 1358 LAGKPGPDGLRGLPGSVGQQGRPGATGQAGPPGPVGPPGLPGLRGDAGAKGEKGHPGLIG 1417
Query: 198 PPVPAGS----GSDLFPGPGAGMYPSRGDHDFGGSM----------LVGPNDPRWFSGGT 243
P G G PGP G +G+ G+ L GP+ P+ G T
Sbjct: 1418 LIGPTGEQGEKGDRGLPGP-QGSPGQKGETGIPGASGPIGPGGPPGLPGPSGPKGAKGAT 1476
Query: 244 GGGIGGGPGFIGGLPG--APPGARFDPI 269
G G + G PG PPG P+
Sbjct: 1477 GPAGPKGEKGVQGPPGHPGPPGEVIQPL 1504
Score = 30.8 bits (68), Expect = 4.5
Identities = 33/99 (33%), Positives = 36/99 (36%), Gaps = 6/99 (6%)
Query: 180 PGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLV-GPNDPRW 238
PG PAGPN G P G +L P G G G V GP R
Sbjct: 1182 PGPRGPAGPN----GADGPQGSPGGVGNLGPPGEKGEPGESGSPGVQGEPGVKGPRGERG 1237
Query: 239 FSGGTG-GGIGGGPGFIGGLPGAPPGARFDPIGPPGVPG 276
G +G G G PG G P P+G PG PG
Sbjct: 1238 EKGESGQAGEAGPPGPKGPTGDNGPKGNPGPVGFPGDPG 1276
>CA15_HUMAN (P20908) Collagen alpha 1(V) chain precursor
Length = 1838
Score = 48.1 bits (113), Expect = 3e-05
Identities = 43/119 (36%), Positives = 55/119 (46%), Gaps = 16/119 (13%)
Query: 166 KTSDSTRPETSEPVP-GFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHD 224
K D +T P P G P+GP PG PP PAG P G ++G+
Sbjct: 1365 KGDDGEPGQTGSPGPTGEPGPSGP----PGKRGPPGPAG------PEGRQGEKGAKGEAG 1414
Query: 225 FGGSM-LVGPNDPRWFSGGTG-GGIGGGPGFIG--GLPGAP-PGARFDPIGPPGVPGFE 278
G GP P+ G G G+ G PG +G GLPG+P P P+GPPG+PG +
Sbjct: 1415 LEGPPGKTGPIGPQGAPGKPGPDGLRGIPGPVGEQGLPGSPGPDGPPGPMGPPGLPGLK 1473
Score = 37.7 bits (86), Expect = 0.036
Identities = 44/123 (35%), Positives = 51/123 (40%), Gaps = 29/123 (23%)
Query: 185 PAGPNDYHPGFIVPPVPAGSGSDLFPG-PGAGMYPSRGDHDFGGSM-------LVGPNDP 236
P GP + PG PP P G D PG PG RG+ F G +VGP P
Sbjct: 953 PQGPTGF-PGPKGPPGPPGK--DGLPGHPG-----QRGETGFQGKTGPPGPPGVVGPQGP 1004
Query: 237 RWFSGGTGG-GIGGGPGFIG--GLPG--APPGARFDP--------IGPPGVPGFEPNRFA 283
+G G G G PG G GLPG G + DP GPPG+ GF +R
Sbjct: 1005 TGETGPMGERGHPGPPGPPGEQGLPGLAGKEGTKGDPGPAGLPGKDGPPGLRGFPGDRGL 1064
Query: 284 RNP 286
P
Sbjct: 1065 PGP 1067
Score = 36.6 bits (83), Expect = 0.081
Identities = 44/130 (33%), Positives = 48/130 (36%), Gaps = 27/130 (20%)
Query: 185 PAGPNDYHPGFIVPPVPA------GSGSDLFPG-PGAGMYPSRGDHDFGGSMLVGPNDPR 237
P GP VP +P GS FPG PGA + + GG G PR
Sbjct: 857 PLGPPGEKGKLGVPGLPGYPGRQGPKGSIGFPGFPGA-------NGEKGGRGTPGKPGPR 909
Query: 238 WFSGGTGG-------GIGGGPGFIGGL----PGAPPGARFDPIGPPGVPGFE-PNRFARN 285
G TG GI G PG G P PPG R P GP G GF P
Sbjct: 910 GQRGPTGPRGERGPRGITGKPGPKGNSGGDGPAGPPGER-GPNGPQGPTGFPGPKGPPGP 968
Query: 286 PRRPGFDAHP 295
P + G HP
Sbjct: 969 PGKDGLPGHP 978
Score = 35.8 bits (81), Expect = 0.14
Identities = 49/157 (31%), Positives = 61/157 (38%), Gaps = 49/157 (31%)
Query: 179 VPGFGEPAGPNDYHPGFIVPP-------VPAGSGSDLFPGP----GA----GMYPSRGDH 223
+PG P G HPG PP P G +PGP GA G+ ++G+
Sbjct: 755 MPGADGPPG----HPGKEGPPGEKGGQGPPGPQGPIGYPGPRGVKGADGIRGLKGTKGEK 810
Query: 224 D------FGGSM-------LVGPNDPRWFSGGTG----GGIGGGPGFIG--------GLP 258
F G M +GP PR G G GG G PG +G G+P
Sbjct: 811 GEDGFPGFKGDMGIKGDRGEIGPPGPRGEDGPEGPKGRGGPNGDPGPLGPPGEKGKLGVP 870
Query: 259 GAP--PGAR--FDPIGPPGVPGFEPNRFAR-NPRRPG 290
G P PG + IG PG PG + R P +PG
Sbjct: 871 GLPGYPGRQGPKGSIGFPGFPGANGEKGGRGTPGKPG 907
Score = 35.8 bits (81), Expect = 0.14
Identities = 34/109 (31%), Positives = 40/109 (36%), Gaps = 10/109 (9%)
Query: 179 VPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPND--- 235
+PG P G + G P G D PGP P D + G VGP
Sbjct: 620 MPGQTGPKGDRGFD-GLAGLPGEKGHRGD--PGPSGPPGPPGDDGERGDDGEVGPRGLPG 676
Query: 236 ---PRWFSGGTGGGIGGGPGFIGGLPGAP-PGARFDPIGPPGVPGFEPN 280
PR G G GP + G+ G P P P G PG PG + N
Sbjct: 677 KPGPRGLLGPKGPPGPPGPPGVTGMDGQPGPKGNVGPQGEPGPPGQQGN 725
Score = 33.9 bits (76), Expect = 0.53
Identities = 34/101 (33%), Positives = 40/101 (38%), Gaps = 19/101 (18%)
