

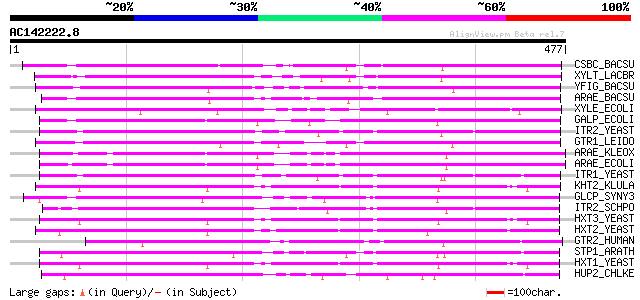
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142222.8 - phase: 0
(477 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CSBC_BACSU (P46333) Probable metabolite transport protein csbC 238 2e-62
XYLT_LACBR (O52733) D-xylose-proton symporter (D-xylose transpor... 237 6e-62
YFIG_BACSU (P54723) Hypothetical metabolite transport protein yfiG 207 5e-53
ARAE_BACSU (P96710) Arabinose-proton symporter (Arabinose transp... 207 5e-53
XYLE_ECOLI (P09098) D-xylose-proton symporter (D-xylose transpor... 200 6e-51
GALP_ECOLI (P37021) Galactose-proton symporter (Galactose transp... 193 9e-49
ITR2_YEAST (P30606) Myo-inositol transporter 2 185 3e-46
GTR1_LEIDO (Q01440) Membrane transporter D1 184 3e-46
ARAE_KLEOX (P45598) Arabinose-proton symporter (Arabinose transp... 184 4e-46
ARAE_ECOLI (P09830) Arabinose-proton symporter (Arabinose transp... 183 7e-46
ITR1_YEAST (P30605) Myo-inositol transporter 1 180 6e-45
KHT2_KLULA (P53387) Hexose transporter 2 179 2e-44
GLCP_SYNY3 (P15729) Glucose transport protein 178 2e-44
ITR2_SCHPO (P87110) Myo-inositol transporter 2 176 2e-43
HXT3_YEAST (P32466) Low-affinity glucose transporter HXT3 175 2e-43
HXT2_YEAST (P23585) High-affinity glucose transporter HXT2 173 8e-43
GTR2_HUMAN (P11168) Solute carrier family 2, facilitated glucose... 171 5e-42
STP1_ARATH (P23586) Glucose transporter (Sugar carrier) 169 1e-41
HXT1_YEAST (P32465) Low-affinity glucose transporter HXT1 169 1e-41
HUP2_CHLKE (Q39524) H(+)/hexose cotransporter 2 (Galactose-H+ sy... 167 4e-41
>CSBC_BACSU (P46333) Probable metabolite transport protein csbC
Length = 461
Score = 238 bits (608), Expect = 2e-62
Identities = 153/470 (32%), Positives = 251/470 (52%), Gaps = 38/470 (8%)
Query: 12 LPKTCLKNLRFLFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGS 71
+ K K + + F A GGLL+GYD G S A + I + L + GL+ S
Sbjct: 1 MKKDTRKYMIYFFGALGGLLYGYDTGVISGALLFINND-------IPLTTLTEGLVVSML 53
Query: 72 LYGALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGL 131
L GA+ GS L+ +D GRR+ + V ++++++GAL AF+ +L+ R++ G+ +G
Sbjct: 54 LLGAIFGSALSGTCSDRWGRRKVVFVLSIIFIIGALACAFSQTIGMLIASRVILGLAVGG 113
Query: 132 AMHAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPV 191
+ P+Y++E APT IRG L ++ IV GI+ Y + L A WR+M G+++
Sbjct: 114 STALVPVYLSEMAPTKIRGTLGTMNNLMIVTGILLAYIVNYLFTPFEA-WRWMVGLAAVP 172
Query: 192 AVIMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIM 251
AV++ G+ ++P SPRW++ R +++ R + + HD P+ ++ +
Sbjct: 173 AVLLLIGIAFMPESPRWLVKRGSEEEA----------RRIMNIT----HD--PKDIEMEL 216
Query: 252 AEFSY-LGEENDVTLGEMFRGKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCIN 307
AE E+ + TLG + R L+I GL +FQQ I TV + PT
Sbjct: 217 AEMKQGEAEKKETTLGVLKAKWIRPMLLIGVGLAIFQQAVGINTVIYYAPT------IFT 270
Query: 308 PTGFSLAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYI 367
G +A A ++ +G+ +IM A++++DR+GR+ LL+ G GI +SL L +
Sbjct: 271 KAGLGTSASALG-TMGIGILNVIMCITAMILIDRVGRKKLLIWGSVGITLSLAALSGVLL 329
Query: 368 FLD---NAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANA 424
L + A + VV L +Y+ YQ ++GP+ W+++ E+FP + RG LV AAN
Sbjct: 330 TLGLSASTAWMTVVFLGVYIVFYQATWGPVVWVLMPELFPSKARGAATGFTTLVLSAANL 389
Query: 425 LVTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEA 474
+V+ F + +G +F +FS I + S F +++VPETKG +LEEIEA
Sbjct: 390 IVSLVFPLMLSAMGIAWVFMVFSVICLLSFFFAFYMVPETKGKSLEEIEA 439
>XYLT_LACBR (O52733) D-xylose-proton symporter (D-xylose
transporter)
Length = 457
Score = 237 bits (604), Expect = 6e-62
Identities = 153/462 (33%), Positives = 243/462 (52%), Gaps = 40/462 (8%)
Query: 22 FLFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVL 81
+ F A GGLLFGYD G S A + IQ G +W G + S L GA++G+ +
Sbjct: 10 YFFGALGGLLFGYDTGVISGAILFIQKQMNLG-SWQQ------GWVVSAVLLGAILGAAI 62
Query: 82 AFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIA 141
+D GRR+ LL++A+++ VGAL +AF+P F L+I R++ G+ +G A P Y+A
Sbjct: 63 IGPSSDRFGRRKLLLLSAIIFFVGALGSAFSPEFWTLIISRIILGMAVGAASALIPTYLA 122
Query: 142 ETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWW 201
E AP+ RG + SL + ++ GI+ Y GWR+M G ++ A ++ G
Sbjct: 123 ELAPSDKRGTVSSLFQLMVMTGILLAYITNYSFSGFYTGWRWMLGFAAIPAALLFLGGLI 182
Query: 202 LPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEEN 261
LP SPR+++ K G L + L HD + E + + E
Sbjct: 183 LPESPRFLV-----KSGHLDEARHV-------LDTMNKHDQVA-----VNKEINDIQESA 225
Query: 262 DVTLG---EMFRGKCRKALVISAGLVLFQQIQ---TVTDHGPTKCSLLRCINPTGFSLAA 315
+ G E+F R +L+I GL +FQQ+ TV + PT GF ++A
Sbjct: 226 KIVSGGWSELFGKMVRPSLIIGIGLAIFQQVMGCNTVLYYAPT------IFTDVGFGVSA 279
Query: 316 DATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFL---DNA 372
A I +G+F +I+T +AV ++D++ R+ ++ G G+ ISLF++ F A
Sbjct: 280 -ALLAHIGIGIFNVIVTAIAVAIMDKIDRKKIVNIGAVGMGISLFVMSIGMKFSGGSQTA 338
