

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142222.7 + phase: 0
(555 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
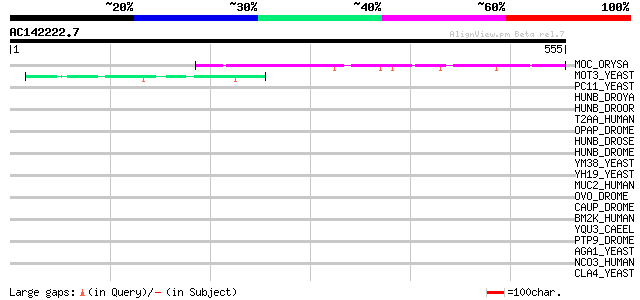
Score E
Sequences producing significant alignments: (bits) Value
MOC_ORYSA (Q84MM9) Protein MONOCULM 1 161 5e-39
MOT3_YEAST (P54785) Zinc finger protein MOT3/HMS1 44 8e-04
PC11_YEAST (P39081) PCF11 protein 44 0.001
HUNB_DROYA (O62541) Hunchback protein 44 0.001
HUNB_DROOR (O62537) Hunchback protein 44 0.001
T2AA_HUMAN (P52655) Transcription initiation factor IIA alpha an... 44 0.001
OPAP_DROME (P23488) DROMSOPA protein 44 0.001
HUNB_DROSE (O62538) Hunchback protein 44 0.001
HUNB_DROME (P05084) Hunchback protein 44 0.001
YM38_YEAST (Q03825) Hypothetical 85.0 kDa protein in HLJ1-SMP2 i... 43 0.002
YH19_YEAST (P38900) Hypothetical 70.1 kDa protein in PUR5 3'region 43 0.002
MUC2_HUMAN (Q02817) Mucin 2 precursor (Intestinal mucin 2) 43 0.002
OVO_DROME (P51521) Ovo protein (Shaven baby protein) 42 0.003
CAUP_DROME (P54269) Homeobox protein caupolican 42 0.003
BM2K_HUMAN (Q9NSY1) BMP-2 inducible protein kinase (EC 2.7.1.37)... 42 0.003
YQU3_CAEEL (Q09550) Hypothetical protein F26C11.3 in chromosome II 41 0.009
PTP9_DROME (P35832) Protein-tyrosine phosphatase 99A precursor (... 41 0.009
AGA1_YEAST (P32323) A-agglutinin attachment subunit precursor 41 0.009
NCO3_HUMAN (Q9Y6Q9) Nuclear receptor coactivator 3 (EC 2.3.1.48)... 40 0.012
CLA4_YEAST (P48562) Serine/threonine-protein kinase CLA4 (EC 2.7... 40 0.012
>MOC_ORYSA (Q84MM9) Protein MONOCULM 1
Length = 441
Score = 161 bits (407), Expect = 5e-39
Identities = 127/406 (31%), Positives = 194/406 (47%), Gaps = 53/406 (13%)
Query: 186 STNPLLKTLTEIASLIETQKPNQAIETLTHLNKSISQNGNPNQRVSFYFSQALTNKITAQ 245
ST LL ++ + +A E + L + S G+ R++++F++AL ++ A+
Sbjct: 49 STRDLLLACADLLQRGDLPAARRAAEIV--LAAAASPRGDAADRLAYHFARALALRVDAK 106
Query: 246 SSIASSNSSSTTWEELTL-SYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIV 304
+ + +Y A N P+ +FAHLTANQAILEA +G+ +HI+D V
Sbjct: 107 AGHGHVVVGGGAARPASSGAYLAFNQIAPFLRFAHLTANQAILEAVDGARRVHILDLDAV 166
Query: 305 QGIQWAALLQAFATRSSGK--PNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLN 362
G+QW LLQA A R+ P VR++G A ++ TGNRL FA+ + L
Sbjct: 167 HGVQWPPLLQAIAERADPALGPPEVRVTGAGA------DRDTLLRTGNRLRAFARSIHLP 220
Query: 363 FEFTPIL------TPIELLDESSFC--------------IQPDEALAVNFMLQLYNLLDE 402
F FTP+L P + S+ PDE LAVN ++ L+NL
Sbjct: 221 FHFTPLLLSCATTAPHHVAGTSTGAAAAASTAAAATGLEFHPDETLAVNCVMFLHNLAGH 280
Query: 403 NTNSVEKALRLAKSLNPKIVTLGEYEA-----------SLTTRVGFVERFETAFNYFAAF 451
+ + L+ K+++P +VT+ E EA L RVG A ++++A
Sbjct: 281 --DELAAFLKWVKAMSPAVVTIAEREAGGGGGGGDHIDDLPRRVG------VAMDHYSAV 332
Query: 452 FESLEPNMALDSPERFQVESLLLGRRIDGVIGVR--ERMEDKEQWKVLMENCGFESVGLS 509
FE+LE + S ER VE +LGR I+ +G E+W GF + LS
