

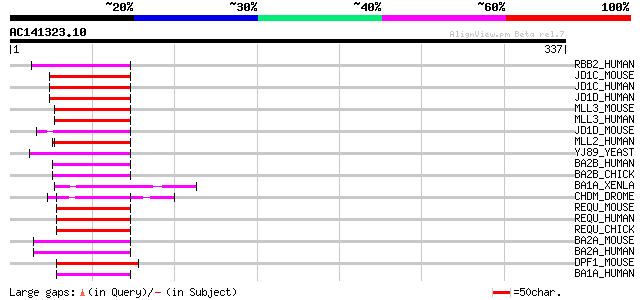
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141323.10 + phase: 0 /pseudo
(337 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RBB2_HUMAN (P29375) Retinoblastoma-binding protein 2 (RBBP-2) 70 6e-12
JD1C_MOUSE (P41230) Jumonji/ARID domain-containing protein 1C (S... 70 7e-12
JD1C_HUMAN (P41229) Jumonji/ARID domain-containing protein 1C (S... 70 7e-12
JD1D_HUMAN (Q9BY66) Jumonji/ARID domain-containing protein 1D (S... 70 1e-11
MLL3_MOUSE (Q8BRH4) Myeloid/lymphoid or mixed-lineage leukemia p... 65 2e-10
MLL3_HUMAN (Q8NEZ4) Myeloid/lymphoid or mixed-lineage leukemia p... 65 2e-10
JD1D_MOUSE (Q62240) Jumonji/ARID domain-containing protein 1D (S... 64 7e-10
MLL2_HUMAN (O14686) Myeloid/lymphoid or mixed-lineage leukemia p... 62 3e-09
YJ89_YEAST (P47156) Hypothetical 85.0 kDa protein in STE24-ATP2 ... 61 5e-09
BA2B_HUMAN (Q9UIF8) Bromodomain adjacent to zinc finger domain 2... 60 8e-09
BA2B_CHICK (Q9DE13) Bromodomain adjacent to zinc finger domain 2... 60 8e-09
BA1A_XENLA (Q8UVR5) ATP-utilizing chromatin assembly and remodel... 60 1e-08
CHDM_DROME (O97159) Chromodomain helicase-DNA-binding protein Mi... 59 1e-08
REQU_MOUSE (Q61103) Zinc-finger protein ubi-d4 (Requiem) (Apopto... 59 2e-08
REQU_HUMAN (Q92785) Zinc-finger protein ubi-d4 (Requiem) (Apopto... 58 3e-08
REQU_CHICK (P58268) Zinc-finger protein ubi-d4 (Requiem) (Apopto... 58 4e-08
BA2A_MOUSE (Q91YE5) Bromodomain adjacent to zinc finger domain 2... 57 5e-08
BA2A_HUMAN (Q9UIF9) Bromodomain adjacent to zinc finger domain 2... 57 5e-08
DPF1_MOUSE (Q9QX66) Zinc-finger protein neuro-d4 (D4, zinc and d... 57 7e-08
BA1A_HUMAN (Q9NRL2) Bromodomain adjacent to zinc finger domain p... 57 7e-08
>RBB2_HUMAN (P29375) Retinoblastoma-binding protein 2 (RBBP-2)
Length = 1722
Score = 70.5 bits (171), Expect = 6e-12
Identities = 30/60 (50%), Positives = 35/60 (58%)
Query: 14 KKQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
+++ T N D VC C G + KLLLCD CD+ YH FCL P LP VPK W CP C
Sbjct: 281 QRKGTLSVNFVDLYVCMFCGRGNNEDKLLLCDGCDDSYHTFCLIPPLPDVPKGDWRCPKC 340
>JD1C_MOUSE (P41230) Jumonji/ARID domain-containing protein 1C (SmcX
protein) (Xe169 protein)
Length = 1554
Score = 70.1 bits (170), Expect = 7e-12
Identities = 26/49 (53%), Positives = 32/49 (65%)
Query: 25 DDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
+ VC+ C+ G KLLLCD CD+ YH+FCL P LP +PK W CP C
Sbjct: 323 ESYVCRMCSRGDEDDKLLLCDGCDDNYHIFCLLPPLPEIPKGVWRCPKC 371
>JD1C_HUMAN (P41229) Jumonji/ARID domain-containing protein 1C (SmcX
protein) (Xe169 protein)
Length = 1560
Score = 70.1 bits (170), Expect = 7e-12
Identities = 26/49 (53%), Positives = 32/49 (65%)
Query: 25 DDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
+ VC+ C+ G KLLLCD CD+ YH+FCL P LP +PK W CP C
Sbjct: 323 ESYVCRMCSRGDEDDKLLLCDGCDDNYHIFCLLPPLPEIPKGVWRCPKC 371
>JD1D_HUMAN (Q9BY66) Jumonji/ARID domain-containing protein 1D (SmcY
