

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141863.4 + phase: 0 /partial
(1196 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
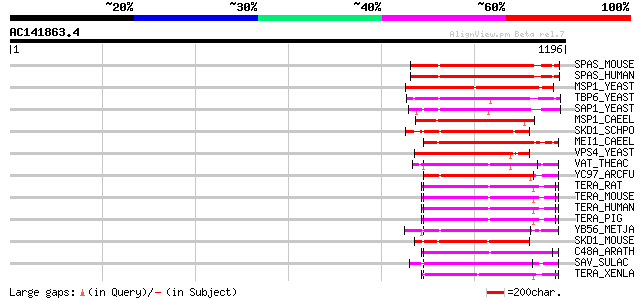
Score E
Sequences producing significant alignments: (bits) Value
SPAS_MOUSE (Q9QYY8) Spastin 246 3e-64
SPAS_HUMAN (Q9UBP0) Spastin 245 6e-64
MSP1_YEAST (P28737) MSP1 protein (TAT-binding homolog 4) 237 2e-61
TBP6_YEAST (P40328) Probable 26S protease subunit YTA6 (TAT-bind... 222 5e-57
SAP1_YEAST (P39955) SAP1 protein 218 1e-55
MSP1_CAEEL (P54815) MSP1 protein homolog 206 2e-52
SKD1_SCHPO (Q09803) Suppressor protein of bem1/bed5 double mutants 206 3e-52
MEI1_CAEEL (P34808) Meiotic spindle formation protein mei-1 200 2e-50
VPS4_YEAST (P52917) Vacuolar protein sorting-associated protein ... 190 2e-47
VAT_THEAC (O05209) VCP-like ATPase 190 2e-47
YC97_ARCFU (O28972) Cell division cycle protein 48 homolog AF1297 187 1e-46
TERA_RAT (P46462) Transitional endoplasmic reticulum ATPase (TER... 186 4e-46
TERA_MOUSE (Q01853) Transitional endoplasmic reticulum ATPase (T... 186 4e-46
TERA_HUMAN (P55072) Transitional endoplasmic reticulum ATPase (T... 186 4e-46
TERA_PIG (P03974) Transitional endoplasmic reticulum ATPase (TER... 184 9e-46
YB56_METJA (Q58556) Cell division cycle protein 48 homolog MJ1156 184 1e-45
SKD1_MOUSE (P46467) SKD1 protein (Vacuolar sorting protein 4b) 184 2e-45
C48A_ARATH (P54609) Cell division control protein 48 homolog A (... 183 2e-45
SAV_SULAC (Q07590) SAV protein 183 3e-45
TERA_XENLA (P23787) Transitional endoplasmic reticulum ATPase (T... 182 4e-45
>SPAS_MOUSE (Q9QYY8) Spastin
Length = 614
Score = 246 bits (628), Expect = 3e-64
Identities = 136/322 (42%), Positives = 207/322 (64%), Gaps = 20/322 (6%)
Query: 864 KKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCK 923
KK LK+ + L+ + I N V FDDI E K L+E+V+LP RPELF
Sbjct: 309 KKDLKNFRNVDSNLANLIMNEIVDNGTAVKFDDIAGQELAKQALQEIVILPSLRPELFTG 368
Query: 924 GQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFS 983
L P +G+LLFGPPG GKTMLAKAVA ++ A F NIS +S+TSK+ GEGEK V+A+F+
Sbjct: 369 --LRAPARGLLLFGPPGNGKTMLAKAVAAESNATFFNISAASLTSKYVGEGEKLVRALFA 426
Query: 984 LASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATN 1043
+A ++ PS+IF+DEVDS+L R GEH+A R++K EF++ +DG+++ +RV+V+ ATN
Sbjct: 427 VARELQPSIIFIDEVDSLLCERRE-GEHDASRRLKTEFLIEFDGVQSAGDDRVLVMGATN 485
Query: 1044 RPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLS-SDVDLGAIANMTDGYSGS 1102
RP +LDEAV+RR +R+ V+LP+ R +LK +L K+ + +L +A MTDGYSGS
Sbjct: 486 RPQELDEAVLRRFIKRVYVSLPNEETRLLLLKNLLCKQGSPLTQKELAQLARMTDGYSGS 545
Query: 1103 DLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVC 1162
DL L AA PI+E+ ++ K ++A+ ++R++ + DF + +++
Sbjct: 546 DLTALAKDAALGPIRELKPEQVKNMSAS--------------EMRNIRLSDFTESLKKIK 591
Query: 1163 ASVSSESVNMTELVQWNELYGE 1184
SVS ++ + ++WN+ +G+
Sbjct: 592 RSVSPQT--LEAYIRWNKDFGD 611
>SPAS_HUMAN (Q9UBP0) Spastin
Length = 616
Score = 245 bits (625), Expect = 6e-64
Identities = 135/322 (41%), Positives = 207/322 (63%), Gaps = 20/322 (6%)
Query: 864 KKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCK 923
KK LK+ + L+ + I N V FDDI + K L+E+V+LP RPELF
Sbjct: 311 KKDLKNFRNVDSNLANLIMNEIVDNGTAVKFDDIAGQDLAKQALQEIVILPSLRPELFTG 370
Query: 924 GQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFS 983
L P +G+LLFGPPG GKTMLAKAVA ++ A F NIS +S+TSK+ GEGEK V+A+F+
Sbjct: 371 --LRAPARGLLLFGPPGNGKTMLAKAVAAESNATFFNISAASLTSKYVGEGEKLVRALFA 428
Query: 984 LASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATN 1043
+A ++ PS+IF+DEVDS+L R GEH+A R++K EF++ +DG+++ +RV+V+ ATN
Sbjct: 429 VARELQPSIIFIDEVDSLLCERRE-GEHDASRRLKTEFLIEFDGVQSAGDDRVLVMGATN 487
Query: 1044 RPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLS-SDVDLGAIANMTDGYSGS 1102
RP +LDEAV+RR +R+ V+LP+ R +LK +L K+ + +L +A MTDGYSGS
Sbjct: 488 RPQELDEAVLRRFIKRVYVSLPNEETRLLLLKNLLCKQGSPLTQKELAQLARMTDGYSGS 547
Query: 1103 DLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVC 1162
DL L AA PI+E+ ++ K ++A+ ++R++ + DF + +++
Sbjct: 548 DLTALAKDAALGPIRELKPEQVKNMSAS--------------EMRNIRLSDFTESLKKIK 593
Query: 1163 ASVSSESVNMTELVQWNELYGE 1184
SVS ++ + ++WN+ +G+
Sbjct: 594 RSVSPQT--LEAYIRWNKDFGD 613
>MSP1_YEAST (P28737) MSP1 protein (TAT-binding homolog 4)
Length = 362
Score = 237 bits (604), Expect = 2e-61
Identities = 132/320 (41%), Positives = 198/320 (61%), Gaps = 9/320 (2%)
Query: 854 QAIQNESKSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVML 913
Q++Q E + VT + +E+ +L ++ P++I +TF DIG L+ + L E V+
Sbjct: 50 QSLQWEKLVKRSPALAEVTLDAYERTILSSIVTPDEINITFQDIGGLDPLISDLHESVIY 109
Query: 914 PLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGE 973
PL PE++ L + G+LL+GPPG GKTMLAKA+A ++GANFI+I MSSI KW+GE
Sbjct: 110 PLMMPEVYSNSPLLQAPSGVLLYGPPGCGKTMLAKALAKESGANFISIRMSSIMDKWYGE 169
