

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142222.5 + phase: 0
(378 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
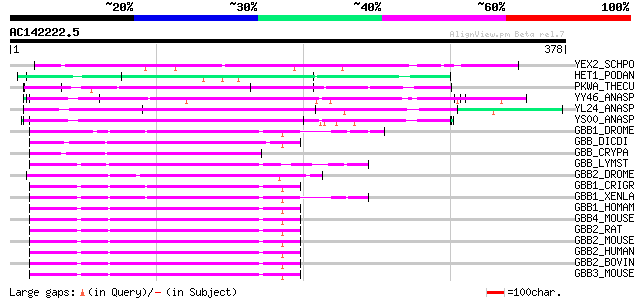
YEX2_SCHPO (O13856) Hypothetical WD-repeat protein C1A6.02 in ch... 107 6e-23
HET1_PODAN (Q00808) Vegetatible incompatibility protein HET-E-1 85 3e-16
PKWA_THECU (P49695) Probable serine/threonine-protein kinase pkw... 81 5e-15
YY46_ANASP (Q8YRI1) Hypothetical WD-repeat protein alr3466 80 8e-15
YL24_ANASP (Q8YV57) Hypothetical WD-repeat protein all2124 70 7e-12
YS00_ANASP (Q8YTC2) Hypothetical WD-repeat protein alr2800 69 2e-11
GBB1_DROME (P26308) Guanine nucleotide-binding protein beta subu... 68 4e-11
GBB_DICDI (P36408) Guanine nucleotide-binding protein beta subunit 67 6e-11
GBB_CRYPA (O14435) Guanine nucleotide-binding protein beta subunit 67 1e-10
GBB_LYMST (Q08706) Guanine nucleotide-binding protein beta subunit 66 1e-10
GBB2_DROME (P29829) Guanine nucleotide-binding protein beta subu... 66 1e-10
GBB1_CRIGR (Q6TMK6) Guanine nucleotide-binding protein G(I)/G(S)... 66 1e-10
GBB1_XENLA (P79959) Guanine nucleotide-binding protein G(I)/G(S)... 66 2e-10
GBB1_HOMAM (O45040) Guanine nucleotide-binding protein G(I)/G(S)... 66 2e-10
GBB4_MOUSE (P29387) Guanine nucleotide-binding protein beta subu... 65 2e-10
GBB2_RAT (P54313) Guanine nucleotide-binding protein G(I)/G(S)/G... 65 4e-10
GBB2_MOUSE (P62880) Guanine nucleotide-binding protein G(I)/G(S)... 65 4e-10
GBB2_HUMAN (P62879) Guanine nucleotide-binding protein G(I)/G(S)... 65 4e-10
GBB2_BOVIN (P11017) Guanine nucleotide-binding protein G(I)/G(S)... 65 4e-10
GBB3_MOUSE (Q61011) Guanine nucleotide-binding protein G(I)/G(S)... 64 6e-10
>YEX2_SCHPO (O13856) Hypothetical WD-repeat protein C1A6.02 in
chromosome I
Length = 361
Score = 107 bits (266), Expect = 6e-23
Identities = 89/340 (26%), Positives = 152/340 (44%), Gaps = 24/340 (7%)
Query: 18 DNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGSPDFS 77
+N V G +G + Y+Y D V H +SCR G ++ D
Sbjct: 25 ENQVLLGFSNGRVSSYQY--DVAQISLVEQWSTKRHKKSCRNISVNESGTEFISVGSDGV 82
Query: 78 ILATDVETGSTIAR--LDNAHEAAVNRLVNLTESTV--ASGDDDGCIKVWDTR-ERSCCN 132
+ D TG ++ +D E + +V E+ + A+GDD+GC+ VWD R E +
Sbjct: 83 LKIADTSTGRVSSKWIVDKNKEISPYSVVQWIENDMVFATGDDNGCVSVWDKRTEGGIIH 142
Query: 133 SFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNG 192
+ H DYIS I+ + +ATSGDG LSV + R K SE ++E+ ++
Sbjct: 143 THNDHIDYISSIS-PFEERYFVATSGDGVLSVIDARNFKKPILSEEQDEEMTCGAFTRDQ 201
Query: 193 ---RKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSN---SIDTMLKLDEDRIITGSENGM 246
+K G+ +G++ L++ G + D +DR + + SI+T+ + D D + G +G
Sbjct: 202 HSKKKFAVGTASGVITLFTKGDWGDHTDRILSPIRSHDFSIETITRADSDSLYVGGSDGC 261
Query: 247 INLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDLDNILQ 306
I L+ ILPN+ + + +HS V + + FL S + + L W +D
Sbjct: 262 IRLLHILPNKYERIIGQHSSRSTVDA-----VDVTTEGNFLVSCSGTE-LAFWPVDQ--- 312
Query: 307 GSRNTQRNENGGVDNDVDSDNEDEMDVDNNTSKFTKGNKR 346
+ + + +D+D DS ++ E K K+
Sbjct: 313 -KEGDESSSSDNLDSDEDSSSDSEFSSPKKKKKVGNQGKK 351
>HET1_PODAN (Q00808) Vegetatible incompatibility protein HET-E-1
Length = 1356
Score = 84.7 bits (208), Expect = 3e-16
Identities = 66/289 (22%), Positives = 117/289 (39%), Gaps = 14/289 (4%)
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLT 71
+ F P VA+G D + ++ +S + H S + F G+ + +
Sbjct: 931 VAFSPDGQRVASGSDDHTIKIWDAASGTCTQT------LEGHGSSVLSVAFSPDGQRVAS 984
Query: 72 GSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCC 131
GS D +I D +G+ L+ + + + VASG DD IK+WDT +C
Sbjct: 985 GSGDKTIKIWDTASGTCTQTLEGHGGSVWSVAFSPDGQRVASGSDDKTIKIWDTASGTCT 1044
Query: 132 NSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKN 191
+ E H ++ + F+ D ++ + S D T+ + + E D + SV +
Sbjct: 1045 QTLEGHGGWVQSVVFSPDGQRVASGSDDHTIKIWDAVSGTCTQTLEGHGDSVWSVAFSPD 1104
Query: 192 GRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENGMINLVG 251
G++V GS G + ++ C+ D R+ +GS +G I +
Sbjct: 1105 GQRVASGSIDGTIKIWD-AASGTCTQTLEGHGGWVHSVAFSPDGQRVASGSIDGTIKIWD 1163
Query: 252 ILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWD 300
Q + H G + VAFS D + + S + D+ +K+WD
Sbjct: 1164 AASGTCTQTLEGHG-------GWVQSVAFSPDGQRVASGSSDKTIKIWD 1205
Score = 79.0 bits (193), Expect = 2e-14
Identities = 70/296 (23%), Positives = 119/296 (39%), Gaps = 16/296 (5%)
Query: 6 GKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFING 65
G + F P VA+G D + ++ D + + LE H S + F
Sbjct: 967 GSSVLSVAFSPDGQRVASGSGDKTIKIW----DTASGTCTQTLE--GHGGSVWSVAFSPD 1020
Query: 66 GRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST-VASGDDDGCIKVWD 124
G+ + +GS D +I D +G+ L+ H V +V + VASG DD IK+WD
Sbjct: 1021 GQRVASGSDDKTIKIWDTASGTCTQTLEG-HGGWVQSVVFSPDGQRVASGSDDHTIKIWD 1079
Query: 125 TRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELL 184
+C + E H D + + F+ D ++ + S DGT+ + + E +
Sbjct: 1080 AVSGTCTQTLEGHGDSVWSVAFSPDGQRVASGSIDGTIKIWDAASGTCTQTLEGHGGWVH 1139
Query: 185 SVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSEN 244
SV +G++V GS G + ++ C+ D R+ +GS +
Sbjct: 1140 SVAFSPDGQRVASGSIDGTIKIWD-AASGTCTQTLEGHGGWVQSVAFSPDGQRVASGSSD 1198
Query: 245 GMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWD 300
I + Q + H + + VAFS D + + S + D +K+WD
Sbjct: 1199 KTIKIWDTASGTCTQTLEGHGGWV-------QSVAFSPDGQRVASGSSDNTIKIWD 1247
Score = 78.6 bits (192), Expect = 2e-14
Identities = 65/296 (21%), Positives = 119/296 (39%), Gaps = 16/296 (5%)
Query: 6 GKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFING 65
G + + F P VA+G D + ++ +S + H ++ F
Sbjct: 883 GGSVWSVAFSPDRERVASGSDDKTIKIWDAASGTCTQT------LEGHGGRVQSVAFSPD 936
Query: 66 GRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDT 125
G+ + +GS D +I D +G+ L+ + ++ + VASG D IK+WDT
Sbjct: 937 GQRVASGSDDHTIKIWDAASGTCTQTLEGHGSSVLSVAFSPDGQRVASGSGDKTIKIWDT 996
Query: 126 RERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLS 185
+C + E H + + F+ D ++ + S D T+ + + E + S
Sbjct: 997 ASGTCTQTLEGHGGSVWSVAFSPDGQRVASGSDDKTIKIWDTASGTCTQTLEGHGGWVQS 1056
Query: 186 VVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSI-DTMLKLDEDRIITGSEN 244
VV +G++V GS + + W + ++ +S+ D R+ +GS +
Sbjct: 1057 VVFSPDGQRVASGSDDHTIKI--WDAVSGTCTQTLEGHGDSVWSVAFSPDGQRVASGSID 1114