Query: 178 PVPGFGEPAGPNDY-HPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDH-DFGGSMLVGPND 235
P G G P + PG P P+G PGP RG+ + G S GP
Sbjct: 1271 PPGGIGNPGAVGEKGEPGEAGEPGPSGRSGP--PGPKG----ERGEKGESGPSGAAGPPG 1324
Query: 236 PRWFSGGTGGGIGGGPGFIGGLPGAPPGARFDPIGPPGVPG 276
P+ G G G PG + G PG P GPPG PG
Sbjct: 1325 PKGPPGDDGP--KGSPGPV-GFPGDP--------GPPGEPG 1354
Score = 33.9 bits (76), Expect = 0.53
Identities = 33/94 (35%), Positives = 39/94 (41%), Gaps = 27/94 (28%)
Query: 185 PAGPNDYHPGFIVPPVPAG-SGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGT 243
P GP PG PP P G +G D PGP + P G P G
Sbjct: 686 PKGP----PG---PPGPPGVTGMDGQPGPKGNVGPQ------------GEPGPPGQQGNP 726
Query: 244 GG-GIGGGPGFIGGLPGAPPGARFDPIGPPGVPG 276
G G+ G G IG PPG + P+G PG+PG
Sbjct: 727 GAQGLPGPQGAIG-----PPGEK-GPLGKPGLPG 754
Score = 33.1 bits (74), Expect = 0.90
Identities = 47/167 (28%), Positives = 57/167 (33%), Gaps = 38/167 (22%)
Query: 173 PETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPG-----PGA-------GMYPSR 220
P + +PG P GP PG + PP G D P PG G +
Sbjct: 1445 PVGEQGLPGSPGPDGP----PGPMGPPGLPGLKGDSGPKGEKGHPGLIGLIGPPGEQGEK 1500
Query: 221 GDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPGAPPGARF-------------- 266
GD G G + P+ G TG GP GLPG PPG +
Sbjct: 1501 GDRGLPGPQ--GSSGPKGEQGITGPSGPIGPPGPPGLPG-PPGPKGAKGSSGPTGPRGEA 1557
Query: 267 ---DPIGPPGVPG--FEPNRFARNPRRPGFDAHPDLQHFRRDTDSDY 308
P GPPG PG +P + R DA L + DY
Sbjct: 1558 GHPGPPGPPGPPGEVIQPLPIQASRTRRNIDASQLLDDGNGENYVDY 1604
Score = 32.7 bits (73), Expect = 1.2
Identities = 35/106 (33%), Positives = 39/106 (36%), Gaps = 15/106 (14%)
Query: 173 PETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVG 232
P PG P GP PG I P G + PG PS G S G
Sbjct: 1256 PRGPSGAPGADGPQGP----PGGIGNPGAVGEKGE--PGEAGEPGPS------GRSGPPG 1303
Query: 233 PNDPRWFSGGTGGGIGGGPGFIGGLPG--APPGARFDPIGPPGVPG 276
P R G +G GP G PG P G+ P+G PG PG
Sbjct: 1304 PKGERGEKGESGPSGAAGPPGPKGPPGDDGPKGSP-GPVGFPGDPG 1348
Score = 32.7 bits (73), Expect = 1.2
Identities = 40/121 (33%), Positives = 46/121 (37%), Gaps = 32/121 (26%)
Query: 180 PGFGEPAGPNDY--------HPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLV 231
PG P GP HPG PP P G PG AG ++GD G + L
Sbjct: 996 PGVVGPQGPTGETGPMGERGHPG---PPGPPGEQG--LPGL-AGKEGTKGDP--GPAGLP 1047
Query: 232 GPNDPRWFSGGTGG-GIGGGPGFIG-----GLPGAP----------PGARFDPIGPPGVP 275
G + P G G G+ G G +G G PG P P PIG PG P
Sbjct: 1048 GKDGPPGLRGFPGDRGLPGPVGALGLKGNEGPPGPPGPAGSPGERGPAGAAGPIGIPGRP 1107
Query: 276 G 276
G
Sbjct: 1108 G 1108
Score = 32.7 bits (73), Expect = 1.2
Identities = 36/98 (36%), Positives = 39/98 (39%), Gaps = 22/98 (22%)
Query: 194 GFIVPPVPAGSGSDLFPGPGAGMYPSRGDH-DFGGSMLVGPNDPRWFSGGTG-------- 244
GF PP P G PGP +G+ D G GP PR SG G
Sbjct: 1219 GFPGPPGPVGLQG--LPGPPG----EKGETGDVGQMGPPGPPGPRGPSGAPGADGPQGPP 1272
Query: 245 GGIGGGPGFIG--GLPGAP----PGARFDPIGPPGVPG 276
GGI G PG +G G PG P R P GP G G
Sbjct: 1273 GGI-GNPGAVGEKGEPGEAGEPGPSGRSGPPGPKGERG 1309
Score = 32.7 bits (73), Expect = 1.2
Identities = 36/105 (34%), Positives = 41/105 (38%), Gaps = 9/105 (8%)
Query: 185 PAGPNDY--HPGFIVPPVPAG---SGSDLFP-GPGAGMYPSRGDHDFGGSMLVGPNDPRW 238
PAGP PG + PP G D+ P GP P G G + R
Sbjct: 560 PAGPMGLTGRPGPVGPPGSGGLKGEPGDVGPQGPRGVQGPPGPAGKPGRRGRAGSDGARG 619
Query: 239 FSGGTGGGIGGGPGFIGGLPGAPPGARFDP--IGPPGVPGFEPNR 281
G TG G + GLPG G R DP GPPG PG + R
Sbjct: 620 MPGQTGPKGDRGFDGLAGLPGEK-GHRGDPGPSGPPGPPGDDGER 663
Score = 32.3 bits (72), Expect = 1.5
Identities = 28/72 (38%), Positives = 31/72 (42%), Gaps = 9/72 (12%)
Query: 210 PGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPGAPPGARFD-- 267
PGP S GD G GPN P+ +G G GP GLPG PG R +
Sbjct: 930 PGPKGN---SGGDGPAGPPGERGPNGPQGPTGFPGPKGPPGPPGKDGLPG-HPGQRGETG 985
Query: 268 ---PIGPPGVPG 276