Query: 974 GEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDT 1033
K V A+FSLA+K+ P +IF+DE+DS L R + +HE +K EFM WDGL +
Sbjct: 170 SNKIVDAMFSLANKLQPCIIFIDEIDSFL-RERSSTDHEVTATLKAEFMTLWDGL--LNN 226
Query: 1034 ERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSD-VDLGAI 1092
RV+++ ATNR D+D+A +RRLP+R +V+LP + R KIL V+L L D DL I
Sbjct: 227 GRVMIIGATNRINDIDDAFLRRLPKRFLVSLPGSDQRYKILSVLLKDTKLDEDEFDLQLI 286
Query: 1093 ANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNME 1152
A+ T G+SGSDLK LC AA KE ++++++ + + + +L+ IR L +
Sbjct: 287 ADNTKGFSGSDLKELCREAALDAAKEYIKQKRQLIDSGTIDVNDTSSLK----IRPLKTK 342
Query: 1153 DF-KHAHQQVCASVSSESVN 1171
DF K +++SS+ ++
Sbjct: 343 DFTKKLRMDATSTLSSQPLD 362
>TBP6_YEAST (P40328) Probable 26S protease subunit YTA6 (TAT-binding
homolog 6)
Length = 754
Score = 222 bits (565), Expect = 5e-57
Identities = 133/344 (38%), Positives = 208/344 (59%), Gaps = 34/344 (9%)
Query: 855 AIQNESKSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLP 914
++ + + + KS++ V + +++L +++ D V ++DI L N K++LKE V+ P
Sbjct: 434 SLDSRKEDILKSVQGV--DRNACEQILNEILV-TDEKVYWEDIAGLRNAKNSLKEAVVYP 490
Query: 915 LQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEG 974
RP+LF KG L +P +G+LLFGPPGTGKTM+AKAVAT++ + F ++S SS+ SK+ GE
Sbjct: 491 FLRPDLF-KG-LREPVRGMLLFGPPGTGKTMIAKAVATESNSTFFSVSASSLLSKYLGES 548
Query: 975 EKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTE 1034
EK V+A+F +A K++PS+IF+DE+DSML R + E+E+ R++K E ++ W L + +
Sbjct: 549 EKLVRALFYMAKKLSPSIIFIDEIDSMLTARSD-NENESSRRIKTELLIQWSSLSSATAQ 607
Query: 1035 ----------RVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLS 1084
RV+VL ATN P+ +D+A RR R+L + LPD R LK ++AK+ S
Sbjct: 608 SEDRNNTLDSRVLVLGATNLPWAIDDAARRRFSRKLYIPLPDYETRLYHLKRLMAKQKNS 667
Query: 1085 -SDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGS 1143
D+D I MT+G+SGSDL +L AA PI+++ +K
Sbjct: 668 LQDLDYELITEMTEGFSGSDLTSLAKEAAMEPIRDLGDK---------------LMFADF 712
Query: 1144 DDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYGEGGS 1187
D IR + ++DF++A + SVSSES+ E +W+ +G GS
Sbjct: 713 DKIRGIEIKDFQNALLTIKKSVSSESLQKYE--EWSSKFGSNGS 754
>SAP1_YEAST (P39955) SAP1 protein
Length = 897
Score = 218 bits (554), Expect = 1e-55
Identities = 131/358 (36%), Positives = 209/358 (57%), Gaps = 53/358 (14%)
Query: 859 ESKSLKKSLKDVVTEN------EFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVM 912
+ K L++ L+D + ++ + K++ +++ D V +DDI LE+ K +LKE V+
Sbjct: 564 DKKVLREILEDEIIDSLQGVDRQAAKQIFAEIVVHGD-EVHWDDIAGLESAKYSLKEAVV 622
Query: 913 LPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFG 972
P RP+LF +G L +P +G+LLFGPPGTGKTMLA+AVAT++ + F +IS SS+TSK+ G
Sbjct: 623 YPFLRPDLF-RG-LREPVRGMLLFGPPGTGKTMLARAVATESHSTFFSISASSLTSKYLG 680
Query: 973 EGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRT-- 1030
E EK V+A+F++A K++PS+IFVDE+DS++G R N E+E+ R++KNEF+V W L +
Sbjct: 681 ESEKLVRALFAIAKKLSPSIIFVDEIDSIMGSRNNENENESSRRIKNEFLVQWSSLSSAA 740
Query: 1031 ----------------KDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLP-DAPNRAKI 1073
+D RV+VLAATN P+ +DEA RR RR + LP D +
Sbjct: 741 AGSNKSNTNNSDTNGDEDDTRVLVLAATNLPWSIDEAARRRFVRRQYIPLPEDQTRHVQF 800
Query: 1074 LKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKE----ILEKEKKELAA 1129
K++ ++ ++ D + +T+GYSGSD+ +L AA P+++ +LE E+
Sbjct: 801 KKLLSHQKHTLTESDFDELVKITEGYSGSDITSLAKDAAMGPLRDLGDKLLETER----- 855
Query: 1130 AVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYGEGGS 1187
+ IR + + DFK++ + SVS + + + +W +G GS
Sbjct: 856 --------------EMIRPIGLVDFKNSLVYIKPSVSQD--GLVKYEKWASQFGSSGS 897
>MSP1_CAEEL (P54815) MSP1 protein homolog
Length = 342
Score = 206 bits (525), Expect = 2e-52
Identities = 110/268 (41%), Positives = 166/268 (61%), Gaps = 13/268 (4%)
Query: 874 NEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGI 933
+E E R+ + D+G +D+IG E + LK+ ++LPL+ L P +GI
Sbjct: 62 SEHEIRIATQFVGGEDVGADWDEIGGCEELVAELKDRIILPLRFASQ-SGSHLLSPPRGI 120
Query: 934 LLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVI 993
LL+GPPG GKT+LAKAVA AG FIN+ +S++T KW+GE +K AVFS+A K P++I
Sbjct: 121 LLYGPPGCGKTLLAKAVARAAGCRFINLQVSNLTDKWYGESQKLAAAVFSVAQKFQPTII 180
Query: 994 FVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVI 1053
F+DE+DS L R++ +HE+ MK +FM WDG + +++IV+ ATNRP D+D A++
Sbjct: 181 FIDEIDSFLRDRQS-HDHESTAMMKAQFMTLWDGF-SSSGDQIIVMGATNRPRDVDAAIL 238
Query: 1054 RRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLC----- 1108
RR+ R V +P+A R++IL VIL E +++ V+LG IA +G SGSDLK +C
Sbjct: 239 RRMTARFQVPVPNAKQRSQILNVILRNEKINNTVNLGEIAQAAEGLSGSDLKEVCRLALL 298
Query: 1109 -----VTAAHRPIKEILEKEKKELAAAV 1131
A+ + ++L E+ + +AV
Sbjct: 299 ARAKATVASGGSVNQLLPLEQSDFESAV 326
>SKD1_SCHPO (Q09803) Suppressor protein of bem1/bed5 double mutants
Length = 432
Score = 206 bits (524), Expect = 3e-52