Query: 373 AVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSP 432
A+++V+ L +Y+ + ++GP+ W+MI E+FPL +RG G S A ++N+ AN +V+ F
Sbjct: 339 AIISVIALTVYIAFFSATWGPVMWVMIGEVFPLNIRGLGNSFASVINWTANMIVSLTFPS 398
Query: 433 LKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEA 474
L D G G LF + + AS+ F+ V ET+ +LE+IEA
Sbjct: 399 LLDFFGTGSLFIGYGILCFASIWFVQKKVFETRNRSLEDIEA 440
>YFIG_BACSU (P54723) Hypothetical metabolite transport protein yfiG
Length = 482
Score = 207 bits (527), Expect = 5e-53
Identities = 145/458 (31%), Positives = 234/458 (50%), Gaps = 30/458 (6%)
Query: 23 LFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLA 82
L FGGLLFGYD G + A + ++ +T V GL+ S L GA G++
Sbjct: 26 LVSTFGGLLFGYDTGVINGALPFMATAGQLNLT-----PVTEGLVASSLLLGAAFGAMFG 80
Query: 83 FNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAE 142
++D GRR+ +L AL+++ L F+PN +++ R + G+ +G A P ++AE
Sbjct: 81 GRLSDRHGRRKTILYLALLFIAATLGCTFSPNASVMIAFRFLLGLAVGCASVTVPTFLAE 140
Query: 143 TAPTPIRGQLVSLKEFFIVIGIVAGYG----LGSLLVDTVAGWRYMFGISSPVAVIMGFG 198
+P RG++V+ E IVIG + Y +GS + ++ WRYM I++ AV++ FG
Sbjct: 141 ISPAERRGRIVTQNELMIVIGQLLAYTFNAIIGSTMGESANVWRYMLVIATLPAVVLWFG 200
Query: 199 MWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLG 258
M +P SPRW L A + GD A+R L Q++ A Q++ EI + + G
Sbjct: 201 MLIVPESPRW--LAAKGRMGD-------ALRVLRQIRE---DSQAQQEIKEI--KHAIEG 246
Query: 259 EENDVTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADAT 318
+ R+ L I G+ + QQI V +LR GF A A
Sbjct: 247 TAKKAGFHDFQEPWIRRILFIGIGIAIVQQITGVNSIMYYGTEILR---EAGFQTEA-AL 302
Query: 319 RVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAAVLAVV 378
+I GV +I + ++ ++ RRP+L+ G G + +L L+G I L+ L V
Sbjct: 303 IGNIANGVISVIAVIFGIWLLGKVRRRPMLIIGQIGTMTALLLIGILSIVLEGTPALPYV 362
Query: 379 GL---LLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKD 435
L +L++ Q + + WLM++EIFP+ +RG G+ I+ + AN L+ F F L +
Sbjct: 363 VLSLTILFLAFQQTAISTVTWLMLSEIFPMHVRGLGMGISTFCLWTANFLIGFTFPILLN 422
Query: 436 LLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
+G F+IF A+ + +++F+ VPETKG +LE++E
Sbjct: 423 HIGMSATFFIFVAMNILAILFVKKYVPETKGRSLEQLE 460
>ARAE_BACSU (P96710) Arabinose-proton symporter (Arabinose
transporter)
Length = 464
Score = 207 bits (527), Expect = 5e-53
Identities = 136/456 (29%), Positives = 234/456 (50%), Gaps = 39/456 (8%)
Query: 28 GGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIAD 87
GGLL+GYD S A ++ Y L GL+ S + G ++G ++ ++D
Sbjct: 33 GGLLYGYDTAVISGAIGFLKDL-------YSLSPFMEGLVISSIMIGGVVGVGISGFLSD 85
Query: 88 FLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTP 147
GRR+ L+ AAL++ + A+++A + + L+I R++ G+GIG+ + YI E AP
Sbjct: 86 RFGRRKILMTAALLFAISAIVSALSQDVSTLIIARIIGGLGIGMGSSLSVTYITEAAPPA 145
Query: 148 IRGQLVSLKEFFIVIGIVAGYGL-------GSLLVDTVAGWRYMFGISSPVAVIMGFGMW 200
IRG L SL + F ++GI A Y + G+ GWR+M +VI +
Sbjct: 146 IRGSLSSLYQLFTILGISATYFINLAVQRSGTYEWGVHTGWRWMLAYGMVPSVIFFLVLL 205
Query: 201 WLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEE 260
+P SPRW+ K G + A++ L ++ G T + ++ + + +G
Sbjct: 206 VVPESPRWLA-----KAGKT----NEALKILTRINGETVAKEELKNIENSL-KIEQMG-- 253
Query: 261 NDVTLGEMFRGKCRKALVISAGLVLFQQI---QTVTDHGPTKCSLLRCINPTGFSLAADA 317
+L ++F+ RKALVI L LF Q+ +T +GP ++ GF
Sbjct: 254 ---SLSQLFKPGLRKALVIGILLALFNQVIGMNAITYYGPEIFKMMGFGQNAGFV----- 305
Query: 318 TRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAAVLAV 377
+ ++GV ++I T +AV+++D++GR+ L+ G + + I + L+G+ + F + ++ +
Sbjct: 306 --TTCIVGVVEVIFTVIAVLLIDKVGRKKLMSIGSAFMAIFMILIGTSFYFELTSGIMMI 363
Query: 378 VGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLL 437
V +L +V + +S GP+ W+MI+EIFP LR + IA + + AN + + D
Sbjct: 364 VLILGFVAAFCVSVGPITWIMISEIFPNHLRARAAGIATIFLWGANWAIGQFVPMMIDSF 423
Query: 438 GAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
G F+IF+ I + +F+ I PETK +LEEIE
Sbjct: 424 GLAYTFWIFAVINILCFLFVVTICPETKNKSLEEIE 459
>XYLE_ECOLI (P09098) D-xylose-proton symporter (D-xylose
transporter)
Length = 491
Score = 200 bits (509), Expect = 6e-51
Identities = 147/489 (30%), Positives = 241/489 (49%), Gaps = 64/489 (13%)
Query: 23 LFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEI-GLLTSGSLYGALIGSVL 81
L GGLLFGYD S S+ + ++ + A + G + +L G +IG L
Sbjct: 15 LVATLGGLLFGYDTAVISGTVESLNTVFVAPQNLSESAANSLLGFCVASALIGCIIGGAL 74
Query: 82 AFNIADFLGRRRELLVAALMYLVGALITAF------------------APNFPLLVIGRL 123
++ GRR L +AA+++ + + +A+ A P VI R+
Sbjct: 75 GGYCSNRFGRRDSLKIAAVLFFISGVGSAWPELGFTSINPDNTVPVYLAGYVPEFVIYRI 134
Query: 124 VFGIGIGLAMHAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDT------ 177
+ GIG+GLA +PMYIAE AP IRG+LVS +F I+ G + Y + + +
Sbjct: 135 IGGIGVGLASMLSPMYIAELAPAHIRGKLVSFNQFAIIFGQLLVYCVNYFIARSGDASWL 194
