

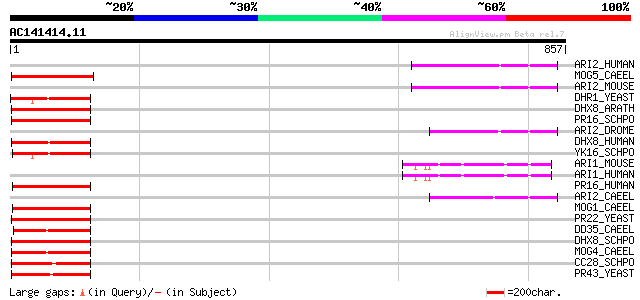
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141414.11 - phase: 0
(857 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ARI2_HUMAN (O95376) Ariadne-2 protein homolog (ARI-2) (Triad1 pr... 106 2e-22
MOG5_CAEEL (Q09530) Probable pre-mRNA splicing factor ATP-depend... 106 3e-22
ARI2_MOUSE (Q9Z1K6) Ariadne-2 protein homolog (ARI-2) (Triad1 pr... 104 1e-21
DHR1_YEAST (Q04217) Probable ATP-dependent RNA helicase DHR1 (DE... 103 1e-21
DHX8_ARATH (Q38953) Putative pre-mRNA splicing factor ATP-depend... 103 3e-21
PR16_SCHPO (Q9P774) Pre-mRNA splicing factor RNA helicase prp16 ... 102 3e-21
ARI2_DROME (O76924) Ariadne-2 protein (Ari-2) 101 1e-20
DHX8_HUMAN (Q14562) ATP-dependent helicase DHX8 (RNA helicase HR... 100 1e-20
YK16_SCHPO (Q9HDY4) Putative ATP-dependent RNA helicase PB1A10.0... 100 2e-20
ARI1_MOUSE (Q9Z1K5) Ariadne-1 protein homolog (ARI-1) (Ubiquitin... 98 8e-20
ARI1_HUMAN (Q9Y4X5) Ariadne-1 protein homolog (ARI-1) (Ubiquitin... 98 8e-20
PR16_HUMAN (Q92620) Pre-mRNA splicing factor ATP-dependent RNA h... 96 3e-19
ARI2_CAEEL (Q22431) Probable ariadne-2 protein (Ari-2) 96 3e-19
MOG1_CAEEL (P34498) Probable pre-mRNA splicing factor ATP-depend... 96 5e-19
PR22_YEAST (P24384) Pre-mRNA splicing factor RNA helicase PRP22 95 7e-19
DD35_CAEEL (Q9BKQ8) Probable ATP-dependent helicase DHX35 homolog 94 1e-18
DHX8_SCHPO (O42643) Putative pre-mRNA splicing factor ATP-depend... 94 2e-18
MOG4_CAEEL (O45244) Probable pre-mRNA splicing factor ATP-depend... 90 2e-17
CC28_SCHPO (Q10752) Putative ATP-dependent RNA helicase cdc28 90 2e-17
PR43_YEAST (P53131) Pre-mRNA splicing factor RNA helicase PRP43 ... 90 3e-17
>ARI2_HUMAN (O95376) Ariadne-2 protein homolog (ARI-2) (Triad1
protein) (HT005)
Length = 493
Score = 106 bits (265), Expect = 2e-22
Identities = 70/232 (30%), Positives = 96/232 (41%), Gaps = 10/232 (4%)
Query: 621 KSRVEEITFEIARLSNPSSERFDTGPS--CPICLCEVEDGYQLE-GCGHLFCQSCMVEQC 677
KS ++ E NPS + P C +C+ V L C H FC+SC + C
Sbjct: 110 KSNSAQLLVEARVQPNPSKHVPTSHPPHHCAVCMQFVRKENLLSLACQHQFCRSCWEQHC 169
Query: 678 ESAIKNQGSFPIRCAHQGCGNHILLVDFRTLLSNDKLEELFRASLGAFVASSSGTYRFCP 737
+K+ + C Q C LL N++L E +R L S + CP
Sbjct: 170 SVLVKDGVGVGVSCMAQDCPLRTPEDFVFPLLPNEELREKYRRYLFRDYVESHYQLQLCP 229
Query: 738 SPDCPSIYRVADPDTASAPFVCGACYSETCTRCHIEYHPYVSCERYRQF--KDDPDSSLR 795
DCP + RV +P C C C +C YH C R++ K DS
Sbjct: 230 GADCPMVIRVQEPRARRVQ--CNRCNEVFCFKCRQMYHAPTDCATIRKWLTKCADDSETA 287
Query: 796 DWCKGKEQVKNCPACGHVIEKVDGCNHIEC-KCGKHICWVCLEFFTTSGECY 846