GPPG PG
Sbjct: 986 FQGKTGPPGPPG 997
Score = 31.2 bits (69), Expect = 3.4
Identities = 41/127 (32%), Positives = 44/127 (34%), Gaps = 35/127 (27%)
Query: 177 EPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGA--GMYPSRGDHDFGGSMLVGPN 234
E PG P GP P P G PGP G+ S G G M GP
Sbjct: 1416 EGPPGKTGPIGPQG------APGKPGPDGLRGIPGPVGEQGLPGSPGPDGPPGPM--GPP 1467
Query: 235 DPRWFSGGTGG-GIGGGPGFIG--------------GLPG-----APPGAR-----FDPI 269
G +G G G PG IG GLPG P G + PI
Sbjct: 1468 GLPGLKGDSGPKGEKGHPGLIGLIGPPGEQGEKGDRGLPGPQGSSGPKGEQGITGPSGPI 1527
Query: 270 GPPGVPG 276
GPPG PG
Sbjct: 1528 GPPGPPG 1534
Score = 30.0 bits (66), Expect = 7.6
Identities = 39/123 (31%), Positives = 43/123 (34%), Gaps = 13/123 (10%)
Query: 174 ETSEPVP-GFGEPAGPNDYH--------PGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHD 224
E EP P G P GP G PP P G D P G GD
Sbjct: 1289 EAGEPGPSGRSGPPGPKGERGEKGESGPSGAAGPPGPKGPPGDDGPKGSPGPVGFPGDPG 1348
Query: 225 FGGSM-LVGPNDPRWFSGGTGG-GIGGGPGFIGGL-PGAPPGARFDPIGPPGVPGFEPNR 281
G G + P G G G G PG G P PPG R P GP G G + +
Sbjct: 1349 PPGEPGPAGQDGPPGDKGDDGEPGQTGSPGPTGEPGPSGPPGKRGPP-GPAGPEGRQGEK 1407
Query: 282 FAR 284
A+
Sbjct: 1408 GAK 1410
>CA1A_CHICK (P08125) Collagen alpha 1(X) chain precursor
Length = 674
Score = 47.8 bits (112), Expect = 4e-05
Identities = 50/139 (35%), Positives = 57/139 (40%), Gaps = 32/139 (23%)
Query: 183 GEPAGPNDYHPGFIVPPVPAG-SGSDLFPGPGAGMYPSRGDHDFGGSMLVG------PND 235
GEP PG + PP PAG G+ PGP AG+ S G FG L G P
Sbjct: 249 GEPGEVGIGKPGPMGPPGPAGIPGAKGLPGP-AGLPGSPGLPGFGKPGLPGMKGHRGPEG 307
Query: 236 PRWFSGGTGG-GIGGGPGFIG-----------GLPGAP-----PGARFD--PIGPPGVPG 276
P F G G G G PG +G GL G P PG + D P+GP G PG
Sbjct: 308 PPGFPGPKGDQGPAGVPGELGPAGPQGNMGPQGLKGLPGENGLPGPKGDMGPVGPAGFPG 367
Query: 277 FEPNRFARNPRRPGFDAHP 295
+ R PG D P
Sbjct: 368 AKGER-----GLPGLDGKP 381
Score = 42.0 bits (97), Expect = 0.002
Identities = 49/153 (32%), Positives = 60/153 (39%), Gaps = 39/153 (25%)
Query: 176 SEPVPGFGEPAGPN-------DYHPGFIVPPV---PAGSGSDLFP-GPGAGMYPSR---- 220
S +PGFG+P P + PGF P PAG +L P GP M P
Sbjct: 285 SPGLPGFGKPGLPGMKGHRGPEGPPGFPGPKGDQGPAGVPGELGPAGPQGNMGPQGLKGL 344
Query: 221 -GDHDFGGSML-VGPNDPRWFSGGTGG----GIGGGPGFIG--GLPGAP-----PGARFD 267
G++ G +GP P F G G G+ G PG+ G GLPG PG + D
Sbjct: 345 PGENGLPGPKGDMGPVGPAGFPGAKGERGLPGLDGKPGYPGEQGLPGPKGHPGLPGQKGD 404
Query: 268 -----------PIGPPGVPGFEPNRFARNPRRP 289
P+GP GV G PR P
Sbjct: 405 TGHAGHPGLPGPVGPQGVKGVPGINGEPGPRGP 437
Score = 40.0 bits (92), Expect = 0.007
Identities = 32/79 (40%), Positives = 36/79 (45%), Gaps = 8/79 (10%)
Query: 201 PAGSGSDLFPGPGA--GMYPSRGDHDFGGSM-LVGPNDPRWFSGGTGGGIGGGPGFIGGL 257
P G PGP G+ +GD G L GP P+ G G I G PG G
Sbjct: 381 PGYPGEQGLPGPKGHPGLPGQKGDTGHAGHPGLPGPVGPQGVKGVPG--INGEPGPRG-- 436
Query: 258 PGAPPGARFDPIGPPGVPG 276
P PG R PIGPPG+PG
Sbjct: 437 PSGIPGVR-GPIGPPGMPG 454
Score = 38.1 bits (87), Expect = 0.028
Identities = 43/131 (32%), Positives = 51/131 (38%), Gaps = 24/131 (18%)
Query: 181 GFGEPAGPNDYHPGFIVPPVPAG-SGSDLFPGPGAGMYPS----RGDHDFGGSM----LV 231
GFG P P + G P P G GS PG P +GD G+ +
Sbjct: 185 GFGVPGRPGNR--GLPGPQGPQGLPGSAGIGKPGENGLPGQPGMKGDRGLPGARGEAGIP 242
Query: 232 GPNDPRWFSGGTGGGIGG--GPGFIGGLPGA-----PPGARFDP----IGPPGVPGFEPN 280
GP P G G G G GP G+PGA P G P G PG+PG + +
Sbjct: 243 GPQGPPGEPGEVGIGKPGPMGPPGPAGIPGAKGLPGPAGLPGSPGLPGFGKPGLPGMKGH 302
Query: 281 RFARNPRRPGF 291
R P PGF
Sbjct: 303 RGPEGP--PGF 311
Score = 37.0 bits (84), Expect = 0.062
Identities = 45/147 (30%), Positives = 56/147 (37%), Gaps = 48/147 (32%)
Query: 185 PAGPNDYHPGFIVPPVPAGSGSDLFPGPGA-GMYPSRGDHDFG------GSMLVGPNDPR 237
P GP PG P P G + PG G+ +G+ FG L GP P+
Sbjct: 153 PQGP----PGAQGPRGPPGEKGE----PGVPGINGQKGEMGFGVPGRPGNRGLPGPQGPQ 204