Identities = 116/268 (43%), Positives = 172/268 (63%), Gaps = 21/268 (7%)
Query: 854 QAIQNESKSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVML 913
+A+ +++K L+ +L + V PN V +DDI LEN K+ LKE V+L
Sbjct: 102 EALDSDAKKLRSALTSAIL-----------VEKPN---VRWDDIAGLENAKEALKETVLL 147
Query: 914 PLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGE 973
P++ P+LF G+ KP GILL+GPPGTGK+ LAKAVAT+AG+ F +IS S + SKW GE
Sbjct: 148 PIKLPQLFSHGR--KPWSGILLYGPPGTGKSYLAKAVATEAGSTFFSISSSDLVSKWMGE 205
Query: 974 GEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDT 1033
E+ V+ +F +A + PS+IF+DE+DS+ G R + GE E+ R++K EF+V +G+ KD
Sbjct: 206 SERLVRQLFEMAREQKPSIIFIDEIDSLCGSR-SEGESESSRRIKTEFLVQMNGV-GKDE 263
Query: 1034 ERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAK--EDLSSDVDLGA 1091
V+VL ATN P+ LD A+ RR +R+ + LP+A RA++ ++ + K +L+S D
Sbjct: 264 SGVLVLGATNIPWTLDSAIRRRFEKRIYIPLPNAHARARMFELNVGKIPSELTSQ-DFKE 322
Query: 1092 IANMTDGYSGSDLKNLCVTAAHRPIKEI 1119
+A MTDGYSGSD+ + A P++ I
Sbjct: 323 LAKMTDGYSGSDISIVVRDAIMEPVRRI 350
>MEI1_CAEEL (P34808) Meiotic spindle formation protein mei-1
Length = 472
Score = 200 bits (509), Expect = 2e-50
Identities = 116/295 (39%), Positives = 180/295 (60%), Gaps = 17/295 (5%)
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
++ DDI + +VK L E V LPL PE F L P K ++L GPPGTGKT++A+A+A
Sbjct: 190 MSLDDIIGMHDVKQVLHEAVTLPLLVPEFF--QGLRSPWKAMVLAGPPGTGKTLIARAIA 247
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEH 1011
+++ + F +S + ++SKW G+ EK V+ +F LA APS+IF+DE+D++ G+R N GEH
Sbjct: 248 SESSSTFFTVSSTDLSSKWRGDSEKIVRLLFELARFYAPSIIFIDEIDTLGGQRGNSGEH 307
Query: 1012 EAMRKMKNEFMVNWDGLRTK-DTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNR 1070
EA R++K+EF+V DG + K D+ RV VLAATN P++LDEA+ RR +R+ + LPD R
Sbjct: 308 EASRRVKSEFLVQMDGSQNKFDSRRVFVLAATNIPWELDEALRRRFEKRIFIPLPDIDAR 367
Query: 1071 AKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKK--ELA 1128
K+++ + S +++ +A T+G+SG+D+ +LC TAA ++ K + EL
Sbjct: 368 KKLIEKSMEGTPKSDEINYDDLAARTEGFSGADVVSLCRTAAINVLRRYDTKSLRGGELT 427
Query: 1129 AAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYG 1183
AA+ E A +R DF+ A Q V S ++ M + +W + +G
Sbjct: 428 AAM-ESLKAELVRNI---------DFEAALQAVSPSAGPDT--MLKCKEWCDSFG 470
>VPS4_YEAST (P52917) Vacuolar protein sorting-associated protein VPS4
(END13 protein)
Length = 437
Score = 190 bits (483), Expect = 2e-47
Identities = 111/254 (43%), Positives = 159/254 (61%), Gaps = 17/254 (6%)
Query: 873 ENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKG 932
+N+ + L I V ++D+ LE K+ LKE V+LP++ P LF KG KP G
Sbjct: 111 DNKKLRGALSSAILSEKPNVKWEDVAGLEGAKEALKEAVILPVKFPHLF-KGN-RKPTSG 168
Query: 933 ILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSV 992
ILL+GPPGTGK+ LAKAVAT+A + F ++S S + SKW GE EK VK +F++A + PS+
Sbjct: 169 ILLYGPPGTGKSYLAKAVATEANSTFFSVSSSDLVSKWMGESEKLVKQLFAMARENKPSI 228
Query: 993 IFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAV 1052
IF+DEVD++ G R GE EA R++K E +V +G+ D++ V+VL ATN P+ LD A+
Sbjct: 229 IFIDEVDALTGTR-GEGESEASRRIKTELLVQMNGV-GNDSQGVLVLGATNIPWQLDSAI 286
Query: 1053 IRRLPRRLMVNLPDAPNRAKILKV-------ILAKEDLSSDVDLGAIANMTDGYSGSDLK 1105
RR RR+ + LPD R + ++ +L KED + LGA MT+GYSGSD+
Sbjct: 287 RRRFERRIYIPLPDLAARTTMFEINVGDTPCVLTKEDYRT---LGA---MTEGYSGSDIA 340
Query: 1106 NLCVTAAHRPIKEI 1119
+ A +PI++I
Sbjct: 341 VVVKDALMQPIRKI 354
>VAT_THEAC (O05209) VCP-like ATPase
Length = 745
Score = 190 bits (483), Expect = 2e-47
Identities = 113/283 (39%), Positives = 167/283 (58%), Gaps = 18/283 (6%)
Query: 868 KDVVTENEFEKRLLGDVIPPNDI--------GVTFDDIGALENVKDTLKELVMLPLQRPE 919
K VVTE++F+ L I P+ + V +DDIG LE+VK +KE V LPL +P+
Sbjct: 434 KMVVTEDDFKNALKS--IEPSSLREVMVEVPNVHWDDIGGLEDVKREIKETVELPLLKPD 491
Query: 920 LFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVK 979
+F K +P KG LL+GPPG GKT+LAKAVAT++ ANFI+I + SKW GE EK ++
Sbjct: 492 VF-KRLGIRPSKGFLLYGPPGVGKTLLAKAVATESNANFISIKGPEVLSKWVGESEKAIR 550
Query: 980 AVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVL 1039
+F A ++AP+++F+DE+DS+ RR + ++ N+ + + DG+ + V+V+
Sbjct: 551 EIFKKAKQVAPAIVFLDEIDSIAPRRGTTSDSGVTERIVNQLLTSLDGIEVMN--GVVVI 608
Query: 1040 AATNRPYDLDEAVIR--RLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTD 1097
ATNRP +D A++R R + + + PD R ILKV L+ DVDL IA T+
Sbjct: 609 GATNRPDIMDPALLRAGRFDKLIYIPPPDKEARLSILKVHTKNMPLAPDVDLNDIAQRTE 668
Query: 1098 GYSGSDLKNLCVTA---AHRPIKEILEKEKKELAAAVAEGRPA 1137
GY G+DL+NLC A A+R + +K A+ RP+
Sbjct: 669 GYVGADLENLCREAGMNAYRENPDATSVSQKNFLDALKTIRPS 711
Score = 151 bits (382), Expect = 9e-36
Identities = 97/299 (32%), Positives = 169/299 (56%), Gaps = 21/299 (7%)
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
++++DIG L ++E++ LPL+ PELF + +T P KG++L+GPPGTGKT++A+AVA
Sbjct: 187 ISYEDIGGLSEQLGKIREMIELPLKHPELFERLGITPP-KGVILYGPPGTGKTLIARAVA 245