Query: 245 GMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWD 300
G I + Q + H G VAFS D + + S + D +K+WD
Sbjct: 1115 GTIKIWDAASGTCTQTLEGHG-------GWVHSVAFSPDGQRVASGSIDGTIKIWD 1163
Score = 62.8 bits (151), Expect = 1e-09
Identities = 48/224 (21%), Positives = 90/224 (39%), Gaps = 8/224 (3%)
Query: 77 SILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNSFEA 136
+I + E + L+ + ++ + VASG DD IK+WDT + + E
Sbjct: 822 TISVVEAEWNACTQTLEGHGSSVLSVAFSADGQRVASGSDDKTIKIWDTASGTGTQTLEG 881
Query: 137 HEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVV 196
H + + F+ D ++ + S D T+ + + E + SV +G++V
Sbjct: 882 HGGSVWSVAFSPDRERVASGSDDKTIKIWDAASGTCTQTLEGHGGRVQSVAFSPDGQRVA 941
Query: 197 CGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENGMINLVGILPNR 256
GS + ++ C+ S+ + D R+ +GS + I +
Sbjct: 942 SGSDDHTIKIWD-AASGTCTQTLEGHGSSVLSVAFSPDGQRVASGSGDKTIKIWDTASGT 1000
Query: 257 VIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWD 300
Q + H G+ VAFS D + + S + D+ +K+WD
Sbjct: 1001 CTQTLEGHG-------GSVWSVAFSPDGQRVASGSDDKTIKIWD 1037
Score = 48.9 bits (115), Expect = 2e-05
Identities = 52/228 (22%), Positives = 86/228 (36%), Gaps = 38/228 (16%)
Query: 6 GKLAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFING 65
G + F P VA+G IDG + ++ +S + H ++ F
Sbjct: 1135 GGWVHSVAFSPDGQRVASGSIDGTIKIWDAASGTCTQT------LEGHGGWVQSVAFSPD 1188
Query: 66 GRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDT 125
G+ + +GS D +I D +G+ L+ + + VASG D IK+WDT
Sbjct: 1189 GQRVASGSSDKTIKIWDTASGTCTQTLEGHGGWVQSVAFSPDGQRVASGSSDNTIKIWDT 1248
Query: 126 RERSC-----------CNSFEAHEDYISD----ITFASDAMKLL-----------ATSGD 159
+C C SF+ YI+ I A+ M+ L D
Sbjct: 1249 ASGTCTQTLNVGSTATCLSFDYTNAYINTNIGRIQIATATMESLNQLSSPVCYSYGLGQD 1308
Query: 160 GTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLY 207
CN +N + E+ + GRK+V GS +G ++++
Sbjct: 1309 HRWITCN-NQNVLWLPPEYH-----TSAFTMQGRKIVLGSYSGRIIIF 1350
>PKWA_THECU (P49695) Probable serine/threonine-protein kinase pkwA
(EC 2.7.1.37)
Length = 742
Score = 80.9 bits (198), Expect = 5e-15
Identities = 67/268 (25%), Positives = 114/268 (42%), Gaps = 11/268 (4%)
Query: 36 SSDNTNSDPVRLLEIHAHT--ESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLD 93
++ + ++DP L E T A F GG L GS D I DV +G + L+
Sbjct: 439 TAPSESADPHELNEPRILTTDREAVAVAFSPGGSLLAGGSGDKLIHVWDVASGDELHTLE 498
Query: 94 NAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKL 153
+ + + +ASG DD +++WD FE H Y+ DI F+ D +
Sbjct: 499 GHTDWVRAVAFSPDGALLASGSDDATVRLWDVAAAEERAVFEGHTHYVLDIAFSPDGSMV 558
Query: 154 LATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFK 213
+ S DGT + N+ A + D + +V +G V GS+ G + L+ K
Sbjct: 559 ASGSRDGTARLWNVATGTEHAVLKGHTDYVYAVAFSPDGSMVASGSRDGTIRLWDVATGK 618
Query: 214 DCSDRFVDLSSNSIDTMLKLDEDRIITGSENGMINLVGILPNRVIQPVAEHSEYPVERLG 273
+ D + N + D ++ GS++ ++L + + H+++
Sbjct: 619 E-RDVLQAPAENVVSLAFSPDGSMLVHGSDS-TVHLWDVASGEALHTFEGHTDWV----- 671
Query: 274 N*RFVAFSHDRKFLGSIAHDQMLKLWDL 301
R VAFS D L S + D+ ++LWD+
Sbjct: 672 --RAVAFSPDGALLASGSDDRTIRLWDV 697
Score = 65.9 bits (159), Expect = 2e-10
Identities = 47/197 (23%), Positives = 84/197 (41%), Gaps = 7/197 (3%)
Query: 11 DIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALL 70
DI F P ++VA+G DG L+ ++ ++ + HT+ A F G +
Sbjct: 548 DIAFSPDGSMVASGSRDGTARLWNVATGTEHA------VLKGHTDYVYAVAFSPDGSMVA 601
Query: 71 TGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSC 130
+GS D +I DV TG L E V+ + S + G D + +WD
Sbjct: 602 SGSRDGTIRLWDVATGKERDVLQAPAENVVSLAFSPDGSMLVHGSDS-TVHLWDVASGEA 660
Query: 131 CNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMK 190
++FE H D++ + F+ D L + S D T+ + ++ + E + + SV
Sbjct: 661 LHTFEGHTDWVRAVAFSPDGALLASGSDDRTIRLWDVAAQEEHTTLEGHTEPVHSVAFHP 720
Query: 191 NGRKVVCGSQTGIMLLY 207
G + S+ G + ++
Sbjct: 721 EGTTLASASEDGTIRIW 737
Score = 50.8 bits (120), Expect = 6e-06
Identities = 35/155 (22%), Positives = 68/155 (43%), Gaps = 7/155 (4%)
Query: 10 FDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRAL 69
+ + F P ++VA+G DG + L+ ++ + A E+ + F G L
Sbjct: 589 YAVAFSPDGSMVASGSRDGTIRLWDVATGKERD------VLQAPAENVVSLAFSPDGSML 642
Query: 70 LTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERS 129
+ GS D ++ DV +G + + + + + +ASG DD I++WD +
Sbjct: 643 VHGS-DSTVHLWDVASGEALHTFEGHTDWVRAVAFSPDGALLASGSDDRTIRLWDVAAQE 701
Query: 130 CCNSFEAHEDYISDITFASDAMKLLATSGDGTLSV 164
+ E H + + + F + L + S DGT+ +
Sbjct: 702 EHTTLEGHTEPVHSVAFHPEGTTLASASEDGTIRI 736
>YY46_ANASP (Q8YRI1) Hypothetical WD-repeat protein alr3466
Length = 1526
Score = 80.1 bits (196), Expect = 8e-15
Identities = 73/303 (24%), Positives = 126/303 (41%), Gaps = 21/303 (6%)
Query: 14 FHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGS 73
F P+ +A G D + L+ SS L + HT A F G L +GS
Sbjct: 1124 FSPNGVTLANGSSDQIVRLWDISSKKC------LYTLQGHTNWVNAVAFSPDGATLASGS 1177
Query: 74 PDFSILATDVETGSTIARLDNAHEAAVNRLV-NLTESTVASGDDDGCIKVWDTRERSCCN 132
D ++ D+ + + L H + VN +V N ST+ASG D +++W+ C
Sbjct: 1178 GDQTVRLWDISSSKCLYILQG-HTSWVNSVVFNPDGSTLASGSSDQTVRLWEINSSKCLC 1236
Query: 133 SFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNG 192
+F+ H +++ + F D L + S D T+ + ++ +K + + + SV +G
Sbjct: 1237 TFQGHTSWVNSVVFNPDGSMLASGSSDKTVRLWDISSSKCLHTFQGHTNWVNSVAFNPDG 1296
Query: 193 RKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENGMINLVGI 252
+ GS + L+ K C F +S D + +GS++ + L I
Sbjct: 1297 SMLASGSGDQTVRLWEISSSK-CLHTFQGHTSWVSSVTFSPDGTMLASGSDDQTVRLWSI 1355
Query: 253 LPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDLDN-----ILQG 307
+ H+ + +G+ V FS D L S + DQ ++LW + + LQG
Sbjct: 1356 SSGECLYTFLGHTNW----VGS---VIFSPDGAILASGSGDQTVRLWSISSGKCLYTLQG 1408
Query: 308 SRN 310
N
Sbjct: 1409 HNN 1411
Score = 79.7 bits (195), Expect = 1e-14
Identities = 76/350 (21%), Positives = 147/350 (41%), Gaps = 24/350 (6%)
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLT 71
+ F P +A+G D + L+ SS L + HT + F G L +
Sbjct: 1164 VAFSPDGATLASGSGDQTVRLWDISSSKC------LYILQGHTSWVNSVVFNPDGSTLAS 1217
Query: 72 GSPDFSILATDVETGSTIARLDNAHEAAVNRLV-NLTESTVASGDDDGCIKVWDTRERSC 130
GS D ++ ++ + + H + VN +V N S +ASG D +++WD C
Sbjct: 1218 GSSDQTVRLWEINSSKCLCTFQG-HTSWVNSVVFNPDGSMLASGSSDKTVRLWDISSSKC 1276