++ K+CP C IEK GCNH++C KC CW+CL + T G Y
Sbjct: 288 NYISA--HTKDCPKCNICIEKNGGCNHMQCSKCKHDFCWMCLGDWKTHGSEY 337
>MOG5_CAEEL (Q09530) Probable pre-mRNA splicing factor ATP-dependent
RNA helicase mog-5 (Sex determination protein mog-5)
(Masculinization of germ line protein 5)
Length = 1200
Score = 106 bits (264), Expect = 3e-22
Identities = 56/126 (44%), Positives = 83/126 (65%), Gaps = 1/126 (0%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
QI V++GETGSGK+TQ+ Q+ ++G+G I CTQPRR+AA S+A+RV EE GC +
Sbjct: 557 QILVVVGETGSGKTTQMTQYAIEAGLGRRGKIGCTQPRRVAAMSVAKRVAEEY-GCKLGT 615
Query: 64 SIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLAL 123
+ F D+ I +MTD LL+ + D + +G S I++DEAHER+I+TD+L L
Sbjct: 616 DVGYTIRFEDCTSQDTIIKYMTDGMLLRECLIDPDLSGYSLIMLDEAHERTIHTDVLFGL 675
Query: 124 VPSSNR 129
+ ++ R
Sbjct: 676 LKAAAR 681
>ARI2_MOUSE (Q9Z1K6) Ariadne-2 protein homolog (ARI-2) (Triad1
protein) (UbcM4-interacting protein 48)
Length = 492
Score = 104 bits (259), Expect = 1e-21
Identities = 69/232 (29%), Positives = 95/232 (40%), Gaps = 10/232 (4%)
Query: 621 KSRVEEITFEIARLSNPSSERFDTGPS--CPICLCEVEDGYQLE-GCGHLFCQSCMVEQC 677
+S ++ E NPS P C +C+ V L C H FC+SC + C
Sbjct: 109 RSNSAQLLVEARVQPNPSKHVPTAHPPHHCAVCMQFVRKENLLSLACQHQFCRSCWEQHC 168
Query: 678 ESAIKNQGSFPIRCAHQGCGNHILLVDFRTLLSNDKLEELFRASLGAFVASSSGTYRFCP 737
+K+ I C Q C LL N++L + +R L S + CP
Sbjct: 169 SVLVKDGVGVGISCMAQDCPLRTPEDFVFPLLPNEELRDKYRRYLFRDYVESHFQLQLCP 228
Query: 738 SPDCPSIYRVADPDTASAPFVCGACYSETCTRCHIEYHPYVSCERYRQF--KDDPDSSLR 795
DCP + RV +P C C C +C YH C R++ K DS
Sbjct: 229 GADCPMVIRVQEPRARRVQ--CNRCSEVFCFKCRQMYHAPTDCATIRKWLTKCADDSETA 286
Query: 796 DWCKGKEQVKNCPACGHVIEKVDGCNHIEC-KCGKHICWVCLEFFTTSGECY 846
++ K+CP C IEK GCNH++C KC CW+CL + T G Y
Sbjct: 287 NYISA--HTKDCPKCNICIEKNGGCNHMQCSKCKHDFCWMCLGDWKTHGSEY 336
>DHR1_YEAST (Q04217) Probable ATP-dependent RNA helicase DHR1
(DEAH-box RNA helicase DHR1)
Length = 1267
Score = 103 bits (258), Expect = 1e-21
Identities = 58/129 (44%), Positives = 80/129 (61%), Gaps = 8/129 (6%)
Query: 2 HVQITVLIGETGSGKSTQLVQFLADSGVGANES------IVCTQPRRIAAKSLAERVREE 55
H + ++ GETGSGK+TQ+ QFL ++G GA +S + TQPRR+AA S+AERV E
Sbjct: 406 HNDVVIICGETGSGKTTQVPQFLYEAGFGAEDSPDYPGMVGITQPRRVAAVSMAERVANE 465
Query: 56 SGGCYEDSSIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSI 115
G + F S K D+++ FMTD LL+ M D T S II+DEAHER+I
Sbjct: 466 LGD--HGHKVGYQIRFDSTAKEDTKVKFMTDGVLLREMMHDFKLTKYSSIIIDEAHERNI 523
Query: 116 NTDLLLALV 124
NTD+L+ ++
Sbjct: 524 NTDILIGML 532