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEH 1011
++GANF++I+ I SK++G+ E+ ++ +FS A + APS+IF+DE+DS+ +RE +
Sbjct: 246 NESGANFLSINGPEIMSKYYGQSEQKLREIFSKAEETAPSIIFIDEIDSIAPKREEV-QG 304
Query: 1012 EAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDAPN 1069
E R++ + + DG+ K+ VIV+ ATNR +D A+ R R R + + +PD
Sbjct: 305 EVERRVVAQLLTLMDGM--KERGHVIVIGATNRIDAIDPALRRPGRFDREIEIGVPDRNG 362
Query: 1070 RAKILKV-----ILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEK 1124
R +IL + L + + L +A+ T G+ G+DL L +A ++ L +
Sbjct: 363 RKEILMIHTRNMPLGMSEEEKNKFLEEMADYTYGFVGADLAALVRESAMNALRRYLPE-- 420
Query: 1125 KELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYG 1183
+ +P P + + +DFK+A + + S E + V W+++ G
Sbjct: 421 ------IDLDKPIPTEILEKMV--VTEDDFKNALKSIEPSSLREVMVEVPNVHWDDIGG 471
>YC97_ARCFU (O28972) Cell division cycle protein 48 homolog AF1297
Length = 733
Score = 187 bits (476), Expect = 1e-46
Identities = 103/244 (42%), Positives = 156/244 (63%), Gaps = 10/244 (4%)
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
V ++DIG LE+ K L E V PL+ PE+F + KP +GILLFGPPGTGKT+LAKAVA
Sbjct: 452 VKWEDIGGLEHAKQELMEAVEWPLKYPEVFRAANI-KPPRGILLFGPPGTGKTLLAKAVA 510
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEH 1011
++ ANFI++ + SKW GE EK+V+ +F A ++AP VIF DE+DS+ RR G+
Sbjct: 511 NESNANFISVKGPELLSKWVGESEKHVREMFRKARQVAPCVIFFDEIDSLAPRRGGIGDS 570
Query: 1012 EAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDAPN 1069
++ ++ + DGL ++ + V+V+AATNRP +D A++R RL R + + PD
Sbjct: 571 HVTERVVSQLLTELDGL--EELKDVVVIAATNRPDMIDPALLRPGRLERHIYIPPPDKKA 628
Query: 1070 RAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILE-----KEK 1124
R +I K+ L + L+ DV++ +A T+GYSG+D++ +C A I+E+++ +E
Sbjct: 629 RVEIFKIHLRGKPLADDVNIEELAEKTEGYSGADIEAVCREAGMLAIRELIKPGMTREEA 688
Query: 1125 KELA 1128
KE A
Sbjct: 689 KEAA 692
Score = 183 bits (464), Expect = 3e-45
Identities = 113/298 (37%), Positives = 175/298 (57%), Gaps = 23/298 (7%)
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
VT++DIG L+ ++E++ LPL+ PELF + + P KG+LL+GPPGTGKT++AKAVA
Sbjct: 179 VTYEDIGGLKRELRLVREMIELPLKHPELFQRLGIEPP-KGVLLYGPPGTGKTLIAKAVA 237
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRREN-PGE 1010
+ A+FI IS I SK++GE E+ ++ +F A + APS+IF+DE+DS+ +RE GE
Sbjct: 238 NEVDAHFIPISGPEIMSKYYGESEQRLREIFEEAKENAPSIIFIDEIDSIAPKREEVTGE 297
Query: 1011 HEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDAP 1068
E R++ + + DGL + VIV+AATNRP +D A+ R R R + + +PD
Sbjct: 298 VE--RRVVAQLLALMDGLEARGD--VIVIAATNRPDAIDPALRRPGRFDREIEIGVPDKE 353
Query: 1069 NRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEK---EKK 1125
R +IL++ K L+ DVDL +A +T+G+ G+DL+ LC AA ++ +L + E +
Sbjct: 354 GRKEILEIHTRKMPLAEDVDLEELAELTNGFVGADLEALCKEAAMHALRRVLPEIDIEAE 413
Query: 1126 ELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYG 1183
E+ A V E + EDF A + + S E + V+W ++ G
Sbjct: 414 EIPAEVIEN------------LKVTREDFMEALKNIEPSAMREVLVEVPNVKWEDIGG 459
>TERA_RAT (P46462) Transitional endoplasmic reticulum ATPase (TER
ATPase) (15S Mg(2+)-ATPase p97 subunit)
(Valosin-containing protein) (VCP)
Length = 806
Score = 186 bits (471), Expect = 4e-46
Identities = 114/296 (38%), Positives = 168/296 (56%), Gaps = 24/296 (8%)
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
VT++DIG LE+VK L+ELV P++ P+ F K +T P KG+L +GPPG GKT+LAKA+A
Sbjct: 474 VTWEDIGGLEDVKRELQELVQYPVEHPDKFLKFGMT-PSKGVLFYGPPGCGKTLLAKAIA 532
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSML-GRRENPGE 1010
+ ANFI+I + + WFGE E V+ +F A + AP V+F DE+DS+ R N G+
Sbjct: 533 NECQANFISIKGPELLTMWFGESEANVREIFDKARQAAPCVLFFDELDSIAKARGGNIGD 592
Query: 1011 -HEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDA 1067
A ++ N+ + DG+ TK + V ++ ATNRP +D A++R RL + + + LPD
Sbjct: 593 GGGAADRVINQILTEMDGMSTK--KNVFIIGATNRPDIIDPAILRPGRLDQLIYIPLPDE 650
Query: 1068 PNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKE- 1126
+R ILK L K ++ DVDL +A MT+G+SG+DL +C A I+E +E E +
Sbjct: 651 KSRVAILKANLRKSPVAKDVDLEFLAKMTNGFSGADLTEICQRACKLAIRESIESEIRRE 710
Query: 1127 -------LAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTEL 1175
A V E P P +R + F+ A + SVS + E+
Sbjct: 711 RERQTNPSAMEVEEDDPVPEIR---------RDHFEEAMRFARRSVSDNDIRKYEM 757
Score = 174 bits (441), Expect = 1e-42
Identities = 113/301 (37%), Positives = 168/301 (55%), Gaps = 23/301 (7%)
Query: 888 NDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLA 947
N++G +DDIG +KE+V LPL+ P LF K KP +GILL+GPPGTGKT++A
Sbjct: 199 NEVG--YDDIGGCRKQLAQIKEMVELPLRHPALF-KAIGVKPPRGILLYGPPGTGKTLIA 255
Query: 948 KAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRREN 1007
+AVA + GA F I+ I SK GE E ++ F A K AP++IF+DE+D++ +RE
Sbjct: 256 RAVANETGAFFFLINGPEIMSKLAGESESNLRKAFEEAEKNAPAIIFIDELDAIAPKREK 315