Query: 131 CNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMK 190
++F+ H ++++ + F D L + SGD T+ + + +K + + SV
Sbjct: 1277 LHTFQGHTNWVNSVAFNPDGSMLASGSGDQTVRLWEISSSKCLHTFQGHTSWVSSVTFSP 1336
Query: 191 NGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENGMINLV 250
+G + GS + L+S +C F+ ++ + D + +GS + + L
Sbjct: 1337 DGTMLASGSDDQTVRLWSISS-GECLYTFLGHTNWVGSVIFSPDGAILASGSGDQTVRLW 1395
Query: 251 GILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDLDN-----IL 305
I + + + H+ + +G+ + FS D L S + DQ ++LW++ + L
Sbjct: 1396 SISSGKCLYTLQGHNNW----VGS---IVFSPDGTLLASGSDDQTVRLWNISSGECLYTL 1448
Query: 306 QGSRNTQRNENGGVDNDVDSDNEDEMDV---DNNTSKFTKGNKRKNASKG 352
G N+ R+ D + + D+ + D T + K K + +G
Sbjct: 1449 HGHINSVRSVAFSSDGLILASGSDDETIKLWDVKTGECIKTLKSEKIYEG 1498
Score = 73.9 bits (180), Expect = 6e-13
Identities = 70/309 (22%), Positives = 120/309 (38%), Gaps = 35/309 (11%)
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLT 71
+ F ++A+G D + L+ SS L HT R+ F L +
Sbjct: 912 VGFSQDGKMLASGSDDQTVRLWDISSGQC------LKTFKGHTSRVRSVVFSPNSLMLAS 965
Query: 72 GSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCC 131
GS D ++ D+ +G + + NL S +A+G D +++WD C
Sbjct: 966 GSSDQTVRLWDISSGECLYIFQGHTGWVYSVAFNLDGSMLATGSGDQTVRLWDISSSQCF 1025
Query: 132 NSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKN 191
F+ H + + F+SD L + S D T+ + ++ + + SVV +
Sbjct: 1026 YIFQGHTSCVRSVVFSSDGAMLASGSDDQTVRLWDISSGNCLYTLQGHTSCVRSVVFSPD 1085
Query: 192 GRKVVCGSQTGIMLLY---SWGCFKDCSD-----RFVDLSSNSIDTMLKLDEDRIITGSE 243
G + G I+ L+ S C RF+ S N + + GS
Sbjct: 1086 GAMLASGGDDQIVRLWDISSGNCLYTLQGYTSWVRFLVFSPNGV---------TLANGSS 1136
Query: 244 NGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDLDN 303
+ ++ L I + + + H+ + VAFS D L S + DQ ++LWD+ +
Sbjct: 1137 DQIVRLWDISSKKCLYTLQGHTNWV-------NAVAFSPDGATLASGSGDQTVRLWDISS 1189
Query: 304 -----ILQG 307
ILQG
Sbjct: 1190 SKCLYILQG 1198
Score = 66.2 bits (160), Expect = 1e-10
Identities = 67/337 (19%), Positives = 130/337 (37%), Gaps = 58/337 (17%)
Query: 10 FDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRAL 69
+ + F+ +++ATG D + L+ SS HT R+ F + G L
Sbjct: 994 YSVAFNLDGSMLATGSGDQTVRLWDISSSQC------FYIFQGHTSCVRSVVFSSDGAML 1047
Query: 70 LTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTV-ASGDDDGC--------- 119
+GS D ++ D+ +G+ + L H + V +V + + ASG DD
Sbjct: 1048 ASGSDDQTVRLWDISSGNCLYTLQG-HTSCVRSVVFSPDGAMLASGGDDQIVRLWDISSG 1106
Query: 120 ---------------------------------IKVWDTRERSCCNSFEAHEDYISDITF 146
+++WD + C + + H ++++ + F
Sbjct: 1107 NCLYTLQGYTSWVRFLVFSPNGVTLANGSSDQIVRLWDISSKKCLYTLQGHTNWVNAVAF 1166
Query: 147 ASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLL 206
+ D L + SGD T+ + ++ +K + + SVV +G + GS + L
Sbjct: 1167 SPDGATLASGSGDQTVRLWDISSSKCLYILQGHTSWVNSVVFNPDGSTLASGSSDQTVRL 1226
Query: 207 YSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENGMINLVGILPNRVIQPVAEHSE 266
+ K C F +S + D + +GS + + L I ++ + H+
Sbjct: 1227 WEINSSK-CLCTFQGHTSWVNSVVFNPDGSMLASGSSDKTVRLWDISSSKCLHTFQGHTN 1285
Query: 267 YPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDLDN 303
+ VAF+ D L S + DQ ++LW++ +
Sbjct: 1286 WV-------NSVAFNPDGSMLASGSGDQTVRLWEISS 1315
Score = 58.9 bits (141), Expect = 2e-08
Identities = 51/243 (20%), Positives = 97/243 (38%), Gaps = 10/243 (4%)
Query: 62 FINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRL-VNLTESTVASGDDDGCI 120
F G+ TG + + TG + H + VN + + +ASG DD +
Sbjct: 872 FSPDGKLFATGDSGGIVRFWEAATGKELLTC-KGHNSWVNSVGFSQDGKMLASGSDDQTV 930
Query: 121 KVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSE 180
++WD C +F+ H + + F+ +++ L + S D T+ + ++ + +
Sbjct: 931 RLWDISSGQCLKTFKGHTSRVRSVVFSPNSLMLASGSSDQTVRLWDISSGECLYIFQGHT 990
Query: 181 DELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIIT 240
+ SV +G + GS + L+ C F +S + D + +
Sbjct: 991 GWVYSVAFNLDGSMLATGSGDQTVRLWDISS-SQCFYIFQGHTSCVRSVVFSSDGAMLAS 1049
Query: 241 GSENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWD 300
GS++ + L I + + H+ R V FS D L S DQ+++LWD
Sbjct: 1050 GSDDQTVRLWDISSGNCLYTLQGHTSCV-------RSVVFSPDGAMLASGGDDQIVRLWD 1102
Query: 301 LDN 303
+ +
Sbjct: 1103 ISS 1105
>YL24_ANASP (Q8YV57) Hypothetical WD-repeat protein all2124
Length = 1683
Score = 70.5 bits (171), Expect = 7e-12
Identities = 41/199 (20%), Positives = 85/199 (42%), Gaps = 6/199 (3%)
Query: 10 FDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRAL 69
+ + F P +A+G D + L++ SD L I H ++ F G+ L
Sbjct: 1117 YSVSFSPDGQTIASGGSDKTIKLWQ------TSDGTLLKTITGHEQTVNNVYFSPDGKNL 1170
Query: 70 LTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERS 129
+ S D SI D +G + L + + T+A+G +D +K+W ++
Sbjct: 1171 ASASSDHSIKLWDTTSGQLLMTLTGHSAGVITVRFSPDGQTIAAGSEDKTVKLWHRQDGK 1230
Query: 130 CCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLM 189
+ H+D+++ ++F+ D L + S D T+ + + K+ + D + V
Sbjct: 1231 LLKTLNGHQDWVNSLSFSPDGKTLASASADKTIKLWRIADGKLVKTLKGHNDSVWDVNFS 1290
Query: 190 KNGRKVVCGSQTGIMLLYS 208
+G+ + S+ + L++
Sbjct: 1291 SDGKAIASASRDNTIKLWN 1309
Score = 67.4 bits (163), Expect = 6e-11
Identities = 56/297 (18%), Positives = 122/297 (40%), Gaps = 18/297 (6%)
Query: 10 FDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRAL 69
+ ++F P N++A+ +D + L++ + LE+ A A F++ G +
Sbjct: 1326 YAVNFLPDSNIIASASLDNTIRLWQRPL-------ISPLEVLAGNSGVYAVSFLHDGSII 1378
Query: 70 LTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERS 129
T D +I + GS + L ++A +AS + D +K+W R+
Sbjct: 1379 ATAGADGNIQLWHSQDGSLLKTLPG-NKAIYGISFTPQGDLIASANADKTVKIWRVRDGK 1437
Query: 130 CCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLM 189
+ H++ ++ + F+ D L + S D T+ + N+ K + + DE+ V
Sbjct: 1438 ALKTLIGHDNEVNKVNFSPDGKTLASASRDNTVKLWNVSDGKFKKTLKGHTDEVFWVSFS 1497
Query: 190 KNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTM-LKLDEDRIITGSENGMIN 248
+G+ + S + L W F + + ++ + ++ D + + S + +
Sbjct: 1498 PDGKIIASASADKTIRL--WDSFSGNLIKSLPAHNDLVYSVNFNPDGSMLASTSADKTVK 1555
Query: 249 LVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDLDNIL 305
L ++ + HS +FS D +++ S + D+ +K+W +D L
Sbjct: 1556 LWRSHDGHLLHTFSGHSNVVYSS-------SFSPDGRYIASASEDKTVKIWQIDGHL 1605
Score = 52.8 bits (125), Expect = 1e-06
