

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141114.7 + phase: 0
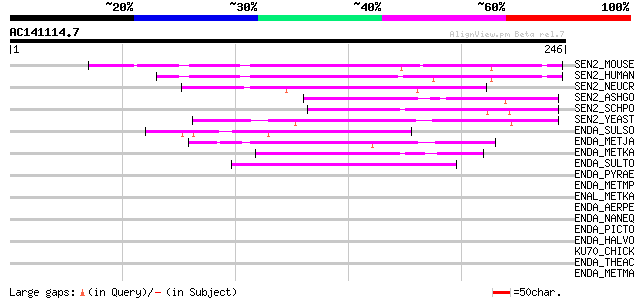
(246 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SEN2_MOUSE (Q6P7W5) tRNA-splicing endonuclease subunit Sen2 (EC ... 100 4e-21
SEN2_HUMAN (Q8NCE0) tRNA-splicing endonuclease subunit SEN2 (EC ... 94 4e-19
SEN2_NEUCR (Q9P6Y2) Probable tRNA-splicing endonuclease subunit ... 70 5e-12
SEN2_ASHGO (Q74ZY5) tRNA-splicing endonuclease subunit SEN2 (EC ... 66 7e-11
SEN2_SCHPO (Q8TFH7) Probable tRNA-splicing endonuclease subunit ... 62 1e-09
SEN2_YEAST (P16658) tRNA-splicing endonuclease subunit SEN2 (EC ... 59 1e-08
ENDA_SULSO (Q97ZY3) tRNA-splicing endonuclease (EC 3.1.27.9) (tR... 55 2e-07
ENDA_METJA (Q58819) tRNA-splicing endonuclease (EC 3.1.27.9) (tR... 54 5e-07
ENDA_METKA (Q8TGZ7) tRNA-splicing endonuclease (EC 3.1.27.9) (tR... 50 4e-06
ENDA_SULTO (Q975R3) tRNA-splicing endonuclease (EC 3.1.27.9) (tR... 47 6e-05
ENDA_PYRAE (Q8ZVI1) tRNA-splicing endonuclease (EC 3.1.27.9) (tR... 42 0.001
ENDA_METMP (Q6LY59) tRNA-splicing endonuclease (EC 3.1.27.9) (tR... 41 0.002
ENAL_METKA (Q8TGZ5) EndA-like protein 40 0.004
ENDA_AERPE (Q9YBF1) tRNA-splicing endonuclease (EC 3.1.27.9) (tR... 37 0.034
ENDA_NANEQ (Q74MP4) tRNA-splicing endonuclease (EC 3.1.27.9) (tR... 36 0.075
ENDA_PICTO (Q6L1P9) tRNA-splicing endonuclease (EC 3.1.27.9) (tR... 35 0.22
ENDA_HALVO (O07118) tRNA-splicing endonuclease (EC 3.1.27.9) (tR... 33 0.49
KU70_CHICK (O93257) ATP-dependent DNA helicase II, 70 kDa subuni... 32 1.4
ENDA_THEAC (Q9HIY5) tRNA-splicing endonuclease (EC 3.1.27.9) (tR... 32 1.4
ENDA_METMA (Q8PZI1) tRNA-splicing endonuclease (EC 3.1.27.9) (tR... 32 1.8
>SEN2_MOUSE (Q6P7W5) tRNA-splicing endonuclease subunit Sen2 (EC
3.1.27.9) (tRNA-intron endonuclease Sen2)
Length = 460
Score = 100 bits (248), Expect = 4e-21
Identities = 69/216 (31%), Positives = 106/216 (48%), Gaps = 18/216 (8%)
Query: 36 LVQSNTCGFLSDNCVHLTVQAEQLDLLDKACFGRPVRIVEKDMYWFQLTVEEAFYLCYSL 95
L++ CG + + + + L C P RI E + QL++EEAF+L Y+L
Sbjct: 252 LIEEELCGAQEEEAAAASDE-KLLKRKKLVCRRNPYRIFE----YLQLSLEEAFFLAYAL 306
Query: 96 KCLKIKINVGADTGPLNDEELWHYFRSKKETFPYFYKAYSHLRMKNWVVRSGAQYGVDLI 155
CL I + PL +LW F + + TF Y AY + R K WV + G +YG DL+
Sbjct: 307 GCLSIYY----EKEPLTIVKLWQAFTAVQPTFRTTYMAYHYFRSKGWVPKVGLKYGTDLL 362
Query: 156 VYRHHPARVHSEYGVLV--LSHDKDGDLNGRLRVWSDVHCTTRLLGGVAKTLLVLYVNK- 212