>DHX8_ARATH (Q38953) Putative pre-mRNA splicing factor ATP-dependent
RNA helicase
Length = 1168
Score = 103 bits (256), Expect = 3e-21
Identities = 56/121 (46%), Positives = 79/121 (65%), Gaps = 1/121 (0%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
Q+ V+IGETGSGK+TQ+ Q+LA++G I CTQPRR+AA S+A+RV EE GC
Sbjct: 532 QVLVVIGETGSGKTTQVTQYLAEAGYTTKGKIGCTQPRRVAAMSVAKRVAEEF-GCRLGE 590
Query: 64 SIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLAL 123
+ F D+ I +MTD LL+ + D+N + S I++DEAHER+I+TD+L L
Sbjct: 591 EVGYAIRFEDCTGPDTVIKYMTDGMLLREILIDENLSQYSVIMLDEAHERTIHTDVLFGL 650
Query: 124 V 124
+
Sbjct: 651 L 651
>PR16_SCHPO (Q9P774) Pre-mRNA splicing factor RNA helicase prp16 (EC
3.6.1.-)
Length = 1173
Score = 102 bits (255), Expect = 3e-21
Identities = 57/121 (47%), Positives = 77/121 (63%), Gaps = 1/121 (0%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
Q+ +++GETGSGK+TQL QFL + G N I CTQPRR+AA S+A+RV EE G S
Sbjct: 509 QVLIVVGETGSGKTTQLAQFLYEDGYHRNGMIGCTQPRRVAAMSVAKRVSEEM-GVRLGS 567
Query: 64 SIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLAL 123
++ F D+ I +MTD LL+ + N S II+DEAHERS+NTD+L+ L
Sbjct: 568 TVGYSIRFEDVTGPDTVIKYMTDGVLLRESLMQNNLEKYSVIIMDEAHERSLNTDILMGL 627
Query: 124 V 124
+
Sbjct: 628 L 628
>ARI2_DROME (O76924) Ariadne-2 protein (Ari-2)
Length = 509
Score = 101 bits (251), Expect = 1e-20
Identities = 60/203 (29%), Positives = 93/203 (45%), Gaps = 8/203 (3%)
Query: 648 CPICLC-EVEDGYQLEGCGHLFCQSCMVEQCESAIKNQGSFPIRCAHQGCGNHILLVDFR 706
CP+C ++ D + CGH FC+ C E+ I S I C Q C +
Sbjct: 153 CPVCASSQLGDKFYSLACGHSFCKDCWTIYFETQIFQGISTQIGCMAQMCNVRVPEDLVL 212
Query: 707 TLLSNDKLEELFRASLGAFVASSSGTYRFCPSPDCPSIYRVADPDTASAPFVCGACYSET 766
TL++ + + ++ S RFCP P+C I + ++ A +C AC++
Sbjct: 213 TLVTRPVMRDKYQQFAFKDYVKSHPELRFCPGPNCQIIVQSSEISAKRA--ICKACHTGF 270
Query: 767 CTRCHIEYHPYVSCERYRQF--KDDPDSSLRDWCKGKEQVKNCPACGHVIEKVDGCNHIE 824
C RC ++YH C+ +++ K DS ++ K+CP C IEK GCNH++
Sbjct: 271 CFRCGMDYHAPTDCQVIKKWLTKCADDSETANYISA--HTKDCPKCHICIEKNGGCNHMQ 328
Query: 825 C-KCGKHICWVCLEFFTTSGECY 846
C C CW+CL + T G Y
Sbjct: 329 CFNCKHDFCWMCLGDWKTHGSEY 351
>DHX8_HUMAN (Q14562) ATP-dependent helicase DHX8 (RNA helicase HRH1)
(DEAH-box protein 8)
Length = 1220
Score = 100 bits (250), Expect = 1e-20
Identities = 54/121 (44%), Positives = 79/121 (64%), Gaps = 1/121 (0%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
QI ++IGETGSGK+TQ+ Q+LA++G + I CTQPRR+AA S+A+RV EE G C
Sbjct: 582 QILIVIGETGSGKTTQITQYLAEAGYTSRGKIGCTQPRRVAAMSVAKRVSEEFGCCL-GQ 640
Query: 64 SIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLAL 123