Query: 238 WFSGGTG------GGIGGGPGFIG--GLPGA-----------PPG-------ARFDPIGP 271
G G G+ G PG G GLPGA PPG + P+GP
Sbjct: 205 GLPGSAGIGKPGENGLPGQPGMKGDRGLPGARGEAGIPGPQGPPGEPGEVGIGKPGPMGP 264
Query: 272 P---GVPGFE----PNRFARNPRRPGF 291
P G+PG + P +P PGF
Sbjct: 265 PGPAGIPGAKGLPGPAGLPGSPGLPGF 291
Score = 32.0 bits (71), Expect = 2.0
Identities = 33/92 (35%), Positives = 35/92 (37%), Gaps = 23/92 (25%)
Query: 198 PPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIG-- 255
PP P G PGP G FG GP P G + G G PG G
Sbjct: 60 PPGPIGPRGQ--PGPA-------GKPGFGSPGPQGPPGPLGPPGFSTVGKLGMPGLPGKP 110
Query: 256 ---GLPGAP--------PGARFDPIGPPGVPG 276
GL G PGAR P GPPG+PG
Sbjct: 111 GERGLNGEKGEAGPVGLPGAR-GPQGPPGIPG 141
Score = 32.0 bits (71), Expect = 2.0
Identities = 37/112 (33%), Positives = 41/112 (36%), Gaps = 22/112 (19%)
Query: 180 PGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWF 239
PG P GP V VP +G PGP RG G + GP P
Sbjct: 411 PGLPGPVGPQG------VKGVPGINGE---PGP-------RGPSGIPG--VRGPIGPPGM 452
Query: 240 SGGTGG-GIGGGPGFIGGLPGAPPGARFDPIGPPGVPGFEPNRFARNPRRPG 290
G G G G PG G G R P+GP G PG P + P PG
Sbjct: 453 PGAPGAKGEAGAPGLPGPAGIVTKGLR-GPMGPLGPPG--PKGNSGEPGLPG 501
>CCR6_CAEEL (P20784) Cuticle collagen rol-6
Length = 348
Score = 47.4 bits (111), Expect = 5e-05
Identities = 48/148 (32%), Positives = 57/148 (38%), Gaps = 30/148 (20%)
Query: 175 TSEPVPGFG--EPAGPNDYHPGFIVPPVPAGSGSDLFPGPGA--GMYPSRGDHDFGG--- 227
T P G+G +PA P + G +P G FPG G G +P+ G G
Sbjct: 88 TPNPYGGYGASQPAPPEKFPDG-----IPNGGNQPKFPGGGFPDGPFPNGGGPRGGNQCQ 142
Query: 228 -----SMLVGPNDPRWFSGGTG-GGIGGGPGFIG----GLPGAPPGARFD-------PIG 270
S GP P G G G G PGF G + PP F P G
Sbjct: 143 CTVENSCPPGPAGPEGEEGPDGHDGQDGVPGFDGKDAEDVQNTPPTGCFTCPQGPLGPQG 202
Query: 271 PPGVPGFEPNRFAR-NPRRPGFDAHPDL 297
P G PG R AR P RPG D +P +
Sbjct: 203 PNGAPGLRGMRGARGQPGRPGRDGNPGM 230
Score = 33.5 bits (75), Expect = 0.69
Identities = 38/114 (33%), Positives = 42/114 (36%), Gaps = 14/114 (12%)
Query: 183 GEPAGPNDYHPGFIVPPVPAGSGSDLFPG-PGA----GMYPSRGDHDFGGSMLVGPNDPR 237
G P P D P P GSD PG PG G P G GP P+
Sbjct: 226 GNPGMPGDCGP-------PGAPGSDGKPGSPGGKGDDGERPLGRPGPRGPPGEAGPEGPQ 278
Query: 238 WFSGGTGGGIGGGPGFIGGLPGAPPGARFD-PIGPPGVPGFEPNRFARNPRRPG 290
+G GP GL G A D P GPPG PG P + A + PG
Sbjct: 279 GPTGRDAYPGQSGPQGEPGLQGYGGAAGEDGPEGPPGAPGL-PGKDAEYCKCPG 331
>CA1A_MOUSE (Q05306) Collagen alpha 1(X) chain precursor
Length = 680
Score = 47.4 bits (111), Expect = 5e-05
Identities = 44/113 (38%), Positives = 51/113 (44%), Gaps = 22/113 (19%)
Query: 181 GFGEPAGPNDYH----PGFIVPPVPAG---SGSDLFPG-PGAGMYPSRGDHDFGGSM-LV 231
G+G P P + G I PP P+G G + FPG PG +GD F G M
Sbjct: 192 GYGSPGRPGERGLPGPQGPIGPPGPSGVGRRGENGFPGQPGI-----KGDRGFPGEMGPS 246
Query: 232 GPNDPRWFSGGTGGGIGGGPGFIGGLPGAP--PGARFDP-----IGPPGVPGF 277
GP P+ G G G PG IG PG P PG + P GPPG PGF
Sbjct: 247 GPPGPQGPPGKQGREGIGKPGAIGS-PGQPGIPGEKGHPGAPGIAGPPGAPGF 298
Score = 41.2 bits (95), Expect = 0.003
Identities = 43/121 (35%), Positives = 50/121 (40%), Gaps = 20/121 (16%)
Query: 181 GFGEPAGPNDYHPGFIVPPVPAG-SGSDLFPG-PGA-------GMYPSRGDHDFGGSM-L 230
G PAGP PG P G G +PG PG G+ +GD GG+ L
Sbjct: 365 GLVGPAGP----PGARGARGPPGLDGKTGYPGEPGLNGPKGNPGLPGQKGDPGVGGTPGL 420
Query: 231 VGPNDPRWFSGGTGGGIGGGPGFIGGLPGAPPGARFDPIGPPGVPGFEPNR-FARNPRRP 289
GP P G G GP G+PG P GPPGVPGF ++ NP P
Sbjct: 421 RGPVGPVGAKGVPGHNGEAGPRGEPGIPGTR-----GPTGPPGVPGFPGSKGDPGNPGAP 475
Query: 290 G 290
G
Sbjct: 476 G 476
Score = 37.4 bits (85), Expect = 0.048
Identities = 41/122 (33%), Positives = 46/122 (37%), Gaps = 10/122 (8%)
Query: 179 VPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRW 238
+PG P GP HPG PP G GS G G+ G G L GP
Sbjct: 61 IPGPPGPTGPRG-HPGPSGPPGKPGYGSPGLQGE-PGLPGPPGISATGKPGLPGPPGKPG 118
Query: 239 FSGGTGGGIGGGPGFIGGLPGAPPGARFDPIGPPGVPGFEPNRFARNPRRPGFDAHPDLQ 298