+YR P H+ Y V++ L + +G L R W + +R+ G V+K L++ Y+ K
Sbjct: 363 LYRKGPPFYHASYSVIIELLDDNYEGSLR-RPFSWKSLAALSRVSGNVSKELMLCYLIKP 421
Query: 213 ---NGNNDESLLCLTNYTVEERTISRWSPEQCRERS 245
+ E+ C+ V+E +SRW RERS
Sbjct: 422 STMTAEDMETPECMKRIQVQEVILSRW--VSSRERS 455
>SEN2_HUMAN (Q8NCE0) tRNA-splicing endonuclease subunit SEN2 (EC
3.1.27.9) (tRNA-intron endonuclease SEN2) (HsSen2
protein)
Length = 465
Score = 93.6 bits (231), Expect = 4e-19
Identities = 65/187 (34%), Positives = 91/187 (47%), Gaps = 19/187 (10%)
Query: 66 CFGRPVRIVEKDMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYFRSKKE 125
C P RI E + QL++EEAF+L Y+L CL I + PL +LW F +
Sbjct: 286 CRRNPYRIFE----YLQLSLEEAFFLVYALGCLSIYY----EKEPLTIVKLWKAFTVVQP 337
Query: 126 TFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGDLNGRL 185
TF Y AY + R K WV + G +YG DL++YR P H+ Y V++ D G L
Sbjct: 338 TFRTTYMAYHYFRSKGWVPKVGLKYGTDLLLYRKGPPFYHASYSVII--ELVDDHFEGSL 395
Query: 186 R---VWSDVHCTTRLLGGVAKTLLVLYVNK----NGNNDESLLCLTNYTVEERTISRWSP 238
R W + +R+ V+K L++ Y+ K ES C+ V+E +SRW
Sbjct: 396 RRPLSWKSLAALSRVSVNVSKELMLCYLIKPSTMTDKEMESPECMKRIKVQEVILSRW-- 453
Query: 239 EQCRERS 245
RERS
Sbjct: 454 VSSRERS 460
>SEN2_NEUCR (Q9P6Y2) Probable tRNA-splicing endonuclease subunit
sen-2 (EC 3.1.27.9) (tRNA-intron endonuclease sen-2)
Length = 633
Score = 70.1 bits (170), Expect = 5e-12
Identities = 47/153 (30%), Positives = 74/153 (47%), Gaps = 20/153 (13%)
Query: 77 DMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYFR-------------SK 123
D FQL EEAF+L + L L++ V P+++E+L R S
Sbjct: 444 DKEHFQLAPEEAFFLTFGLGALRVVDPV--TEAPISNEQLLIKLRANSYFPPRSVDNLSP 501
Query: 124 KETFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDG---- 179
++ F Y Y H R WV R G ++GVD I+Y+ P HSE+G++V+ D
Sbjct: 502 EDPFLVQYAVYHHFRSLGWVPRHGIKFGVDWIIYQRGPVFDHSEFGIMVVPSFSDPRWSE 561
Query: 180 -DLNGRLRVWSDVHCTTRLLGGVAKTLLVLYVN 211
+ + WS + R+L V K+L+++YV+
Sbjct: 562 FEHEESKKTWSWLMGVNRVLSHVLKSLVLVYVD 594
>SEN2_ASHGO (Q74ZY5) tRNA-splicing endonuclease subunit SEN2 (EC
3.1.27.9) (tRNA-intron endonuclease SEN2)
Length = 356
Score = 66.2 bits (160), Expect = 7e-11
Identities = 39/122 (31%), Positives = 57/122 (45%), Gaps = 16/122 (13%)
Query: 131 YKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGDLNGRLRVWSD 190
Y AY H R W VRSG ++G D ++YR P H+E+ V+VL+ D+ D W
Sbjct: 242 YAAYHHYRSHGWCVRSGIKFGCDFLLYRRGPPFHHAEFSVMVLAPDERHDY-----TWYS 296