protein) (Histocompatibility Y antigen) (H-Y)
Length = 1539
Score = 69.7 bits (169), Expect = 1e-11
Identities = 25/49 (51%), Positives = 31/49 (63%)
Query: 25 DDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
D +CQ C+ G KLL CD CD+ YH+FCL P LP +P+ W CP C
Sbjct: 313 DSYICQVCSRGDEDDKLLFCDGCDDNYHIFCLLPPLPEIPRGIWRCPKC 361
>MLL3_MOUSE (Q8BRH4) Myeloid/lymphoid or mixed-lineage leukemia
protein 3 homolog (Histone-lysine N-methyltransferase,
H3 lysine-4 specific MLL3) (EC 2.1.1.43)
Length = 4903
Score = 65.1 bits (157), Expect = 2e-10
Identities = 23/46 (50%), Positives = 30/46 (65%)
Query: 28 VCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
VCQ C +K+L+CD CD GYH FCL P++ SVP + W C +C
Sbjct: 389 VCQNCKQSGEDSKMLVCDTCDKGYHTFCLQPVMKSVPTNGWKCKNC 434
Score = 60.1 bits (144), Expect = 8e-09
Identities = 23/46 (50%), Positives = 28/46 (60%)
Query: 28 VCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
VC+ C P +LLLCD+CD YH +CL P L +VPK W C C
Sbjct: 1002 VCEACGKATDPGRLLLCDDCDISYHTYCLDPPLQTVPKGGWKCKWC 1047
Score = 36.2 bits (82), Expect = 0.12
Identities = 14/49 (28%), Positives = 20/49 (40%)
Query: 25 DDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
+D C C+S C C YH CL + + ++ W CP C
Sbjct: 339 EDANCAVCDSPGDLLDQFFCTTCGQHYHGMCLDIAVTPLKRAGWQCPEC 387
Score = 30.8 bits (68), Expect = 5.0
Identities = 31/117 (26%), Positives = 53/117 (44%), Gaps = 14/117 (11%)
Query: 213 VEGFTIEADKSIKDLTIITEYVGDV--DFLKNREHDDGDSIMTLLSASNPSQSLVICPDK 270
++G + A + I+ T++ EY+G + + + NR+ +S + VI
Sbjct: 4772 IQGLGLYAARDIEKHTMVIEYIGTIIRNEVANRKEKLYESQNRGVYMFRMDNDHVIDATL 4831
Query: 271 RSNIARFINGINNHTPEGKKKQNLKCVRYNVDGEC--RVLLIANRDIAKGERLYYDY 325
AR+IN ++ P CV V E ++++ +NR I KGE L YDY
Sbjct: 4832 TGGPARYIN--HSCAPN--------CVAEVVTFERGHKIIISSNRRIQKGEELCYDY 4878
Score = 30.0 bits (66), Expect = 8.6
Identities = 16/49 (32%), Positives = 22/49 (44%), Gaps = 2/49 (4%)
Query: 27 VVCQKCNSGKSPTKLLLCDNCDNGYHLFCLT-PILPSVPKSSWFCPSCS 74
VVC G + +LL C C YH +C++ I V W C C+
Sbjct: 954 VVCGSFGQG-AEGRLLACSQCGQCYHPYCVSIKITKVVLSKGWRCLECT 1001
>MLL3_HUMAN (Q8NEZ4) Myeloid/lymphoid or mixed-lineage leukemia
protein 3 homolog (Histone-lysine N-methyltransferase,
H3 lysine-4 specific MLL3) (EC 2.1.1.43) (Homologous to
ALR protein)
Length = 4911
Score = 65.1 bits (157), Expect = 2e-10
Identities = 23/46 (50%), Positives = 30/46 (65%)
Query: 28 VCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
VCQ C +K+L+CD CD GYH FCL P++ SVP + W C +C
Sbjct: 390 VCQNCKQSGEDSKMLVCDTCDKGYHTFCLQPVMKSVPTNGWKCKNC 435
Score = 60.1 bits (144), Expect = 8e-09
Identities = 23/46 (50%), Positives = 28/46 (60%)
Query: 28 VCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
VC+ C P +LLLCD+CD YH +CL P L +VPK W C C
Sbjct: 1009 VCEACGKATDPGRLLLCDDCDISYHTYCLDPPLQTVPKGGWKCKWC 1054
Score = 36.2 bits (82), Expect = 0.12
Identities = 14/49 (28%), Positives = 20/49 (40%)