Query: 1008 PGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLP 1065
E R++ ++ + DGL K VIV+AATNRP +D A+ R R R + + +P
Sbjct: 316 T-HGEVERRIVSQLLTLMDGL--KQRAHVIVMAATNRPNSIDPALRRFGRFDREVDIGIP 372
Query: 1066 DAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIK---EILEK 1122
DA R +IL++ L+ DVDL +AN T G+ G+DL LC AA + I+ ++++
Sbjct: 373 DATGRLEILQIHTKNMKLADDVDLEQVANETHGHVGADLAALCSEAALQAIRKKMDLIDL 432
Query: 1123 EKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELY 1182
E + + A V ++ M+DF+ A Q S E+V V W ++
Sbjct: 433 EDETIDAEVMNS------------LAVTMDDFRWALSQSNPSALRETVVEVPQVTWEDIG 480
Query: 1183 G 1183
G
Sbjct: 481 G 481
>TERA_MOUSE (Q01853) Transitional endoplasmic reticulum ATPase (TER
ATPase) (15S Mg(2+)-ATPase p97 subunit)
(Valosin-containing protein) (VCP)
Length = 806
Score = 186 bits (471), Expect = 4e-46
Identities = 114/296 (38%), Positives = 168/296 (56%), Gaps = 24/296 (8%)
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
VT++DIG LE+VK L+ELV P++ P+ F K +T P KG+L +GPPG GKT+LAKA+A
Sbjct: 474 VTWEDIGGLEDVKRELQELVQYPVEHPDKFLKFGMT-PSKGVLFYGPPGCGKTLLAKAIA 532
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSML-GRRENPGE 1010
+ ANFI+I + + WFGE E V+ +F A + AP V+F DE+DS+ R N G+
Sbjct: 533 NECQANFISIKGPELLTMWFGESEANVREIFDKARQAAPCVLFFDELDSIAKARGGNIGD 592
Query: 1011 -HEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDA 1067
A ++ N+ + DG+ TK + V ++ ATNRP +D A++R RL + + + LPD
Sbjct: 593 GGGAADRVINQILTEMDGMSTK--KNVFIIGATNRPDIIDPAILRPGRLDQLIYIPLPDE 650
Query: 1068 PNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKE- 1126
+R ILK L K ++ DVDL +A MT+G+SG+DL +C A I+E +E E +
Sbjct: 651 KSRVAILKANLRKSPVAKDVDLEFLAKMTNGFSGADLTEICQRACKLAIRESIESEIRRE 710
Query: 1127 -------LAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTEL 1175
A V E P P +R + F+ A + SVS + E+
Sbjct: 711 RERQTNPSAMEVEEDDPVPEIR---------RDHFEEAMRFARRSVSDNDIRKYEM 757
Score = 174 bits (440), Expect = 2e-42
Identities = 112/301 (37%), Positives = 168/301 (55%), Gaps = 23/301 (7%)
Query: 888 NDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLA 947
N++G +DD+G +KE+V LPL+ P LF K KP +GILL+GPPGTGKT++A
Sbjct: 199 NEVG--YDDVGGCRKQLAQIKEMVELPLRHPALF-KAIGVKPPRGILLYGPPGTGKTLIA 255
Query: 948 KAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRREN 1007
+AVA + GA F I+ I SK GE E ++ F A K AP++IF+DE+D++ +RE
Sbjct: 256 RAVANETGAFFFLINGPEIMSKLAGESESNLRKAFEEAEKNAPAIIFIDELDAIAPKREK 315
Query: 1008 PGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLP 1065
E R++ ++ + DGL K VIV+AATNRP +D A+ R R R + + +P
Sbjct: 316 T-HGEVERRIVSQLLTLMDGL--KQRAHVIVMAATNRPNSIDPALRRFGRFDREVDIGIP 372
Query: 1066 DAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIK---EILEK 1122
DA R +IL++ L+ DVDL +AN T G+ G+DL LC AA + I+ ++++
Sbjct: 373 DATGRLEILQIHTKNMKLADDVDLEQVANETHGHVGADLAALCSEAALQAIRKKMDLIDL 432
Query: 1123 EKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELY 1182
E + + A V ++ M+DF+ A Q S E+V V W ++
Sbjct: 433 EDETIDAEVMNS------------LAVTMDDFRWALSQSNPSALRETVVEVPQVTWEDIG 480
Query: 1183 G 1183
G
Sbjct: 481 G 481
>TERA_HUMAN (P55072) Transitional endoplasmic reticulum ATPase (TER
ATPase) (15S Mg(2+)-ATPase p97 subunit)
(Valosin-containing protein) (VCP)
Length = 805
Score = 186 bits (471), Expect = 4e-46
Identities = 114/296 (38%), Positives = 168/296 (56%), Gaps = 24/296 (8%)
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
VT++DIG LE+VK L+ELV P++ P+ F K +T P KG+L +GPPG GKT+LAKA+A
Sbjct: 473 VTWEDIGGLEDVKRELQELVQYPVEHPDKFLKFGMT-PSKGVLFYGPPGCGKTLLAKAIA 531
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSML-GRRENPGE 1010
+ ANFI+I + + WFGE E V+ +F A + AP V+F DE+DS+ R N G+
Sbjct: 532 NECQANFISIKGPELLTMWFGESEANVREIFDKARQAAPCVLFFDELDSIAKARGGNIGD 591
Query: 1011 -HEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDA 1067
A ++ N+ + DG+ TK + V ++ ATNRP +D A++R RL + + + LPD
Sbjct: 592 GGGAADRVINQILTEMDGMSTK--KNVFIIGATNRPDIIDPAILRPGRLDQLIYIPLPDE 649
Query: 1068 PNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKE- 1126
+R ILK L K ++ DVDL +A MT+G+SG+DL +C A I+E +E E +
Sbjct: 650 KSRVAILKANLRKSPVAKDVDLEFLAKMTNGFSGADLTEICQRACKLAIRESIESEIRRE 709
Query: 1127 -------LAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTEL 1175
A V E P P +R + F+ A + SVS + E+
Sbjct: 710 RERQTNPSAMEVEEDDPVPEIR---------RDHFEEAMRFARRSVSDNDIRKYEM 756
Score = 174 bits (441), Expect = 1e-42
Identities = 113/301 (37%), Positives = 168/301 (55%), Gaps = 23/301 (7%)
Query: 888 NDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLA 947
N++G +DDIG +KE+V LPL+ P LF K KP +GILL+GPPGTGKT++A
Sbjct: 198 NEVG--YDDIGGCRKQLAQIKEMVELPLRHPALF-KAIGVKPPRGILLYGPPGTGKTLIA 254
Query: 948 KAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRREN 1007
+AVA + GA F I+ I SK GE E ++ F A K AP++IF+DE+D++ +RE