Query: 178 -VAGWRYMFGISSPVAVIMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQG 236
GWRYMF A++ ++ +P SPRW++ R Q++ + L ++ G
Sbjct: 195 NTDGWRYMFASECIPALLFLMLLYTVPESPRWLMSRGKQEQAE---------GILRKIMG 245
Query: 237 RTFHDSAPQQVDEIMAEFSYLGEENDVTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHG 296
T A Q V EI + G + L G +VI L +FQQ +
Sbjct: 246 NTL---ATQAVQEIKHSLDH-GRKTGGRLLMFGVG----VIVIGVMLSIFQQFVGI---- 293
Query: 297 PTKCSLLRCINPTGFSLAADATRVSIL----LGVFKLIMTGVAVVVVDRLGRRPLLLGGV 352
+++ P F +T +++L +GV L T +A++ VD+ GR+PL + G
Sbjct: 294 ----NVVLYYAPEVFKTLGASTDIALLQTIIVGVINLTFTVLAIMTVDKFGRKPLQIIGA 349
Query: 353 SGIVISLFLLGSYYIFLDNAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGL 412
G+ I +F LG+ + + ++A++ +L YV + +S+GP+ W++++EIFP +RGK L
Sbjct: 350 LGMAIGMFSLGTAF-YTQAPGIVALLSMLFYVAAFAMSWGPVCWVLLSEIFPNAIRGKAL 408
Query: 413 SIAVLVNFAANALVTFAFSPLKDL-------LGAGILFYIFSAIAVASLVFIYFIVPETK 465
+IAV + AN V++ F P+ D G ++I+ + V + +F++ VPETK
Sbjct: 409 AIAVAAQWLANYFVSWTF-PMMDKNSWLVAHFHNGFSYWIYGCMGVLAALFMWKFVPETK 467
Query: 466 GLTLEEIEA 474
G TLEE+EA
Sbjct: 468 GKTLEELEA 476
>GALP_ECOLI (P37021) Galactose-proton symporter (Galactose
transporter)
Length = 464
Score = 193 bits (490), Expect = 9e-49
Identities = 127/459 (27%), Positives = 233/459 (50%), Gaps = 44/459 (9%)
Query: 26 AFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNI 85
A GLLFG DIG + A I + + + + S ++GA +G+V + +
Sbjct: 23 ALAGLLFGLDIGVIAGALPFIADE-------FQITSHTQEWVVSSMMFGAAVGAVGSGWL 75
Query: 86 ADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAP 145
+ LGR++ L++ A++++ G+L +A APN +L++ R++ G+ +G+A + AP+Y++E AP
Sbjct: 76 SFKLGRKKSLMIGAILFVAGSLFSAAAPNVEVLILSRVLLGLAVGVASYTAPLYLSEIAP 135
Query: 146 TPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPAS 205
IRG ++S+ + I IGI+ Y L WR+M G+ A+++ G+++LP S
Sbjct: 136 EKIRGSMISMYQLMITIGILGAY-LSDTAFSYTGAWRWMLGVIIIPAILLLIGVFFLPDS 194
Query: 206 PRWILL--RAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSY------L 257
PRW R + + L L+DT+ A +++DEI L
Sbjct: 195 PRWFAAKRRFVDAERVLLRLRDTSA-------------EAKRELDEIRESLQVKQSGWAL 241
Query: 258 GEENDVTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADA 317
+EN +F G VL Q +Q T + G++ +
Sbjct: 242 FKENSNFRRAVFLG------------VLLQVMQQFTGMNVIMYYAPKIFELAGYTNTTEQ 289
Query: 318 TRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYY---IFLDNAAV 374
++++G+ ++ T +A+ +VDR GR+P L G + + +LG+ I +A
Sbjct: 290 MWGTVIVGLTNVLATFIAIGLVDRWGRKPTLTLGFLVMAAGMGVLGTMMHIGIHSPSAQY 349
Query: 375 LAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLK 434
A+ LL+++ + +S GP+ W++ +EI PL+ R G++ + N+ AN +V F +
Sbjct: 350 FAIAMLLMFIVGFAMSAGPLIWVLCSEIQPLKGRDFGITCSTATNWIANMIVGATFLTML 409
Query: 435 DLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
+ LG F++++A+ V ++ ++VPETK ++LE IE
Sbjct: 410 NTLGNANTFWVYAALNVLFILLTLWLVPETKHVSLEHIE 448
>ITR2_YEAST (P30606) Myo-inositol transporter 2
Length = 612
Score = 185 bits (469), Expect = 3e-46
Identities = 137/470 (29%), Positives = 225/470 (47%), Gaps = 42/470 (8%)
Query: 26 AFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNI 85
+ G +FGYD G SSA ISI + + Y E L+T+ + GALI SV A
Sbjct: 120 SISGFMFGYDTGYISSALISINRDLDNKVLTYG----EKELITAATSLGALITSVGAGTA 175
Query: 86 ADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAP 145
AD GRR L+ + LM+L+GA++ A F + GRL+ G G+G+ +P++I+E AP
Sbjct: 176 ADVFGRRPCLMFSNLMFLIGAILQITAHKFWQMAAGRLIMGFGVGIGSLISPLFISEIAP 235
Query: 146 TPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPAS 205
IRG+L + ++ G + YG G+ L GWR + G+S V+ +LP +
Sbjct: 236 KMIRGRLTVINSLWLTGGQLIAYGCGAGLNHVKNGWRILVGLSLIPTVLQFSFFCFLPDT 295
Query: 206 PRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEENDVT- 264
PR+ ++ KGDL+ K RS T + Q+V+E+ + + +N +T
Sbjct: 296 PRYYVM-----KGDLKRAKMVLKRSYV----NTEDEIIDQKVEELSSLNQSIPGKNPITK 346
Query: 265 ----LGEMFRGKCR-KALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADATR 319
+ E+ +AL+I GL Q IQ T GF +++
Sbjct: 347 FWNMVKELHTVPSNFRALIIGCGL---QAIQQFTGWNSLMYFSGTIFETVGFK---NSSA 400
Query: 320 VSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFL---------- 369
VSI++ + T +A +D++GRR +LL G+ G+ ++L + + FL
Sbjct: 401 VSIIVSGTNFVFTLIAFFCIDKIGRRYILLIGLPGMTVALVICAIAFHFLGIKFNGADAV 460
Query: 370 ------DNAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAAN 423
+ ++ +V +++Y Y + G + W +E+FP +RG G S A N+A +
Sbjct: 461 VASDGFSSWGIVIIVFIIVYAAFYALGIGTVPWQQ-SELFPQNVRGVGTSYATATNWAGS 519
Query: 424 ALVTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
++ F + + F F+ +A S +F YF PE GL LEE++
Sbjct: 520 LVIASTFLTMLQNITPTGTFSFFAGVACLSTIFCYFCYPELSGLELEEVQ 569
>GTR1_LEIDO (Q01440) Membrane transporter D1
Length = 547
Score = 184 bits (468), Expect = 3e-46
Identities = 135/468 (28%), Positives = 218/468 (45%), Gaps = 52/468 (11%)