+ F ++ I +MTD LL+ + D + T + I++DEAHER+I+TD+L L
Sbjct: 641 EVGYTIRFEDCTSPETVIKYMTDGMLLRECLIDPDLTQYAIIMLDEAHERTIHTDVLFGL 700
Query: 124 V 124
+
Sbjct: 701 L 701
>YK16_SCHPO (Q9HDY4) Putative ATP-dependent RNA helicase PB1A10.06c
(EC 3.6.1.-)
Length = 1183
Score = 100 bits (249), Expect = 2e-20
Identities = 60/125 (48%), Positives = 77/125 (61%), Gaps = 7/125 (5%)
Query: 5 ITVLIGETGSGKSTQLVQFLADSGVGANES-----IVCTQPRRIAAKSLAERVREESGGC 59
+ ++ G TGSGK+TQL QFL ++G + ES I TQPRR+AA S+A+RV EE G
Sbjct: 416 VVIICGATGSGKTTQLPQFLFEAGFSSPESENPGMIAITQPRRVAAVSIAKRVSEELTGF 475
Query: 60 YEDSSIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDL 119
S + F S D+ I FMTD LL+ SD T S +IVDEAHERS+NTD+
Sbjct: 476 --SSKVSYQIRFDSTINPDTAIKFMTDGILLRELSSDFLLTAYSAVIVDEAHERSVNTDI 533
Query: 120 LLALV 124
LL L+
Sbjct: 534 LLGLL 538
>ARI1_MOUSE (Q9Z1K5) Ariadne-1 protein homolog (ARI-1)
(Ubiquitin-conjugating enzyme E2-binding protein 1)
(UbcH7-binding protein) (UbcM4-interacting protein 77)
Length = 555
Score = 98.2 bits (243), Expect = 8e-20
Identities = 76/257 (29%), Positives = 111/257 (42%), Gaps = 36/257 (14%)
Query: 607 TRNRTILCHGNSELKSRVE-------EITFEIARLSNPSSE----RFDTGPS-----CPI 650
T R +L H N + + +E E F + NPS + + +T S C I
Sbjct: 127 TITRILLSHFNWDKEKLMERYFDGNLEKLFAECHVINPSKKSRTRQMNTRSSAQDMPCQI 186
Query: 651 CLCEVEDGY--QLEGCGHLFCQSCMVEQCESAIKNQG-SFPIRCAHQGCGNHILLVDFRT 707
C + Y LE CGH FC C E + I +G I C GC +LVD T
Sbjct: 187 CYLNYPNSYFTGLE-CGHKFCMQCWSEYLTTKIMEEGMGQTISCPAHGCD---ILVDDNT 242
Query: 708 ---LLSNDKLEELFRASLGAFVASSSGTYRFCPSPDCPSIYRVADPDTASAPFVCGACYS 764
L+++ K++ ++ + + ++CP+PDC + +V PD CG +
Sbjct: 243 VMRLITDSKVKLKYQHLITNSFVECNRLLKWCPAPDCHHVVKVQYPDAKPVRCKCGRQF- 301
Query: 765 ETCTRCHIEYHPYVSCERYRQF--KDDPDSSLRDWCKGKEQVKNCPACGHVIEKVDGCNH 822
C C +H V C+ +++ K D DS +W K CP C IEK GCNH
Sbjct: 302 --CFNCGENWHDPVKCKWLKKWIKKCDDDSETSNWIAAN--TKECPKCHVTIEKDGGCNH 357
Query: 823 IECK---CGKHICWVCL 836
+ C+ C CWVCL
Sbjct: 358 MVCRNQNCKAEFCWVCL 374
>ARI1_HUMAN (Q9Y4X5) Ariadne-1 protein homolog (ARI-1)
(Ubiquitin-conjugating enzyme E2-binding protein 1)
(UbcH7-binding protein) (UbcM4-interacting protein)
(HHARI) (H7-AP2) (HUSSY-27) (MOP-6)
Length = 557
Score = 98.2 bits (243), Expect = 8e-20
Identities = 76/257 (29%), Positives = 111/257 (42%), Gaps = 36/257 (14%)
Query: 607 TRNRTILCHGNSELKSRVE-------EITFEIARLSNPSSE----RFDTGPS-----CPI 650
T R +L H N + + +E E F + NPS + + +T S C I
Sbjct: 129 TITRILLSHFNWDKEKLMERYFDGNLEKLFAECHVINPSKKSRTRQMNTRSSAQDMPCQI 188