Sbjct: 255 RAVANETGAFFFLINGPEIMSKLAGESESNLRKAFEEAEKNAPAIIFIDELDAIAPKREK 314
Query: 1008 PGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLP 1065
E R++ ++ + DGL K VIV+AATNRP +D A+ R R R + + +P
Sbjct: 315 T-HGEVERRIVSQLLTLMDGL--KQRAHVIVMAATNRPNSIDPALRRFGRFDREVDIGIP 371
Query: 1066 DAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIK---EILEK 1122
DA R +IL++ L+ DVDL +AN T G+ G+DL LC AA + I+ ++++
Sbjct: 372 DATGRLEILQIHTKNMKLADDVDLEQVANETHGHVGADLAALCSEAALQAIRKKMDLIDL 431
Query: 1123 EKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELY 1182
E + + A V ++ M+DF+ A Q S E+V V W ++
Sbjct: 432 EDETIDAEVMNS------------LAVTMDDFRWALSQSNPSALRETVVEVPQVTWEDIG 479
Query: 1183 G 1183
G
Sbjct: 480 G 480
>TERA_PIG (P03974) Transitional endoplasmic reticulum ATPase (TER
ATPase) (15S Mg(2+)-ATPase p97 subunit)
(Valosin-containing protein) (VCP)
Length = 806
Score = 184 bits (468), Expect = 9e-46
Identities = 113/296 (38%), Positives = 168/296 (56%), Gaps = 24/296 (8%)
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
VT++DIG LE+VK L++LV P++ P+ F K +T P KG+L +GPPG GKT+LAKA+A
Sbjct: 474 VTWEDIGGLEDVKRELQDLVQYPVEHPDKFLKFGMT-PSKGVLFYGPPGCGKTLLAKAIA 532
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSML-GRRENPGE 1010
+ ANFI+I + + WFGE E V+ +F A + AP V+F DE+DS+ R N G+
Sbjct: 533 NECQANFISIKGPELLTMWFGESEANVREIFDKARQAAPCVLFFDELDSIAKARGGNIGD 592
Query: 1011 -HEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDA 1067
A ++ N+ + DG+ TK + V ++ ATNRP +D A++R RL + + + LPD
Sbjct: 593 GGGAADRVINQILTEMDGMSTK--KNVFIIGATNRPDIIDPAILRPGRLDQLIYIPLPDE 650
Query: 1068 PNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKE- 1126
+R ILK L K ++ DVDL +A MT+G+SG+DL +C A I+E +E E +
Sbjct: 651 KSRVAILKANLRKSPVAKDVDLEFLAKMTNGFSGADLTEICQRACKLAIRESIESEIRRE 710
Query: 1127 -------LAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTEL 1175
A V E P P +R + F+ A + SVS + E+
Sbjct: 711 RERQTNPSAMEVEEDDPVPEIR---------RDHFEEAMRFARRSVSDNDIRKYEM 757
Score = 174 bits (441), Expect = 1e-42
Identities = 113/301 (37%), Positives = 168/301 (55%), Gaps = 23/301 (7%)
Query: 888 NDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLA 947
N++G +DDIG +KE+V LPL+ P LF K KP +GILL+GPPGTGKT++A
Sbjct: 199 NEVG--YDDIGGCRKQLAQIKEMVELPLRHPALF-KAIGVKPPRGILLYGPPGTGKTLIA 255
Query: 948 KAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRREN 1007
+AVA + GA F I+ I SK GE E ++ F A K AP++IF+DE+D++ +RE
Sbjct: 256 RAVANETGAFFFLINGPEIMSKLAGESESNLRKAFEEAEKNAPAIIFIDELDAIAPKREK 315
Query: 1008 PGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLP 1065
E R++ ++ + DGL K VIV+AATNRP +D A+ R R R + + +P
Sbjct: 316 T-HGEVERRIVSQLLTLMDGL--KQRAHVIVMAATNRPNSIDPALRRFGRFDREVDIGIP 372
Query: 1066 DAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIK---EILEK 1122
DA R +IL++ L+ DVDL +AN T G+ G+DL LC AA + I+ ++++
Sbjct: 373 DATGRLEILQIHTKNMKLADDVDLEQVANETHGHVGADLAALCSEAALQAIRKKMDLIDL 432
Query: 1123 EKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELY 1182
E + + A V ++ M+DF+ A Q S E+V V W ++
Sbjct: 433 EDETIDAEVMNS------------LAVTMDDFRWALSQSNPSALRETVVEVPQVTWEDIG 480
Query: 1183 G 1183
G
Sbjct: 481 G 481
>YB56_METJA (Q58556) Cell division cycle protein 48 homolog MJ1156
Length = 903
Score = 184 bits (467), Expect = 1e-45
Identities = 114/294 (38%), Positives = 172/294 (57%), Gaps = 15/294 (5%)
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
VT++DIG L+ ++E++ LP++ PELF K + P KG+LL GPPGTGKT+LAKAVA
Sbjct: 176 VTYEDIGGLKEEVKKVREMIELPMRHPELFEKLGIEPP-KGVLLVGPPGTGKTLLAKAVA 234
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEH 1011
+AGANF I+ I SK+ GE E+ ++ +F A + APS+IF+DE+D++ +R+
Sbjct: 235 NEAGANFYVINGPEIMSKYVGETEENLRKIFEEAEENAPSIIFIDEIDAIAPKRDE-ATG 293
Query: 1012 EAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDAPN 1069
E R++ + + DGL K +V+V+ ATNRP LD A+ R R R +++ +PD
Sbjct: 294 EVERRLVAQLLTLMDGL--KGRGQVVVIGATNRPNALDPALRRPGRFDREIVIGVPDREG 351
Query: 1070 RAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAA 1129
R +IL++ L+ DVDL +A++T G+ G+DL LC AA R ++ +L E
Sbjct: 352 RKEILQIHTRNMPLAEDVDLDYLADVTHGFVGADLAALCKEAAMRALRRVLPSIDLE--- 408
Query: 1130 AVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYG 1183
AE P L D + M+DFK A + V S E + V+W ++ G
Sbjct: 409 --AEEIPKEVL----DNLKVTMDDFKEALKDVEPSAMREVLVEVPNVKWEDIGG 456
Score = 183 bits (464), Expect = 3e-45
Identities = 113/285 (39%), Positives = 171/285 (59%), Gaps = 21/285 (7%)
Query: 852 IFQAIQNESKSLKKSLKD--VVTENEFEKRLLGDVIP----------PNDIGVTFDDIGA 899
+ +I E++ + K + D VT ++F K L DV P PN V ++DIG
Sbjct: 401 VLPSIDLEAEEIPKEVLDNLKVTMDDF-KEALKDVEPSAMREVLVEVPN---VKWEDIGG 456
Query: 900 LENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFI 959
LE VK L+E V PL+ E+F K + +P KG+LLFGPPGTGKT+LAKAVA ++GANFI