Query: 23 LFPAFGGLLFGYDIGATSSATISIQSS-SLSGITWYDLDAVEIGLLTSGSLYGALIGSVL 81
L A GG LFGYD G ++A ++ S +W + L+ + ++ GA +G+ +
Sbjct: 7 LCAALGGFLFGYDTGVINAALFQMKDHFGFSEHSW------QYALIVAIAIAGAFVGAFI 60
Query: 82 AFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIA 141
+ I+ GRR + VA ++++G+++ APN ++++ R++ G+ IG++ P+Y+A
Sbjct: 61 SGFISAAFGRRPCIAVADALFVIGSVLMGAAPNVEVVLVSRVIVGLAIGISSATIPVYLA 120
Query: 142 ETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVA---GWRYMFGISSPVAVIMGFG 198
E RG + L F+ G G +++V + GWR GI + AV+ F
Sbjct: 121 EVTSPKHRGATIVLNNLFLTGGQFVAAGFTAIMVVFTSKNIGWRVAIGIGALPAVVQAFC 180
Query: 199 M-WWLPASPRWILLRAIQKKGDLQTLKDTAIR---SLCQLQGRTFHDSAPQQVDEIMAEF 254
+ ++LP SPRW+L KG K A + LC+ Q S +MA
Sbjct: 181 LLFFLPESPRWLL-----SKGHADRAKAVADKFEVDLCEFQEGDELPSVRIDYRPLMAR- 234
Query: 255 SYLGEENDVTLGEMFRGKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCINPTGF 311
R +V+S+GL + QQ I T+ + + GF
Sbjct: 235 -----------------DMRFRVVLSSGLQIIQQFSGINTIMYYSSV------ILYDAGF 271
Query: 312 SLAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDN 371
A +SI L + T VA+ VDR GRR +LL V G ++ L ++ F+
Sbjct: 272 RDAIMPVVLSIPLAFMNALFTAVAIFTVDRFGRRRMLLISVFGCLVLLVVIAIIGFFIGT 331
Query: 372 AAVLAVVG------LLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANAL 425
+V G L +++ Y G + W+++ EIFP LR S+A + N+ AN L
Sbjct: 332 RISYSVGGGLFLALLAVFLALYAPGIGCIPWVIMGEIFPTHLRTSAASVATMANWGANVL 391
Query: 426 VTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
V+ F L +G G F I S + +F+YF ETKGLTLE+I+
Sbjct: 392 VSQVFPILMGAIGVGGTFTIISGLMALGCIFVYFFAVETKGLTLEQID 439
>ARAE_KLEOX (P45598) Arabinose-proton symporter (Arabinose
transporter)
Length = 472
Score = 184 bits (467), Expect = 4e-46
Identities = 126/460 (27%), Positives = 232/460 (50%), Gaps = 35/460 (7%)
Query: 26 AFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNI 85
A GLLFG DIG + A I + L + + G+ GAL L+F
Sbjct: 30 AVAGLLFGLDIGVIAGALPFITDHFVLSSR---LQEWVVSSMMLGAAIGALFNGWLSFR- 85
Query: 86 ADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAP 145
LGR+ L+V A++++ G++ +AFA + +L++ R+V G+ +G+A + AP+Y++E A
Sbjct: 86 ---LGRKYSLMVGAVLFVAGSVGSAFATSVEMLLVARIVLGVAVGIASYTAPLYLSEMAS 142
Query: 146 TPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPAS 205
+RG+++S+ + + +GIV + L WR M G+ + AV++ + +LP S
Sbjct: 143 ENVRGKMISMYQLMVTLGIVMAF-LSDTAFSYSGNWRAMLGVLALPAVVLIILVIFLPNS 201
Query: 206 PRWILL--RAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEENDV 263
PRW+ R ++ + L+ L+DT+ + A +++EI ++
Sbjct: 202 PRWLAEKGRHVEAEEVLRMLRDTS-------------EKARDELNEIRESLKL--KQGGW 246
Query: 264 TLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADATRVSIL 323
L ++ R R+A+ + +L Q +Q T R GF+ +++
Sbjct: 247 ALFKVNR-NVRRAVFLG---MLLQAMQQFTGMNIIMYYAPRIFKMAGFTTTEQQMVATLV 302
Query: 324 LGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAAV------LAV 377
+G+ + T +AV VD+ GR+P L G S + I +LG + DN L+V
Sbjct: 303 VGLTFMFATFIAVFTVDKAGRKPALKIGFSVMAIGTLVLGYCLMQFDNGTASSGLSWLSV 362
Query: 378 VGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLL 437
++ + Y +S P+ W++ +EI PL+ R G++ + N+ +N ++ F L D +
Sbjct: 363 GMTMMCIAGYAMSAAPVVWILCSEIQPLKCRDFGITCSTTTNWVSNMIIGATFLTLLDAI 422
Query: 438 GAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEAKCL 477
GA F++++A+ VA + ++++PETK +TLE IE + +
Sbjct: 423 GAAGTFWLYTALNVAFIGVTFWLIPETKNVTLEHIERRLM 462
>ARAE_ECOLI (P09830) Arabinose-proton symporter (Arabinose
transporter)
Length = 472
Score = 183 bits (465), Expect = 7e-46
Identities = 126/460 (27%), Positives = 232/460 (50%), Gaps = 35/460 (7%)
Query: 26 AFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNI 85
A GLLFG DIG + A I + L + + G+ GAL L+F
Sbjct: 30 AVAGLLFGLDIGVIAGALPFITDHF---VLTSRLQEWVVSSMMLGAAIGALFNGWLSFR- 85
Query: 86 ADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAP 145
LGR+ L+ A+++++G++ +AFA + +L+ R+V GI +G+A + AP+Y++E A
Sbjct: 86 ---LGRKYSLMAGAILFVLGSIGSAFATSVEMLIAARVVLGIAVGIASYTAPLYLSEMAS 142
Query: 146 TPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPAS 205
+RG+++S+ + + +GIV + L WR M G+ + AV++ + +LP S
Sbjct: 143 ENVRGKMISMYQLMVTLGIVLAF-LSDTAFSYSGNWRAMLGVLALPAVLLIILVVFLPNS 201
Query: 206 PRWILL--RAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEENDV 263
PRW+ R I+ + L+ L+DT+ + A ++++EI ++
Sbjct: 202 PRWLAEKGRHIEAEEVLRMLRDTS-------------EKAREELNEIRESLKL--KQGGW 246
Query: 264 TLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADATRVSIL 323
L ++ R R+A+ + +L Q +Q T R GF+ +++
Sbjct: 247 ALFKINR-NVRRAVFLG---MLLQAMQQFTGMNIIMYYAPRIFKMAGFTTTEQQMIATLV 302
Query: 324 LGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAAV------LAV 377
+G+ + T +AV VD+ GR+P L G S + + +LG + DN L+V
Sbjct: 303 VGLTFMFATFIAVFTVDKAGRKPALKIGFSVMALGTLVLGYCLMQFDNGTASSGLSWLSV 362