Sbjct: 333 FEALEATVPPGSRERLAVEQEVLGREIEAAVGPSGGRWWRGIERWGGAARAAGFAARPLS 392
Query: 510 HYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSWR 555
+A+SQA++LL Y S LV+ L W+ PLL+VS+W+
Sbjct: 393 AFAVSQARLLL-RLHYPSEGYLVQEARGACFLGWQTRPLLSVSAWQ 437
>MOT3_YEAST (P54785) Zinc finger protein MOT3/HMS1
Length = 490
Score = 44.3 bits (103), Expect = 8e-04
Identities = 57/248 (22%), Positives = 95/248 (37%), Gaps = 37/248 (14%)
Query: 16 QQVINQKQQQEQHEQQQHQHHHHHQNILGITNPLSVSLHPWHNNNIPVSSSTSLPPLSFP 75
QQ Q+ QQ+QH+QQQHQH H Q + N VS + NNI S S P +
Sbjct: 10 QQQQRQQHQQQQHQQQQHQHQHQQQQHTILQN---VS----NTNNIGSDSLASQPFNTTT 62
Query: 76 PTDFTDPFQVGSGPENTDPAFNFPSPLDPHSTTFRFSDFDSDDWMDTLMSAADSHSD--- 132
+ D V SG A P PL + + + S++ + ++A ++ S
Sbjct: 63 VSSNKDDVMVNSG------ARELPMPLHQQQYIYPYYQYTSNNSNNNNVTAGNNMSASPI 116
Query: 133 FHLNPFSSCPTRLSPQPDLLLPATAPLQIPTQPPTPQRKNDNTTTETTSSFSSSTNPLLK 192
H N +S + +S + + + N+N +++ N +
Sbjct: 117 VHNNSNNSNNSNIS---------ASDYTVANNSTSNNNNNNNNN-------NNNNNNIHP 160
Query: 193 TLTEIASLIETQKPNQAIETLTHLNKSISQNG-----NPNQRVSFYFSQALTNKITAQSS 247
A+ + + A + N I Q NP +S Y+S +N ++
Sbjct: 161 NQFTAAANMNSNAAAAAYYSFPTANMPIPQQDQQYMFNPASYISHYYSAVNSNNNGNNAA 220
Query: 248 IASSNSSS 255
SN+SS
Sbjct: 221 NNGSNNSS 228
>PC11_YEAST (P39081) PCF11 protein
Length = 626
Score = 43.9 bits (102), Expect = 0.001
Identities = 52/246 (21%), Positives = 102/246 (41%), Gaps = 11/246 (4%)
Query: 16 QQVINQKQQQEQHEQQQHQHHHHHQNILGITNPLSVSLHP-WHNNNIPVSSSTSLPPLSF 74
QQ Q+QQQ+Q +QQQ Q +H ++++G S S P + NN+ + SL F
Sbjct: 234 QQQQQQQQQQQQQQQQQQQQYHETKDMVGSYTQNSNSAIPLFGNNSDTTNQQNSLSSSLF 293
Query: 75 PPTDFTDPFQVGSGPENTDPAFNFPSPLDPHSTTFRFSDFDSDDWMDTLMSAADSHSDFH 134
+ FQ ++ + N + L + + + TL + HS++
Sbjct: 294 GNISGVESFQEIEKKKSLNKINNLYASLKAEGLIYT----PPKESIVTLYKKLNGHSNYS 349
Query: 135 LNPFSSCPTRLSPQPDLLLPATAPLQIPTQPPTPQRKNDNTTTETTSSFSSSTNPLLKTL 194
L+ + P+ LL + + N N + + + NP+++
Sbjct: 350 LDSHEKQLMKNLPKIPLLNDILSDCKAYFATVNIDVLN-NPSLQLSEQTLLQENPIVQ-- 406
Query: 195 TEIASLIETQKPNQAIETLTHLNKSISQNGNPNQRVSFYF---SQALTNKITAQSSIASS 251
+ L+ KPN+ S S+ N+ + ++F ++ ++ TA + I++S
Sbjct: 407 NNLIHLLYRSKPNKCSVCGKRFGNSESEKLLQNEHLDWHFRINTRIKGSQNTANTGISNS 466
Query: 252 NSSSTT 257
N ++TT
Sbjct: 467 NLNTTT 472
>HUNB_DROYA (O62541) Hunchback protein
Length = 759
Score = 43.9 bits (102), Expect = 0.001
Identities = 38/151 (25%), Positives = 62/151 (40%), Gaps = 34/151 (22%)
Query: 13 AIAQQVINQKQQQEQHEQQQHQHHHHHQNILGITNPLSV-----SLHPWHNNNI------ 61
A QQ + Q+QQ +QH Q Q HHHH +++G NPL+ + ++ N+
Sbjct: 95 ASLQQQLLQQQQYQQHFQAAQQQHHHHHHLMGGFNPLTPPGLPNPMQHFYGGNLRPSPQP 154
Query: 62 -PVSSSTSLP-----------PLSFPPTDFTDPFQVGSGPENTDPAFNFPSPLDPHSTTF 109
P S+ST P PP D T P PA + S ++P
Sbjct: 155 TPTSASTVAPVAVATGSSEKLQALTPPMDVTPP---------KSPAKSSQSNIEPEKEHD 205
Query: 110 RFSDFDSDDWMDTLMSAADSHSDFHLNPFSS 140