Query: 25 DDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
+D C C+S C C YH CL + + ++ W CP C
Sbjct: 340 EDANCAVCDSPGDLLDQFFCTTCGQHYHGMCLDIAVTPLKRAGWQCPEC 388
Score = 32.3 bits (72), Expect = 1.7
Identities = 18/54 (33%), Positives = 24/54 (44%), Gaps = 2/54 (3%)
Query: 22 NNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLT-PILPSVPKSSWFCPSCS 74
N D VVC G + +LL C C YH +C++ I V W C C+
Sbjct: 956 NQDMCVVCGSFGQG-AEGRLLACSQCGQCYHPYCVSIKITKVVLSKGWRCLECT 1008
>JD1D_MOUSE (Q62240) Jumonji/ARID domain-containing protein 1D (SmcY
protein) (Histocompatibility Y antigen) (H-Y)
Length = 1548
Score = 63.5 bits (153), Expect = 7e-10
Identities = 28/57 (49%), Positives = 33/57 (57%), Gaps = 3/57 (5%)
Query: 17 STTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
ST F N+ VC+ C+ G K LLCD C + YH+FCL P L VPK W CP C
Sbjct: 317 STQFMNS---YVCRICSRGDEVDKFLLCDGCSDNYHIFCLLPPLSEVPKGVWRCPKC 370
>MLL2_HUMAN (O14686) Myeloid/lymphoid or mixed-lineage leukemia
protein 2 (ALL1-related protein)
Length = 5262
Score = 61.6 bits (148), Expect = 3e-09
Identities = 23/47 (48%), Positives = 30/47 (62%)
Query: 27 VVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
+VC+ C P++LLLCD+CD YH +CL P L +VPK W C C
Sbjct: 1153 IVCEVCGQASDPSRLLLCDDCDISYHTYCLDPPLLTVPKGGWKCKWC 1199
Score = 58.5 bits (140), Expect = 2e-08
Identities = 21/46 (45%), Positives = 29/46 (62%)
Query: 28 VCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
VCQ C + +K+L+C+ CD GYH FCL P + +P SW C +C
Sbjct: 275 VCQACRKPGNDSKMLVCETCDKGYHTFCLKPPMEELPAHSWKCKAC 320
Score = 36.2 bits (82), Expect = 0.12
Identities = 14/49 (28%), Positives = 21/49 (42%)
Query: 25 DDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
++ C C L C +C + YH CL L + ++ W CP C
Sbjct: 225 EEARCAVCEGPGELCDLFFCTSCGHHYHGACLDTALTARKRAGWQCPEC 273
Score = 33.9 bits (76), Expect = 0.60
Identities = 35/132 (26%), Positives = 62/132 (46%), Gaps = 24/132 (18%)
Query: 213 VEGFTIEADKSIKDLTIITEYVGDV---DFLKNRE--HDDGDSIMTLLSASNPSQSLVIC 267
++G + A K ++ T++ EY+G + + RE +++ + + + +N VI
Sbjct: 5131 IQGLGLYAAKDLEKHTMVIEYIGTIIRNEVANRREKIYEEQNRGIYMFRINNEH---VID 5187
Query: 268 PDKRSNIARFINGINNHTPEGKKKQNLKCVRYNV--DGECRVLLIANRDIAKGERLYYDY 325
AR+IN ++ P CV V D E ++++I++R I KGE L YDY
Sbjct: 5188 ATLTGGPARYIN--HSCAPN--------CVAEVVTFDKEDKIIIISSRRIPKGEELTYDY 5237
Query: 326 ----NGLEHEYP 333
+HE P
Sbjct: 5238 QFDFEDDQHEIP 5249
>YJ89_YEAST (P47156) Hypothetical 85.0 kDa protein in STE24-ATP2
intergenic region
Length = 728
Score = 60.8 bits (146), Expect = 5e-09
Identities = 23/61 (37%), Positives = 32/61 (51%)
Query: 13 PKKQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPS 72
P S + DDD C C P + +LCD+CD +H++CL+P L VP W C +
Sbjct: 222 PDSNSGSDFEEDDDDACIVCRKTNDPKRTILCDSCDKPFHIYCLSPPLERVPSGDWICNT 281
Query: 73 C 73
C
Sbjct: 282 C 282
>BA2B_HUMAN (Q9UIF8) Bromodomain adjacent to zinc finger domain 2B
(hWALp4)
Length = 1972
Score = 60.1 bits (144), Expect = 8e-09
