

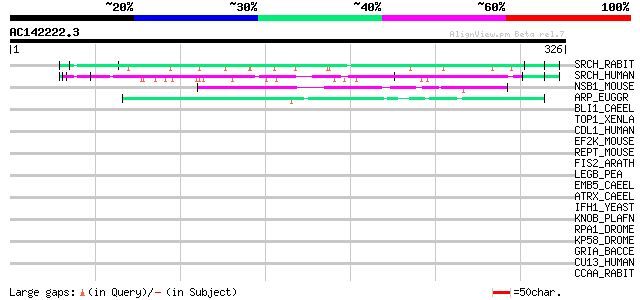
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC142222.3 + phase: 0
(326 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SRCH_RABIT (P16230) Sarcoplasmic reticulum histidine-rich calciu... 98 3e-20
SRCH_HUMAN (P23327) Sarcoplasmic reticulum histidine-rich calciu... 65 3e-10
NSB1_MOUSE (Q9JL35) Nucleosome binding protein 1 (Nucleosome bin... 48 3e-05
ARP_EUGGR (Q04732) Calcium-binding acidic-repeat protein precurs... 44 7e-04
BLI1_CAEEL (Q09457) Cuticle collagen bli-1 41 0.005
TOP1_XENLA (P41512) DNA topoisomerase I (EC 5.99.1.2) 40 0.008
CDL1_HUMAN (P21127) PITSLRE serine/threonine-protein kinase CDC2... 40 0.008
EF2K_MOUSE (O08796) Elongation factor 2 kinase (EC 2.7.1.-) (eEF... 40 0.010
REPT_MOUSE (P97347) Repetin 39 0.014
FIS2_ARATH (Q9ZNT9) Polycomb group protein FERTILIZATION-INDEPEN... 39 0.018
LEGB_PEA (P14594) Legumin B [Contains: Legumin B alpha chain (Le... 39 0.023
EMB5_CAEEL (P34703) Abnormal embryogenesis protein 5 38 0.030
ATRX_CAEEL (Q9U7E0) Transcriptional regulator ATRX homolog (X-li... 38 0.030
IFH1_YEAST (P39520) IFH1 protein (RRP3 protein) 38 0.040
KNOB_PLAFN (P06719) Knob-associated histidine-rich protein precu... 37 0.067
RPA1_DROME (P91875) DNA-directed RNA polymerase I largest subuni... 37 0.088
KP58_DROME (Q9VPC0) Serine/threonine-protein kinase PITSLRE (EC ... 37 0.088
GRIA_BACCE (O85467) Spore germination protein gerIA 37 0.088
CU13_HUMAN (O95447) Protein C21orf13 37 0.088
CCAA_RABIT (P27884) Voltage-dependent P/Q-type calcium channel a... 37 0.088
>SRCH_RABIT (P16230) Sarcoplasmic reticulum histidine-rich
calcium-binding protein precursor
Length = 852
Score = 97.8 bits (242), Expect = 3e-20
Identities = 85/315 (26%), Positives = 125/315 (38%), Gaps = 50/315 (15%)
Query: 30 SVSQARRGHPRYDDDHSLDEFLADRRRKKHLALRRHPHYDDDYSLDEIFPNRRQKHQAHK 89
S RGH + D++ D+ R + H D+D DE HQAH+
Sbjct: 196 SPEHRHRGHGKEDEEDEDDDSTESDRHQAHRHRGHREEEDEDDDDDEGDSTESDHHQAHR 255
Query: 90 RHYH------YDDEDSLDSLLPKQHQA--ERGHTRYDDEDSLDE---VLPKQRQAKRGRS 138
H DD+D DS +HQA RGH +DED DE + QA R R
Sbjct: 256 HRGHEEEEDEEDDDDEGDSTESDRHQAHRHRGHREEEDEDDDDEGDSTESDRHQAHRHRG 315
Query: 139 HY------DDDDSLDEVLPKQH--HAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHY---- 186
H DDDD D +H H RGH +D+DD +G + + H H
Sbjct: 316 HREEEDDDDDDDEGDSTESDRHQAHRHRGHREEEDEDDDDEGDSTESDRHQAHRHRGHRE 375
Query: 187 ---DNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPHYDNDGDSLHE 243
++D+D HQA R R H + ED D++GDS
Sbjct: 376 EEDEDDDDEGDSTESDRHQAHRHRGHREEEDE-----------------DDDDEGDSTES 418
Query: 244 DLPKNHQAER----GQPRYDDDDDLPDGVLPKQRQAKKLHPHYDNDEDSLHEDLPKNHQA 299
D +HQA R + ++DDD D + QA + H + +++ ++ +H
Sbjct: 419 D---HHQAHRHRGHREEEDEEDDDEGDSTESDRHQAHRHRGHGEEEDEDDDDEGEHHHVP 475
Query: 300 KRGRPLYDDTDSLDE 314
RG +++ D D+
Sbjct: 476 HRGHRGHEEDDGGDD 490
Score = 83.6 bits (205), Expect = 6e-16
Identities = 81/309 (26%), Positives = 119/309 (38%), Gaps = 45/309 (14%)
Query: 36 RGHPRYDDDHSLDEFLADRRRKKHLALRRHPHY---DDDYSLDEIFPNRRQKHQAHKRHY 92
RGH R ++D D+ +H A R H DDD DE +HQAH+
Sbjct: 286 RGH-REEEDEDDDDEGDSTESDRHQAHRHRGHREEEDDDDDDDEGDSTESDRHQAHRHRG 344
Query: 93 H-----YDDEDSLDSLLPKQHQA--ERGHTRYDDEDSLDE---VLPKQRQAKRGRSH--- 139
H DD+D DS +HQA RGH +DED DE + QA R R H
Sbjct: 345 HREEEDEDDDDEGDSTESDRHQAHRHRGHREEEDEDDDDEGDSTESDRHQAHRHRGHREE 404
Query: 140 --YDDDDSLDEVLPKQH--HAKRGHPHYDDDDDLPDGVLPKQQAKKRHPH--------YD 187
DDDD D H H RGH +D++D +G + + H H D
Sbjct: 405 EDEDDDDEGDSTESDHHQAHRHRGHREEEDEEDDDEGDSTESDRHQAHRHRGHGEEEDED 464
Query: 188 NDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPHYDNDGDSLHEDLPK 247
+D++ H +P H+ RG D D+ + RH + + + D
Sbjct: 465 DDDEGEHHHVP--HRGHRGHEEDDGGDDDDGDDSTENGHQAHRHQGHGKEEAEVTSDEHH 522
Query: 248 NHQAERGQPRYDDDDDLPDGVLPKQRQAKKLHPHY--DNDEDSLHEDL------------ 293
+H + G + D + +GV H H+ +E+ E+L
Sbjct: 523 HHVPDHGHQGHGDKEGEEEGVSTDHWHQVPRHAHHGPGGEEEGGEEELTVKAGHHVASHP 582
Query: 294 PKNHQAKRG 302
P H+++ G
Sbjct: 583 PPGHRSREG 591
Score = 63.2 bits (152), Expect = 9e-10
Identities = 69/282 (24%), Positives = 107/282 (37%), Gaps = 30/282 (10%)
Query: 65 HPHYDDDYSLDEIFPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLD 124
H H +++ +H+ H DDE S + Q Q R H+ ++S+
Sbjct: 63 HSHRSPGEENEDVSMENGHHFWSHRDHGETDDEVSRE--YGHQPQGHRYHSPEAGDESVS 120