Identities = 62/294 (21%), Positives = 115/294 (39%), Gaps = 23/294 (7%)
Query: 91 RLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDA 150
RL+ + ++ ++ T+ASG D IK+W +R+ + HED + ++F+ D
Sbjct: 1067 RLEGHKDGVISISISRDGQTIASGSLDKTIKLW-SRDGRLFRTLNGHEDAVYSVSFSPDG 1125
Query: 151 MKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLYSWG 210
+ + D T+ + + E + +V +G+ + S + L+
Sbjct: 1126 QTIASGGSDKTIKLWQTSDGTLLKTITGHEQTVNNVYFSPDGKNLASASSDHSIKLWD-- 1183
Query: 211 CFKDCSDRFVDLSSNS---IDTMLKLDEDRIITGSENGMINLVGILPNRVIQPVAEHSEY 267
+ L+ +S I D I GSE+ + L ++++ + H ++
Sbjct: 1184 --TTSGQLLMTLTGHSAGVITVRFSPDGQTIAAGSEDKTVKLWHRQDGKLLKTLNGHQDW 1241
Query: 268 PVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDLDNILQGSR-NTQRNENGGV-DNDVDS 325
++FS D K L S + D+ +KLW I G T + N V D + S
Sbjct: 1242 V-------NSLSFSPDGKTLASASADKTIKLW---RIADGKLVKTLKGHNDSVWDVNFSS 1291
Query: 326 DNE--DEMDVDNNTSKFTKGNKRKNASKGHAAG-DSNNFFADL*EIMAATFSTT 376
D + DN + + GH+ G + NF D I +A+ T
Sbjct: 1292 DGKAIASASRDNTIKLWNRHGIELETFTGHSGGVYAVNFLPDSNIIASASLDNT 1345
Score = 32.3 bits (72), Expect = 2.0
Identities = 23/116 (19%), Positives = 50/116 (42%), Gaps = 7/116 (6%)
Query: 8 LAFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGR 67
L + ++F+P +++A+ D + L+R + D L H+ ++ F GR
Sbjct: 1532 LVYSVNFNPDGSMLASTSADKTVKLWR------SHDGHLLHTFSGHSNVVYSSSFSPDGR 1585
Query: 68 ALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVW 123
+ + S D ++ ++ G + L ++ + + T+ SG D K+W
Sbjct: 1586 YIASASEDKTVKIWQID-GHLLTTLPQHQAGVMSAIFSPDGKTLISGSLDTTTKIW 1640
>YS00_ANASP (Q8YTC2) Hypothetical WD-repeat protein alr2800
Length = 1258
Score = 68.9 bits (167), Expect = 2e-11
Identities = 59/287 (20%), Positives = 117/287 (40%), Gaps = 14/287 (4%)
Query: 14 FHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGS 73
F P L+AT D + ++ S LL H+ R F G L +
Sbjct: 650 FSPEGQLLATCDTDCHVRVWEVKSGKL------LLICRGHSNWVRFVVFSPDGEILASCG 703
Query: 74 PDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNS 133
D ++ V G I L + + T+AS D IK+WD ++ +C +
Sbjct: 704 ADENVKLWSVRDGVCIKTLTGHEHEVFSVAFHPDGETLASASGDKTIKLWDIQDGTCLQT 763
Query: 134 FEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGR 193
H D++ + F+ D L +++ D T+ + ++ + K + + SV +G+
Sbjct: 764 LTGHTDWVRCVAFSPDGNTLASSAADHTIKLWDVSQGKCLRTLKSHTGWVRSVAFSADGQ 823
Query: 194 KVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENGMINLVGIL 253
+ GS + ++++ +C ++ +++ D +++GS + I L
Sbjct: 824 TLASGSGDRTIKIWNYHT-GECLKTYIGHTNSVYSIAYSPDSKILVSGSGDRTIKLWDCQ 882
Query: 254 PNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWD 300
+ I+ + H+ VAFS D + L ++ DQ ++LW+
Sbjct: 883 THICIKTLHGHTNEVCS-------VAFSPDGQTLACVSLDQSVRLWN 922
Score = 68.2 bits (165), Expect = 3e-11
Identities = 68/334 (20%), Positives = 134/334 (39%), Gaps = 54/334 (16%)
Query: 10 FDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRAL 69
+ I + P ++ +G D + L+ + + + +H HT + F G+ L
Sbjct: 856 YSIAYSPDSKILVSGSGDRTIKLWDCQTH------ICIKTLHGHTNEVCSVAFSPDGQTL 909
Query: 70 LTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERS 129
S D S+ + TG + + A+ + +ASG +D +K+WD +
Sbjct: 910 ACVSLDQSVRLWNCRTGQCLKAWYGNTDWALPVAFSPDRQILASGSNDKTVKLWDWQTGK 969
Query: 130 CCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLM 189
+S E H D+I I F+ D+ L + S D ++ + N+ + D + +VV
Sbjct: 970 YISSLEGHTDFIYGIAFSPDSQTLASASTDSSVRLWNISTGQCFQILLEHTDWVYAVVFH 1029
Query: 190 KNGRKVVCGSQTGIMLLYSWG---CFK---DCSDRFVD---------LSSNSIDTMLKL- 233
G+ + GS + L++ C K + SD+ + L+S S D ++L
Sbjct: 1030 PQGKIIATGSADCTVKLWNISTGQCLKTLSEHSDKILGMAWSPDGQLLASASADQSVRLW 1089
Query: 234 -------------------------DEDRIITGSENGMINLVGILPNRVIQPVAEHSEYP 268
+ + I T S + + + + ++ + H+ +
Sbjct: 1090 DCCTGRCVGILRGHSNRVYSAIFSPNGEIIATCSTDQTVKIWDWQQGKCLKTLTGHTNWV 1149
Query: 269 VERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDLD 302
+ +AFS D K L S +HDQ +++WD++
Sbjct: 1150 FD-------IAFSPDGKILASASHDQTVRIWDVN 1176
Score = 65.1 bits (157), Expect = 3e-10
Identities = 43/191 (22%), Positives = 79/191 (40%), Gaps = 6/191 (3%)
Query: 10 FDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRAL 69
+ + FHP ++ATG D + L+ S+ L + H++ + G+ L
Sbjct: 1024 YAVVFHPQGKIIATGSADCTVKLWNISTGQC------LKTLSEHSDKILGMAWSPDGQLL 1077
Query: 70 LTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERS 129
+ S D S+ D TG + L + + + +A+ D +K+WD ++
Sbjct: 1078 ASASADQSVRLWDCCTGRCVGILRGHSNRVYSAIFSPNGEIIATCSTDQTVKIWDWQQGK 1137
Query: 130 CCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLM 189
C + H +++ DI F+ D L + S D T+ + ++ K + SV
Sbjct: 1138 CLKTLTGHTNWVFDIAFSPDGKILASASHDQTVRIWDVNTGKCHHICIGHTHLVSSVAFS 1197
Query: 190 KNGRKVVCGSQ 200
+G V GSQ
Sbjct: 1198 PDGEVVASGSQ 1208
Score = 53.9 bits (128), Expect = 7e-07
Identities = 51/293 (17%), Positives = 109/293 (36%), Gaps = 14/293 (4%)
Query: 9 AFDIDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRA 68
A + F P ++A+G D + L+ + + S + HT+ F +
Sbjct: 939 ALPVAFSPDRQILASGSNDKTVKLWDWQTGKYISS------LEGHTDFIYGIAFSPDSQT 992
Query: 69 LLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRER 128
L + S D S+ ++ TG L + + + +A+G D +K+W+
Sbjct: 993 LASASTDSSVRLWNISTGQCFQILLEHTDWVYAVVFHPQGKIIATGSADCTVKLWNISTG 1052
Query: 129 SCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVL 188
C + H D I + ++ D L + S D ++ + + + + + S +
Sbjct: 1053 QCLKTLSEHSDKILGMAWSPDGQLLASASADQSVRLWDCCTGRCVGILRGHSNRVYSAIF 1112
Query: 189 MKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENGMIN 248
NG + S + ++ W K C ++ D D + + S + +
Sbjct: 1113 SPNGEIIATCSTDQTVKIWDWQQGK-CLKTLTGHTNWVFDIAFSPDGKILASASHDQTVR 1171
Query: 249 LVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSIAHDQMLKLWDL 301
+ + + H+ VAFS D + + S + DQ +++W++
Sbjct: 1172 IWDVNTGKCHHICIGHTHLVSS-------VAFSPDGEVVASGSQDQTVRIWNV 1217
Score = 33.1 bits (74), Expect = 1.2
Identities = 39/191 (20%), Positives = 67/191 (34%), Gaps = 50/191 (26%)
Query: 111 VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRN 170
+A+ D D ++VW+ + H +++ + F+ D L + D + + ++R
Sbjct: 657 LATCDTDCHVRVWEVKSGKLLLICRGHSNWVRFVVFSPDGEILASCGADENVKLWSVRDG 716
Query: 171 KVQAQSEFSEDELLSVVLMKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTM 230