Identities = 22/47 (46%), Positives = 28/47 (58%)
Query: 27 VVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
V CQ C G + LLLCD CD G H +C P + ++P WFCP+C
Sbjct: 1736 VYCQICRKGDNEELLLLCDGCDKGCHTYCHRPKITTIPDGDWFCPAC 1782
>BA2B_CHICK (Q9DE13) Bromodomain adjacent to zinc finger domain 2B
(Extracellular matrix protein F22)
Length = 2130
Score = 60.1 bits (144), Expect = 8e-09
Identities = 22/47 (46%), Positives = 28/47 (58%)
Query: 27 VVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
V CQ C G + LLLCD CD G H +C P + ++P WFCP+C
Sbjct: 1896 VYCQICRKGDNEELLLLCDGCDKGCHTYCHRPKITTIPDGDWFCPAC 1942
>BA1A_XENLA (Q8UVR5) ATP-utilizing chromatin assembly and remodely
factor 1 (xACF1) (Fragment)
Length = 627
Score = 59.7 bits (143), Expect = 1e-08
Identities = 27/86 (31%), Positives = 45/86 (51%), Gaps = 8/86 (9%)
Query: 28 VCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSCSHNPKIPKFPLVQT 87
VC+K G+S ++LCD CD G+H++C+ P L VP+ WFCP C + + P
Sbjct: 227 VCRKKGDGES---MVLCDGCDRGHHIYCVRPKLKYVPEGDWFCPECHPKQRSHRLPSRHR 283
Query: 88 KIIDFFKIQRTSDASQILNHEEEKAE 113
+ + + + L+ +EE+ E
Sbjct: 284 -----YSMDSDEEEEEELDQKEEEEE 304
>CHDM_DROME (O97159) Chromodomain helicase-DNA-binding protein Mi-2
homolog (dMi-2)
Length = 1982
Score = 59.3 bits (142), Expect = 1e-08
Identities = 31/77 (40%), Positives = 40/77 (51%), Gaps = 4/77 (5%)
Query: 24 DDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSCSHNPKIPKFP 83
DDD + C K +LL CD+C + YH FCL P L ++P W CP CS P K
Sbjct: 432 DDDEHQEFCRVCKDGGELLCCDSCPSAYHTFCLNPPLDTIPDGDWRCPRCSCPPLTGK-- 489
Query: 84 LVQTKIIDFFKIQRTSD 100
KII + QR++D
Sbjct: 490 --AEKIITWRWAQRSND 504
Score = 49.3 bits (116), Expect = 1e-05
Identities = 19/45 (42%), Positives = 24/45 (53%), Gaps = 3/45 (6%)
Query: 29 CQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
C+ C G +++LCD C YHL CL P L P+ W CP C
Sbjct: 380 CEVCQQGG---EIILCDTCPRAYHLVCLEPELDEPPEGKWSCPHC 421
>REQU_MOUSE (Q61103) Zinc-finger protein ubi-d4 (Requiem) (Apoptosis
response zinc finger protein) (D4, zinc and double PHD
fingers family 2)
Length = 391
Score = 58.5 bits (140), Expect = 2e-08
Identities = 20/45 (44%), Positives = 29/45 (64%)
Query: 29 CQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
C C + ++ +LL CD+CD GYH++CLTP + P+ SW C C
Sbjct: 330 CNLCGTSENDDQLLFCDDCDRGYHMYCLTPSMSEPPEGSWSCHLC 374
>REQU_HUMAN (Q92785) Zinc-finger protein ubi-d4 (Requiem) (Apoptosis
response zinc finger protein) (D4, zinc and double PHD
fingers family 2)
Length = 391
Score = 58.2 bits (139), Expect = 3e-08
Identities = 20/45 (44%), Positives = 29/45 (64%)
Query: 29 CQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
C C + ++ +LL CD+CD GYH++CLTP + P+ SW C C
Sbjct: 330 CNICGTSENDDQLLFCDDCDRGYHMYCLTPSMSEPPEGSWSCHLC 374
>REQU_CHICK (P58268) Zinc-finger protein ubi-d4 (Requiem) (Apoptosis
response zinc finger protein)
Length = 405
Score = 57.8 bits (138), Expect = 4e-08