Sbjct: 457 LEEVKQELREAVEWPLKAKEVFEKIGV-RPPKGVLLFGPPGTGKTLLAKAVANESGANFI 515
Query: 960 NISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKN 1019
++ I SKW GE EK ++ +F A + AP +IF DE+D++ +R K+ N
Sbjct: 516 SVKGPEIFSKWVGESEKAIREIFRKARQSAPCIIFFDEIDAIAPKRGRDLSSAVTDKVVN 575
Query: 1020 EFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDAPNRAKILKVI 1077
+ + DG+ ++ + V+V+AATNRP +D A++R RL R ++V +PD R I K+
Sbjct: 576 QLLTELDGM--EEPKDVVVIAATNRPDIIDPALLRPGRLDRVILVPVPDEKARLDIFKIH 633
Query: 1078 LAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEK 1122
+L+ DV+L +A T+GY+G+D++ LC AA ++E + K
Sbjct: 634 TRSMNLAEDVNLEELAKKTEGYTGADIEALCREAAMLAVRESIGK 678
>SKD1_MOUSE (P46467) SKD1 protein (Vacuolar sorting protein 4b)
Length = 444
Score = 184 bits (466), Expect = 2e-45
Identities = 106/251 (42%), Positives = 159/251 (63%), Gaps = 11/251 (4%)
Query: 873 ENEFEKRLLGDVI--PPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPC 930
+ + + +L G ++ PN V + D+ LE K+ LKE V+LP++ P LF G+ T P
Sbjct: 113 KKKLQNQLQGAIVIERPN---VKWSDVAGLEGAKEALKEAVILPIKFPHLFT-GKRT-PW 167
Query: 931 KGILLFGPPGTGKTMLAKAVATDAG-ANFINISMSSITSKWFGEGEKYVKAVFSLASKIA 989
+GILLFGPPGTGK+ LAKAVAT+A + F +IS S + SKW GE EK VK +F LA +
Sbjct: 168 RGILLFGPPGTGKSYLAKAVATEANNSTFFSISSSDLVSKWLGESEKLVKNLFQLARENK 227
Query: 990 PSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLD 1049
PS+IF+DE+DS+ G R + E EA R++K EF+V G+ D + ++VL ATN P+ LD
Sbjct: 228 PSIIFIDEIDSLCGSR-SENESEAARRIKTEFLVQMQGVGV-DNDGILVLGATNIPWVLD 285
Query: 1050 EAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLS-SDVDLGAIANMTDGYSGSDLKNLC 1108
A+ RR +R+ + LP+A RA + ++ L S ++ D + TDGYSG+D+ +
Sbjct: 286 SAIRRRFEKRIYIPLPEAHARAAMFRLHLGSTQNSLTEADFQELGRKTDGYSGADISIIV 345
Query: 1109 VTAAHRPIKEI 1119
A +P++++
Sbjct: 346 RDALMQPVRKV 356
>C48A_ARATH (P54609) Cell division control protein 48 homolog A
(AtCDC48a)
Length = 809
Score = 183 bits (465), Expect = 2e-45
Identities = 105/284 (36%), Positives = 163/284 (56%), Gaps = 9/284 (3%)
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
V+++DIG LENVK L+E V P++ PE F K ++ P KG+L +GPPG GKT+LAKA+A
Sbjct: 477 VSWNDIGGLENVKRELQETVQYPVEHPEKFEKFGMS-PSKGVLFYGPPGCGKTLLAKAIA 535
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEH 1011
+ ANFI++ + + WFGE E V+ +F A + AP V+F DE+DS+ +R
Sbjct: 536 NECQANFISVKGPELLTMWFGESEANVREIFDKARQSAPCVLFFDELDSIATQRGGGSGG 595
Query: 1012 E---AMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPD 1066
+ A ++ N+ + DG+ K T V ++ ATNRP +D A++R RL + + + LPD
Sbjct: 596 DGGGAADRVLNQLLTEMDGMNAKKT--VFIIGATNRPDIIDSALLRPGRLDQLIYIPLPD 653
Query: 1067 APNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKE 1126
+R I K L K ++ DVD+GA+A T G+SG+D+ +C A I+E +EK+ E
Sbjct: 654 EDSRLNIFKAALRKSPIAKDVDIGALAKYTQGFSGADITEICQRACKYAIRENIEKD-IE 712
Query: 1127 LAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESV 1170
+E A G D++ + F+ + + SVS +
Sbjct: 713 KEKRRSENPEAMEEDGVDEVSEIKAAHFEESMKYARRSVSDADI 756
Score = 168 bits (425), Expect = 9e-41
Identities = 109/301 (36%), Positives = 165/301 (54%), Gaps = 23/301 (7%)
Query: 888 NDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLA 947
+D+G +DD+G + ++ELV LPL+ P+LF K KP KGILL+GPPG+GKT++A
Sbjct: 202 DDVG--YDDVGGVRKQMAQIRELVELPLRHPQLF-KSIGVKPPKGILLYGPPGSGKTLIA 258
Query: 948 KAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRREN 1007
+AVA + GA F I+ I SK GE E ++ F A K APS+IF+DE+DS+ +RE
Sbjct: 259 RAVANETGAFFFCINGPEIMSKLAGESESNLRKAFEEAEKNAPSIIFIDEIDSIAPKREK 318
Query: 1008 PGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLP 1065
E R++ ++ + DGL+++ VIV+ ATNRP +D A+ R R R + + +P
Sbjct: 319 T-NGEVERRIVSQLLTLMDGLKSR--AHVIVMGATNRPNSIDPALRRFGRFDREIDIGVP 375
Query: 1066 DAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKE---ILEK 1122
D R ++L++ L+ DVDL I+ T GY G+DL LC AA + I+E +++
Sbjct: 376 DEIGRLEVLRIHTKNMKLAEDVDLERISKDTHGYVGADLAALCTEAALQCIREKMDVIDL 435
Query: 1123 EKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELY 1182
E + A + ++ E F A S E+V V WN++
Sbjct: 436 EDDSIDAEILNS------------MAVTNEHFHTALGNSNPSALRETVVEVPNVSWNDIG 483
Query: 1183 G 1183
G
Sbjct: 484 G 484
>SAV_SULAC (Q07590) SAV protein
Length = 780
Score = 183 bits (464), Expect = 3e-45
Identities = 117/275 (42%), Positives = 162/275 (58%), Gaps = 16/275 (5%)
Query: 861 KSLKKSLKDVVTE-NEFEKRLLGDV---IPPNDIGVTFDDIGALENVKDTLKELVMLPLQ 916
K LK S+ D + + LL +V +P V ++DIG L+NVK L+E V PL+
Sbjct: 453 KELKVSMNDFLNALKSIQPSLLREVYVEVPK----VNWNDIGGLDNVKQQLREAVEWPLR 508
Query: 917 RPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEK 976
PELF K +T P KGILLFGPPGTGKTMLAKAVAT++GANFI + I SKW GE EK
Sbjct: 509 FPELFTKSGVTPP-KGILLFGPPGTGKTMLAKAVATESGANFIAVRGPEILSKWVGESEK 567
Query: 977 YVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERV 1036