E E+ SV +G L+S S D
Sbjct: 717 VCIKTLTGHEHEVFSVAFHPDGET---------------------------LASASGDKT 749
Query: 231 LKLDEDRIITGSENGMINLVGILPNRVIQPVAEHSEYPVERLGN*RFVAFSHDRKFLGSI 290
+KL + I +Q + H+++ R VAFS D L S
Sbjct: 750 IKLWD----------------IQDGTCLQTLTGHTDWV-------RCVAFSPDGNTLASS 786
Query: 291 AHDQMLKLWDL 301
A D +KLWD+
Sbjct: 787 AADHTIKLWDV 797
Score = 33.1 bits (74), Expect = 1.2
Identities = 28/128 (21%), Positives = 49/128 (37%), Gaps = 8/128 (6%)
Query: 14 FHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGS 73
F P+ ++AT D + ++ + L + HT F G+ L + S
Sbjct: 1112 FSPNGEIIATCSTDQTVKIWDWQQGKC------LKTLTGHTNWVFDIAFSPDGKILASAS 1165
Query: 74 PDFSILATDVETGSTIARLDNAHEAAVNRLVNLTES-TVASGDDDGCIKVWDTRERSCCN 132
D ++ DV TG + H V+ + + VASG D +++W+ + C
Sbjct: 1166 HDQTVRIWDVNTGKC-HHICIGHTHLVSSVAFSPDGEVVASGSQDQTVRIWNVKTGECLQ 1224
Query: 133 SFEAHEDY 140
A Y
Sbjct: 1225 ILRAKRLY 1232
>GBB1_DROME (P26308) Guanine nucleotide-binding protein beta subunit
1
Length = 340
Score = 67.8 bits (164), Expect = 4e-11
Identities = 66/247 (26%), Positives = 104/247 (41%), Gaps = 33/247 (13%)
Query: 14 FHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGS 73
+ PS + VA G +D +Y + N R L H SC RF++ + ++T S
Sbjct: 105 YAPSGSYVACGGLDNMCSIYNLKTREGNVRVSRELPGHGGYLSC--CRFLDDNQ-IVTSS 161
Query: 74 PDFSILATDVETGSTIARLDNAHEAAVNRLVNLTE-STVASGDDDGCIKVWDTRERSCCN 132
D S D+ETG + H V L + T SG D K+WD RE C
Sbjct: 162 GDMSCGLWDIETGLQVTSF-LGHTGDVMALSLAPQCKTFVSGACDASAKLWDIREGVCKQ 220
Query: 133 SFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELL----SVVL 188
+F HE I+ +TF + S D T + ++R + Q + +S D ++ SV
Sbjct: 221 TFPGHESDINAVTFFPNGQAFATGSDDATCRLFDIRAD--QELAMYSHDNIICGITSVAF 278
Query: 189 MKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSENGMIN 248
K+GR ++ G + D + N DTM K + I+ G +N ++
Sbjct: 279 SKSGRLLLAG--------------------YDDFNCNVWDTM-KAERSGILAGHDN-RVS 316
Query: 249 LVGILPN 255
+G+ N
Sbjct: 317 CLGVTEN 323
Score = 38.5 bits (88), Expect = 0.028
Identities = 28/118 (23%), Positives = 47/118 (39%), Gaps = 6/118 (5%)
Query: 53 HTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST-- 110
H A F G+A TGS D + D+ +A +H+ + + ++ S
Sbjct: 225 HESDINAVTFFPNGQAFATGSDDATCRLFDIRADQELAMY--SHDNIICGITSVAFSKSG 282
Query: 111 --VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCN 166
+ +G DD VWDT + H++ +S + + M + S D L V N
Sbjct: 283 RLLLAGYDDFNCNVWDTMKAERSGILAGHDNRVSCLGVTENGMAVATGSWDSFLRVWN 340
>GBB_DICDI (P36408) Guanine nucleotide-binding protein beta subunit
Length = 347
Score = 67.4 bits (163), Expect = 6e-11
Identities = 49/190 (25%), Positives = 89/190 (46%), Gaps = 11/190 (5%)
Query: 14 FHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLL-EIHAHTESCRAARFINGGRALLTG 72
+ P+ N VA G +D +Y S P+R+ E+++HT RF+N R ++T
Sbjct: 112 YSPTANFVACGGLDNICSIYNLRS---REQPIRVCRELNSHTGYLSCCRFLND-RQIVTS 167
Query: 73 SPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCN 132
S D + + DVE G+ I + + ++ V+ ++ SG D K+WD R C
Sbjct: 168 SGDMTCILWDVENGTKITEFSDHNGDVMSVSVSPDKNYFISGACDATAKLWDLRSGKCVQ 227
Query: 133 SFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELL----SVVL 188
+F HE I+ + + + + S D + + ++R ++ Q ++ D +L SV
Sbjct: 228 TFTGHEADINAVQYFPNGLSFGTGSDDASCRLFDIRADRELMQ--YTHDNILCGITSVGF 285
Query: 189 MKNGRKVVCG 198
+GR + G
Sbjct: 286 SFSGRFLFAG 295
Score = 34.7 bits (78), Expect = 0.41
Identities = 33/155 (21%), Positives = 54/155 (34%), Gaps = 8/155 (5%)
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLT 71
+ P N +G D L+ S + H A ++ G + T
Sbjct: 197 VSVSPDKNYFISGACDATAKLWDLRSGKC------VQTFTGHEADINAVQYFPNGLSFGT 250
Query: 72 GSPDFSILATDVETGSTIARL--DNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERS 129
GS D S D+ + + DN + + + + +G DD VWDT +
Sbjct: 251 GSDDASCRLFDIRADRELMQYTHDNILCGITSVGFSFSGRFLFAGYDDFTCNVWDTLKGE 310
Query: 130 CCNSFEAHEDYISDITFASDAMKLLATSGDGTLSV 164
S H + +S + +D M L S D L +
Sbjct: 311 RVLSLTGHGNRVSCLGVPTDGMALCTGSWDSLLKI 345
>GBB_CRYPA (O14435) Guanine nucleotide-binding protein beta subunit
Length = 359
Score = 66.6 bits (161), Expect = 1e-10
Identities = 50/161 (31%), Positives = 74/161 (45%), Gaps = 7/161 (4%)
Query: 14 FHPSDNLVATGLIDGDLHLYRYSSDNTNSD-PVRLL-EIHAHTESCRAARFINGGRALLT 71
+ PS N VA G +D +Y N N D P R+ E+ H RFIN R++LT
Sbjct: 121 YAPSGNYVACGGLDNICSIYNL---NQNRDGPTRVARELSGHAGYLSCCRFIND-RSILT 176
Query: 72 GSPDFSILATDVETGST-IARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSC 130
S D + + D+ETG+ I D+ + L ++T SG D K+WD R
Sbjct: 177 SSGDMTCMKWDIETGTKQIEFADHLGDVMSISLNPTNQNTFISGACDAFAKLWDIRAGKA 236
Query: 131 CNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNK 171
+F HE I+ I F D + S D T + ++R ++
Sbjct: 237 VQTFAGHESDINAIQFFPDGHSFVTGSDDATCRLFDIRADR 277
Score = 36.6 bits (83), Expect = 0.11
Identities = 27/116 (23%), Positives = 48/116 (41%), Gaps = 6/116 (5%)
Query: 53 HTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST-- 110
H A +F G + +TGS D + D+ + + E+ + + ++ S
Sbjct: 243 HESDINAIQFFPDGHSFVTGSDDATCRLFDIRADRELNVYGS--ESILCGITSVATSVSG 300
Query: 111 --VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSV 164
+ +G DD KVWD S HE+ +S + ++D + L S D L +
Sbjct: 301 RLLFAGYDDFECKVWDVTRGEKVGSLVGHENRVSCLGVSNDGISLCTGSWDSLLKI 356
>GBB_LYMST (Q08706) Guanine nucleotide-binding protein beta subunit
Length = 341
Score = 66.2 bits (160), Expect = 1e-10
Identities = 60/231 (25%), Positives = 96/231 (40%), Gaps = 22/231 (9%)
Query: 14 FHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGS 73
+ PS N VA G +D +Y + N R E+ HT RFI+ +++T S
Sbjct: 106 YAPSGNFVACGGLDNICSIYSLKTREGNVRVSR--ELPGHTGYLSCCRFIDDN-SIVTSS 162
Query: 74 PDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNS 133
D S D+ETG ++ + T SG D K+WD R+ C +
Sbjct: 163 GDMSCALWDIETGQQTTSFTGHTGDVMSLSTSPDFRTFVSGACDASAKLWDVRDGMCKQT 222
Query: 134 FEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELLSVVLMKNGR 193
F HE I+ IT+ + S D T + ++R + Q +S D ++ G
Sbjct: 223 FSGHESDINAITYFPNGHAFATGSDDATCRLFDIRAD--QEIGMYSHDNIIC------GI 274
Query: 194 KVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSEN 244
V S++G +LL R+ D + N D +LK + ++ G +N
Sbjct: 275 TSVAFSKSGRLLL----------GRYDDFNCNVWD-VLKQETHGVLAGHDN 314