Query: 125 EVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDD---DDLPDGVLPKQQAKK 181
E + QA++ H ++ E L + GH D+D + P VL ++A +
Sbjct: 121 EEGVHREQARQAPGHGGHGEAGAEDLAEHGSHGHGHEEEDEDVISSERPRHVL--RRAPR 178
Query: 182 RHPHYDNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPHY------- 234
H + E+ E+ RG D ED D + QA + H
Sbjct: 179 GHGGEEEGEEEEEEEEVSPEHRHRGHGKEDEEDEDDDSTESDRHQAHRHRGHREEEDEDD 238
Query: 235 -DNDGDSLHEDLPKNHQAER-----GQPRYDDDDDLPDGVLPKQRQAKKLHPHY------ 282
D++GDS D +HQA R + +DDDD D + QA + H
Sbjct: 239 DDDEGDSTESD---HHQAHRHRGHEEEEDEEDDDDEGDSTESDRHQAHRHRGHREEEDED 295
Query: 283 -DNDEDSLHEDLPKNHQAKRGRPLYDDTDSLDEVLPARRRRH 323
D++ DS D + H+ + R DD D DE RH
Sbjct: 296 DDDEGDSTESDRHQAHRHRGHREEEDDDDDDDEGDSTESDRH 337
Score = 38.5 bits (88), Expect = 0.023
Identities = 41/185 (22%), Positives = 73/185 (39%), Gaps = 10/185 (5%)
Query: 136 GRSHYDDDDSLDEV-LPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHYDNDEDSLH 194
G SH + ++V + HH H + DD++ + Q + H DE S+
Sbjct: 62 GHSHRSPGEENEDVSMENGHHFWSHRDHGETDDEVSREYGHQPQGHRYHSPEAGDE-SVS 120
Query: 195 EDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPHYDNDGDSLHEDLPKN--HQAE 252
E+ QA++ H + G H H + D D + + P++ +A
Sbjct: 121 EEGVHREQARQAPGHGGHGE---AGAEDLAEHGSHGHGHEEEDEDVISSERPRHVLRRAP 177
Query: 253 RGQ--PRYDDDDDLPDGVLPKQR-QAKKLHPHYDNDEDSLHEDLPKNHQAKRGRPLYDDT 309
RG ++++ + V P+ R + D D+DS D + H+ + R D+
Sbjct: 178 RGHGGEEEGEEEEEEEEVSPEHRHRGHGKEDEEDEDDDSTESDRHQAHRHRGHREEEDED 237
Query: 310 DSLDE 314
D DE
Sbjct: 238 DDDDE 242
>SRCH_HUMAN (P23327) Sarcoplasmic reticulum histidine-rich
calcium-binding protein precursor
Length = 699
Score = 64.7 bits (156), Expect = 3e-10
Identities = 70/287 (24%), Positives = 120/287 (41%), Gaps = 50/287 (17%)
Query: 48 DEFLADRRRKKHLALRRHPHYDDDYSLDE-----IFPNRRQKHQAHKRHYHY-------- 94
+E A+ R H + R HP + D S + P+R ++ + + Y +
Sbjct: 51 EEASAELRHHLH-SPRDHPDENKDVSTENGHHFWSHPDREKEDEDVSKEYGHLLPGHRSQ 109
Query: 95 -----DDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQR-QAKRGRSHYDDDDSLDE 148
D+ S + + + RGH + ED+ D + + R SH D+D+ DE
Sbjct: 110 DHKVGDEGVSGEEVFAEHGGQARGHRGHGSEDTEDSAEHRHHLPSHRSHSHQDEDE--DE 167
Query: 149 VLPKQHH---AKRGHPHYDDDDDLPDGVLPKQQAKKRHPHYDNDEDSLHEDLPKNHQAKR 205
V+ +HH + GH +D +DD + ++ + +E++ E HQA R
Sbjct: 168 VVSSEHHHHILRHGHRGHDGEDD---------EGEEEEEEEEEEEEASTE---YGHQAHR 215
Query: 206 GRSHYDVED-NLLDGVLPKQPQAKKRHPHYDNDGDSLHED--------LPKNHQAERGQP 256
R H ED ++ DG P + + D+D D +D + HQA R Q
Sbjct: 216 HRGHGSEEDEDVSDGHHHHGPSHRHQGHEEDDDDDDDDDDDDDDDDVSIEYRHQAHRHQG 275
Query: 257 R-YDDDDDLPDGVLPKQRQAK-KLHPHYDNDEDSLHEDLPKNHQAKR 301
++D+D+ DG + + + H DND+D + + HQA R
Sbjct: 276 HGIEEDEDVSDGHHHRDPSHRHRSHEEDDNDDDDVSTEY--GHQAHR 320
Score = 60.8 bits (146), Expect = 4e-09
Identities = 66/312 (21%), Positives = 112/312 (35%), Gaps = 42/312 (13%)
Query: 34 ARRGHPRYDDDHSLDEFLADRRRKKHLALRRHPHYDDDYSLDEIFPNRRQK--------- 84
+ R H D+D DE ++ H + RH H D DE ++
Sbjct: 154 SHRSHSHQDEDE--DEVVSSEH---HHHILRHGHRGHDGEDDEGEEEEEEEEEEEEASTE 208
Query: 85 --HQAHKRHYHYDDEDSLDSLLPKQH---QAERGHTRYDDEDSLDEVLPKQRQAKRGRSH 139
HQAH+ H +ED S H +GH DD+D D+
Sbjct: 209 YGHQAHRHRGHGSEEDEDVSDGHHHHGPSHRHQGHEEDDDDDDDDD-------------- 254
Query: 140 YDDDDSLDEVLPKQHHAKRGHPH-YDDDDDLPDGVLPKQQAKKRHPHYDNDEDSLHEDLP 198
DDDD D + +H A R H ++D+D+ DG + + + H ++D D
Sbjct: 255 -DDDDDDDVSIEYRHQAHRHQGHGIEEDEDVSDGHHHRDPSHRHRSHEEDDNDDDDVSTE 313
Query: 199 KNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPHYDNDGDS------LHEDLPKNHQAE 252
HQA R + H E + G + H D + D H+ H
Sbjct: 314 YGHQAHRHQDHRKEEVEAVSGEHHHHVPDHRHQGHRDEEEDEDVSTERWHQGPQHVHHGL 373
Query: 253 RGQPRYDDDDDLPDGVLPKQRQAKKLHPHYDNDEDSLHEDLPKNHQAKRGRPLYDDTDS- 311
+ +++ + G Q + ++ +D ++P +H + R ++ +
Sbjct: 374 VDEEEEEEEITVQFGHYVASHQPRGHKSDEEDFQDEYKTEVPHHHHHRVPREEDEEVSAE 433
Query: 312 LDEVLPARRRRH 323
L P+ R+ H
Sbjct: 434 LGHQAPSHRQSH 445
Score = 52.4 bits (124), Expect = 2e-06
Identities = 67/325 (20%), Positives = 129/325 (39%), Gaps = 56/325 (17%)
Query: 30 SVSQARRGHPRYDDDHSLDEFLADRRRKKHLALRRHPHYDDDYSLDEI---FPNRRQKHQ 86
S+ + H DE ++D + + R H +DD D++ + ++ +HQ
Sbjct: 263 SIEYRHQAHRHQGHGIEEDEDVSDGHHHRDPSHRHRSHEEDDNDDDDVSTEYGHQAHRHQ 322
Query: 87 AHKR---------HYHY----------DDEDSLDSLLPKQHQAER--GHTRYDDEDSLDE 125
H++ H+H+ D+E+ D + HQ + H D+E+ +E
Sbjct: 323 DHRKEEVEAVSGEHHHHVPDHRHQGHRDEEEDEDVSTERWHQGPQHVHHGLVDEEEEEEE 382