+ S+ D M + + D ++ + ++S
Sbjct: 206 QMSNSSED--MKYMAESEDDDTNIRMPIYNS 234
>HUNB_DROOR (O62537) Hunchback protein
Length = 767
Score = 43.9 bits (102), Expect = 0.001
Identities = 38/149 (25%), Positives = 65/149 (43%), Gaps = 36/149 (24%)
Query: 16 QQVINQKQQQEQHEQQQHQHHHHHQNILGITNPLSV-----SLHPWHNNNI-------PV 63
QQ + Q+QQ +QH Q Q HHHH +++G NPL+ + ++ N+ P+
Sbjct: 99 QQQLLQQQQYQQHFQAAQQQHHHHHHLMGGFNPLTPPGLPNPMQHFYGGNLRPSPQPTPI 158
Query: 64 SSST------------SLPPLSFPPTDFTDPFQVGSGPENTDPAFNFPSPLDPHSTTFRF 111
S+ST L L+ PP D T P PA + S ++P +
Sbjct: 159 SASTVASVAVATGSSEKLQALT-PPMDVTPP---------KSPAKSSQSNIEPEKEHDQM 208
Query: 112 SDFDSDDWMDTLMSAADSHSDFHLNPFSS 140
S+ D M ++ + D ++ + ++S
Sbjct: 209 SNSSED--MKYMVESEDDDTNIRMPIYNS 235
>T2AA_HUMAN (P52655) Transcription initiation factor IIA alpha and
beta chains (TFIIA p35 and p19 subunits) (TFIIA-42)
(TFIIAL)
Length = 376
Score = 43.5 bits (101), Expect = 0.001
Identities = 17/25 (68%), Positives = 20/25 (80%)
Query: 16 QQVINQKQQQEQHEQQQHQHHHHHQ 40
QQ++ Q QQQ Q +QQQH HHHHHQ
Sbjct: 64 QQLLLQVQQQHQPQQQQHHHHHHHQ 88
Score = 35.4 bits (80), Expect = 0.39
Identities = 12/19 (63%), Positives = 15/19 (78%)
Query: 23 QQQEQHEQQQHQHHHHHQN 41
Q Q+QH+ QQ QHHHHH +
Sbjct: 69 QVQQQHQPQQQQHHHHHHH 87
>OPAP_DROME (P23488) DROMSOPA protein
Length = 69
Score = 43.5 bits (101), Expect = 0.001
Identities = 19/35 (54%), Positives = 23/35 (65%)
Query: 8 SGNLMAIAQQVINQKQQQEQHEQQQHQHHHHHQNI 42
SGNL A QQ +Q+QQQ Q +QQQH HH H +
Sbjct: 24 SGNLPAPEQQQHHQQQQQHQQQQQQHHQHHQHHQL 58
>HUNB_DROSE (O62538) Hunchback protein
Length = 757
Score = 43.5 bits (101), Expect = 0.001
Identities = 37/148 (25%), Positives = 61/148 (41%), Gaps = 34/148 (22%)
Query: 16 QQVINQKQQQEQHEQQQHQHHHHHQNILGITNPLSV-----SLHPWHNNNI-------PV 63
QQ + Q+QQ +QH Q Q HHHH +++G NPL+ + ++ N+ P
Sbjct: 96 QQQLLQQQQYQQHFQAAQQQHHHHHHLMGGFNPLTPPGLPNPMQHFYGGNLRPSPQPTPT 155
Query: 64 SSSTSLP-----------PLSFPPTDFTDPFQVGSGPENTDPAFNFPSPLDPHSTTFRFS 112
S+ST P PP D T P PA + S ++P + S
Sbjct: 156 SASTIAPVAVATGSSEKLQALTPPMDVTPP---------KSPAKSSQSNIEPEKEHDQMS 206
Query: 113 DFDSDDWMDTLMSAADSHSDFHLNPFSS 140
+ D M + + D ++ + ++S
Sbjct: 207 NSSED--MKYMAESEDDDTNIRMPIYNS 232
>HUNB_DROME (P05084) Hunchback protein
Length = 758
Score = 43.5 bits (101), Expect = 0.001
Identities = 37/148 (25%), Positives = 61/148 (41%), Gaps = 34/148 (22%)
Query: 16 QQVINQKQQQEQHEQQQHQHHHHHQNILGITNPLSV-----SLHPWHNNNI-------PV 63
QQ + Q+QQ +QH Q Q HHHH +++G NPL+ + ++ N+ P
Sbjct: 97 QQQLLQQQQYQQHFQAAQQQHHHHHHLMGGFNPLTPPGLPNPMQHFYGGNLRPSPQPTPT 156
Query: 64 SSSTSLP-----------PLSFPPTDFTDPFQVGSGPENTDPAFNFPSPLDPHSTTFRFS 112
S+ST P PP D T P PA + S ++P + S
Sbjct: 157 SASTIAPVAVATGSSEKLQALTPPMDVTPP---------KSPAKSSQSNIEPEKEHDQMS 207
Query: 113 DFDSDDWMDTLMSAADSHSDFHLNPFSS 140
+ D M + + D ++ + ++S