Score = 31.2 bits (69), Expect = 4.5
Identities = 25/119 (21%), Positives = 43/119 (36%), Gaps = 8/119 (6%)
Query: 53 HTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST-- 110
H A + G A TGS D + D+ I +H+ + + ++ S
Sbjct: 226 HESDINAITYFPNGHAFATGSDDATCRLFDIRADQEIGMY--SHDNIICGITSVAFSKSG 283
Query: 111 ---VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCN 166
+ DD C VWD ++ H++ +S + D + S D L + N
Sbjct: 284 RLLLGRYDDFNC-NVWDVLKQETHGVLAGHDNRVSCLGVTEDGSAVATGSWDSFLKIWN 341
>GBB2_DROME (P29829) Guanine nucleotide-binding protein beta subunit
2
Length = 346
Score = 66.2 bits (160), Expect = 1e-10
Identities = 55/206 (26%), Positives = 92/206 (43%), Gaps = 9/206 (4%)
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLT 71
+ F PS N VA G +D +Y ++ + + + E+ + + RF++ G L+T
Sbjct: 107 VAFSPSGNFVACGGMDNQCTVYDVNNRDASGVAKMVKELMGYEGFLSSCRFLDDGH-LIT 165
Query: 72 GSPDFSILATDVETGSTIARLD-NAHEAAVNRL-VNLTESTVASGDDDGCIKVWDTRERS 129
GS D I D+E G + +D N H + L ++ T +G D K+WD RE
Sbjct: 166 GSGDMKICHWDLEKG--VKTMDFNGHAGDIAGLSLSPDMKTYITGSVDKTAKLWDVREEG 223
Query: 130 CCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDE--LLSVV 187
F H+ +S + + + S D T + +LR ++ AQ E + S
Sbjct: 224 HKQMFFGHDMDVSSVCYHPSGFGFASCSEDQTARMYDLRADQQIAQYEPPQKNTGFTSCA 283
Query: 188 LMKNGRKVVCGSQTGIMLLYSWGCFK 213
L +GR ++CG G ++SW K
Sbjct: 284 LSTSGRYLMCGGIEG--NVHSWDTMK 307
>GBB1_CRIGR (Q6TMK6) Guanine nucleotide-binding protein
G(I)/G(S)/G(T) beta subunit 1 (Transducin beta chain 1)
Length = 340
Score = 66.2 bits (160), Expect = 1e-10
Identities = 55/190 (28%), Positives = 82/190 (42%), Gaps = 11/190 (5%)
Query: 14 FHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGS 73
+ PS N VA G +D +Y + N R E+ HT RF+N + ++T S
Sbjct: 105 YAPSGNYVACGGLDNICSIYNLKTREGNVRVSR--ELAGHTGYLSCCRFLNDNQ-IVTSS 161
Query: 74 PDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTV-ASGDDDGCIKVWDTRERSCCN 132
D + D+ETG H V L ++ + SG D K+WD RE C
Sbjct: 162 EDTTCALWDIETGQQTTTF-TGHTGDVMSLSLAPDTRLFVSGACDASAKLWDVREGMCRQ 220
Query: 133 SFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELL----SVVL 188
+F HE I+ I F + S D T + +LR + Q +S D ++ SV
Sbjct: 221 TFTGHESDINAICFFPNGNAFATGSDDATCRLFDLRAD--QELMTYSHDNIICGITSVSF 278
Query: 189 MKNGRKVVCG 198
K+GR ++ G
Sbjct: 279 SKSGRLLLAG 288
Score = 35.4 bits (80), Expect = 0.24
Identities = 26/118 (22%), Positives = 46/118 (38%), Gaps = 6/118 (5%)
Query: 53 HTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST-- 110
H A F G A TGS D + D+ + + +H+ + + +++ S
Sbjct: 225 HESDINAICFFPNGNAFATGSDDATCRLFDLRADQEL--MTYSHDNIICGITSVSFSKSG 282
Query: 111 --VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCN 166
+ +G DD VWD + H++ +S + D M + S D L + N
Sbjct: 283 RLLLAGYDDFNCNVWDALKADRAGVLAGHDNRVSCLGVTDDGMAVATGSWDSFLKIWN 340
Score = 31.6 bits (70), Expect = 3.5
Identities = 32/117 (27%), Positives = 45/117 (38%), Gaps = 12/117 (10%)
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTE---SCRAARFINGGRA 68
I F P+ N ATG D L+ +D L ++H + F GR
Sbjct: 232 ICFFPNGNAFATGSDDATCRLFDLRADQE-------LMTYSHDNIICGITSVSFSKSGRL 284
Query: 69 LLTGSPDFSILATDVETGSTIARLDNAHEAAVNRL-VNLTESTVASGDDDGCIKVWD 124
LL G DF+ D L H+ V+ L V VA+G D +K+W+
Sbjct: 285 LLAGYDDFNCNVWDALKADRAGVL-AGHDNRVSCLGVTDDGMAVATGSWDSFLKIWN 340
>GBB1_XENLA (P79959) Guanine nucleotide-binding protein
G(I)/G(S)/G(T) beta subunit 1 (Transducin beta chain 1)
(XGbeta1)
Length = 340
Score = 65.9 bits (159), Expect = 2e-10
Identities = 64/236 (27%), Positives = 95/236 (40%), Gaps = 32/236 (13%)
Query: 14 FHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGS 73
+ PS N VA G +D +Y + N R E+ HT RF++ + ++T S
Sbjct: 105 YAPSGNYVACGGLDNICPIYNLKTREGNVRVSR--ELAGHTGYLSCCRFLDDNQ-IITSS 161
Query: 74 PDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTV-ASGDDDGCIKVWDTRERSCCN 132
D + D+ETG H V L +S SG D K+WD RE C
Sbjct: 162 GDTTCALWDIETGQQTTTF-TGHTGDVMSLSLAPDSRCFVSGACDASAKLWDVREGMCRQ 220
Query: 133 SFEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELL----SVVL 188
+F HE I+ I F + S D T + +LR + Q +S D ++ SV
Sbjct: 221 TFTGHESDINAICFFPNGNAFATGSDDATCRLFDLRAD--QELMVYSHDNIICGITSVAF 278
Query: 189 MKNGRKVVCGSQTGIMLLYSWGCFKDCSDRFVDLSSNSIDTMLKLDEDRIITGSEN 244
K+GR ++ G + D + N DT LK D ++ G +N
Sbjct: 279 SKSGRLLLAG--------------------YDDFNCNVWDT-LKADRAGVLAGHDN 313
Score = 36.2 bits (82), Expect = 0.14
Identities = 27/118 (22%), Positives = 46/118 (38%), Gaps = 6/118 (5%)
Query: 53 HTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST-- 110
H A F G A TGS D + D+ + + +H+ + + ++ S
Sbjct: 225 HESDINAICFFPNGNAFATGSDDATCRLFDLRADQEL--MVYSHDNIICGITSVAFSKSG 282
Query: 111 --VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCN 166
+ +G DD VWDT + H++ +S + D M + S D L + N
Sbjct: 283 RLLLAGYDDFNCNVWDTLKADRAGVLAGHDNRVSCLGVTDDGMAVATGSWDSFLKIWN 340
Score = 33.1 bits (74), Expect = 1.2
Identities = 32/117 (27%), Positives = 46/117 (38%), Gaps = 12/117 (10%)
Query: 12 IDFHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTE---SCRAARFINGGRA 68
I F P+ N ATG D L+ +D L +++H + F GR
Sbjct: 232 ICFFPNGNAFATGSDDATCRLFDLRADQE-------LMVYSHDNIICGITSVAFSKSGRL 284
Query: 69 LLTGSPDFSILATDVETGSTIARLDNAHEAAVNRL-VNLTESTVASGDDDGCIKVWD 124
LL G DF+ D L H+ V+ L V VA+G D +K+W+
Sbjct: 285 LLAGYDDFNCNVWDTLKADRAGVL-AGHDNRVSCLGVTDDGMAVATGSWDSFLKIWN 340
>GBB1_HOMAM (O45040) Guanine nucleotide-binding protein
G(I)/G(S)/G(T) beta subunit 1 (Transducin beta chain 1)
Length = 340
Score = 65.9 bits (159), Expect = 2e-10
Identities = 48/189 (25%), Positives = 84/189 (44%), Gaps = 9/189 (4%)
Query: 14 FHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGS 73
+ PS + VA G +D +Y + N R E+ HT RF++ + ++T S
Sbjct: 105 YAPSGSYVACGGLDNICSIYSLKTREGNVRVSR--ELPGHTGYLSCCRFVDDNQ-IVTSS 161
Query: 74 PDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNS 133
D + D+ETG + ++ ++ T SG D K+WD R+ C +
Sbjct: 162 GDMTCALWDIETGQQCTQFTGHTGDVMSLSLSPNMRTFTSGACDASAKLWDIRDGMCRQT 221
Query: 134 FEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELL----SVVLM 189
F HE I+ +TF + S D T + ++R + Q + +S D ++ SV
Sbjct: 222 FPGHESDINAVTFFPNGHAFATGSDDATCRLFDIRAD--QELAMYSHDNIICGITSVAFS 279
Query: 190 KNGRKVVCG 198