Query: 126 VLPK---QRQAKRGRSH-YDDDDSLDEV---LPKQHHAKRGHPHYDDDDDLPDGVLPKQQ 178
+ + + + R H D++D DE +P HH H ++D+ L Q
Sbjct: 383 ITVQFGHYVASHQPRGHKSDEEDFQDEYKTEVPHHHH----HRVPREEDEEVSAELGHQA 438
Query: 179 AKKRHPHYDND-----EDSLHE--DLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRH 231
R H D + S+ E P H + RSH +D+ ++ + K+
Sbjct: 439 PSHRQSHQDEETGHGQRGSIKEMSHHPPGHTVVKDRSHLRKDDS-------EEEKEKEED 491
Query: 232 P-HYDNDGDSLHEDLPKNHQAERGQPRYDDDDDLPDGVLPKQRQAKKLHPHYDNDEDSLH 290
P ++ D +S + H R Q +D+++ L ++ + ++ D +E+
Sbjct: 492 PGSHEEDDESSEQGEKGTHHGSRDQEDEEDEEEGHGLSLNQEEEEEE-----DKEEEEEE 546
Query: 291 EDLPKNHQ-AKRGRPLYDDTDSLDE 314
ED + + A+ G PL D +E
Sbjct: 547 EDEERREERAEVGAPLSPDHSEEEE 571
Score = 44.7 bits (104), Expect = 3e-04
Identities = 45/202 (22%), Positives = 82/202 (40%), Gaps = 18/202 (8%)
Query: 32 SQARRGHPRYDDDHSLDEFLADRRRKKHLALRRHPHYDDDYSLDEIFPNRRQKHQAHKRH 91
S RGH + D++ DE+ + H + R + L P+ RQ HQ +
Sbjct: 393 SHQPRGH-KSDEEDFQDEYKTEVPHHHHHRVPREEDEEVSAELGHQAPSHRQSHQDEETG 451
Query: 92 YHYDDEDSLDSLLPKQHQA--ERGHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDDSLDEV 149
+ S P H +R H R DD + K+++ G SH +DD+S ++
Sbjct: 452 HGQRGSIKEMSHHPPGHTVVKDRSHLRKDDSEE-----EKEKEEDPG-SHEEDDESSEQG 505
Query: 150 LPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHYDNDEDSLHEDLPKNHQ-----AK 204
HH R +D+++ G+ Q+ ++ D +E+ ED + + A
Sbjct: 506 EKGTHHGSRDQEDEEDEEE-GHGLSLNQEEEEEE---DKEEEEEEEDEERREERAEVGAP 561
Query: 205 RGRSHYDVEDNLLDGVLPKQPQ 226
H + E+ +G+ +P+
Sbjct: 562 LSPDHSEEEEEEEEGLEEDEPR 583
>NSB1_MOUSE (Q9JL35) Nucleosome binding protein 1 (Nucleosome
binding protein 45) (NBP-45) (GARP45 protein)
Length = 406
Score = 48.1 bits (113), Expect = 3e-05
Identities = 42/184 (22%), Positives = 76/184 (40%), Gaps = 25/184 (13%)
Query: 111 ERGHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLP 170
E G + +++D + +++ K ++DD + + K G P D +DL
Sbjct: 190 EEGDEKEEEKDDKEGDTGTEKEVKEQNKEAEEDDGKCKEEENKEVGKEGQPEEDGKEDL- 248
Query: 171 DGVLPKQQAKKRHPHYDNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKR 230
H + ++ LHE+ K Q + E++ +G QP+ +
Sbjct: 249 --------------HEEVGKEDLHEEDGKEGQPEEDGKEIHHEEDGKEG----QPEEDGK 290
Query: 231 HPHYDNDGDSLHEDLPKNHQAERGQPRYDDDDDLP--DGVLPKQRQAKKLHPHYDNDEDS 288
++ DG+ E PK Q E GQP D +D P DG + ++ K H + ++
Sbjct: 291 EYLHEEDGE---EGQPKEDQKE-GQPEEDGKEDQPEEDGKEGQCKEDGKEGHHEEGGKED 346
Query: 289 LHED 292
LHE+
Sbjct: 347 LHEE 350
>ARP_EUGGR (Q04732) Calcium-binding acidic-repeat protein precursor
(ARP)
Length = 695
Score = 43.5 bits (101), Expect = 7e-04
Identities = 60/254 (23%), Positives = 99/254 (38%), Gaps = 19/254 (7%)
Query: 67 HYDDDYSLDEIFPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEV 126
++ D Y+ D + ++ H D+D+ D + + T D DS D+
Sbjct: 40 YFTDPYNADSDQDGLTDGLEVNRHQTHPQDKDTDDDSIGDGVEVNNLGTNPKDPDSDDDG 99
Query: 127 LPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYD---DDDDLPDGVLPKQQAKKRH 183
L + R+ D DS P A+ H + DDD L DG + + +
Sbjct: 100 LTDGAEVNLYRTDPLDADSTTTGCPMGGGAEVRHRPQNGDTDDDGLTDGA--EVNVHRTN 157
Query: 184 PH-YDNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPHYDNDGDSLH 242
P D+D+D L + N + +D + DG P+ K D+DGD L
Sbjct: 158 PQDGDSDDDGLSDGAEVNTYHSNPKDGDSDDDGVSDGA-EVNPKLK------DSDGDGLT 210
Query: 243 EDLPKNHQAERGQPRYDDDDDLPDGVLP-KQRQAKKLHP-HYDNDEDSLHEDLPKNHQAK 300
++ + + R P D D DG+L ++ + K +P D+D+D L + H
Sbjct: 211 DE--EEIKLYRTDPFCADSDF--DGLLDGEEVKVHKTNPLDGDSDDDGLGDGAEVTHFNT 266
Query: 301 RGRPLYDDTDSLDE 314
D D LD+
Sbjct: 267 NPLDADSDNDGLDD 280
Score = 38.5 bits (88), Expect = 0.023
Identities = 52/204 (25%), Positives = 81/204 (39%), Gaps = 23/204 (11%)
Query: 95 DDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAKRGRSHY----DDDDSLDEVL 150
+DED+ D L + T +D D+ D+ L + R++ D D L +
Sbjct: 468 NDEDTDDDGLTDGAEVNLHRTDPEDADTDDDGLTDGAEVNTYRTNPKLADSDGDGLSDGA 527
Query: 151 PKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHYDNDEDSLHEDLPKNHQAKRGRSHY 210
H + P+ D DD DGV +AK + D+D D L + + +
Sbjct: 528 EVNTH--KSDPNDGDSDD--DGVPDAAEAKVK----DSDGDGLSDTDEVRFRTNPKLADT 579
Query: 211 DVEDNLLDG--VLPKQPQAKKRHPHYDNDGDSLHEDLPKNHQAERGQPRYDDDDDLPDGV 268
D D L DG +L + + R D DGD + + L N + DDD L DG
Sbjct: 580 DF-DGLTDGAEILKHKTDPRNR----DTDGDGVADGLEVNTYGSDPKDADTDDDGLTDGA 634
Query: 269 LPKQRQAKKLHP-HYDNDEDSLHE 291
+ +P D+D+D L +
Sbjct: 635 ---EINVHDTNPTDADSDDDGLSD 655