Query: 378 VGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLL 437
++ + Y +S P+ W++ +EI PL+ R G++ + N+ +N ++ F L D +
Sbjct: 363 GMTMMCIAGYAMSAAPVVWILCSEIQPLKCRDFGITCSTTTNWVSNMIIGATFLTLLDSI 422
Query: 438 GAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEAKCL 477
GA F++++A+ +A + ++++PETK +TLE IE K +
Sbjct: 423 GAAGTFWLYTALNIAFVGITFWLIPETKNVTLEHIERKLM 462
>ITR1_YEAST (P30605) Myo-inositol transporter 1
Length = 584
Score = 180 bits (457), Expect = 6e-45
Identities = 137/471 (29%), Positives = 231/471 (48%), Gaps = 44/471 (9%)
Query: 26 AFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNI 85
+ G +FGYD G SSA ISI + + Y E ++T+ + GALI S+ A
Sbjct: 94 SISGFMFGYDTGYISSALISIGTDLDHKVLTYG----EKEIVTAATSLGALITSIFAGTA 149
Query: 86 ADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAP 145
AD GR+R L+ + LM+++GA++ A F + +GRL+ G G+G+ AP++I+E AP
Sbjct: 150 ADIFGRKRCLMGSNLMFVIGAILQVSAHTFWQMAVGRLIMGFGVGIGSLIAPLFISEIAP 209
Query: 146 TPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPAS 205
IRG+L + ++ G + YG G+ L GWR + G+S + + +LP +
Sbjct: 210 KMIRGRLTVINSLWLTGGQLVAYGCGAGLNYVNNGWRILVGLSLIPTAVQFTCLCFLPDT 269
Query: 206 PRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEENDV-- 263
PR+ ++ KGDL + RS T + ++V+E++ + +N
Sbjct: 270 PRYYVM-----KGDLARATEVLKRSYTD----TSEEIIERKVEELVTLNQSIPGKNVPEK 320
Query: 264 ---TLGEMFRGKCR-KALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADATR 319
T+ E+ +AL+I GL Q IQ T GF +++
Sbjct: 321 VWNTIKELHTVPSNLRALIIGCGL---QAIQQFTGWNSLMYFSGTIFETVGFK---NSSA 374
Query: 320 VSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFL----DNA--- 372
VSI++ I T VA +D++GRR +LL G+ G+ ++L + + FL D A
Sbjct: 375 VSIIVSGTNFIFTLVAFFSIDKIGRRTILLIGLPGMTMALVVCSIAFHFLGIKFDGAVAV 434
Query: 373 ---------AVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAAN 423
++ +V ++++ Y + G + W +E+FP +RG G S A N+A +
Sbjct: 435 VVSSGFSSWGIVIIVFIIVFAAFYALGIGTVPWQQ-SELFPQNVRGIGTSYATATNWAGS 493
Query: 424 ALVTFAF-SPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
++ F + L+++ AG F F+ ++ S +F YF PE GL LEE++
Sbjct: 494 LVIASTFLTMLQNITPAG-TFAFFAGLSCLSTIFCYFCYPELSGLELEEVQ 543
>KHT2_KLULA (P53387) Hexose transporter 2
Length = 566
Score = 179 bits (453), Expect = 2e-44
Identities = 144/475 (30%), Positives = 220/475 (46%), Gaps = 41/475 (8%)
Query: 23 LFPAFGGLLFGYDIGATSSATISIQSSSLSGITWYD----LDAVEIGLLTSGSLYGALIG 78
L AFGG +FG+D G S G D L V GL+ S G IG
Sbjct: 67 LMVAFGGFVFGWDTGTISGFVNQTDFIRRFGQEKADGSHYLSNVRTGLIVSIFNIGCAIG 126
Query: 79 SVLAFNIADFLGRRRELLVAALMYLVGALIT-AFAPNFPLLVIGRLVFGIGIGLAMHAAP 137
++ + D GRR L++ L+Y+VG +I A + IGR++ G+G+G +P
Sbjct: 127 GIILSKLGDMYGRRIGLMIVVLIYVVGIIIQIASIDKWYQYFIGRIISGLGVGGISVLSP 186
Query: 138 MYIAETAPTPIRGQLVSLKEFFIVIGIVAGY--GLGSLLVDTVAGWRYMFGISSPVAVIM 195
M I+ETAP IRG LVS + I GI GY G+ WR G+ A+ M
Sbjct: 187 MLISETAPKHIRGTLVSFYQLMITFGIFLGYCTNYGTKTYSNSVQWRVPLGLCFAWAIFM 246
Query: 196 GFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQ-QVDEIMAEF 254
GM ++P SPR+++ +K + D A RS+ + ++ D A Q +VD I A
Sbjct: 247 ITGMLFVPESPRFLV-----EKDRI----DEAKRSIAKSNKVSYEDPAVQAEVDLICAGV 297
Query: 255 SYLGEENDVTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLA 314
++ E+F K + + G+ L Q Q +T + N G
Sbjct: 298 EAERLAGSASIKELFSTKTKVFQRLIMGM-LIQSFQQLTGNNYFFYYGTTIFNSVGMD-- 354
Query: 315 ADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYI------- 367
D+ SI+LG+ T VA+ VVD+ GRR LL G + + + + S +
Sbjct: 355 -DSFETSIVLGIVNFASTFVAIYVVDKFGRRKCLLWGAAAMTACMVVFASVGVTRLWPDG 413
Query: 368 ------FLDNAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFA 421
A +V Y+ C+ S+ P+ ++++AE +PLR++ K ++IA N+
Sbjct: 414 ANHPETASKGAGNCMIVFACFYIFCFATSWAPIAYVVVAESYPLRVKAKCMAIATASNWI 473
Query: 422 ANALVTFAFSPLKDLLGAGILF---YIFSAIAVASLVFIYFIVPETKGLTLEEIE 473
L F F+P + + I F Y+F VA +++F VPETKGLTLEE++
Sbjct: 474 WGFLNGF-FTP---FITSAIHFYYGYVFMGCLVAMFFYVFFFVPETKGLTLEEVQ 524
>GLCP_SYNY3 (P15729) Glucose transport protein
Length = 468
Score = 178 bits (452), Expect = 2e-44
Identities = 137/489 (28%), Positives = 238/489 (48%), Gaps = 56/489 (11%)
Query: 13 PKTCLKNLRFLF-----PAFGGLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLL 67
P N++F+ A GG LFG+D + A ++Q + D++ GL
Sbjct: 7 PSQSTANVKFVLLISGVAALGGFLFGFDTAVINGAVAALQKH-------FQTDSLLTGLS 59
Query: 68 TSGSLYGALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGI 127
S +L G+ +G+ A IAD GR + +++AA+++ + ++ + + R++ GI
Sbjct: 60 VSLALLGSALGAFGAGPIADRHGRIKTMILAAVLFTLSSIGSGLPFTIWDFIFWRVLGGI 119
Query: 128 GIGLAMHAAPMYIAETAPTPIRGQLVSLKEFFIVIGI------------VAGYGLGSLLV 175
G+G A AP YIAE +P +RG+L SL++ IV GI +AG + +
Sbjct: 120 GVGAASVIAPAYIAEVSPAHLRGRLGSLQQLAIVSGIFIALLSNWFIALMAGGSAQNPWL 179
Query: 176 DTVAGWRYMFGISSPVAVIMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQ 235