Query: 651 CLCEVEDGY--QLEGCGHLFCQSCMVEQCESAIKNQG-SFPIRCAHQGCGNHILLVDFRT 707
C + Y LE CGH FC C E + I +G I C GC +LVD T
Sbjct: 189 CYLNYPNSYFTGLE-CGHKFCMQCWSEYLTTKIMEEGMGQTISCPAHGCD---ILVDDNT 244
Query: 708 ---LLSNDKLEELFRASLGAFVASSSGTYRFCPSPDCPSIYRVADPDTASAPFVCGACYS 764
L+++ K++ ++ + + ++CP+PDC + +V PD CG +
Sbjct: 245 VMRLITDSKVKLKYQHLITNSFVECNRLLKWCPAPDCHHVVKVQYPDAKPVRCKCGRQF- 303
Query: 765 ETCTRCHIEYHPYVSCERYRQF--KDDPDSSLRDWCKGKEQVKNCPACGHVIEKVDGCNH 822
C C +H V C+ +++ K D DS +W K CP C IEK GCNH
Sbjct: 304 --CFNCGENWHDPVKCKWLKKWIKKCDDDSETSNWIAAN--TKECPKCHVTIEKDGGCNH 359
Query: 823 IECK---CGKHICWVCL 836
+ C+ C CWVCL
Sbjct: 360 MVCRNQNCKAEFCWVCL 376
>PR16_HUMAN (Q92620) Pre-mRNA splicing factor ATP-dependent RNA
helicase PRP16 (EC 3.6.1.-) (ATP-dependent RNA helicase
DHX38) (DEAH-box protein 38)
Length = 1227
Score = 96.3 bits (238), Expect = 3e-19
Identities = 53/120 (44%), Positives = 75/120 (62%), Gaps = 1/120 (0%)
Query: 5 ITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDSS 64
I +++GETGSGK+TQL Q+L + G I CTQPRR+AA S+A+RV EE GG +
Sbjct: 550 IVIVVGETGSGKTTQLTQYLHEDGYTDYGMIGCTQPRRVAAMSVAKRVSEEMGGNLGE-E 608
Query: 65 IKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLALV 124
+ F ++ I +MTD LL+ + + + S II+DEAHERS+NTD+L L+
Sbjct: 609 VGYAIRFEDCTSENTLIKYMTDGILLRESLREADLDHYSAIIMDEAHERSLNTDVLFGLL 668
>ARI2_CAEEL (Q22431) Probable ariadne-2 protein (Ari-2)
Length = 482
Score = 96.3 bits (238), Expect = 3e-19
Identities = 56/203 (27%), Positives = 87/203 (42%), Gaps = 8/203 (3%)
Query: 648 CPICLCEVEDGYQLEGCGHLFCQSCMVEQCESAIKNQGSFPIRCAHQGCGNHILLVDFRT 707
C +C + CGH FC+ C ES + + I C C + +
Sbjct: 129 CSVCAMDGYTELPHLTCGHCFCEHCWKSHVESRLSEGVASRIECMESECEVYAPSEFVLS 188
Query: 708 LLSNDKLEEL-FRASLGAFVASSSGTYRFCPSPDCPSIYRVADPDTASAPFVCGACYSET 766
++ N + +L + L + +S +FC +CP I R + C C++
Sbjct: 189 IIKNSPVIKLKYERFLLRDMVNSHPHLKFCVGNECPVIIR--STEVKPKRVTCMQCHTSF 246
Query: 767 CTRCHIEYHPYVSCERYRQF--KDDPDSSLRDWCKGKEQVKNCPACGHVIEKVDGCNHIE 824
C +C +YH SCE +Q+ K DS ++ K+CP C IEK GCNHI+
Sbjct: 247 CVKCGADYHAPTSCETIKQWMTKCADDSETANYISA--HTKDCPQCHSCIEKAGGCNHIQ 304
Query: 825 C-KCGKHICWVCLEFFTTSGECY 846
C +C H CW+C + + G Y
Sbjct: 305 CTRCRHHFCWMCFGDWKSHGSEY 327
>MOG1_CAEEL (P34498) Probable pre-mRNA splicing factor ATP-dependent
RNA helicase mog-1 (EC 3.6.1.-) (Sex determination
protein mog-1) (Masculinization of germ line protein 1)
Length = 1131
Score = 95.5 bits (236), Expect = 5e-19
Identities = 52/120 (43%), Positives = 73/120 (60%), Gaps = 1/120 (0%)
Query: 5 ITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDSS 64