Sbjct: 208 NSSED--MKYMAESEDDDTNIRMPIYNS 233
>YM38_YEAST (Q03825) Hypothetical 85.0 kDa protein in HLJ1-SMP2
intergenic region
Length = 758
Score = 42.7 bits (99), Expect = 0.002
Identities = 28/63 (44%), Positives = 32/63 (50%), Gaps = 6/63 (9%)
Query: 16 QQVINQKQQQEQHEQQQHQHHHHHQNILGITNPLSVSLHPWHNNNIPVSSSTSLPPLSFP 75
QQ Q+QQQ+Q +QQQ QH Q I NP V P N S ST L P S P
Sbjct: 305 QQQQQQQQQQQQQQQQQQQHQQQQQTPYPIVNPQMVPHIPSEN-----SHSTGLMP-SVP 358
Query: 76 PTD 78
PT+
Sbjct: 359 PTN 361
>YH19_YEAST (P38900) Hypothetical 70.1 kDa protein in PUR5 3'region
Length = 624
Score = 42.7 bits (99), Expect = 0.002
Identities = 45/162 (27%), Positives = 69/162 (41%), Gaps = 11/162 (6%)
Query: 112 SDFDSDDWMDTLMSAADSHSDFHLNPFSSCPTRLSPQPDLLLPATAPLQIPTQPPTPQRK 171
SD DSD + +A++ ++ N ++ T S TA + + T T +
Sbjct: 70 SDEDSDT---CIHGSANASTNATTNSSTNATTTASINVRTSATTTASINVRTSATTTEST 126
Query: 172 NDNTTTETTSSFSSSTNPLLKTLTEI---ASLIETQKPNQAIETL--THLNKSISQNGNP 226
N NT TT S +SSTN + A+ E+ N + T T N S + +
Sbjct: 127 NSNTNATTTESTNSSTNATTTASINVRTSATTTESTNSNTSATTTESTDSNTSATTTEST 186
Query: 227 NQRVSFYFSQALTNKITAQSSIASSNSS--STTWEELTLSYK 266
+ S + A TN T ++ AS+NSS +TT E S K
Sbjct: 187 DSNTS-ATTTASTNSSTNATTTASTNSSTNATTTESTNASAK 227
>MUC2_HUMAN (Q02817) Mucin 2 precursor (Intestinal mucin 2)
Length = 5179
Score = 42.7 bits (99), Expect = 0.002
Identities = 39/139 (28%), Positives = 54/139 (38%), Gaps = 5/139 (3%)
Query: 62 PVSSSTSLPPL---SFPPTDFTDP--FQVGSGPENTDPAFNFPSPLDPHSTTFRFSDFDS 116
P +S+T+LPP S PPT T P S P T P+ + P TT S +
Sbjct: 1628 PPTSTTTLPPTTTPSPPPTTTTTPPPTTTPSPPTTTTPSPPITTTTTPPPTTTPSSPITT 1687
Query: 117 DDWMDTLMSAADSHSDFHLNPFSSCPTRLSPQPDLLLPATAPLQIPTQPPTPQRKNDNTT 176
T S + +P ++ T S P T PT P+P T
Sbjct: 1688 TPSPPTTTMTTPSPTTTPSSPITTTTTPSSTTTPSPPPTTMTTPSPTTTPSPPTTTMTTL 1747
Query: 177 TETTSSFSSSTNPLLKTLT 195
TT+S +T PL ++T
Sbjct: 1748 PPTTTSSPLTTTPLPPSIT 1766
Score = 39.3 bits (90), Expect = 0.027
Identities = 42/162 (25%), Positives = 59/162 (35%), Gaps = 9/162 (5%)
Query: 62 PVSSSTSLPPLSFPP----TDFTDPFQVGSGPENTDPAFNFPS--PLDPHSTTFRFSDFD 115
P +++T+LPP + P T T P P T P+ P P STT
Sbjct: 1409 PPTTTTTLPPTTTPSPPTTTTTTPPPTTTPSPPITTTTTPLPTTTPSPPISTTTTPPPTT 1468
Query: 116 SDDWMDTLMSAADSHSDFHLNPFSSCPTRLSPQPDLLLPATAPLQIPTQPPTPQRKNDNT 175
+ T S + ++ P +P P + P T P T PPT T
Sbjct: 1469 TPSPPTTTPSPPTTTPSPPTTTTTTPPPTTTPSPPMTTPITPPASTTTLPPTTTPSPPTT 1528
Query: 176 TTET---TSSFSSSTNPLLKTLTEIASLIETQKPNQAIETLT 214
TT T T++ S T + T +L T P+ T T
Sbjct: 1529 TTTTPPPTTTPSPPTTTPITPPTSTTTLPPTTTPSPPPTTTT 1570
Score = 38.1 bits (87), Expect = 0.061
Identities = 42/180 (23%), Positives = 64/180 (35%), Gaps = 12/180 (6%)
Query: 45 ITNPLSVSLHPWHNNNIPVSSSTSLPPLSFPP-----TDFTDPFQVGSGPENTDPAFNFP 99
IT P S + P P +++T+ PP + P T T P + P T P+
Sbjct: 1508 ITPPASTTTLPPTTTPSPPTTTTTTPPPTTTPSPPTTTPITPPTSTTTLPPTTTPSPPPT 1567