G G GP GLPG P G P GPPG+PG P + G P +
Sbjct: 119 ERGPYGHKGDIGPA---GLPG-PRG----PPGPPGIPGPAGISVPGKPGQQGLTGAPGPR 170
Query: 299 HF 300
F
Sbjct: 171 GF 172
Score = 33.5 bits (75), Expect = 0.69
Identities = 28/64 (43%), Positives = 31/64 (47%), Gaps = 10/64 (15%)
Query: 218 PSRGDHDFGGSMLVGPNDPRWFSGGTG--GGIG-GGPGFIG--GLPGAPPGARFDPIGPP 272
P RG+ G GP PR G +G G G G PG G GLPG PPG G P
Sbjct: 54 PVRGEQGIPGPP--GPTGPRGHPGPSGPPGKPGYGSPGLQGEPGLPG-PPG--ISATGKP 108
Query: 273 GVPG 276
G+PG
Sbjct: 109 GLPG 112
Score = 29.6 bits (65), Expect = 9.9
Identities = 19/45 (42%), Positives = 20/45 (44%), Gaps = 6/45 (13%)
Query: 246 GIGGGPGFIGGLPGAPPGARFDPIGPPGVPGFEPNRFARNPRRPG 290
GI G PG G P PG P GPPG PG+ P PG
Sbjct: 60 GIPGPPGPTG--PRGHPG----PSGPPGKPGYGSPGLQGEPGLPG 98
>CA11_CHICK (P02457) Collagen alpha 1(I) chain precursor
Length = 1453
Score = 47.4 bits (111), Expect = 5e-05
Identities = 50/130 (38%), Positives = 55/130 (41%), Gaps = 21/130 (16%)
Query: 180 PGFGEPAGP--NDYHPGFIVPPVPAG-SGSDLFPGPG-----AGMYPSRGDHDFGGSM-L 230
PG PAG ND PG PP P G +G FPG G +RG GS
Sbjct: 302 PGPSGPAGARGNDGAPGAAGPPGPTGPAGPPGFPGAAGAKGETGPQGARGSEGPQGSRGE 361
Query: 231 VGPNDPRWFSGGTGG-GIGGGPGFIG-----GLPGAP--PGAR--FDPIGPPGVPGFEPN 280
GP P +G G G G PG G G+ GAP PGAR P GP G PG P
Sbjct: 362 PGPPGPAGAAGPAGNPGADGQPGAKGATGAPGIAGAPGFPGARGPSGPQGPSGAPG--PK 419
Query: 281 RFARNPRRPG 290
+ P PG
Sbjct: 420 GNSGEPGAPG 429
Score = 39.7 bits (91), Expect = 0.010
Identities = 47/135 (34%), Positives = 51/135 (36%), Gaps = 19/135 (14%)
Query: 173 PETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVG 232
P + +PG GP PG PP P G G + P G G M G
Sbjct: 121 PPGRDGIPGQPGLPGP----PG---PPGPPGLGGNFAPQMSYGYDEKSAGVAVPGPM--G 171
Query: 233 PNDPRWFSGGTGG----GIGGGPGFIGGLPGA--PPGARFDPIGPPGVPGFEPNRFARNP 286
P PR G G G G PG G PGA P G R P GPPG G + A P
Sbjct: 172 PAGPRGLPGPPGAPGPQGFQGPPGEPGE-PGASGPMGPR-GPAGPPGKNGDDGE--AGKP 227
Query: 287 RRPGFDAHPDLQHFR 301
RPG P Q R
Sbjct: 228 GRPGQRGPPGPQGAR 242
Score = 39.3 bits (90), Expect = 0.013
Identities = 51/157 (32%), Positives = 59/157 (37%), Gaps = 46/157 (29%)
Query: 185 PAGPNDYHPGFIVPPVPAGSGSDLFP-GPGA--GMYPSRGDHDFGG----SMLVGPNDPR 237
PAGP PP PAG+ P GP G +GD G S L GP P
Sbjct: 1060 PAGPAG-------PPGPAGARGPAGPQGPRGDKGETGEQGDRGMKGHRGFSGLQGPPGPP 1112
Query: 238 WFSGGTG-------GGIGGGPGFIG--------GLPG--APPGAR-----FDPIGPPGVP 275
G G G G PG G GLPG PPG R P+GPPG P
Sbjct: 1113 GAPGEQGPSGASGPAGPRGPPGSAGAAGKDGLNGLPGPIGPPGPRGRTGEVGPVGPPGPP 1172
Query: 276 GFEPN---------RFARNPRRPGFDAHPDLQHFRRD 303
G P F+ P+ P AH +++R D
Sbjct: 1173 G-PPGPPGPPSGGFDFSFLPQPPQEKAHDGGRYYRAD 1208
Score = 38.9 bits (89), Expect = 0.016
Identities = 39/107 (36%), Positives = 41/107 (37%), Gaps = 17/107 (15%)
Query: 185 PAGPNDYH--PGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGG 242
PAGP PG P PAG D AG P+ G GP PR G
Sbjct: 1030 PAGPPGAPGAPGAPGPVGPAGKNGDRGETGPAG--PAGPPGPAGARGPAGPQGPRGDKGE 1087
Query: 243 TGG----GIGGGPGFIG-----GLPGAP----PGARFDPIGPPGVPG 276
TG G+ G GF G G PGAP P P GP G PG
Sbjct: 1088 TGEQGDRGMKGHRGFSGLQGPPGPPGAPGEQGPSGASGPAGPRGPPG 1134
Score = 37.0 bits (84), Expect = 0.062
Identities = 44/136 (32%), Positives = 48/136 (34%), Gaps = 15/136 (11%)
Query: 152 LDGTAHASSSTKSSKTSDSTRPETSEPVPGFGEPAGPNDYHP-----GFIVPPVPAGS-- 204
L G + S S T P G PAGP GF P AG
Sbjct: 523 LPGAKGLTGSPGSPGPDGKTGPPGPAGQDGRPGPAGPPGARGQAGVMGFPGPKGAAGEPG 582
Query: 205 --GSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPG--A 260
G PGP + + D + G GP P G G G PGF GLPG
Sbjct: 583 KPGERGAPGPPGAVGAAGKDGEAGAQGPPGPTGPAGERGEQGPA--GAPGF-QGLPGPAG 639
Query: 261 PPGARFDPIGPPGVPG 276
PPG P G GVPG
Sbjct: 640 PPGEAGKP-GEQGVPG 654
Score = 36.6 bits (83), Expect = 0.081