+ +++GETGSGK+TQL Q+L + G G + I CTQPRR+AA S+A RV +E G
Sbjct: 459 VVIIVGETGSGKTTQLAQYLLEDGFGDSGLIGCTQPRRVAAMSVARRVADEM-GVDLGQD 517
Query: 65 IKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLALV 124
+ F + I +MTD LL+ + D + S II+DEAHERS+NTD+L L+
Sbjct: 518 VGYAIRFEDCTSEKTIIKYMTDGILLRECLGDGSLDQYSAIIMDEAHERSLNTDVLFGLL 577
>PR22_YEAST (P24384) Pre-mRNA splicing factor RNA helicase PRP22
Length = 1145
Score = 95.1 bits (235), Expect = 7e-19
Identities = 53/121 (43%), Positives = 74/121 (60%), Gaps = 1/121 (0%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
Q V++GETGSGK+TQ+ Q+L + G I CTQPRR+AA S+A+RV EE GC
Sbjct: 500 QFLVIVGETGSGKTTQITQYLDEEGFSNYGMIGCTQPRRVAAVSVAKRVAEEV-GCKVGH 558
Query: 64 SIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLAL 123
+ F D+RI +MTD L + + D + S I++DEAHER++ TD+L AL
Sbjct: 559 DVGYTIRFEDVTGPDTRIKYMTDGMLQREALLDPEMSKYSVIMLDEAHERTVATDVLFAL 618
Query: 124 V 124
+
Sbjct: 619 L 619
>DD35_CAEEL (Q9BKQ8) Probable ATP-dependent helicase DHX35 homolog
Length = 732
Score = 94.4 bits (233), Expect = 1e-18
Identities = 53/119 (44%), Positives = 75/119 (62%), Gaps = 2/119 (1%)
Query: 7 VLIGETGSGKSTQLVQFLADSGVGAN-ESIVCTQPRRIAAKSLAERVREESGGCYEDSSI 65
+++GETG GKSTQ+ QFL ++G A+ IV TQPRR+A +LA RV EE C +
Sbjct: 97 IIVGETGCGKSTQVPQFLLEAGWAADGRQIVITQPRRVAVVTLATRVAEEKD-CILGHDV 155
Query: 66 KCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLALV 124
F + D+++ FMTD LL+ ++D + S I++DEAHERS NTD+LL L+
Sbjct: 156 GYTVRFDDVSDKDTKVKFMTDGLLLREILADPLLSKYSIIMIDEAHERSCNTDILLGLL 214
>DHX8_SCHPO (O42643) Putative pre-mRNA splicing factor ATP-dependent
RNA helicase C10F6.02c
Length = 1168
Score = 94.0 bits (232), Expect = 2e-18
Identities = 53/122 (43%), Positives = 77/122 (62%), Gaps = 2/122 (1%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIV-CTQPRRIAAKSLAERVREESGGCYED 62
QI VL+GETGSGK+TQ+ Q+LA+ G ++ ++ CTQPRR+AA S+A+RV EE GC
Sbjct: 527 QILVLLGETGSGKTTQITQYLAEEGYTSDSKMIGCTQPRRVAAMSVAKRVAEEV-GCRVG 585
Query: 63 SSIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLA 122
+ F ++I +MTD L + + D + S II+DEAHER++ TD+L
Sbjct: 586 EEVGYTIRFEDKTSRMTQIKYMTDGMLQRECLVDPLLSKYSVIILDEAHERTVATDVLFG 645
Query: 123 LV 124
L+
Sbjct: 646 LL 647
>MOG4_CAEEL (O45244) Probable pre-mRNA splicing factor ATP-dependent
RNA helicase mog-4 (Sex determination protein mog-4)
(Masculinization of germ line protein 4)
Length = 1008
Score = 90.1 bits (222), Expect = 2e-17
Identities = 48/122 (39%), Positives = 77/122 (62%), Gaps = 2/122 (1%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGV-GANESIVCTQPRRIAAKSLAERVREESGGCYED 62