++ +F A + AP+VIF DE+DS+ R + ++ N+ + DG+ +V
Sbjct: 568 AIREIFRKARQAAPTVIFFDEIDSIAPIRGLSTDSGVTERIVNQLLAEMDGI--VPLNKV 625
Query: 1037 IVLAATNRPYDLDEAVIR--RLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIAN 1094
+++AATNRP LD A++R R R + V PD R +ILKV L+ DV L IA
Sbjct: 626 VIIAATNRPDILDPALLRPGRFDRLIYVPPPDKTARFEILKVHTKNVPLAEDVSLEDIAE 685
Query: 1095 MTDGYSGSDLKNLCVTA---AHRPIKEILEKEKKE 1126
+GY+G+DL+ L A A R I + +K+ ++
Sbjct: 686 KAEGYTGADLEALVREATINAMRSIYSMCDKQSRD 720
Score = 174 bits (441), Expect = 1e-42
Identities = 107/295 (36%), Positives = 170/295 (57%), Gaps = 15/295 (5%)
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
V+++DIG LE K ++E+V P++ PELF + + P KGILL+GPPGTGKT+LA+A+
Sbjct: 209 VSWEDIGDLEEAKQKIREIVEWPMRHPELFQRLGIDPP-KGILLYGPPGTGKTLLARALR 267
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRREN-PGE 1010
+ GA FI ++ I SK++GE E+ ++ +F A + APS+IF+DE+D++ +RE+ GE
Sbjct: 268 NEIGAYFITVNGPEIMSKFYGESEQRIREIFKEAEENAPSIIFIDEIDAIAPKREDVTGE 327
Query: 1011 HEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDAP 1068
E +++ + + DG+ K RVIV+ ATNRP +D A+ R R R + + PD
Sbjct: 328 VE--KRVVAQLLTLMDGI--KGRGRVIVIGATNRPDAIDPALRRPGRFDREIEIRPPDTK 383
Query: 1069 NRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELA 1128
R IL+V ++ DVDL +A MT GY+G+DL L AA ++ ++++K L
Sbjct: 384 GRKDILQVHTRNMPITDDVDLDKLAEMTYGYTGADLAALAKEAAIYALRRFVDEKKLNLD 443
Query: 1129 AAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYG 1183
L+ ++M DF +A + + S+ E V WN++ G
Sbjct: 444 QPTIPAEIIKELK-------VSMNDFLNALKSIQPSLLREVYVEVPKVNWNDIGG 491
>TERA_XENLA (P23787) Transitional endoplasmic reticulum ATPase (TER
ATPase) (15S Mg(2+)-ATPase p97 subunit)
Length = 805
Score = 182 bits (463), Expect = 4e-45
Identities = 112/296 (37%), Positives = 167/296 (55%), Gaps = 24/296 (8%)
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
VT++DIG LE+VK L+ELV P++ P+ F K +T P KG+L +GPPG GKT+LAKA+A
Sbjct: 474 VTWEDIGGLEDVKRELQELVQYPVEHPDKFLKFGMT-PSKGVLFYGPPGCGKTLLAKAIA 532
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSML-GRRENPGE 1010
+ ANFI+I + + WFGE E V+ +F A + AP V+F DE+DS+ R N G+
Sbjct: 533 NECQANFISIKGPELLTMWFGESEANVREIFDKARQAAPCVLFFDELDSIAKARGGNIGD 592
Query: 1011 -HEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDA 1067
A ++ N+ + DG+ K + V ++ ATNRP +D A++R RL + + + LPD
Sbjct: 593 GGGAADRVINQILTEMDGMSIK--KNVFIIGATNRPDIIDPAILRPGRLDQLIYIPLPDE 650
Query: 1068 PNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKE- 1126
+R ILK L K ++ DVD+ +A MT+G+SG+DL +C A I+E +E E +
Sbjct: 651 KSRMAILKANLRKSPVAKDVDVDFLAKMTNGFSGADLTEICQRACKLAIRESIENEIRRE 710
Query: 1127 -------LAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTEL 1175
A V E P P +R + F+ A + SVS + E+
Sbjct: 711 RDRQTNPSAMEVEEDDPVPEIR---------RDHFEEAMRLARRSVSDNDIRKYEM 757
Score = 172 bits (437), Expect = 4e-42
Identities = 112/301 (37%), Positives = 167/301 (55%), Gaps = 23/301 (7%)
Query: 888 NDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLA 947
N++G +DDIG +KE+V LPL+ P LF K KP +GILL+GPPGTGKT++A
Sbjct: 199 NEVG--YDDIGGCRKQLAQIKEMVELPLRHPALF-KAIGVKPPRGILLYGPPGTGKTLIA 255
Query: 948 KAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRREN 1007
+AVA + GA F I+ I SK GE E ++ F A K AP++IF+DE+D++ +RE
Sbjct: 256 RAVANETGAFFFLINGPEIMSKLAGESESNLRKAFEEAEKNAPAIIFIDELDAIAPKREK 315
Query: 1008 PGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLP 1065
E R++ ++ + DGL K VIV+AATNRP +D A+ R R R + + +P
Sbjct: 316 T-HGEVERRIVSQLLTLMDGL--KQRAHVIVMAATNRPNSIDPALRRFGRFDREVDIGIP 372
Query: 1066 DAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIK---EILEK 1122
D+ R +IL++ LS DVDL +AN T G+ G+DL LC AA + I+ ++++
Sbjct: 373 DSTGRLEILQIHTKNMKLSDDVDLEQVANETHGHVGADLAALCSEAALQAIRKKMDLIDL 432
Query: 1123 EKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELY 1182
E + + A V ++ M+DF+ Q S E+V V W ++
Sbjct: 433 EDETIDAEVMNS------------LAVTMDDFRWGLSQSNPSALRETVVEVPQVTWEDIG 480
Query: 1183 G 1183
G
Sbjct: 481 G 481
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.313 0.131 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 143,267,027
Number of Sequences: 164201
Number of extensions: 6344665
Number of successful extensions: 18982
Number of sequences better than 10.0: 778
Number of HSP's better than 10.0 without gapping: 416
Number of HSP's successfully gapped in prelim test: 363
Number of HSP's that attempted gapping in prelim test: 17331
Number of HSP's gapped (non-prelim): 1294
length of query: 1196
length of database: 59,974,054
effective HSP length: 122
effective length of query: 1074
effective length of database: 39,941,532
effective search space: 42897205368
effective search space used: 42897205368
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 72 (32.3 bits)
Medicago: description of AC141863.4