Identities = 53/188 (28%), Positives = 64/188 (33%), Gaps = 62/188 (32%)
Query: 153 DGTAHASSSTKSSKTSDSTRPETSEPVPGFGE---PAGPNDYHPGFIVPP----VPAGSG 205
DG A A + + + PGF PAGP PG P VP +G
Sbjct: 602 DGEAGAQGPPGPTGPAGERGEQGPAGAPGFQGLPGPAGP----PGEAGKPGEQGVPGNAG 657
Query: 206 SDLFPGPGAGMYPSRGDHDFGGSMLV----GPNDPRWFSGGTGG----GIGGGPGFIG-- 255
+ PGP +RG+ F G V GP PR +G G G G PG G
Sbjct: 658 A---PGPAG----ARGERGFPGERGVQGPPGPQGPRGANGAPGNDGAKGDAGAPGAPGNE 710
Query: 256 ---GLPGAP--------PGARFD-----------------------PIGPPGVPGFEPNR 281
GL G P PGA+ D PIGPPG G ++
Sbjct: 711 GPPGLEGMPGERGAAGLPGAKGDRGDPGPKGADGAPGKDGLRGLTGPIGPPGPAGAPGDK 770
Query: 282 FARNPRRP 289
P P
Sbjct: 771 GEAGPPGP 778
Score = 35.4 bits (80), Expect = 0.18
Identities = 45/140 (32%), Positives = 52/140 (37%), Gaps = 52/140 (37%)
Query: 180 PGFGEPAGPNDYHPGFIVPPVPAGS-------GSDLFPGPGA--------GMYPSRGDHD 224
PG PAGP PG PP GS G+ PGP G+ RG+
Sbjct: 908 PGEPGPAGP----PG---PPGEKGSPGADGPIGAPGTPGPQGIAGQRGVVGLPGQRGERG 960
Query: 225 FGGSMLVGPNDPRWFSGGTGG-------------GIGGGPGFIG--GLPGA--------- 260
F G L GP+ G +G G+ G PG G G PGA
Sbjct: 961 FPG--LPGPSGEPGKQGPSGASGERGPPGPMGPPGLAGPPGEAGREGAPGAEGAPGRDGA 1018
Query: 261 --PPGARFD--PIGPPGVPG 276
P G R + P GPPG PG
Sbjct: 1019 AGPKGDRGETGPAGPPGAPG 1038
Score = 34.7 bits (78), Expect = 0.31
Identities = 40/122 (32%), Positives = 46/122 (36%), Gaps = 35/122 (28%)
Query: 183 GEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGG----SMLVGPNDPRW 238
GE P PG PP P G+ PG AG+ +G F G GP P+
Sbjct: 222 GEAGKPG--RPGQRGPPGPQGARG--LPGT-AGLPGMKGHRGFSGLDGAKGQPGPAGPKG 276
Query: 239 FSGGTGGGIGGGPGFIG--GLPG--------APPGARFD--------------PIGPPGV 274
G G G PG +G GLPG P GAR + P GPPG
Sbjct: 277 EPGSPGEN--GAPGQMGPRGLPGERGRPGPSGPAGARGNDGAPGAAGPPGPTGPAGPPGF 334
Query: 275 PG 276
PG
Sbjct: 335 PG 336
Score = 34.3 bits (77), Expect = 0.40
Identities = 40/125 (32%), Positives = 44/125 (35%), Gaps = 23/125 (18%)
Query: 172 RPETSEPVP-GFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSML 230
R E P P GF P G D PG AG+ D P AG P+ G
Sbjct: 791 RGEPGPPGPAGFAGPPGA-DGQPGAKGETGDAGAKGDAGPPGPAG--PTGAPGPAGZVGA 847
Query: 231 VGPNDPRWFSGGTGG-GIGGGPGFIG--------GLPGAP----------PGARFDPIGP 271
GP R +G G G G G +G GLPG P P P G
Sbjct: 848 PGPKGARGSAGPPGATGFPGAAGRVGPPGPSGNIGLPGPPGPAGKZGSKGPRGETGPAGR 907
Query: 272 PGVPG 276
PG PG
Sbjct: 908 PGEPG 912
Score = 34.3 bits (77), Expect = 0.40
Identities = 35/96 (36%), Positives = 39/96 (40%), Gaps = 11/96 (11%)
Query: 185 PAGPNDYH--PGFIVPPVPAGSGSDLFPGPGAGMYPSRGDH-DFGGSMLVGPNDPRWFSG 241
PAGP PG P P G P G ++G+ D G GP P +G
Sbjct: 778 PAGPTGARGAPGDRGEPGPPGPAGFAGPPGADGQPGAKGETGDAGAKGDAGPPGP---AG 834
Query: 242 GTGGGIGGGPGFIGGLPGAPPGARFDPIGPPGVPGF 277
TG GP G PG P GAR GPPG GF
Sbjct: 835 PTGAP---GPAGZVGAPG-PKGAR-GSAGPPGATGF 865
Score = 33.9 bits (76), Expect = 0.53
Identities = 41/123 (33%), Positives = 47/123 (37%), Gaps = 26/123 (21%)
Query: 193 PGFIVPPVPAGS----GSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTG-GGI 247
PG P PAG GS FPG P + G VGP +G G G+
Sbjct: 464 PGPAGLPGPAGERGAPGSRGFPGADGIAGPKGPPGERGSPGAVGPKGSPGEAGRPGEAGL 523
Query: 248 GGGPGFIGGLPGA--------PPG-----ARFDPIGPP------GVPGFE-PNRFARNPR 287
G G + G PG+ PPG R P GPP GV GF P A P
Sbjct: 524 PGAKG-LTGSPGSPGPDGKTGPPGPAGQDGRPGPAGPPGARGQAGVMGFPGPKGAAGEPG 582
Query: 288 RPG 290
+PG
Sbjct: 583 KPG 585
Score = 33.9 bits (76), Expect = 0.53
Identities = 35/109 (32%), Positives = 40/109 (36%), Gaps = 16/109 (14%)
Query: 185 PAGPNDYH--PGFIVPPVPAGSGSDLFPGPGAGMYPSRGDH-----DFGGSMLVGPNDPR 237
PAGP G + P P G+ P PGA +P G L GP P
Sbjct: 832 PAGPTGAPGPAGZVGAPGPKGARGSAGP-PGATGFPGAAGRVGPPGPSGNIGLPGPPGPA 890
Query: 238 WFSGGTGGGIGGGPGFIGGLPGAPPGARFDPIGPPGVPGFEPNRFARNP 286
G G GP G PG P P GPPG PG + + A P