Query: 100 SPLDPHSTTFRFSDFDSDDWMDTLMSAADSHSDFHLNPFSSC---PTRLSPQPDLLLPAT 156
+ P TT + T+ + + P ++ P +P P P T
Sbjct: 1568 TTTTPPPTTTPSPPTTTTPSPPTITTTTPPPTTTPSPPTTTTTTPPPTTTPSPPTTTPIT 1627
Query: 157 APLQIPTQPPTPQRKNDNTTTETTSSFSSSTNPLLKT----LTEIASLIETQKPNQAIET 212
P T PPT TTT T ++ + P T +T + T P+ I T
Sbjct: 1628 PPTSTTTLPPTTTPSPPPTTTTTPPPTTTPSPPTTTTPSPPITTTTTPPPTTTPSSPITT 1687
Score = 34.7 bits (78), Expect = 0.67
Identities = 37/159 (23%), Positives = 56/159 (34%), Gaps = 17/159 (10%)
Query: 48 PLSVSLHPWHNNNIPVSSSTSLPPLSFPPTDFTDPFQVGSGPENTDPAFNFPSPLDPHST 107
P + P + P +++T+ PP + P T P T P PSP P +T
Sbjct: 1473 PTTTPSPPTTTPSPPTTTTTTPPPTTTPSPPMTTPI-TPPASTTTLPPTTTPSP--PTTT 1529
Query: 108 TFRFSDFDSDDWMDTLMSAADSHSDFHLNPFSSCPTRLSPQPDLLLPATAPLQIPTQPPT 167
T T + S P + PT + P P+ P T PPT
Sbjct: 1530 T-------------TTPPPTTTPSPPTTTPITP-PTSTTTLPPTTTPSPPPTTTTTPPPT 1575
Query: 168 PQRKNDNTTTETTSSFSSSTNPLLKTLTEIASLIETQKP 206
TTT + + +++T P T + + T P
Sbjct: 1576 TTPSPPTTTTPSPPTITTTTPPPTTTPSPPTTTTTTPPP 1614
Score = 34.3 bits (77), Expect = 0.87
Identities = 23/78 (29%), Positives = 31/78 (39%), Gaps = 6/78 (7%)
Query: 137 PFSSCPTRLSPQPDLLLPATAPLQIPTQPPTPQRKNDNTTTETTSSFSSSTNPLLKTLTE 196
P C T +P P P+ P T PPT TTT T ++ + P+ T T
Sbjct: 1391 PMDKCIT--TPSPPTTTPSPPPTTTTTLPPTTTPSPPTTTTTTPPPTTTPSPPITTTTTP 1448
Query: 197 IASLIETQKPNQAIETLT 214
+ T P+ I T T
Sbjct: 1449 ----LPTTTPSPPISTTT 1462
Score = 32.3 bits (72), Expect = 3.3
Identities = 33/148 (22%), Positives = 57/148 (38%), Gaps = 14/148 (9%)
Query: 62 PVSSSTSLPPLSFP--PTDFTDPFQVGSGPENTDPAFNFPSPLDPHSTTFRFSDFDSDDW 119
P ++T+ PP + P PT T + P T P+ +P+ P ++T +
Sbjct: 1589 PTITTTTPPPTTTPSPPTTTTT-----TPPPTTTPSPPTTTPITPPTSTTTLPPTTTPSP 1643
Query: 120 MDTLMSAADSHSDFHLNPFSSCPTRLSPQPDLLLPAT-APLQIPTQPPTPQRKNDNTTTE 178
T + + S PT +P P + T P P+ P T TT
Sbjct: 1644 PPTTTTTPPPTTT------PSPPTTTTPSPPITTTTTPPPTTTPSSPITTTPSPPTTTMT 1697
Query: 179 TTSSFSSSTNPLLKTLTEIASLIETQKP 206
T S ++ ++P+ T T ++ + P
Sbjct: 1698 TPSPTTTPSSPITTTTTPSSTTTPSPPP 1725
Score = 32.0 bits (71), Expect = 4.3
Identities = 41/171 (23%), Positives = 60/171 (34%), Gaps = 5/171 (2%)
Query: 45 ITNPLSVS-LHPWHNNNIPVSSSTSLPPLSFPPTDFTDPFQVGSGPENTDPAFNFPSPLD 103
IT P S + L P + P +++T+ PP + P T + T P PSP
Sbjct: 1547 ITPPTSTTTLPPTTTPSPPPTTTTTPPPTTTPSPPTTTTPSPPTITTTTPPPTTTPSPPT 1606
Query: 104 PHSTTFRFSDFDSDDWMDTLMSAADSHSDFHLNPFSSCPTRLSPQPDLLLPATAPLQIPT 163
+TT + S + + + S PT + P P T P T
Sbjct: 1607 TTTTTPPPTTTPSPPTTTPITPPTSTTTLPPTTTPSPPPTTTTTPP----PTTTPSPPTT 1662
Query: 164 QPPTPQRKNDNTTTETTSSFSSSTNPLLKTLTEIASLIETQKPNQAIETLT 214
P+P T TT+ S T T + + T P+ I T T
Sbjct: 1663 TTPSPPITTTTTPPPTTTPSSPITTTPSPPTTTMTTPSPTTTPSSPITTTT 1713
>OVO_DROME (P51521) Ovo protein (Shaven baby protein)
Length = 1028
Score = 42.4 bits (98), Expect = 0.003