A WR+MF A++ G + +P SPR+++ + +K A L +++
Sbjct: 180 FGAAAWRWMFWTELIPALLYGVCAFLIPESPRYLVAQGQGEK---------AAAILWKVE 230
Query: 236 GRTFHDSAPQQVDEIMAEFSYLGEENDVTLGEMF--RGKCRKALVISAGLVLFQQIQTVT 293
G P +++EI A S ++ ++ RG + I GL QQ +
Sbjct: 231 G----GDVPSRIEEIQATVSL---DHKPRFSDLLSRRGGLLPIVWIGMGLSALQQFVGIN 283
Query: 294 DHGPTKCSLLRCINPTGFSLAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVS 353
L R + GF+ + ++++ G ++ T VA+ VD+ GR+PLLL G
Sbjct: 284 VIFYYSSVLWRSV---GFT-EEKSLLITVITGFINILTTLVAIAFVDKFGRKPLLLMGSI 339
Query: 354 GIVISLFLLGSYY----------IFLDNAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIF 403
G+ I+L +L + A ++A+V LYV + S+GP+ W+++ E+F
Sbjct: 340 GMTITLGILSVVFGGATVVNGQPTLTGAAGIIALVTANLYVFSFGFSWGPIVWVLLGEMF 399
Query: 404 PLRLRGKGLSIAVLVNFAANALVTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPE 463
++R LS+A V + AN +++ F PL D +G G + +++ A S+ FI+F V E
Sbjct: 400 NNKIRAAALSVAAGVQWIANFIISTTFPPLLDTVGLGPAYGLYATSAAISIFFIWFFVKE 459
Query: 464 TKGLTLEEI 472
TKG TLE++
Sbjct: 460 TKGKTLEQM 468
>ITR2_SCHPO (P87110) Myo-inositol transporter 2
Length = 557
Score = 176 bits (445), Expect = 2e-43
Identities = 126/465 (27%), Positives = 217/465 (46%), Gaps = 47/465 (10%)
Query: 29 GLLFGYDIGATSSATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIGSVLAFNIADF 88
GLLFGYD G S A +++ S L + L + + L+TS + + ALI + + +AD+
Sbjct: 92 GLLFGYDTGVISGA-LAVLGSDLGHV----LSSGQKELITSATSFAALISATTSGWLADW 146
Query: 89 LGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAMHAAPMYIAETAPTPI 148
+GR+R LL A ++++G++I A + N ++V+GR + G GIGL PMYI E AP +
Sbjct: 147 VGRKRLLLCADAIFVIGSVIMAASRNVAMMVVGRFIVGYGIGLTSLIVPMYITELAPARL 206
Query: 149 RGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAVIMGFGMWWLPASPRW 208
RG+LV + FI G + Y L + GWR MFGI + A+ ++W P SPR+
Sbjct: 207 RGRLVIIYVVFITGGQLIAYSLNAAFEHVHQGWRIMFGIGAAPALGQLISLFWTPESPRY 266
Query: 209 ILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAEFSYLGEENDVTLGEM 268
+L + + + ++ R ++ P ++ G + D G
Sbjct: 267 LL-------------RHNHVEKVYKILSRIHPEAKPAEI-AYKVSLIQEGVKVDFPEGNK 312
Query: 269 FRG------------KCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAAD 316
F+ R++L I L FQQ + + + GF +
Sbjct: 313 FQHFFHSLKVLFTVPSNRRSLFIGCFLQWFQQFSGTNAIQYFSAIIFQSV---GFK---N 366
Query: 317 ATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYIFLDNAAV-- 374
+ VSI++G + T VA + +DR+GRR +LL + ++ L L Y FL
Sbjct: 367 SISVSIVVGATNFVFTIVAFMFIDRIGRRRILLCTSAVMIAGLALCAIAYHFLPADTTQN 426
Query: 375 -------LAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALVT 427
+ + +++++ Y G + W AE+FP+ +R G + +N+ N +++
Sbjct: 427 TNSGWQYVVLASIIIFLASYASGIGNIPWQQ-AELFPMEVRALGAGFSTAINWVGNLIIS 485
Query: 428 FAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEI 472
+F + + + F +F+ LV YF PE G+++E I
Sbjct: 486 ASFLTMMESITPTGTFALFAGFCFVGLVTSYFTYPELAGMSIENI 530
>HXT3_YEAST (P32466) Low-affinity glucose transporter HXT3
Length = 567
Score = 175 bits (444), Expect = 2e-43
Identities = 138/468 (29%), Positives = 216/468 (45%), Gaps = 37/468 (7%)
Query: 26 AFGGLLFGYDIGATSSATISIQSSSLSGITWYD----LDAVEIGLLTSGSLYGALIGSVL 81
AFGG +FG+D G S G+ D L V GL+ S G IG ++
Sbjct: 70 AFGGFVFGWDTGTISGFVAQTDFLRRFGMKHKDGSYYLSKVRTGLIVSIFNIGCAIGGII 129
Query: 82 AFNIADFLGRRRELLVAALMYLVGALITAFAPN-FPLLVIGRLVFGIGIGLAMHAAPMYI 140
+ D GR+ L+V ++Y++G +I + N + IGR++ G+G+G +PM I
Sbjct: 130 LAKLGDMYGRKMGLIVVVVIYIIGIIIQIASINKWYQYFIGRIISGLGVGGIAVLSPMLI 189
Query: 141 AETAPTPIRGQLVSLKEFFIVIGIVAGY--GLGSLLVDTVAGWRYMFGISSPVAVIMGFG 198
+E AP +RG LVS + I +GI GY G+ WR G+ A+ M G
Sbjct: 190 SEVAPKEMRGTLVSCYQLMITLGIFLGYCTNFGTKNYSNSVQWRVPLGLCFAWALFMIGG 249
Query: 199 MWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQL-QGRTFHDSAPQQVDEIMAEFSYL 257
M ++P SPR+++ + G + D A SL ++ + H Q+++ I A
Sbjct: 250 MTFVPESPRYLV-----EAGQI----DEARASLSKVNKVAPDHPFIQQELEVIEASVEEA 300
Query: 258 GEENDVTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADA 317
+ GE+F GK G ++ Q +Q +T N G S D+
Sbjct: 301 RAAGSASWGELFTGKPAMFKRTMMG-IMIQSLQQLTGDNYFFYYGTTVFNAVGMS---DS 356
Query: 318 TRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYI---------- 367
SI+ GV T ++ VDR GRR LL G G+V + S +
Sbjct: 357 FETSIVFGVVNFFSTCCSLYTVDRFGRRNCLLYGAIGMVCCYVVYASVGVTRLWPNGEGN 416
Query: 368 -FLDNAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALV 426
A +V Y+ C+ ++ P+ +++I+E FPLR++ K +SIA N+ L+
Sbjct: 417 GSSKGAGNCMIVFACFYIFCFATTWAPIAYVVISETFPLRVKSKAMSIATAANWLWGFLI 476
Query: 427 TFAFSPLKDLLGAGILF--YIFSAIAVASLVFIYFIVPETKGLTLEEI 472
F F+P + GA + Y+F V + +++F VPETKGLTLEE+
Sbjct: 477 GF-FTPF--ITGAINFYYGYVFMGCMVFAYFYVFFFVPETKGLTLEEV 521
>HXT2_YEAST (P23585) High-affinity glucose transporter HXT2
Length = 541
Score = 173 bits (439), Expect = 8e-43