K+G+ ++ G
Sbjct: 280 KSGKLLLAG 288
Score = 38.9 bits (89), Expect = 0.022
Identities = 27/116 (23%), Positives = 42/116 (35%), Gaps = 2/116 (1%)
Query: 53 HTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARL--DNAHEAAVNRLVNLTEST 110
H A F G A TGS D + D+ +A DN + + +
Sbjct: 225 HESDINAVTFFPNGHAFATGSDDATCRLFDIRADQELAMYSHDNIICGITSVAFSKSGKL 284
Query: 111 VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCN 166
+ +G DD VWD+ H++ +S + D M + S D L + N
Sbjct: 285 LLAGYDDFNCNVWDSMRTERAGVLAGHDNRVSCLGVTEDGMAVATGSWDSFLKIWN 340
>GBB4_MOUSE (P29387) Guanine nucleotide-binding protein beta subunit
4 (Transducin beta chain 4)
Length = 340
Score = 65.5 bits (158), Expect = 2e-10
Identities = 51/189 (26%), Positives = 81/189 (41%), Gaps = 9/189 (4%)
Query: 14 FHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGS 73
+ PS N VA G +D +Y + N R E+ HT RF++ G+ ++T S
Sbjct: 105 YAPSGNYVACGGLDNICSIYNLKTREGNVRVSR--ELPGHTGYLSCCRFLDDGQ-IITSS 161
Query: 74 PDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNS 133
D + D+ETG ++ ++ T SG D K+WD R+ C S
Sbjct: 162 GDTTCALWDIETGQQTTTFTGHSGDVMSLSLSPDLKTFVSGACDASSKLWDIRDGMCRQS 221
Query: 134 FEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELL----SVVLM 189
F H I+ ++F S D T + +LR + Q +S D ++ SV
Sbjct: 222 FTGHISDINAVSFFPSGYAFATGSDDATCRLFDLRAD--QELLLYSHDNIICGITSVAFS 279
Query: 190 KNGRKVVCG 198
K+GR ++ G
Sbjct: 280 KSGRLLLAG 288
Score = 33.9 bits (76), Expect = 0.70
Identities = 27/118 (22%), Positives = 45/118 (37%), Gaps = 6/118 (5%)
Query: 53 HTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST-- 110
H A F G A TGS D + D+ + L +H+ + + ++ S
Sbjct: 225 HISDINAVSFFPSGYAFATGSDDATCRLFDLRADQEL--LLYSHDNIICGITSVAFSKSG 282
Query: 111 --VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCN 166
+ +G DD VWD + H++ +S + D M + S D L + N
Sbjct: 283 RLLLAGYDDFNCSVWDALKGGRSGVLAGHDNRVSCLGVTDDGMAVATGSWDSFLRIWN 340
>GBB2_RAT (P54313) Guanine nucleotide-binding protein G(I)/G(S)/G(T)
beta subunit 2 (Transducin beta chain 2) (G protein beta
2 subunit)
Length = 340
Score = 64.7 bits (156), Expect = 4e-10
Identities = 50/189 (26%), Positives = 81/189 (42%), Gaps = 9/189 (4%)
Query: 14 FHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGS 73
+ PS N VA G +D +Y + N R E+ HT RF++ + ++T S
Sbjct: 105 YAPSGNFVACGGLDNICSIYSLKTREGNVRVSR--ELPGHTGYLSCCRFLDDNQ-IITSS 161
Query: 74 PDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNS 133
D + D+ETG ++ + T SG D IK+WD R+ C +
Sbjct: 162 GDTTCALWDIETGQQTVGFAGHSGDVMSLSLAPDGRTFVSGACDASIKLWDVRDSMCRQT 221
Query: 134 FEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELL----SVVLM 189
F HE I+ + F + S D T + +LR + Q +S D ++ SV
Sbjct: 222 FIGHESDINAVAFFPNGYAFTTGSDDATCRLFDLRAD--QELLMYSHDNIICGITSVAFS 279
Query: 190 KNGRKVVCG 198
++GR ++ G
Sbjct: 280 RSGRLLLAG 288
Score = 35.0 bits (79), Expect = 0.31
Identities = 26/118 (22%), Positives = 45/118 (38%), Gaps = 6/118 (5%)
Query: 53 HTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST-- 110
H A F G A TGS D + D+ + L +H+ + + ++ S
Sbjct: 225 HESDINAVAFFPNGYAFTTGSDDATCRLFDLRADQEL--LMYSHDNIICGITSVAFSRSG 282
Query: 111 --VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCN 166
+ +G DD +WD + H++ +S + D M + S D L + N
Sbjct: 283 RLLLAGYDDFNCNIWDAMKGDRAGVLAGHDNRVSCLGVTDDGMAVATGSWDSFLKIWN 340
Score = 30.4 bits (67), Expect = 7.7
Identities = 28/154 (18%), Positives = 59/154 (38%), Gaps = 5/154 (3%)
Query: 50 IHAHTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTES 109
+ H A + R L++ S D ++ D T + + + + + +
Sbjct: 51 LRGHLAKIYAMHWGTDSRLLVSASQDGKLIIWDSYTTNKVHAIPLRSSWVMTCAYAPSGN 110
Query: 110 TVASGDDDGCIKVWDTRERS----CCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVC 165
VA G D ++ + R H Y+S F D +++ +SGD T ++
Sbjct: 111 FVACGGLDNICSIYSLKTREGNVRVSRELPGHTGYLSCCRFLDDN-QIITSSGDTTCALW 169
Query: 166 NLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGS 199
++ + +++S+ L +GR V G+
Sbjct: 170 DIETGQQTVGFAGHSGDVMSLSLAPDGRTFVSGA 203
>GBB2_MOUSE (P62880) Guanine nucleotide-binding protein
G(I)/G(S)/G(T) beta subunit 2 (Transducin beta chain 2)
(G protein beta 2 subunit)
Length = 340
Score = 64.7 bits (156), Expect = 4e-10
Identities = 50/189 (26%), Positives = 81/189 (42%), Gaps = 9/189 (4%)
Query: 14 FHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGS 73
+ PS N VA G +D +Y + N R E+ HT RF++ + ++T S
Sbjct: 105 YAPSGNFVACGGLDNICSIYSLKTREGNVRVSR--ELPGHTGYLSCCRFLDDNQ-IITSS 161
Query: 74 PDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNS 133
D + D+ETG ++ + T SG D IK+WD R+ C +
Sbjct: 162 GDTTCALWDIETGQQTVGFAGHSGDVMSLSLAPDGRTFVSGACDASIKLWDVRDSMCRQT 221
Query: 134 FEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELL----SVVLM 189
F HE I+ + F + S D T + +LR + Q +S D ++ SV
Sbjct: 222 FIGHESDINAVAFFPNGYAFTTGSDDATCRLFDLRAD--QELLMYSHDNIICGITSVAFS 279
Query: 190 KNGRKVVCG 198
++GR ++ G
Sbjct: 280 RSGRLLLAG 288
Score = 35.0 bits (79), Expect = 0.31
Identities = 26/118 (22%), Positives = 45/118 (38%), Gaps = 6/118 (5%)
Query: 53 HTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST-- 110
H A F G A TGS D + D+ + L +H+ + + ++ S
Sbjct: 225 HESDINAVAFFPNGYAFTTGSDDATCRLFDLRADQEL--LMYSHDNIICGITSVAFSRSG 282
Query: 111 --VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCN 166
+ +G DD +WD + H++ +S + D M + S D L + N
Sbjct: 283 RLLLAGYDDFNCNIWDAMKGDRAGVLAGHDNRVSCLGVTDDGMAVATGSWDSFLKIWN 340
Score = 30.4 bits (67), Expect = 7.7
Identities = 28/154 (18%), Positives = 59/154 (38%), Gaps = 5/154 (3%)
Query: 50 IHAHTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTES 109
+ H A + R L++ S D ++ D T + + + + + +
Sbjct: 51 LRGHLAKIYAMHWGTDSRLLVSASQDGKLIIWDSYTTNKVHAIPLRSSWVMTCAYAPSGN 110
Query: 110 TVASGDDDGCIKVWDTRERS----CCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVC 165
VA G D ++ + R H Y+S F D +++ +SGD T ++
Sbjct: 111 FVACGGLDNICSIYSLKTREGNVRVSRELPGHTGYLSCCRFLDDN-QIITSSGDTTCALW 169
Query: 166 NLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGS 199
++ + +++S+ L +GR V G+
Sbjct: 170 DIETGQQTVGFAGHSGDVMSLSLAPDGRTFVSGA 203
>GBB2_HUMAN (P62879) Guanine nucleotide-binding protein
G(I)/G(S)/G(T) beta subunit 2 (Transducin beta chain 2)