Identities = 22/50 (44%), Positives = 28/50 (56%), Gaps = 3/50 (6%)
Query: 21 QKQQQEQHEQQQHQHHHHHQNILGITNPLSVSLHPWHNNNIPVSSSTSLP 70
Q+QQQ+QH QQQ HH HHQ P S H H+ + +S+ SLP
Sbjct: 447 QQQQQQQHHQQQQHHHFHHQQ---QQQPQPQSHHSHHHGHGHDNSNMSLP 493
Score = 34.7 bits (78), Expect = 0.67
Identities = 17/38 (44%), Positives = 21/38 (54%), Gaps = 3/38 (7%)
Query: 16 QQVINQK---QQQEQHEQQQHQHHHHHQNILGITNPLS 50
QQ + Q+ QQQ +QQQ QHH HH N +N S
Sbjct: 811 QQSVQQQSVQQQQSLQQQQQQQHHQHHSNSSASSNASS 848
>CAUP_DROME (P54269) Homeobox protein caupolican
Length = 693
Score = 42.4 bits (98), Expect = 0.003
Identities = 46/187 (24%), Positives = 68/187 (35%), Gaps = 55/187 (29%)
Query: 16 QQVINQKQQQEQHEQQQHQHHHHHQNILGITNPLSVSLHPWHNNNIPVSSSTSLPPLSFP 75
QQ + Q QQQ+QH H HH HH LG +PLS+ +SS P S
Sbjct: 505 QQQLQQLQQQQQHHHHPHHHHPHHSMELG--SPLSM-----------MSSYAGGSPYSRI 551
Query: 76 PTDFTDPFQVGSGPENTDPAFNFPSPLDPHSTTFRFSDFDSDDWMDTLMSAADSHSDFHL 135
PT +T+ + PS S+T +
Sbjct: 552 PTAYTEAM-----------GMHLPSSSSSSSSTGKLP----------------------- 577
Query: 136 NPFSSCPTRLSPQPDLL-LPATAPLQIPTQPPTPQRKNDNTTTETTSSFSSSTNPLLKTL 194
PT + P P + P P P PPT + N + + ++S S S++ T
Sbjct: 578 ------PTHIHPAPQRVGFPEIQPDTPPQTPPT-MKLNSSGGSSSSSGSSHSSSMHSVTP 630
Query: 195 TEIASLI 201
+AS++
Sbjct: 631 VTVASMV 637
>BM2K_HUMAN (Q9NSY1) BMP-2 inducible protein kinase (EC 2.7.1.37)
(BIKe) (HRIHFB2017)
Length = 1161
Score = 42.4 bits (98), Expect = 0.003
Identities = 16/24 (66%), Positives = 19/24 (78%)
Query: 16 QQVINQKQQQEQHEQQQHQHHHHH 39
QQ Q+QQQ+Q +QQQH HHHHH
Sbjct: 470 QQQQQQQQQQQQQQQQQHHHHHHH 493
Score = 42.0 bits (97), Expect = 0.004
Identities = 17/28 (60%), Positives = 21/28 (74%)
Query: 16 QQVINQKQQQEQHEQQQHQHHHHHQNIL 43
QQ Q+QQQ+Q +QQQ QHHHHH + L
Sbjct: 468 QQQQQQQQQQQQQQQQQQQHHHHHHHHL 495
Score = 42.0 bits (97), Expect = 0.004
Identities = 16/28 (57%), Positives = 21/28 (74%)
Query: 16 QQVINQKQQQEQHEQQQHQHHHHHQNIL 43
QQ Q+QQQ+Q +QQQ HHHHH ++L
Sbjct: 469 QQQQQQQQQQQQQQQQQQHHHHHHHHLL 496
>YQU3_CAEEL (Q09550) Hypothetical protein F26C11.3 in chromosome II
Length = 1240
Score = 40.8 bits (94), Expect = 0.009
Identities = 66/300 (22%), Positives = 111/300 (37%), Gaps = 50/300 (16%)
Query: 43 LGITNPLSVSLHPWHN--------------NNIPVSSSTSLPPLSFPPTDFTDPFQVG-- 86
L +T LS W N + + SSST + F +F F
Sbjct: 53 LKVTECLSKQYEDWRNVTLIKGYERELEKCDCVLTSSSTETGTVKFLDREFFRVFSSAEI 112
Query: 87 ----SGPENTDPAFN---------FPSPLDPHSTTFRFSD----------FDSDDWMDTL 123
S P T P+ N F +D T R++ +S D +
Sbjct: 113 ASTTSLPTTTSPSLNCYWLSEPSNFSEWIDGKQTNLRYNGGCCSETSIQVLNSSDSTRWI 172
Query: 124 MSAADSHSDFH-LNPFSSCPTRLSPQPDLLLPATAPLQIPTQPPTPQRKNDNTTTETTSS 182
++ +DS + + L C PQ +L + L T T TTET +
Sbjct: 173 LTTSDSWNKANALINLLYCTPNACPQQSMLWTNCSNLSTTTSSSTMLSSTTLLTTETETR 232
Query: 183 FSSSTNPLLKTL--TEIASLIETQKPNQAIETLTHLNKSISQNGNPNQRVSFYFSQALTN 240