Sbjct: 891 GKZGSKGPRGETGPA---GRPGEP-----GPAGPPGPPGEKGSPGADGP 931
Score = 33.9 bits (76), Expect = 0.53
Identities = 38/115 (33%), Positives = 46/115 (39%), Gaps = 12/115 (10%)
Query: 183 GEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGG 242
G P P G PP PAG D PGP AG +RG + ++G P+ +G
Sbjct: 531 GSPGSPGP--DGKTGPPGPAGQ--DGRPGP-AGPPGARGQ-----AGVMGFPGPKGAAGE 580
Query: 243 TGG-GIGGGPGFIGGLPGAPPGARFDPIGPPGVPGFEPNRFARNPR-RPGFDAHP 295
G G G PG G + A GPPG G R + P PGF P
Sbjct: 581 PGKPGERGAPGPPGAVGAAGKDGEAGAQGPPGPTGPAGERGEQGPAGAPGFQGLP 635
>HRP_PLABE (Q08168) 58 kDa phosphoprotein (Heat shock-related
protein) (HRP)
Length = 423
Score = 46.6 bits (109), Expect = 8e-05
Identities = 42/146 (28%), Positives = 59/146 (39%), Gaps = 22/146 (15%)
Query: 135 KNLDKLVKKIDADILSKLDGTAHASSSTKSSKTSDSTRPETSEPVPGFGEPAGPNDYHPG 194
K +K K + ++ KL A K + ++ ++S+ + EP D+ G
Sbjct: 240 KEEEKQRLKREKELKKKLAAKKKAEKMYKENNKRENYDSDSSDS--SYSEPDFSGDFPGG 297
Query: 195 FIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFI 254
+P G PG GM G M P P F G GG GG PG +
Sbjct: 298 -----MPGG-----MPGMPGGM----------GGMGGMPGMPGGFPGMPGGMPGGMPGGM 337
Query: 255 GGLPGAPPGARFDPIGPPGVPGFEPN 280
GG+PG P G G PG+PG P+
Sbjct: 338 GGMPGMPGGMPGGMGGMPGMPGGMPD 363
Score = 45.1 bits (105), Expect = 2e-04
Identities = 47/158 (29%), Positives = 62/158 (38%), Gaps = 27/158 (17%)
Query: 132 QQFKNLDKLVKKIDADILSKLDGTAHASSSTKSSKTSDST--RPETSEPVPGFGEPAGPN 189
Q+ K +L KK+ A ++ + S +SDS+ P+ S PG G P G
Sbjct: 245 QRLKREKELKKKLAAKKKAEKMYKENNKRENYDSDSSDSSYSEPDFSGDFPG-GMPGG-- 301
Query: 190 DYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGG 249
+P +P G G G G P GG P P GG GG+GG
Sbjct: 302 -------MPGMPGGMG-------GMGGMPGMP----GGF----PGMPGGMPGGMPGGMGG 339
Query: 250 GPGFIGGLPGAPPGARFDPIGPPGVPGFEPNRFARNPR 287
PG GG+PG G P G P + E NP+
Sbjct: 340 MPGMPGGMPGGMGGMPGMPGGMPDLNSPEMKELFNNPQ 377
Score = 42.0 bits (97), Expect = 0.002
Identities = 32/77 (41%), Positives = 35/77 (44%), Gaps = 8/77 (10%)
Query: 224 DFGGSMLVGPNDPRWFSGGTGG--GIGGG-PGFIGGLPGAPPGARFDPIGPPGVPGFEPN 280
DF G M G GG GG G+ GG PG GG+PG PG G PG+PG P
Sbjct: 293 DFPGGMPGGMPGMPGGMGGMGGMPGMPGGFPGMPGGMPGGMPGGMG---GMPGMPGGMPG 349
Query: 281 RFARNPRRPGFDAHPDL 297
P PG PDL
Sbjct: 350 GMGGMPGMPG--GMPDL 364
Score = 39.7 bits (91), Expect = 0.010
Identities = 25/69 (36%), Positives = 30/69 (43%)
Query: 222 DHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGFIGGLPGAPPGARFDPIGPPGVPGFEPNR 281
D D S P+ F GG GG+ G PG +GG+ G P P P G+PG P
Sbjct: 277 DSDSSDSSYSEPDFSGDFPGGMPGGMPGMPGGMGGMGGMPGMPGGFPGMPGGMPGGMPGG 336
Query: 282 FARNPRRPG 290
P PG
Sbjct: 337 MGGMPGMPG 345
>ELS_MOUSE (P54320) Elastin precursor (Tropoelastin)
Length = 860
Score = 46.6 bits (109), Expect = 8e-05
Identities = 40/99 (40%), Positives = 47/99 (47%), Gaps = 12/99 (12%)
Query: 179 VPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRW 238
VPGFG AG + G VP AG+G F G GAG+ P G GS+
Sbjct: 616 VPGFGAGAGVPGFGAGAGVPGFGAGAGVPGF-GAGAGV-PGFGAGAVPGSLAASKAAKYG 673
Query: 239 FSGGTGGGIG-GGPGFIGGLPGAPPGARFDPIGPPGVPG 276
+GG GG G GGPG +GG PG +G GVPG
Sbjct: 674 AAGGLGGPGGLGGPGGLGG-PGG--------LGGAGVPG 703
Score = 33.9 bits (76), Expect = 0.53
Identities = 30/77 (38%), Positives = 33/77 (41%), Gaps = 10/77 (12%)
Query: 193 PGFIVPPVPAGSGSDLFPG--PGAGMYPSRGDHDFGGSM-LVGPND-PRWFSGGTGGGIG 248
PG + VP G PG PG YP G GG +GP P G G G
Sbjct: 27 PGGVPGAVPGG-----LPGGVPGGVYYPGAGIGGLGGGGGALGPGGKPPKPGAGLLGTFG 81
Query: 249 GGPGFIGGL-PGAPPGA 264
GPG +GG PGA GA
Sbjct: 82 AGPGGLGGAGPGAGLGA 98
Score = 32.7 bits (73), Expect = 1.2
Identities = 27/94 (28%), Positives = 35/94 (36%), Gaps = 22/94 (23%)
Query: 205 GSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFS-----------GGTGGGIGGGPGF 253