Query: 191 VHCTTRLLGGVAKTLLVLYVNKNGNNDE---------SLLCLTNYTVEERTISRWSPEQC 241
R++ G KTL++ YV++ D+ + Y V E RW P +
Sbjct: 297 T--VDRVVAGAKKTLVLAYVDRRATTDQLTALWHARRYMEAFALYKVHELVYRRWLPGKN 354
Query: 242 RE 243
RE
Sbjct: 355 RE 356
>SEN2_SCHPO (Q8TFH7) Probable tRNA-splicing endonuclease subunit
sen2 (EC 3.1.27.9) (tRNA-intron endonuclease sen2)
Length = 380
Score = 62.4 bits (150), Expect = 1e-09
Identities = 36/126 (28%), Positives = 62/126 (48%), Gaps = 17/126 (13%)
Query: 133 AYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGDLNGRLRVWSDVH 192
AY + R + WVV++G ++ VD ++Y+ P H+E+ +L++ G+ W +VH
Sbjct: 257 AYFYFRQQGWVVKNGTKFSVDFLLYKKGPVFSHAEFAILLI--PCVGNKQKYNMQWHEVH 314
Query: 193 CTTRLLGGVAKTLLVLYV--------NKNGNNDESL-------LCLTNYTVEERTISRWS 237
C R++ V K+L++ YV NK N S+ L Y + T+ RW
Sbjct: 315 CLNRVIAQVKKSLILCYVQCPSIEDFNKIWKNQASMNEWDWAESVLRQYLIRCVTLRRWV 374
Query: 238 PEQCRE 243
P + R+
Sbjct: 375 PSRNRD 380
>SEN2_YEAST (P16658) tRNA-splicing endonuclease subunit SEN2 (EC
3.1.27.9) (tRNA-intron endonuclease SEN2) (Splicing
endonuclease protein 2)
Length = 377
Score = 58.5 bits (140), Expect = 1e-08
Identities = 45/173 (26%), Positives = 76/173 (43%), Gaps = 25/173 (14%)
Query: 82 QLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYFRSKKE--TFPYFYKAYSHLRM 139
+L EA +L ++L L I A +L+ + K+ +F Y Y H R
Sbjct: 219 ELMPVEAMFLTFALPVLDISPACLAG-------KLFQFDAKYKDIHSFVRSYVIYHHYRS 271
Query: 140 KNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGDLNGRLRVWSDVHCTTRLLG 199
W VRSG ++G D ++Y+ P H+E+ V+ L HD D ++ R++G
Sbjct: 272 HGWCVRSGIKFGCDYLLYKRGPPFQHAEFCVMGLDHDVSKD-------YTWYSSIARVVG 324
Query: 200 GVAKTLLVLYVNKNGNNDESLL---------CLTNYTVEERTISRWSPEQCRE 243
G KT ++ YV + + E++ ++ V E RW P + R+
Sbjct: 325 GAKKTFVLCYVERLISEQEAIALWKSNNFTKLFNSFQVGEVLYKRWVPGRNRD 377
>ENDA_SULSO (Q97ZY3) tRNA-splicing endonuclease (EC 3.1.27.9)
(tRNA-intron endonuclease)
Length = 182
Score = 54.7 bits (130), Expect = 2e-07
Identities = 40/124 (32%), Positives = 61/124 (48%), Gaps = 11/124 (8%)
Query: 61 LLDKACFGRPVRIVE----KDMYW-FQLTVEEAFYLCYSLKCLKIKINVGADTGPLND-E 114
L +G+P+ I + KD+ +L++ E+ YL K INV G L + +
Sbjct: 22 LYSNGFYGKPIGISKPKGPKDIVRPLELSLIESVYLTK-----KGLINVVDKNGDLLEYK 76
Query: 115 ELWHYFRSKKETFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLS 174
+L+ Y K F YK Y LR K ++VRSG +YG D VY P H+ Y V+ +