Score = 33.1 bits (74), Expect = 0.97
Identities = 59/264 (22%), Positives = 89/264 (33%), Gaps = 51/264 (19%)
Query: 86 QAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDDS 145
+ + H + DEDS + +P + T +DEDS D+ + + S D
Sbjct: 329 EVNVHHTNPKDEDSDNDGIPDGAEINTHKTDPNDEDSDDDGIADGAEVTLTDSDGDGLPD 388
Query: 146 LDEVLPKQHHAKRGHPHYD--------------------DDDDLPDGV------------ 173
DEV + YD DDD L DGV
Sbjct: 389 EDEVALYNTNPANADSDYDGLTDGAEVKRYQSNPLDKDTDDDGLGDGVEVTVGTDPHDAT 448
Query: 174 ---LPKQQAKKRHPHYD--NDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLP-KQPQA 227
+ A + + H NDED+ + L + R+ + D DG+ +
Sbjct: 449 VTTTGSRTAVEINVHGSDPNDEDTDDDGLTDGAEVNLHRTDPEDADTDDDGLTDGAEVNT 508
Query: 228 KKRHPHY-DNDGDSLHEDLPKNHQAERGQPRYDDDDDLPDGVLPKQRQAKKLHPHYDNDE 286
+ +P D+DGD L + N DDD +PD K + D+D
Sbjct: 509 YRTNPKLADSDGDGLSDGAEVNTHKSDPNDGDSDDDGVPDAAEAKVK---------DSDG 559
Query: 287 DSLHEDLPKNHQAKRGRPLYDDTD 310
D L + + R P DTD
Sbjct: 560 DGLSD---TDEVRFRTNPKLADTD 580
>BLI1_CAEEL (Q09457) Cuticle collagen bli-1
Length = 963
Score = 40.8 bits (94), Expect = 0.005
Identities = 43/226 (19%), Positives = 89/226 (39%), Gaps = 25/226 (11%)
Query: 92 YHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLP 151
Y+ D++ L+ P+ + +ER P+ RQ R ++ ++ + + P
Sbjct: 586 YNDDEKRGLEEHRPRGYDSERAEE------------PRPRQTVRTNTYDENSGAEHQRRP 633
Query: 152 KQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHYDNDEDSLHEDLP--KNHQAKRGRSH 209
+ P D + + V + PH + E+ P + + R +
Sbjct: 634 NYEPSAEVAPPRQDRYEDEERVREPPPKRPPPPHRQTPHELYPEEQPYVRRPPPPQNRGN 693
Query: 210 YDVEDNLLDGVLPKQPQAKKRHPHYDNDGDSLHEDLPKNHQAER-GQPRYDDDDDLPDGV 268
Y+VE + +P+ P+ ++ + H G ++ PK ++ +PRY+ D P
Sbjct: 694 YEVERSR--EYVPEAPRPRQGYEHSSGYGGDDRKEQPKYMESRPVDEPRYET--DAPSRS 749
Query: 269 LPKQRQAKKLHPH--YDNDEDSLHEDLPKNHQAKR----GRPLYDD 308
P ++ HP YD + S + P+ + +R P Y++
Sbjct: 750 RPLKKVEIIRHPERGYDRRQPSYEDSKPRQEEPRRYETEAEPRYEE 795
>TOP1_XENLA (P41512) DNA topoisomerase I (EC 5.99.1.2)
Length = 829
Score = 40.0 bits (92), Expect = 0.008
Identities = 58/250 (23%), Positives = 99/250 (39%), Gaps = 45/250 (18%)
Query: 67 HYDDDYSLDEIFP-NRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDE 125
H +D ++ +F N KH+ K H H + + D K + +E +
Sbjct: 5 HVQNDSQIEAVFRVNDSHKHKKDKEHRHKEHKKDKDREKSKHNNSEHRDPSEKKHKDKHK 64
Query: 126 VLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPH 185
K R+ K G H + D ++H K G H D + +H
Sbjct: 65 NNDKHRE-KDGEKHRERDG-------EKHRDKNGEKHRDGE---------------KHKE 101
Query: 186 YDNDEDSLHEDLPKNHQAKRGRSH--YDVEDNLLDGVLPKQPQAKKRHPHYDNDGDSLHE 243
D ++ H+++ K H+ K G H DVE + V ++ + ++H H D D + E
Sbjct: 102 KDIEK---HKEVEK-HRVKDGEKHKEKDVEKHKEKDV--EKHRDGEKHKHRDKDREKKKE 155
Query: 244 DLPKNH------QAERGQPR----YDDDDDLPDGVLPKQRQAKKLHPHYDNDEDSLHEDL 293
+ K+ + E G D+ +D V PK+ +A K P D DED + +
Sbjct: 156 EKMKSSSGGVKVKKENGFSSPVRVKDEPEDQGFYVSPKENKAMK-RPRED-DEDYKPKKI 213
Query: 294 PKNHQAKRGR 303
K+ K+G+
Sbjct: 214 -KSEDDKKGK 222
>CDL1_HUMAN (P21127) PITSLRE serine/threonine-protein kinase CDC2L1
(EC 2.7.1.37) (Galactosyltransferase associated protein
kinase p58/GTA) (Cell division cycle 2-like 1) (CLK-1)
(CDK11) (p58 CLK-1)
Length = 795
Score = 40.0 bits (92), Expect = 0.008
Identities = 39/191 (20%), Positives = 82/191 (42%), Gaps = 29/191 (15%)
Query: 46 SLDEFLADRRRKKH----LALRRHPHYDD-DYSLDEIFPNRRQKHQAHK--RHYHYDDED 98
+LDE L +++R+K ++R + DD D D + + H+ R+ Y ED
Sbjct: 12 TLDEILQEKKRRKEQEEKAEIKRLKNSDDRDSKRDSLEEGELRDHRMEITIRNSPYRRED 71
Query: 99 SLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKR 158
S++ R +++DSL + P Q+ +++ + H+ D+ E + H+
Sbjct: 72 SMED-------------RGEEDDSL-AIKPPQQMSRKEKVHHRKDEKRKEKRRHRSHSAE 117
Query: 159 GHPHYDDDDDLPDGVLPKQQAKKRHPHYDNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLD 218
G H V K++ +R + ++D + + + + R H E + L+
Sbjct: 118 GGKH--------ARVKEKEREHERRKRHREEQDKARREWERQKRREMAREHSRRERDRLE 169
Query: 219 GVLPKQPQAKK 229
+ K+ + +K
Sbjct: 170 QLERKRERERK 180
Score = 30.0 bits (66), Expect = 8.2
Identities = 20/92 (21%), Positives = 42/92 (44%), Gaps = 1/92 (1%)
Query: 46 SLDEFLADRRRKKHLALRRHPHYDDDYSLDEIFPNRRQKHQAHKRHYHYDDEDSLDSLLP 105
S E + R+ +K RRH + + + ++H+ KRH D+ +
Sbjct: 92 SRKEKVHHRKDEKRKEKRRHRSHSAEGGKHARVKEKEREHERRKRHREEQDKARREWERQ 151
Query: 106 KQHQAERGHTRYDDEDSLDEVLPKQRQAKRGR 137
K+ + R H+R + D L+++ K+ + ++ R