Identities = 140/469 (29%), Positives = 217/469 (45%), Gaps = 33/469 (7%)
Query: 23 LFPAFGGLLFGYDIGATSS----ATISIQSSSLSGITWYDLDAVEIGLLTSGSLYGALIG 78
L AFGG +FG+D G S + + Y L V GL+ G G
Sbjct: 61 LMIAFGGFVFGWDTGTISGFVNQTDFKRRFGQMKSDGTYYLSDVRTGLIVGIFNIGCAFG 120
Query: 79 SVLAFNIADFLGRRRELLVAALMYLVGALIT-AFAPNFPLLVIGRLVFGIGIGLAMHAAP 137
+ + D GRR L+ L+Y+VG +I A + + IGR++ G+G+G +P
Sbjct: 121 GLTLGRLGDMYGRRIGLMCVVLVYIVGIVIQIASSDKWYQYFIGRIISGMGVGGIAVLSP 180
Query: 138 MYIAETAPTPIRGQLVSLKEFFIVIGIVAGY--GLGSLLVDTVAGWRYMFGISSPVAVIM 195
I+ETAP IRG VS + I +GI GY G+ WR G++ A+ M
Sbjct: 181 TLISETAPKHIRGTCVSFYQLMITLGIFLGYCTNYGTKDYSNSVQWRVPLGLNFAFAIFM 240
Query: 196 GFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHD-SAPQQVDEIMAEF 254
GM +P SPR+++ +KG + K RSL + T D S ++D IMA
Sbjct: 241 IAGMLMVPESPRFLV-----EKGRYEDAK----RSLAKSNKVTIEDPSIVAEMDTIMANV 291
Query: 255 SYLGEENDVTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLA 314
+ + GE+F K + G ++ Q +Q +T + N G
Sbjct: 292 ETERLAGNASWGELFSNKGAILPRVIMG-IMIQSLQQLTGNNYFFYYGTTIFNAVGMK-- 348
Query: 315 ADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVI-----------SLFLLG 363
D+ + SI+LG+ T VA+ VD+ GRR LLGG + + I SL+ G
Sbjct: 349 -DSFQTSIVLGIVNFASTFVALYTVDKFGRRKCLLGGSASMAICFVIFSTVGVTSLYPNG 407
Query: 364 SYYIFLDNAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAAN 423
A + +V L++ + IS+ P+ ++++AE +PLR++ + ++IAV N+
Sbjct: 408 KDQPSSKAAGNVMIVFTCLFIFFFAISWAPIAYVIVAESYPLRVKNRAMAIAVGANWIWG 467
Query: 424 ALVTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEI 472
L+ F F+P Y+F V S +++F V ETKGLTLEE+
Sbjct: 468 FLIGF-FTPFITSAIGFSYGYVFMGCLVFSFFYVFFFVCETKGLTLEEV 515
>GTR2_HUMAN (P11168) Solute carrier family 2, facilitated glucose
transporter, member 2 (Glucose transporter type 2,
liver)
Length = 524
Score = 171 bits (432), Expect = 5e-42
Identities = 118/418 (28%), Positives = 210/418 (50%), Gaps = 26/418 (6%)
Query: 66 LLTSGSLYGALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFA---PNFPLLVIGR 122
L S G + S + D LGR + +LVA ++ LVGAL+ F+ P+ L++ GR
Sbjct: 99 LSVSSFAVGGMTASFFGGWLGDTLGRIKAMLVANILSLVGALLMGFSKLGPSHILIIAGR 158
Query: 123 LVFGIGIGLAMHAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLG-SLLVDTVAGW 181
+ G+ GL PMYI E APT +RG L + + IV GI+ +G ++ W
Sbjct: 159 SISGLYCGLISGLVPMYIGEIAPTALRGALGTFHQLAIVTGILISQIIGLEFILGNYDLW 218
Query: 182 RYMFGISSPVAVIMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHD 241
+ G+S A++ +++ P SPR++ ++ ++ Q+LK +L+G +D
Sbjct: 219 HILLGLSGVRAILQSLLLFFCPESPRYLYIKLDEEVKAKQSLK--------RLRG---YD 267
Query: 242 SAPQQVDEIMAEFSYLGEENDVTLGEMFRGKC-RKALVISAGLVLFQQIQTVTDHGPTKC 300
+ ++E+ E E V++ ++F R+ ++++ L + QQ +
Sbjct: 268 DVTKDINEMRKEREEASSEQKVSIIQLFTNSSYRQPILVALMLHVAQQFSGINGIFYYST 327
Query: 301 SLLRCINPTGFSLAADATRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLF 360
S+ + G S AT I +G ++ T V+V +V++ GRR L L G+SG+ +
Sbjct: 328 SIFQT---AGISKPVYAT---IGVGAVNMVFTAVSVFLVEKAGRRSLFLIGMSGMFVCAI 381
Query: 361 LLGSYYIFLDN---AAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVL 417
+ + L+ + ++++ + L+V ++I GP+ W M+AE F R L+IA
Sbjct: 382 FMSVGLVLLNKFSWMSYVSMIAIFLFVSFFEIGPGPIPWFMVAEFFSQGPRPAALAIAAF 441
Query: 418 VNFAANALVTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEIEAK 475
N+ N +V F + D G +F++F+ + +A +F +F VPETKG + EEI A+
Sbjct: 442 SNWTCNFIVALCFQYIADFCGP-YVFFLFAGVLLAFTLFTFFKVPETKGKSFEEIAAE 498
>STP1_ARATH (P23586) Glucose transporter (Sugar carrier)
Length = 522
Score = 169 bits (428), Expect = 1e-41
Identities = 137/477 (28%), Positives = 221/477 (45%), Gaps = 52/477 (10%)
Query: 26 AFGGLLFGYDIGATSSAT-------------ISIQSSSLSGITWYDLDAVEIGLLTSGSL 72
A GGL+FGYDIG + T Q S + D+ + + TS
Sbjct: 31 AMGGLIFGYDIGISGGVTSMPSFLKRFFPSVYRKQQEDASTNQYCQYDSPTLTMFTSSLY 90
Query: 73 YGALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLA 132
ALI S++A + GRR +L +++ GALI FA + +L++GR++ G GIG A
Sbjct: 91 LAALISSLVASTVTRKFGRRLSMLFGGILFCAGALINGFAKHVWMLIVGRILLGFGIGFA 150
Query: 133 MHAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPV- 191
A P+Y++E AP RG L + I IGI+ L GW + + V
Sbjct: 151 NQAVPLYLSEMAPYKYRGALNIGFQLSITIGILVAEVLNYFFAKIKGGWGWRLSLGGAVV 210
Query: 192 -AVIMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEI 250
A+I+ G LP +P ++ R ++ A L +++G D Q+ D++
Sbjct: 211 PALIITIGSLVLPDTPNSMIERGQHEE---------AKTKLRRIRG---VDDVSQEFDDL 258
Query: 251 MAEFSYLGEENDVTLGEMFRGKCRKALVISAGLVLFQQ---IQTVTDHGPTKCSLLRCIN 307
+A S + + + R K R L ++ + FQQ I + + P N
Sbjct: 259 VAA-SKESQSIEHPWRNLLRRKYRPHLTMAVMIPFFQQLTGINVIMFYAPV------LFN 311
Query: 308 PTGFSLAADATRVS-ILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYY 366
GF+ DA+ +S ++ G + T V++ VDR GRR L L G + ++I ++ +
Sbjct: 312 TIGFT--TDASLMSAVVTGSVNVAATLVSIYGVDRWGRRFLFLEGGTQMLICQAVVAACI 369