Sbjct: 77 KLYEYSAMKINKFEILYKVYEDLREKGFIVRSGVKYGADFAVYTLGPGLEHAPYVVIAVD 136
Query: 175 HDKD 178
D++
Sbjct: 137 IDEE 140
>ENDA_METJA (Q58819) tRNA-splicing endonuclease (EC 3.1.27.9)
(tRNA-intron endonuclease)
Length = 179
Score = 53.5 bits (127), Expect = 5e-07
Identities = 42/138 (30%), Positives = 68/138 (48%), Gaps = 13/138 (9%)
Query: 80 WFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYFRSKKETFPYFYKAYSHLRM 139
+ L++ EA YL +L L++K D PL+ EEL+ Y R+ +E Y Y LR
Sbjct: 42 FLSLSLVEALYLI-NLGWLEVKYK---DNKPLSFEELYEYARNVEERLCLKYLVYKDLRT 97
Query: 140 KNWVVRSGAQYGVDLIVYRH--HPARVHSEYGVLVLSHDKDGDLNGRLRVWSDVHCTTRL 197
+ ++V++G +YG D +Y + + HS Y V V D L S++ R+
Sbjct: 98 RGYIVKTGLKYGADFRLYERGANIDKEHSVYLVKVFPEDSSFLL-------SELTGFVRV 150
Query: 198 LGGVAKTLLVLYVNKNGN 215
V K LL+ V+ +G+
Sbjct: 151 AHSVRKKLLIAIVDADGD 168
>ENDA_METKA (Q8TGZ7) tRNA-splicing endonuclease (EC 3.1.27.9)
(tRNA-intron endonuclease)
Length = 179
Score = 50.4 bits (119), Expect = 4e-06
Identities = 35/101 (34%), Positives = 51/101 (49%), Gaps = 7/101 (6%)
Query: 110 PLNDEELWHYFRSKKETFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYG 169
P++ EEL +F ++ F Y Y L + +VV+SG +YG VY P R HS+Y
Sbjct: 68 PMSVEELLSFFERRRPGFRAGYVVYRDLTERGYVVKSGFKYGGRFRVYEEDPDREHSKYV 127
Query: 170 VLVLSHDKDGDLNGRLRVWSDVHCTTRLLGGVAKTLLVLYV 210
V V+ + D +L+ R DV TRL V K ++ V
Sbjct: 128 VRVV--EPDTELSTR-----DVLRATRLAHSVRKDFVLAVV 161
>ENDA_SULTO (Q975R3) tRNA-splicing endonuclease (EC 3.1.27.9)
(tRNA-intron endonuclease)
Length = 180
Score = 46.6 bits (109), Expect = 6e-05
Identities = 31/100 (31%), Positives = 45/100 (45%)
Query: 99 KIKINVGADTGPLNDEELWHYFRSKKETFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYR 158
K K+ + ++ L+ E L+ ++ F Y Y LR K +VVRSG +YG D VY
Sbjct: 59 KGKLEIVSNGERLDFERLYQIGVTQIPRFRILYSVYEDLREKGYVVRSGIKYGADFAVYT 118
Query: 159 HHPARVHSEYGVLVLSHDKDGDLNGRLRVWSDVHCTTRLL 198
P H+ Y V+ L + N L H T + L
Sbjct: 119 IGPGIEHAPYLVIALDENSQISSNEILGFGRVSHSTRKEL 158
>ENDA_PYRAE (Q8ZVI1) tRNA-splicing endonuclease (EC 3.1.27.9)
(tRNA-intron endonuclease)
Length = 183
Score = 42.4 bits (98), Expect = 0.001
Identities = 31/95 (32%), Positives = 43/95 (44%), Gaps = 4/95 (4%)
Query: 83 LTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYFRSKKETFPYFYKAYSHLRMKNW 142
L + EA YL + LK+ G + P EEL R + F YK Y + R +
Sbjct: 50 LGLYEALYLAEKGR-LKVMGEDGREVAP---EELAALGRERMRNFDEIYKIYKYFRDLGY 105