SSST T TE ++ I T P + T++ + K+ + P+ + S A T+
Sbjct: 233 ESSSTGSTQTTTPSTEPSTTITT--PMEQSSTVSSVQKTRTSEDKPSSSTTVPTS-ASTS 289
Query: 241 KITAQSSIASSNSSSTTWEELTLSYKALND-----ACPYSKFAHLTANQAILEATEGSNN 295
+ + S +A ++SSSTT + S + + + P + L+ N+ + G ++
Sbjct: 290 ESSTSSPMAETSSSSTTSQSSPASTSTVPESSTVGSTPTTGLTTLSTNEQSTSTSSGGHS 349
>PTP9_DROME (P35832) Protein-tyrosine phosphatase 99A precursor (EC
3.1.3.48) (Receptor-linked protein-tyrosine phosphatase
99A)
Length = 1301
Score = 40.8 bits (94), Expect = 0.009
Identities = 57/248 (22%), Positives = 92/248 (36%), Gaps = 37/248 (14%)
Query: 10 NLMAIAQQVIN--QKQQQEQHEQQQHQHHHHHQNILGITNPLSVSLHPWHNNNIPVSSST 67
+++ + QQ + Q+QQQ+Q +QQQ Q ++ + + + SL +P+ S
Sbjct: 1064 HMLQLQQQHLETQQQQQQQQQQQQQQQQTALNETVSTPSTDTNPSL-------LPILS-- 1114
Query: 68 SLPPLSFPPTDFTDPFQVGSGPENTDPAFNFPSPLDPHSTTFRFSDFDSDDWMDTLMSAA 127
LPP P + S T P + P+P P T + S D + S
Sbjct: 1115 LLPPTVAP---------LSSSSSTTPPTPSTPTPQPPQ--TIQLSSHSPSDLSHQISSTV 1163
Query: 128 DSHSDFHLNPFSSCPTRLSPQPDLLLPATAPLQIPTQPPTPQRKNDNTTTETTSSFSSST 187
+ + +P + S P T P PT P P + N+ T T ++F + T
Sbjct: 1164 ANAA----SPVTPATASASAGATPTTPMT-PTVPPTIPTIPSLASQNSLTLTNANFHTVT 1218
Query: 188 NPLLKTLTEIASLIETQKPNQAIETLTHLNKSISQNGNPNQ---RVSFYFSQALTNKITA 244
N A L+E Q+ N +P Q V N TA
Sbjct: 1219 N-------NAADLMEHQQQQMLALMQQQTQLQQQYNTHPQQHHNNVGDLLMNNADNSPTA 1271
Query: 245 QSSIASSN 252
+I ++N
Sbjct: 1272 SPTITNNN 1279
>AGA1_YEAST (P32323) A-agglutinin attachment subunit precursor
Length = 725
Score = 40.8 bits (94), Expect = 0.009
Identities = 48/217 (22%), Positives = 89/217 (40%), Gaps = 13/217 (5%)
Query: 62 PVSSSTSLPPLSFPPTDF-----TDPFQVGSGPENTDPAFNFPSPLDPHSTTFRFSDFDS 116
PV+S+ S S P T T P + P +T + + S ++T S S
Sbjct: 168 PVTSTLSSTTSSNPTTTSLSSTSTSPSSTSTSPSSTSTSSSSTSTSSSSTSTSSSSTSTS 227
Query: 117 DDWMDTLMSAADSHSDFHLNPFSSCPTRLSPQPDLLLPATAPLQIPTQPPTPQRKNDNTT 176
T S + S SS T S P++ + +P K+ + +
Sbjct: 228 PSSTSTSSSLTSTSSSSTSTSQSSTST--SSSSTSTSPSSTSTSSSSTSTSPSSKSTSAS 285
Query: 177 TETTSSFSSSTNPLLKTLTEIASLIETQKPNQA--IETLTHLNKSI-SQNGNPNQRVSFY 233
+ +TSS+S+ST+P +LT + + + P+ T T S+ S + + VS Y
Sbjct: 286 STSTSSYSTSTSP---SLTSSSPTLASTSPSSTSISSTFTDSTSSLGSSIASSSTSVSLY 342
Query: 234 FSQALTNKITAQSSIASSNSSSTTWEELTLSYKALND 270
+ + SS ++ S +++ E T+S ++ ++
Sbjct: 343 SPSTPVYSVPSTSSNVATPSMTSSTVETTVSSQSSSE 379
Score = 39.3 bits (90), Expect = 0.027
Identities = 36/141 (25%), Positives = 65/141 (45%), Gaps = 8/141 (5%)
Query: 139 SSCPTRLSPQPDLLLPATAPLQIPTQPPTPQRKNDNTTTET----TSSFSSSTNPLLKTL 194
SS PT S L +T+P T P + + +T+T + TSS S+ST+P +
Sbjct: 178 SSNPTTTS----LSSTSTSPSSTSTSPSSTSTSSSSTSTSSSSTSTSSSSTSTSPSSTST 233
Query: 195 TEIASLIETQKPNQAIETLTHLNKSISQNGNPNQRVSFYFSQALTNKITAQSSIASSNSS 254
+ + + + + + + + S S + + S S + ++K T+ SS ++S+ S
Sbjct: 234 SSSLTSTSSSSTSTSQSSTSTSSSSTSTSPSSTSTSSSSTSTSPSSKSTSASSTSTSSYS 293