Query: 367 ---IFLDNA--------AVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIA 415
+D A++ V + +YV + S+GP+GWL+ +EIFPL +R SI
Sbjct: 370 GAKFGVDGTPGELPKWYAIVVVTFICIYVAGFAWSWGPLGWLVPSEIFPLEIRSAAQSIT 429
Query: 416 VLVNFAANALVTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEI 472
V VN ++ F + L G LF +F+ V +F+Y +PETKG+ +EE+
Sbjct: 430 VSVNMIFTFIIAQIFLTMLCHLKFG-LFLVFAFFVVVMSIFVYIFLPETKGIPIEEM 485
>HXT1_YEAST (P32465) Low-affinity glucose transporter HXT1
Length = 570
Score = 169 bits (428), Expect = 1e-41
Identities = 135/468 (28%), Positives = 217/468 (45%), Gaps = 37/468 (7%)
Query: 26 AFGGLLFGYDIGATSSATISIQSSSLSGITWYD----LDAVEIGLLTSGSLYGALIGSVL 81
AFGG +FG+D G S G+ +D L V GL+ S G IG ++
Sbjct: 73 AFGGFIFGWDTGTISGFVAQTDFLRRFGMKHHDGSHYLSKVRTGLIVSIFNIGCAIGGIV 132
Query: 82 AFNIADFLGRRRELLVAALMYLVGALITAFAPN-FPLLVIGRLVFGIGIGLAMHAAPMYI 140
+ D GRR L+V ++Y +G +I + N + IGR++ G+G+G +PM I
Sbjct: 133 LAKLGDMYGRRIGLIVVVVIYTIGIIIQIASINKWYQYFIGRIISGLGVGGITVLSPMLI 192
Query: 141 AETAPTPIRGQLVSLKEFFIVIGIVAGY--GLGSLLVDTVAGWRYMFGISSPVAVIMGFG 198
+E AP+ +RG LVS + I +GI GY G+ WR G+ A+ M G
Sbjct: 193 SEVAPSEMRGTLVSCYQVMITLGIFLGYCTNFGTKNYSNSVQWRVPLGLCFAWALFMIGG 252
Query: 199 MWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQL-QGRTFHDSAPQQVDEIMAEFSYL 257
M ++P SPR+++ + G + D A SL ++ + H +++ I A +
Sbjct: 253 MMFVPESPRYLV-----EAGRI----DEARASLAKVNKCPPDHPYIQYELETIEASVEEM 303
Query: 258 GEENDVTLGEMFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPTGFSLAADA 317
+ GE+F GK G ++ Q +Q +T G S D+
Sbjct: 304 RAAGTASWGELFTGKPAMFQRTMMG-IMIQSLQQLTGDNYFFYYGTIVFQAVGLS---DS 359
Query: 318 TRVSILLGVFKLIMTGVAVVVVDRLGRRPLLLGGVSGIVISLFLLGSYYI---------- 367
SI+ GV T ++ VDR GRR L+ G G+V + S +
Sbjct: 360 FETSIVFGVVNFFSTCCSLYTVDRFGRRNCLMWGAVGMVCCYVVYASVGVTRLWPNGQDQ 419
Query: 368 -FLDNAAVLAVVGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAVLVNFAANALV 426
A +V Y+ C+ ++ P+ +++I+E FPLR++ K +SIA N+ L+
Sbjct: 420 PSSKGAGNCMIVFACFYIFCFATTWAPIAYVVISECFPLRVKSKCMSIASAANWIWGFLI 479
Query: 427 TFAFSPLKDLLGAGILF--YIFSAIAVASLVFIYFIVPETKGLTLEEI 472
+F F+P + GA + Y+F V + +++F VPETKGL+LEE+
Sbjct: 480 SF-FTPF--ITGAINFYYGYVFMGCMVFAYFYVFFFVPETKGLSLEEV 524
>HUP2_CHLKE (Q39524) H(+)/hexose cotransporter 2 (Galactose-H+
symporter)
Length = 540
Score = 167 bits (424), Expect = 4e-41
Identities = 135/476 (28%), Positives = 227/476 (47%), Gaps = 55/476 (11%)
Query: 28 GGLLFGYDIGATSSATIS--------------IQSSSLSGITWYDLDAVEIGLLTSGSLY 73
GGLLFGYDIG T T Q S S + D ++ L TS
Sbjct: 39 GGLLFGYDIGVTGGVTSMPEFLQKFFPSIYDRTQQPSDSKDPYCTYDDQKLQLFTSSFFL 98
Query: 74 GALIGSVLAFNIADFLGRRRELLVAALMYLVGALITAFAPNFPLLVIGRLVFGIGIGLAM 133
+ S A ++ GR+ +L+A++++L GA + A A + +LVIGR++ G G+G
Sbjct: 99 AGMFVSFFAGSVVRRWGRKPTMLIASVLFLAGAGLNAGAQDLAMLVIGRVLLGFGVGGGN 158
Query: 134 HAAPMYIAETAPTPIRGQLVSLKEFFIVIGIVAGYGLGSLLVDTVAGWRYMFGISSPVAV 193
+A P+Y++E AP RG L + + + IGI+ + GWR G++ A+
Sbjct: 159 NAVPLYLSECAPPKYRGGLNMMFQLAVTIGIIVAQLVNYGTQTMNNGWRLSLGLAGVPAI 218
Query: 194 IMGFGMWWLPASPRWILLRAIQKKGDLQTLKDTAIRSLCQLQGRTFHDSAPQQVDEIMAE 253
I+ G LP +P ++ R +++G R++ RT ++ + ++I A
Sbjct: 219 ILLIGSLLLPETPNSLIERGHRRRG----------RAVLARLRRT--EAVDTEFEDICAA 266
Query: 254 FSYLGEENDVTLGE----MFRGKCRKALVISAGLVLFQQIQTVTDHGPTKCSLLRCINPT 309
E TL + +F + L++++ + + QQ+ T + + P
Sbjct: 267 AE---ESTRYTLRQSWAALFSRQYSPMLIVTSLIAMLQQL--------TGINAIMFYVPV 315
Query: 310 GFSLAADATRVSIL----LGVFKLIMTGVAVVVVDRLGRRPLLL-GGVS---GIVISLFL 361
FS A ++L +G + T V++ VD+ GRR L L GG+ G V++ +
Sbjct: 316 LFSSFGTARHAALLNTVIIGAVNVAATFVSIFSVDKFGRRGLFLEGGIQMFIGQVVTAAV 375
Query: 362 LG----SYYIFLDNAAVLAV-VGLLLYVGCYQISFGPMGWLMIAEIFPLRLRGKGLSIAV 416
LG Y L ++ V V + +YV + S+GP+GWL+ +EI L RG G+S+AV
Sbjct: 376 LGVELNKYGTNLPSSTAAGVLVVICVYVAAFAWSWGPLGWLVPSEIQTLETRGAGMSMAV 435
Query: 417 LVNFAANALVTFAFSPLKDLLGAGILFYIFSAIAVASLVFIYFIVPETKGLTLEEI 472
+VNF + ++ AF + + G+ F F+ V F+YF +PETKG+ +E +
Sbjct: 436 IVNFLFSFVIGQAFLSMMCAMRWGV-FLFFAGWVVIMTFFVYFCLPETKGVPVETV 490
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.328 0.145 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 52,387,630
Number of Sequences: 164201
Number of extensions: 2188292
Number of successful extensions: 8100
Number of sequences better than 10.0: 351
Number of HSP's better than 10.0 without gapping: 209
Number of HSP's successfully gapped in prelim test: 142
Number of HSP's that attempted gapping in prelim test: 7201
Number of HSP's gapped (non-prelim): 538
length of query: 477
length of database: 59,974,054
effective HSP length: 114
effective length of query: 363
effective length of database: 41,255,140
effective search space: 14975615820
effective search space used: 14975615820
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC142222.8