Sbjct: 152 KRREMAREHSR-RERDRLEQLERKRERERKMR 182
Score = 30.0 bits (66), Expect = 8.2
Identities = 22/81 (27%), Positives = 38/81 (46%), Gaps = 14/81 (17%)
Query: 235 DNDGDSLHEDLPKNHQAE---RGQP---------RYDDDDDLPDGVLPKQRQAKKLHPHY 282
D+ DSL E ++H+ E R P R ++DD L + P Q+ ++K H+
Sbjct: 42 DSKRDSLEEGELRDHRMEITIRNSPYRREDSMEDRGEEDDSL--AIKPPQQMSRKEKVHH 99
Query: 283 DNDEDSLHEDLPKNHQAKRGR 303
DE + ++H A+ G+
Sbjct: 100 RKDEKRKEKRRHRSHSAEGGK 120
>EF2K_MOUSE (O08796) Elongation factor 2 kinase (EC 2.7.1.-) (eEF-2
kinase) (eEF-2K) (Calcium/calmodulin-dependent
eukaryotic elongation factor-2 kinase)
Length = 724
Score = 39.7 bits (91), Expect = 0.010
Identities = 37/161 (22%), Positives = 63/161 (38%), Gaps = 28/161 (17%)
Query: 14 SVSQARRARQSAGAFTSVSQARRGHPRYDDDHSLDEFLADRRRKKHLALRRHPHYDDDYS 73
+V+Q+ R QSA ++ + G PR S + L LR + D+
Sbjct: 334 AVNQSTRLLQSAKTILRGTEEKCGSPRIRTLSS---------SRPPLLLRLSENSGDENM 384
Query: 74 LDEIF------PNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVL 127
D F P+ H H H+ LD++ P+ H H D E+S D
Sbjct: 385 SDVTFDSLPSSPSSATPHSQKLDHLHWPVFGDLDNMGPRDHDRMDNHR--DSENSGDSGY 442
Query: 128 PKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDD 168
P ++++ LD+ P++H G+ ++ D+D
Sbjct: 443 PSEKRS-----------DLDDPEPREHGHSNGNRRHESDED 472
>REPT_MOUSE (P97347) Repetin
Length = 1130
Score = 39.3 bits (90), Expect = 0.014
Identities = 50/245 (20%), Positives = 93/245 (37%), Gaps = 50/245 (20%)
Query: 32 SQARRGHPRYDDDHSLDEFLA--DRRRKKHLALRRHPHYDDDYSLDEIFPNR----RQKH 85
+Q++R H R + + D + K +++ P+ ++Y + +++
Sbjct: 909 TQSQRSHDRREQQIQQQTWKPKEDNQHKLLAQVQQEPYSYEEYDWQSQSSEQDHCGEEEY 968
Query: 86 QAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDDS 145
Q RH D E+ + + H+ E+ H D + +DE Q Q ++ R ++++
Sbjct: 969 QDWDRHSVEDQENLYEMQNWQTHEEEQSHQTSDRQTHVDE----QNQQRQHRQTHEEN-- 1022
Query: 146 LDEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHYDNDEDSLHEDLPKNHQAK- 204
H + G H D+ + +R H+ E HE+ K +
Sbjct: 1023 --------HDHQHGRHHEDEHN------------HRRQDHHQQRERQTHEEKEKYQGGQD 1062
Query: 205 -------RGRSHYDVEDNLLDGVLPKQPQAKKRHPHYDNDGDSLHEDLPKNH---QAERG 254
R +SH ED+ +G PQ ++ HP + G S + NH A
Sbjct: 1063 QSRSFPNREKSHMS-EDDQCEG-----PQGRRFHPTH-GGGKSQRREKSGNHPTKPANYS 1115
Query: 255 QPRYD 259
P YD
Sbjct: 1116 SPLYD 1120
>FIS2_ARATH (Q9ZNT9) Polycomb group protein
FERTILIZATION-INDEPENDENT SEED 2
Length = 692
Score = 38.9 bits (89), Expect = 0.018
Identities = 46/235 (19%), Positives = 98/235 (41%), Gaps = 25/235 (10%)
Query: 93 HYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLPK 152
H +DE+ S P+ H +++ + + D++ PK R +K+ D L P
Sbjct: 338 HVNDENV--SSTPRAHSSKKNKSTRKNVDNVPSP-PKTRSSKK------TSDILTTTQPT 388
Query: 153 QHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHYDNDEDSLHEDLPKNHQAKRGRSHYDV 212
+ + +DD++ P+ + K++ ++D++ PK +K+
Sbjct: 389 IAESSEPKVRHVNDDNVSS--TPRAHSSKKNKSTRKNDDNIPSP-PKTRSSKK------- 438
Query: 213 EDNLLDGVLPKQPQAKKRHPHYDNDGDSLHEDLPKNHQAERGQPRYDDDDDLPDGVLPKQ 272
N+L P +++ + PH ++D S P+ H +++ + + DD+ + PK
Sbjct: 439 TSNILTRTQPAIAESEPKVPHVNDDKVS---STPRAHSSKKNKSTHKKDDNA--SLPPKT 493
Query: 273 RQAKKLHPHYDNDEDSLHE-DLPKNHQAKRGRPLYDDTDSLDEVLPARRRRHYRS 326
R +KK + + E PK + R + L+ + + + R+ Y S
Sbjct: 494 RSSKKTSDILATTQPAKAEPSEPKVTRVSRRKELHAERCEAKRLERLKGRQFYHS 548
Score = 31.6 bits (70), Expect = 2.8
Identities = 21/80 (26%), Positives = 37/80 (46%), Gaps = 6/80 (7%)
Query: 66 PHYDDDYSLDEIFPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDE 125
PH +D + P R H +D+D + S P+ H E+ + + +ED++
Sbjct: 160 PHVNDG---NVSSPPRAHSSAEKNESTHVNDDDDVSSP-PRAHSLEKNESTHVNEDNISS 215
Query: 126 VLPKQRQAKRGRS-HYDDDD 144
PK +K+ S H +D+D
Sbjct: 216 P-PKAHSSKKNESTHMNDED 234
>LEGB_PEA (P14594) Legumin B [Contains: Legumin B alpha chain
(Legumin B acidic chain); Legumin B beta chain (Legumin
B basic chain)] (Fragment)
Length = 338
Score = 38.5 bits (88), Expect = 0.023
Identities = 28/113 (24%), Positives = 54/113 (47%), Gaps = 14/113 (12%)
Query: 97 EDSLDSLLPK-QHQAERGHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHH 155
ED L + P+ Q + E+ H++ +E+ ++ +QR K + +D+D +E +Q H
Sbjct: 41 EDGLHIISPELQEEEEQSHSQRKEEEEEEQ---EQRHRKHSKKEDEDEDEEEEEEREQRH 97
Query: 156 AKRGHPHYDDDDDLPDGVLPKQQ------AKKRHPHYDNDEDSLHEDLPKNHQ 202
K +D+D+ P +++ KKR H +E+ E+L K +
Sbjct: 98 RKHSEKEEEDEDE-PRSYETRRKWKKHTAEKKRESHGQGEEE---EELEKEEE 146
Score = 30.4 bits (67), Expect = 6.3