Query: 255 STTWEELTLSYKALNDACPYS 275
++T LT S L P S
Sbjct: 294 TSTSPSLTSSSPTLASTSPSS 314
Score = 35.8 bits (81), Expect = 0.30
Identities = 42/194 (21%), Positives = 80/194 (40%), Gaps = 21/194 (10%)
Query: 154 PATAPLQIPTQPPTPQRKNDNTTTETTSSFSSSTNPLLKTLTEIASLIETQKPNQAIETL 213
P++A + P + N TT TS S+ST+P + + ++ + + + +
Sbjct: 160 PSSASIISPVTSTLSSTTSSNPTT--TSLSSTSTSPSSTSTSPSSTSTSSSSTSTSSSST 217
Query: 214 THLNKSISQNGNPNQRVSFYFSQALTNKITAQSSIASSNSSSTTWEELTLSYKALNDACP 273
+ + S S + + S S + ++ T+QSS ++S+SS++T T + + P
Sbjct: 218 STSSSSTSTSPSSTSTSSSLTSTSSSSTSTSQSSTSTSSSSTSTSPSSTSTSSSSTSTSP 277
Query: 274 YSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRISGI- 332
SK ++ +T S +L + T +S P+S IS
Sbjct: 278 SSKSTSASSTSTSSYSTSTS----------------PSLTSSSPTLASTSPSSTSISSTF 321
Query: 333 --PAMALGTSPVSS 344
+LG+S SS
Sbjct: 322 TDSTSSLGSSIASS 335
>NCO3_HUMAN (Q9Y6Q9) Nuclear receptor coactivator 3 (EC 2.3.1.48)
(NCoA-3) (Thyroid hormone receptor activator molecule 1)
(TRAM-1) (ACTR) (Receptor-associated coactivator 3)
(RAC-3) (Amplified in breast cancer-1 protein) (AIB-1)
(Steroid receptor coactiv
Length = 1424
Score = 40.4 bits (93), Expect = 0.012
Identities = 26/86 (30%), Positives = 39/86 (45%), Gaps = 6/86 (6%)
Query: 11 LMAIAQQVINQKQQQEQHEQQQHQHHHHHQNILGITNPLSVSLHPWHNNNIPVSSSTSLP 70
+M QQ Q+QQQ+Q +QQQ Q Q + P +V+ P + + + P
Sbjct: 1246 MMQQQQQQQQQQQQQQQQQQQQQQQQQQQQQTQAFSPPPNVTASPSMDGLLAGPTMPQAP 1305
Query: 71 PLSFPPTDFTDPFQVGSGPENTDPAF 96
P FP + + +G P DPAF
Sbjct: 1306 PQQFP---YQPNYGMGQQP---DPAF 1325
>CLA4_YEAST (P48562) Serine/threonine-protein kinase CLA4 (EC
2.7.1.37)
Length = 842
Score = 40.4 bits (93), Expect = 0.012
Identities = 24/66 (36%), Positives = 37/66 (55%), Gaps = 3/66 (4%)
Query: 15 AQQVINQKQQQEQHEQQQHQHH-HHHQNILGITNPLSVSLHPW--HNNNIPVSSSTSLPP 71
+Q+ + Q+QQQ+Q ++QQHQ + +HHQ +P L+P+ H+N I S P
Sbjct: 367 SQRSLQQQQQQQQQQKQQHQQYPYHHQGPSPSPSPSPSPLNPYRPHHNMINPYSKQPQSP 426
Query: 72 LSFPPT 77
LS T
Sbjct: 427 LSSQST 432
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.129 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 64,200,637
Number of Sequences: 164201
Number of extensions: 2792543
Number of successful extensions: 21658
Number of sequences better than 10.0: 319
Number of HSP's better than 10.0 without gapping: 139
Number of HSP's successfully gapped in prelim test: 190
Number of HSP's that attempted gapping in prelim test: 16948
Number of HSP's gapped (non-prelim): 2234
length of query: 555
length of database: 59,974,054
effective HSP length: 115
effective length of query: 440
effective length of database: 41,090,939
effective search space: 18080013160
effective search space used: 18080013160
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 68 (30.8 bits)
Medicago: description of AC142222.7