Q+ ++ GETGSGK+TQL Q+L ++G + I CTQPRR+AA S+A RV +E G C
Sbjct: 381 QVLIIEGETGSGKTTQLPQYLYEAGFCEGGKRIGCTQPRRVAAMSVAARVADEVG-CKLG 439
Query: 63 SSIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLA 122
+ + F + + +MTD LL+ ++++ + S +++DEAHER+++TD+L
Sbjct: 440 TQVGYSIRFEDCTSEKTVLKYMTDGMLLREFLNEPDLASYSVMMIDEAHERTLHTDILFG 499
Query: 123 LV 124
LV
Sbjct: 500 LV 501
>CC28_SCHPO (Q10752) Putative ATP-dependent RNA helicase cdc28
Length = 1055
Score = 90.1 bits (222), Expect = 2e-17
Identities = 52/125 (41%), Positives = 79/125 (62%), Gaps = 8/125 (6%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSG-VGANESIVCTQPRRIAAKSLAERVREESG---GC 59
Q+ +++ ETGSGK+TQL QFL ++G N+ I CTQPRR+AA S+A RV +E G
Sbjct: 435 QVLLIVAETGSGKTTQLPQFLHEAGYTKGNKKICCTQPRRVAAMSVAARVAKEMDVRLGQ 494
Query: 60 YEDSSIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDL 119
SI+ F + + I ++TD LL+ ++++ + S II+DEAHER+++TD+
Sbjct: 495 EVGYSIR----FENATSEKTVIKYLTDGMLLREFLTEPDLASYSVIIIDEAHERTLHTDI 550
Query: 120 LLALV 124
L LV
Sbjct: 551 LFGLV 555
>PR43_YEAST (P53131) Pre-mRNA splicing factor RNA helicase PRP43
(Helicase JA1)
Length = 767
Score = 89.7 bits (221), Expect = 3e-17
Identities = 54/125 (43%), Positives = 77/125 (61%), Gaps = 7/125 (5%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVG--ANESIVCTQPRRIAAKSLAERVREESGGCYE 61
QI V +GETGSGK+TQ+ QF+ + N + CTQPRR+AA S+A+RV EE
Sbjct: 110 QIMVFVGETGSGKTTQIPQFVLFDEMPHLENTQVACTQPRRVAAMSVAQRVAEEMDVKLG 169
Query: 62 DSSIKCYSSFSSWNKFDSRII--FMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDL 119
+ + S NK ++ I +MTD LL+ M D + + SCII+DEAHER++ TD+
Sbjct: 170 E---EVGYSIRFENKTSNKTILKYMTDGMLLREAMEDHDLSRYSCIILDEAHERTLATDI 226
Query: 120 LLALV 124
L+ L+
Sbjct: 227 LMGLL 231
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 100,206,834
Number of Sequences: 164201
Number of extensions: 4288366
Number of successful extensions: 12051
Number of sequences better than 10.0: 161
Number of HSP's better than 10.0 without gapping: 61
Number of HSP's successfully gapped in prelim test: 100
Number of HSP's that attempted gapping in prelim test: 11799
Number of HSP's gapped (non-prelim): 214
length of query: 857
length of database: 59,974,054
effective HSP length: 119
effective length of query: 738
effective length of database: 40,434,135
effective search space: 29840391630
effective search space used: 29840391630
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 70 (31.6 bits)
Medicago: description of AC141414.11