Identities = 12/53 (22%), Positives = 30/53 (55%), Gaps = 3/53 (5%)
Query: 83 QKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVLPKQRQAKR 135
++ Q H++H +DED + ++ + E+ H ++ +++ DE P+ + +R
Sbjct: 69 EQEQRHRKHSKKEDEDEDEE---EEEEREQRHRKHSEKEEEDEDEPRSYETRR 118
>EMB5_CAEEL (P34703) Abnormal embryogenesis protein 5
Length = 1521
Score = 38.1 bits (87), Expect = 0.030
Identities = 62/312 (19%), Positives = 115/312 (35%), Gaps = 51/312 (16%)
Query: 32 SQARRGHPRYDDDHSLDEFLADRRRKKHLALRRHPHYDDDYSLDEIFPNRRQKHQAHKRH 91
S A GH ++ S +A + K+ + D+D DE ++++ +
Sbjct: 11 SDASSGHSDDEEPQSKKMKMAKEKSKRKKKMVASSDEDEDDDDDE------EENRKEMQG 64
Query: 92 YHYDDEDSLDSLLPKQHQAER--GHTRYDDEDS--LDEVLPKQRQAKRGRSHYDDDDSLD 147
+ DD+D + ++ + R G DDED ++E + K+ R D D
Sbjct: 65 FIADDDDEEEDAKSEKSEKSRHSGEDELDDEDLDLINENYDIRETKKQNRVQLGDSSDED 124
Query: 148 EVLPKQHHAKRGHPHYDDDDDL------PDGVLPKQQAKKRHPHYDNDEDSLHEDLPKN- 200
E + + +H +DDDL DG K + + Y ++ + +D ++
Sbjct: 125 EPIRRPNH---------EDDDLLSERGSDDGDRRKDRGRGDRGGYGSESERSEDDFIEDD 175
Query: 201 -HQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPHYDNDGDSLHEDLPKNHQAERGQPRYD 259
+R R + ++NL +G + A+ D + D ++D E +
Sbjct: 176 GDAPRRHRKRHRGDENLPEGA---EDDARDVFGVEDFNLDEFYDDDDGEDGLEDEEEEII 232
Query: 260 DDD------------------DLPDGVLPKQRQAKKLHPHYDNDEDSLHEDLPKNHQAKR 301
+DD L + + P + L P D+ EDLP+ Q +R
Sbjct: 233 EDDGEGGEIKIRRKKDTTKKSTLLESIEPSEIDRGFLLP---GDKKIAKEDLPERFQLRR 289
Query: 302 GRPLYDDTDSLD 313
D D L+
Sbjct: 290 TPVTEADDDELE 301
Score = 37.0 bits (84), Expect = 0.067
Identities = 36/194 (18%), Positives = 76/194 (38%), Gaps = 10/194 (5%)
Query: 106 KQHQAERGHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDD 165
++ A GH+ ++ S + K++ ++ + D+ D+ ++ + K D
Sbjct: 9 EESDASSGHSDDEEPQSKKMKMAKEKSKRKKKMVASSDEDEDDDDDEEENRKEMQGFIAD 68
Query: 166 DDDLPDGVLPKQQAKKRHPHYDNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQP 225
DDD + ++ K RH D +D + + +N+ + + V+ L D +P
Sbjct: 69 DDDEEEDAKSEKSEKSRHSGEDELDDEDLDLINENYDIRETKKQNRVQ--LGDSSDEDEP 126
Query: 226 QAKKRHPHYD-------NDGDSLHEDLPKNHQAERGQPRYDDDDDLPDGVLPKQRQAKKL 278
+ H D +DGD +D + + G +DD + R+ +K
Sbjct: 127 IRRPNHEDDDLLSERGSDDGDR-RKDRGRGDRGGYGSESERSEDDFIEDDGDAPRRHRKR 185
Query: 279 HPHYDNDEDSLHED 292
H +N + +D
Sbjct: 186 HRGDENLPEGAEDD 199
Score = 36.6 bits (83), Expect = 0.088
Identities = 49/211 (23%), Positives = 80/211 (37%), Gaps = 38/211 (18%)
Query: 64 RHPHYDDDYSLDEIFPN----RRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDD 119
R P+++DD L E + R+ + + + Y + E S D + A R H +
Sbjct: 128 RRPNHEDDDLLSERGSDDGDRRKDRGRGDRGGYGSESERSEDDFIEDDGDAPRRHRK--- 184
Query: 120 EDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQA 179
DE LP+ + +D +LDE + DDDD DG+
Sbjct: 185 RHRGDENLPEGAEDDARDVFGVEDFNLDE--------------FYDDDDGEDGL------ 224
Query: 180 KKRHPHYDNDEDSLHEDLPKNHQAK-RGRSHYDVEDNLLDGVLPKQPQAKKRHPHYDNDG 238
+++E+ + ED + + K R + + LL+ + P + P D
Sbjct: 225 -------EDEEEEIIEDDGEGGEIKIRRKKDTTKKSTLLESIEPSEIDRGFLLP---GDK 274
Query: 239 DSLHEDLPKNHQAERGQPRYDDDDDLPDGVL 269
EDLP+ Q R DDD+L L
Sbjct: 275 KIAKEDLPERFQLRRTPVTEADDDELESEAL 305
>ATRX_CAEEL (Q9U7E0) Transcriptional regulator ATRX homolog
(X-linked nuclear protein-1)
Length = 1359
Score = 38.1 bits (87), Expect = 0.030
Identities = 33/194 (17%), Positives = 74/194 (38%), Gaps = 14/194 (7%)
Query: 113 GHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLPDG 172
GH D++ + + +R+ KR + + + + PK+ AK+ ++DD +
Sbjct: 13 GHVIEDEDLEMARQIENERKEKRAQKLKEKREREGKPPPKKRPAKKRKASSSEEDDDDEE 72
Query: 173 VLPKQQAKKRHPHYDNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHP 232
P++ +KK ++ +S D ++ + + + D + K+ KKR
Sbjct: 73 ESPRKSSKKSRKRAKSESESDESDEEEDRKKSKSKKKVDQK--------KKEKSKKKRTT 124
Query: 233 HYDNDGDSLHEDLPKNHQAERGQPRYDDDDDLPDG------VLPKQRQAKKLHPHYDNDE 286
D DS E K+ + + + + + K+ + K + + E
Sbjct: 125 SSSEDEDSDEEREQKSKKKSKKTKKQTSSESSEESEEERKVKKSKKNKEKSVKKRAETSE 184
Query: 287 DSLHEDLPKNHQAK 300
+S ++ P K
Sbjct: 185 ESDEDEKPSKKSKK 198
Score = 35.4 bits (80), Expect = 0.20
Identities = 46/261 (17%), Positives = 101/261 (38%), Gaps = 22/261 (8%)
Query: 65 HPHYDDDYSLDEIFPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLD 124
H D+D + N R++ +A K + E PK+ A++ +ED D
Sbjct: 14 HVIEDEDLEMARQIENERKEKRAQKLKEKREREGKPP---PKKRPAKKRKASSSEEDDDD 70