(G protein beta 2 subunit)
Length = 340
Score = 64.7 bits (156), Expect = 4e-10
Identities = 50/189 (26%), Positives = 81/189 (42%), Gaps = 9/189 (4%)
Query: 14 FHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGS 73
+ PS N VA G +D +Y + N R E+ HT RF++ + ++T S
Sbjct: 105 YAPSGNFVACGGLDNICSIYSLKTREGNVRVSR--ELPGHTGYLSCCRFLDDNQ-IITSS 161
Query: 74 PDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNS 133
D + D+ETG ++ + T SG D IK+WD R+ C +
Sbjct: 162 GDTTCALWDIETGQQTVGFAGHSGDVMSLSLAPDGRTFVSGACDASIKLWDVRDSMCRQT 221
Query: 134 FEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELL----SVVLM 189
F HE I+ + F + S D T + +LR + Q +S D ++ SV
Sbjct: 222 FIGHESDINAVAFFPNGYAFTTGSDDATCRLFDLRAD--QELLMYSHDNIICGITSVAFS 279
Query: 190 KNGRKVVCG 198
++GR ++ G
Sbjct: 280 RSGRLLLAG 288
Score = 35.0 bits (79), Expect = 0.31
Identities = 26/118 (22%), Positives = 45/118 (38%), Gaps = 6/118 (5%)
Query: 53 HTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST-- 110
H A F G A TGS D + D+ + L +H+ + + ++ S
Sbjct: 225 HESDINAVAFFPNGYAFTTGSDDATCRLFDLRADQEL--LMYSHDNIICGITSVAFSRSG 282
Query: 111 --VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCN 166
+ +G DD +WD + H++ +S + D M + S D L + N
Sbjct: 283 RLLLAGYDDFNCNIWDAMKGDRAGVLAGHDNRVSCLGVTDDGMAVATGSWDSFLKIWN 340
Score = 30.4 bits (67), Expect = 7.7
Identities = 28/154 (18%), Positives = 59/154 (38%), Gaps = 5/154 (3%)
Query: 50 IHAHTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTES 109
+ H A + R L++ S D ++ D T + + + + + +
Sbjct: 51 LRGHLAKIYAMHWGTDSRLLVSASQDGKLIIWDSYTTNKVHAIPLRSSWVMTCAYAPSGN 110
Query: 110 TVASGDDDGCIKVWDTRERS----CCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVC 165
VA G D ++ + R H Y+S F D +++ +SGD T ++
Sbjct: 111 FVACGGLDNICSIYSLKTREGNVRVSRELPGHTGYLSCCRFLDDN-QIITSSGDTTCALW 169
Query: 166 NLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGS 199
++ + +++S+ L +GR V G+
Sbjct: 170 DIETGQQTVGFAGHSGDVMSLSLAPDGRTFVSGA 203
>GBB2_BOVIN (P11017) Guanine nucleotide-binding protein
G(I)/G(S)/G(T) beta subunit 2 (Transducin beta chain 2)
(G protein beta 2 subunit) (Fragment)
Length = 326
Score = 64.7 bits (156), Expect = 4e-10
Identities = 50/189 (26%), Positives = 81/189 (42%), Gaps = 9/189 (4%)
Query: 14 FHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGS 73
+ PS N VA G +D +Y + N R E+ HT RF++ + ++T S
Sbjct: 91 YAPSGNFVACGGLDNICSIYSLKTREGNVRVSR--ELPGHTGYLSCCRFLDDNQ-IITSS 147
Query: 74 PDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNS 133
D + D+ETG ++ + T SG D IK+WD R+ C +
Sbjct: 148 GDTTCALWDIETGQQTVGFAGHSGDVMSLSLAPDGRTFVSGACDASIKLWDVRDSMCRQT 207
Query: 134 FEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELL----SVVLM 189
F HE I+ + F + S D T + +LR + Q +S D ++ SV
Sbjct: 208 FIGHESDINAVAFFPNGYAFTTGSDDATCRLFDLRAD--QELLMYSHDNIICGITSVAFS 265
Query: 190 KNGRKVVCG 198
++GR ++ G
Sbjct: 266 RSGRLLLAG 274
Score = 35.0 bits (79), Expect = 0.31
Identities = 26/118 (22%), Positives = 45/118 (38%), Gaps = 6/118 (5%)
Query: 53 HTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST-- 110
H A F G A TGS D + D+ + L +H+ + + ++ S
Sbjct: 211 HESDINAVAFFPNGYAFTTGSDDATCRLFDLRADQEL--LMYSHDNIICGITSVAFSRSG 268
Query: 111 --VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCN 166
+ +G DD +WD + H++ +S + D M + S D L + N
Sbjct: 269 RLLLAGYDDFNCNIWDAMKGDRAGVLAGHDNRVSCLGVTDDGMAVATGSWDSFLKIWN 326
Score = 30.4 bits (67), Expect = 7.7
Identities = 28/154 (18%), Positives = 59/154 (38%), Gaps = 5/154 (3%)
Query: 50 IHAHTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTES 109
+ H A + R L++ S D ++ D T + + + + + +
Sbjct: 37 LRGHLAKIYAMHWGTDSRLLVSASQDGKLIIWDSYTTNKVHAIPLRSSWVMTCAYAPSGN 96
Query: 110 TVASGDDDGCIKVWDTRERS----CCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVC 165
VA G D ++ + R H Y+S F D +++ +SGD T ++
Sbjct: 97 FVACGGLDNICSIYSLKTREGNVRVSRELPGHTGYLSCCRFLDDN-QIITSSGDTTCALW 155
Query: 166 NLRRNKVQAQSEFSEDELLSVVLMKNGRKVVCGS 199
++ + +++S+ L +GR V G+
Sbjct: 156 DIETGQQTVGFAGHSGDVMSLSLAPDGRTFVSGA 189
>GBB3_MOUSE (Q61011) Guanine nucleotide-binding protein
G(I)/G(S)/G(T) beta subunit 3 (Transducin beta chain 3)
Length = 340
Score = 63.9 bits (154), Expect = 6e-10
Identities = 51/189 (26%), Positives = 82/189 (42%), Gaps = 9/189 (4%)
Query: 14 FHPSDNLVATGLIDGDLHLYRYSSDNTNSDPVRLLEIHAHTESCRAARFINGGRALLTGS 73
+ PS N VA G +D +Y S N R E+ AHT RF++ ++T S
Sbjct: 105 YAPSGNFVACGGLDNMCSIYNLKSREGNVKVSR--ELSAHTGYLSCCRFLDDNN-IVTSS 161
Query: 74 PDFSILATDVETGSTIARLDNAHEAAVNRLVNLTESTVASGDDDGCIKVWDTRERSCCNS 133
D + D+ETG ++ V+ SG D K+WD RE +C +
Sbjct: 162 GDTTCALWDIETGQQKTVFVGHTGDCMSLAVSPDYKLFISGACDASAKLWDVREGTCRQT 221
Query: 134 FEAHEDYISDITFASDAMKLLATSGDGTLSVCNLRRNKVQAQSEFSEDELL----SVVLM 189
F HE I+ I F + + S D + + +LR + Q + +S++ ++ SV
Sbjct: 222 FTGHESDINAICFFPNGEAICTGSDDASCRLFDLRAD--QELTAYSQESIICGITSVAFS 279
Query: 190 KNGRKVVCG 198
+GR + G
Sbjct: 280 LSGRLLFAG 288
Score = 35.4 bits (80), Expect = 0.24
Identities = 27/118 (22%), Positives = 47/118 (38%), Gaps = 6/118 (5%)
Query: 53 HTESCRAARFINGGRALLTGSPDFSILATDVETGSTIARLDNAHEAAVNRLVNLTEST-- 110
H A F G A+ TGS D S D+ + + E+ + + ++ S
Sbjct: 225 HESDINAICFFPNGEAICTGSDDASCRLFDLRADQELTAY--SQESIICGITSVAFSLSG 282
Query: 111 --VASGDDDGCIKVWDTRERSCCNSFEAHEDYISDITFASDAMKLLATSGDGTLSVCN 166
+ +G DD VWD+ + H++ +S + +D M + S D L + N
Sbjct: 283 RLLFAGYDDFNCNVWDSLKCERVGILSGHDNRVSCLGVTADGMAVATGSWDSFLKIWN 340
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.135 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,148,048
Number of Sequences: 164201
Number of extensions: 1815999
Number of successful extensions: 8224
Number of sequences better than 10.0: 399
Number of HSP's better than 10.0 without gapping: 238
Number of HSP's successfully gapped in prelim test: 164
Number of HSP's that attempted gapping in prelim test: 6214
Number of HSP's gapped (non-prelim): 1501
length of query: 378
length of database: 59,974,054
effective HSP length: 112
effective length of query: 266
effective length of database: 41,583,542
effective search space: 11061222172
effective search space used: 11061222172
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC142222.5