G+ + PG G G P G + G P + G GG + GGPG
Sbjct: 326 GAGVLPGVGGGGIPGGAGAIPGIGGIAGAGTPAAAAAAKAAAKAAKYGAAGGLVPGGPGV 385
Query: 254 ---------IGGLPGAP--PGARFDPIGPPGVPG 276
+GG+PG PG IG PG+ G
Sbjct: 386 RLPGAGIPGVGGIPGVGGIPGVGGPGIGGPGIVG 419
Score = 32.0 bits (71), Expect = 2.0
Identities = 34/104 (32%), Positives = 41/104 (38%), Gaps = 15/104 (14%)
Query: 194 GFIVPPVPAGSGSDLFPG--PGAGM---YPSRGDHDFGGSMLVGPNDPRWFSGGTG--GG 246
G +VP V AG G+ PG PG G+ YP G G+ G GTG
Sbjct: 183 GAVVPQVGAGIGAGGKPGKVPGVGLPGVYPG-GVLPGTGARFPGVGVLPGVPTGTGVKAK 241
Query: 247 IGGGPGFIGGLPGAPPGARFDPIGPPGVPGFEPNRFARNPRRPG 290
GG G G+PG P P P G P + P+ PG
Sbjct: 242 APGGGGAFSGIPGVGPFGGQQPGVPLGYP-------IKAPKLPG 278
Score = 32.0 bits (71), Expect = 2.0
Identities = 26/84 (30%), Positives = 34/84 (39%), Gaps = 10/84 (11%)
Query: 194 GFIVPPVPAGSGSDLFPG-PGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPG 252
G +VP G+ P PGAG P G + + G GG+ GG G
Sbjct: 494 GGLVPGAVPGALPGAVPAVPGAGGVPGAGTPAAAAAAAAAKAAAKAGLGPGVGGVPGGVG 553
Query: 253 FIGGLPGAPPGARFDPIGPPGVPG 276
+GG+PG +G GVPG
Sbjct: 554 -VGGIPGG--------VGVGGVPG 568
Score = 31.6 bits (70), Expect = 2.6
Identities = 28/83 (33%), Positives = 32/83 (37%), Gaps = 6/83 (7%)
Query: 194 GFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVGPNDPRWFSGGTGGGIGGGPGF 253
G +P G+G FPG G G G G S + G GG G
Sbjct: 444 GVGIPTYGVGAGG--FPGYGVGAGAGLG----GASPAAAAAAAKAAKYGAGGAGALGGLV 497
Query: 254 IGGLPGAPPGARFDPIGPPGVPG 276
G +PGA PGA G GVPG
Sbjct: 498 PGAVPGALPGAVPAVPGAGGVPG 520
Score = 31.6 bits (70), Expect = 2.6
Identities = 35/108 (32%), Positives = 43/108 (39%), Gaps = 18/108 (16%)
Query: 173 PETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSRGDHDFGGSMLVG 232
P P PG G G PG + P G+G FP AG +P G GG+
Sbjct: 65 PGGKPPKPGAGL-LGTFGAGPGGLGGAGP-GAGLGAFP---AGTFPGAGALVPGGAAGAA 119
Query: 233 PNDPRWFSGGTG----GGIGGGPGFIGGLPGAPPGARFDPIGPPGVPG 276
G G GG+ GG G +GG+PG +G GVPG
Sbjct: 120 AAYKAAAKAGAGLGGVGGVPGGVG-VGGVPGG--------VGVGGVPG 158
Score = 30.0 bits (66), Expect = 7.6
Identities = 30/94 (31%), Positives = 36/94 (37%), Gaps = 5/94 (5%)
Query: 173 PETSEPVPGFGEPAGPNDYHPGFIVPPVPAGSGSDLFPGPGAGMYPSR-GDHDFGGSMLV 231
P VPG G G P A + + GPG G P G G + V
Sbjct: 506 PGAVPAVPGAGGVPGAGT--PAAAAAAAAAKAAAKAGLGPGVGGVPGGVGVGGIPGGVGV 563
Query: 232 GPNDPRWFSGGTGGGIGGGPGFIGGLPGAPPGAR 265
G P G GIG GPG +GG G+P A+
Sbjct: 564 G-GVPGGVGPGGVTGIGAGPGGLGG-AGSPAAAK 595
>DAH1_MOUSE (Q9QYB2) Dachshund homolog 1 (Dach1)
Length = 751
Score = 46.2 bits (108), Expect = 1e-04
Identities = 35/104 (33%), Positives = 46/104 (43%), Gaps = 5/104 (4%)
Query: 159 SSSTKSSKTSDSTRPETSEPVPGFGEPA--GPNDYHPGFIVPPVPAGSGSDLFPGPGAGM 216
S+S SS T+ ST TS P P G PA GP + P I + S + PG G
Sbjct: 21 STSASSSGTTTSTSSATSSPAPSIGPPASSGPTLFRPEPIASSA-SSSAAATVTSPGGGG 79
Query: 217 YPSRGDHDFGGSMLVGPN--DPRWFSGGTGGGIGGGPGFIGGLP 258
S G GG+ G + +P +G +GGG+ G G P
Sbjct: 80 GGSGGGGGSGGNGGGGGSNCNPSLAAGSSGGGVSAGGGGASSTP 123
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.138 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 45,776,173
Number of Sequences: 164201
Number of extensions: 2615873
Number of successful extensions: 13648
Number of sequences better than 10.0: 554
Number of HSP's better than 10.0 without gapping: 112
Number of HSP's successfully gapped in prelim test: 464
Number of HSP's that attempted gapping in prelim test: 8655
Number of HSP's gapped (non-prelim): 2887
length of query: 309
length of database: 59,974,054
effective HSP length: 110
effective length of query: 199
effective length of database: 41,911,944
effective search space: 8340476856
effective search space used: 8340476856
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 65 (29.6 bits)
Medicago: description of AC142224.1