Query: 125 EVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRH- 183
E ++ +K+ R + DE ++ K D K+++KK+
Sbjct: 71 EEESPRKSSKKSRKRAKSESESDESDEEEDRKKSKSKKKVDQK-------KKEKSKKKRT 123
Query: 184 ----PHYDNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAK----KRHPHYD 235
D+DE+ + K+ + K+ S E++ + + K + K K+
Sbjct: 124 TSSSEDEDSDEEREQKSKKKSKKTKKQTSSESSEESEEERKVKKSKKNKEKSVKKRAETS 183
Query: 236 NDGDSLHEDLPKNHQAERGQPRYDDDDDLPD--GVLPKQRQAKKLHPHYDNDEDSLHEDL 293
+ D + K+ + + + + + + + D V ++++KK+ ED E
Sbjct: 184 EESDEDEKPSKKSKKGLKKKAKSESESESEDEKEVKKSKKKSKKVVKKESESEDEAPEK- 242
Query: 294 PKNHQAKRGRPLYDDTDSLDE 314
K + KR + +++ ++
Sbjct: 243 KKTEKRKRSKTSSEESSESEK 263
>IFH1_YEAST (P39520) IFH1 protein (RRP3 protein)
Length = 1085
Score = 37.7 bits (86), Expect = 0.040
Identities = 51/218 (23%), Positives = 85/218 (38%), Gaps = 38/218 (17%)
Query: 97 EDSLDSLLPKQHQAERGHTRYDDEDSLDEV-----LPKQRQAKRGRSHYDDDDSLDEVLP 151
ED+ + L ++ E G+ DDE+ DE+ +P K +Y D S ++
Sbjct: 386 EDAENKFLQNEYNQENGYDEEDDEE--DEIMSDFDMPFYEDPKFANLYYYGDGSEPKL-- 441
Query: 152 KQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHYDNDEDSLHEDLPKNHQAKRGRSHYD 211
P +D+ L L K++AKKR L++ K + R S+ D
Sbjct: 442 ---SLSTSLPLMLNDEKL--SKLKKKEAKKREQEERKQRRKLYKKTQK--PSTRTTSNVD 494
Query: 212 VEDNLLD----------GVLPKQPQAK--KRHPHYDNDGDSL---HEDL-PKNHQAERGQ 255
++ + + G K+ + K K H + N G +L ++DL P H
Sbjct: 495 NDEYIFNVFFQSDDENSGHKSKKGRHKSGKSHIEHKNKGSNLIKSNDDLEPSTHSTVLNS 554
Query: 256 PRYDDDDDLPDGVL------PKQRQAKKLHPHYDNDED 287
+YD DD D +L P + + +D D D
Sbjct: 555 GKYDSSDDEYDNILLDVAHMPSDDECSESETSHDADTD 592
>KNOB_PLAFN (P06719) Knob-associated histidine-rich protein
precursor (KAHRP)
Length = 657
Score = 37.0 bits (84), Expect = 0.067
Identities = 43/181 (23%), Positives = 75/181 (40%), Gaps = 52/181 (28%)
Query: 68 YDDDYSLDEIFPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDEVL 127
Y S+++ + +KH + K+H D E ++ + + H +D E
Sbjct: 346 YSSFSSVNKYGKHGDEKHHSSKKHEGNDGEG-------EKKKKSKKHKDHDGEK------ 392
Query: 128 PKQRQAKRGRSHYDDDDSLDEVLPKQHHA-----KRGHPHYDDDDDLPDGVLPKQQAKKR 182
K+ + H D++D+ + V K+H + K+ H D++D + V K+ KKR
Sbjct: 393 ------KKSKKHKDNEDA-ESVKSKKHKSHDCEKKKSKKHKDNED--AESVKSKKVLKKR 443
Query: 183 HPHYDNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQAKKRHPHYDNDGDSLH 242
E S+ E KNH AK+ L K+ + KK+ + +DG H
Sbjct: 444 -------EKSIME---KNHAAKK---------------LTKKIKIKKKTNNSKSDGSKAH 478
Query: 243 E 243
E
Sbjct: 479 E 479
Score = 30.8 bits (68), Expect = 4.8
Identities = 28/127 (22%), Positives = 56/127 (44%), Gaps = 23/127 (18%)
Query: 40 RYDDDHSLDEFLADRRRKKHLALRRHPHYDDDYSLDEIFPNRRQKHQAH-------KRHY 92
RY S++++ +KH + ++H D + + + +KH+ H K+H
Sbjct: 345 RYSSFSSVNKY-GKHGDEKHHSSKKHEGNDGEGEKKK----KSKKHKDHDGEKKKSKKHK 399
Query: 93 HYDDEDSLDSLLPKQHQAERGHT-RYDDEDSLDEVLPKQRQAKRGRSHYDDDDSLDEVLP 151
+D +S+ S K H E+ + ++ D + + V K+ KR +S ++
Sbjct: 400 DNEDAESVKSKKHKSHDCEKKKSKKHKDNEDAESVKSKKVLKKREKS----------IME 449
Query: 152 KQHHAKR 158
K H AK+
Sbjct: 450 KNHAAKK 456
>RPA1_DROME (P91875) DNA-directed RNA polymerase I largest subunit (EC
2.7.7.6)
Length = 1644
Score = 36.6 bits (83), Expect = 0.088
Identities = 37/135 (27%), Positives = 56/135 (41%), Gaps = 27/135 (20%)
Query: 89 KRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDSLDE------VLPKQRQAKRGRSHYDD 142
K+ DD++ LD+ + R + +D+DS D+ K +Q K YDD
Sbjct: 1337 KKDADKDDDNDLDN----GDEVGRSKAKANDDDSSDDNDDDDATGVKLKQRKTDEKDYDD 1392
Query: 143 DDSLDEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKRHPHYDND-EDSLHEDLPKNH 201
D ++E+ H DDDD+ D + +K DND +D E L N
Sbjct: 1393 PDDVEEL----------HDANDDDDEAED----EDDEEKGQDGNDNDGDDKAVERLLSND 1438
Query: 202 QAKRGRSHYDVEDNL 216
K YD E++L
Sbjct: 1439 MVK--AYTYDKENHL 1451
>KP58_DROME (Q9VPC0) Serine/threonine-protein kinase PITSLRE (EC
2.7.1.37) (Cell division cycle 2-like)
Length = 952
Score = 36.6 bits (83), Expect = 0.088
Identities = 36/170 (21%), Positives = 60/170 (35%), Gaps = 12/170 (7%)
Query: 54 RRRKKHLALRRHPHYDDDYSLDEIFPNR--RQKHQAHKRHYHYDDEDSLDSLLPKQHQAE 111
R +KKH RR DD + R R +++ + HYH+ + + +
Sbjct: 53 REKKKHSRERRRHKERDDVGGAALALERDHRYDYRSREEHYHHHQRERSSNAAAAYAKHH 112
Query: 112 RGHTRYDDEDSLDEVLPKQRQAKRGRSHYDDDDSLD--EVLPKQHHAKRGHPHYDDDDDL 169
GH + + + P HY L P+++H+ H
Sbjct: 113 LGHAYHYPQPPQQQQQPLPPAPSYAAHHYHHHQHLSGARAAPREYHSYPSGYH----SGS 168
Query: 170 PDGVLPKQQAKKRHPHY--DNDEDSLHEDLPKNHQAKRGR--SHYDVEDN 215