Identities = 20/45 (44%), Positives = 29/45 (64%)
Query: 29 CQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
C C + ++ +LL CD+CD GYH++CLTP + P+ SW C C
Sbjct: 344 CNICGTSENDDQLLFCDDCDRGYHMYCLTPPMSEPPEGSWSCHLC 388
>BA2A_MOUSE (Q91YE5) Bromodomain adjacent to zinc finger domain 2A
(Transcription termination factor-I interacting protein
5) (TTF-I interacting protein 5) (Tip5)
Length = 1850
Score = 57.4 bits (137), Expect = 5e-08
Identities = 22/59 (37%), Positives = 33/59 (55%)
Query: 15 KQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
++S + + + V C C G + LLLCD CD G H++C P + +VP+ WFC C
Sbjct: 1612 ERSIAWEKSVNKVTCLVCRKGDNDEFLLLCDGCDRGCHIYCHRPKMEAVPEGDWFCAVC 1670
>BA2A_HUMAN (Q9UIF9) Bromodomain adjacent to zinc finger domain 2A
(Transcription termination factor-I interacting protein
5) (TTF-I interacting protein 5) (Tip5) (hWALp3)
Length = 1878
Score = 57.4 bits (137), Expect = 5e-08
Identities = 22/59 (37%), Positives = 33/59 (55%)
Query: 15 KQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
++S + + + V C C G + LLLCD CD G H++C P + +VP+ WFC C
Sbjct: 1638 ERSIAWEKSVNKVTCLVCRKGDNDEFLLLCDGCDRGCHIYCHRPKMEAVPEGDWFCTVC 1696
>DPF1_MOUSE (Q9QX66) Zinc-finger protein neuro-d4 (D4, zinc and
double PHD fingers family 1)
Length = 387
Score = 57.0 bits (136), Expect = 7e-08
Identities = 20/50 (40%), Positives = 31/50 (62%)
Query: 29 CQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSCSHNPK 78
C C + ++ +LL CD+CD GYH++CL+P + P+ SW C C + K
Sbjct: 328 CSLCGTSENDDQLLFCDDCDRGYHMYCLSPPMAEPPEGSWSCHLCLRHLK 377
>BA1A_HUMAN (Q9NRL2) Bromodomain adjacent to zinc finger domain
protein 1A (ATP-utilizing chromatin assembly and
remodeling factor 1) (hACF1) (ATP-dependent chromatin
remodelling protein) (Williams syndrome transcription
factor-related chromatin remodelin
Length = 1556
Score = 57.0 bits (136), Expect = 7e-08
Identities = 19/45 (42%), Positives = 27/45 (59%)
Query: 29 CQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
C+ C ++LCD CD G+H +C+ P L +VP+ WFCP C
Sbjct: 1151 CKICRKKGDAENMVLCDGCDRGHHTYCVRPKLKTVPEGDWFCPEC 1195
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.333 0.146 0.480
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,650,770
Number of Sequences: 164201
Number of extensions: 1678808
Number of successful extensions: 4326
Number of sequences better than 10.0: 100
Number of HSP's better than 10.0 without gapping: 56
Number of HSP's successfully gapped in prelim test: 44
Number of HSP's that attempted gapping in prelim test: 4195
Number of HSP's gapped (non-prelim): 152
length of query: 337
length of database: 59,974,054
effective HSP length: 111
effective length of query: 226
effective length of database: 41,747,743
effective search space: 9434989918
effective search space used: 9434989918
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 66 (30.0 bits)
Medicago: description of AC141323.10