G P ++ +R Y D +SL +DL KR Y+ E+N
Sbjct: 169 RHGDYPMEEPTRRSSKYAESKDAESLEQDLRSRLLKKRHNYVKDYETEEN 218
>GRIA_BACCE (O85467) Spore germination protein gerIA
Length = 663
Score = 36.6 bits (83), Expect = 0.088
Identities = 37/170 (21%), Positives = 68/170 (39%), Gaps = 16/170 (9%)
Query: 54 RRRKKHLALRRHPHYDDDYSLDEIFPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERG 113
+++K + + + +S ++ N+ Q +H + + DS KQ A++G
Sbjct: 8 KKKKSNTSKLNETDNQEQHSNNQEDDNKEQTRSM--KHNKGKNNEQKDSSQDKQQSAKQG 65
Query: 114 HTRYDDEDSL---DEVLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDD-- 168
+ D + + D KQ+ K+G S D S + P Q K+ +P +D
Sbjct: 66 DSSQDKQQNPKQEDSSQDKQQNPKQGDSSQDKQQSAKQKDPSQD--KQQNPKQEDSSQDK 123
Query: 169 -----LPDGVLPKQQAKKRHPHYDNDEDSLHEDLP--KNHQAKRGRSHYD 211
D KQQ+ K+ + + + +D P Q+ G S YD
Sbjct: 124 QQSAKQGDSSQDKQQSAKQGDSSQDKQQNAKQDEPSQSKQQSSGGNSIYD 173
Score = 32.3 bits (72), Expect = 1.7
Identities = 32/128 (25%), Positives = 49/128 (38%), Gaps = 12/128 (9%)
Query: 187 DNDEDSLHEDLPKNHQAKRGRSHYDVEDN-LLDGVLPKQPQAKKRHPHYDNDGDSLHEDL 245
DN E + N + R H ++N D KQ AK+ D + ED
Sbjct: 21 DNQEQHSNNQEDDNKEQTRSMKHNKGKNNEQKDSSQDKQQSAKQGDSSQDKQQNPKQED- 79
Query: 246 PKNHQAERGQPRYDDDDDLPDGVLPKQRQAKKLHPHYDNDEDSLHEDLPKNHQ--AKRGR 303
+ Q ++ P+ D KQ+ AK+ P D ++ ED ++ Q AK+G
Sbjct: 80 --SSQDKQQNPKQGDSSQ------DKQQSAKQKDPSQDKQQNPKQEDSSQDKQQSAKQGD 131
Query: 304 PLYDDTDS 311
D S
Sbjct: 132 SSQDKQQS 139
>CU13_HUMAN (O95447) Protein C21orf13
Length = 670
Score = 36.6 bits (83), Expect = 0.088
Identities = 36/140 (25%), Positives = 55/140 (38%), Gaps = 12/140 (8%)
Query: 127 LPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKR---- 182
LPKQ +KR ++ EV + R +D + V +Q+ +
Sbjct: 416 LPKQEDSKRKYEDLSGEEKHLEVQILLENTGRQKDKKEDQEKKNIFVKEEQELPPKIIEV 475
Query: 183 -HPHYDNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLP--KQPQAKKRHPHYDNDGD 239
HP + +S ED+ + KR V+D L G P K P ++RH + +
Sbjct: 476 IHP----ERESNQEDVLVREKFKRSMQRNGVDDTLGKGTAPYTKGPLRQRRHYSFTEATE 531
Query: 240 SLHEDLP-KNHQAERGQPRY 258
+LH LP A G RY
Sbjct: 532 NLHHGLPASGGPANAGNMRY 551
Score = 30.0 bits (66), Expect = 8.2
Identities = 32/130 (24%), Positives = 52/130 (39%), Gaps = 16/130 (12%)
Query: 174 LPKQQ-AKKRHPHYDNDEDSLHEDLPKNHQAKRGRSHYDVEDNLLDGVLPKQPQA----- 227
LPKQ+ +K+++ +E L + + GR ED + K+ Q
Sbjct: 416 LPKQEDSKRKYEDLSGEEKHLEVQILLENT---GRQKDKKEDQEKKNIFVKEEQELPPKI 472
Query: 228 -KKRHPHYDNDGDSLHEDLPKNHQAERGQPRYDDDDDLPDGVLPKQRQAKKLHPHYDNDE 286
+ HP + +S ED+ + +R R DD L G P + + HY E
Sbjct: 473 IEVIHP----ERESNQEDVLVREKFKRSMQRNGVDDTLGKGTAPYTKGPLRQRRHYSFTE 528
Query: 287 --DSLHEDLP 294
++LH LP
Sbjct: 529 ATENLHHGLP 538
>CCAA_RABIT (P27884) Voltage-dependent P/Q-type calcium channel
alpha-1A subunit (Voltage-gated calcium channel alpha
subunit Cav2.1) (Calcium channel, L type, alpha-1
polypeptide isoform 4) (Brain calcium channel I) (BI)
Length = 2424
Score = 36.6 bits (83), Expect = 0.088
Identities = 41/177 (23%), Positives = 66/177 (37%), Gaps = 38/177 (21%)
Query: 17 QARRARQSAGAFTSVSQARRGHPRYDDDHSLDEFLADRRRKKHLALRRHPHYDDDYSLDE 76
QAR A S + +Q RRG PR D ++ + +R L + DD YSL+
Sbjct: 2095 QARAA--SMPRLPAENQRRRGRPRGSDLSTICDTSPMKRSASVLGPKASRRLDD-YSLER 2151
Query: 77 IFPNRRQKHQAHKRHYHYDDEDSLDSLLPKQHQAERGHTRYDDEDS-----------LDE 125
+ P Q+H +R + +ER RY D D+ +
Sbjct: 2152 VPPEENQRHHPRRRERAH-------------RTSERSLGRYTDVDTGLGTDLSMTTQSGD 2198
Query: 126 VLPKQRQAKRGRSHYDDDDSLDEVLPKQHHAKRGHPHYDDDDDLPDGVLPKQQAKKR 182
+ ++R+ +RGR ++H H H+ P V P A++R
Sbjct: 2199 LPSREREQERGRPK-----------DRKHRPHHHHHHHHHPGRGPGRVSPGVSARRR 2244
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.135 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 46,948,436
Number of Sequences: 164201
Number of extensions: 2433652
Number of successful extensions: 5849
Number of sequences better than 10.0: 164
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 155
Number of HSP's that attempted gapping in prelim test: 5273
Number of HSP's gapped (non-prelim): 428
length of query: 326
length of database: 59,974,054
effective HSP length: 110
effective length of query: 216
effective length of database: 41,911,944
effective search space: 9052979904
effective search space used: 9052979904
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 66 (30.0 bits)
Medicago: description of AC142222.3