Query: 143 VVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDK 177
VV+SG ++G VY P H+ V+ L DK
Sbjct: 106 VVKSGLKFGALFSVYEKGPGIDHAPMVVVFLEPDK 140
>ENDA_METMP (Q6LY59) tRNA-splicing endonuclease (EC 3.1.27.9)
(tRNA-intron endonuclease)
Length = 177
Score = 41.2 bits (95), Expect = 0.002
Identities = 30/106 (28%), Positives = 52/106 (48%), Gaps = 8/106 (7%)
Query: 111 LNDEELWHYFRSKKETFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVY-RHHPARVHSEYG 169
L+ EEL+ ++ Y AY LR + + VR+G +YG D +Y R + +HS Y
Sbjct: 68 LSFEELFDVAQNIDRKLCIRYLAYKDLRNRGYTVRTGLKYGSDFRLYERSNIDEIHSRYL 127
Query: 170 VLVLSHDKDGDLNGRLRVWSDVHCTTRLLGGVAKTLLVLYVNKNGN 215
V V S + ++ S++ R+ V K L++ V+ +G+
Sbjct: 128 VKVFSEEIPCEI-------SEITGFVRVAHSVRKELIIAIVDADGS 166
>ENAL_METKA (Q8TGZ5) EndA-like protein
Length = 166
Score = 40.4 bits (93), Expect = 0.004
Identities = 31/117 (26%), Positives = 50/117 (42%), Gaps = 15/117 (12%)
Query: 62 LDKACFGRPVRIVEKDMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYFR 121
L + +G+P R QL EEA +LC + + V + ++ EEL F
Sbjct: 23 LYSSMYGKPSR------RGLQLWPEEALFLCEIGR-----LEVRSGNVRISPEELMDRFV 71
Query: 122 SKKETFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKD 178
+ FP Y Y+ LR + W + G ++G + +R R+ V VL + D
Sbjct: 72 EEDPRFPVRYAVYADLRRRGWKPKPGRKFGTEFRAFRGEDERI----AVKVLQEELD 124
>ENDA_AERPE (Q9YBF1) tRNA-splicing endonuclease (EC 3.1.27.9)
(tRNA-intron endonuclease)
Length = 187
Score = 37.4 bits (85), Expect = 0.034
Identities = 20/54 (37%), Positives = 28/54 (51%)
Query: 127 FPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGD 180
F Y Y LR + +VVRSG ++G D VYR P H+ + V S + + D
Sbjct: 96 FSMLYNIYRDLRERGFVVRSGLKFGSDFAVYRLGPGIDHAPFIVHAYSPEDNID 149
>ENDA_NANEQ (Q74MP4) tRNA-splicing endonuclease (EC 3.1.27.9)
(tRNA-intron endonuclease)
Length = 154
Score = 36.2 bits (82), Expect = 0.075
Identities = 34/122 (27%), Positives = 53/122 (42%), Gaps = 18/122 (14%)
Query: 67 FGRPVRIVEKDMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYFRSKKET 126
FG V + +K+ L + EA+YL K ++ V D L+ EE + E
Sbjct: 6 FGNRVLVDDKE-----LPLIEAYYLLD-----KGELEVYEDDKKLSKEEFLKKCLTYDER 55
Query: 127 FPYFYKAYSHLRMKNWVVRSGAQYGVDLIVY--------RHHPARVHSEYGVLVLSHDKD 178
F YKAY LR K + + + ++G D VY R HS++ + +S D+
Sbjct: 56 FLIRYKAYKELRDKGYTLGTALKFGADFRVYDIGVIPKKGKRSEREHSKWVLYPVSKDET 115
Query: 179 GD 180
D
Sbjct: 116 FD 117
>ENDA_PICTO (Q6L1P9) tRNA-splicing endonuclease (EC 3.1.27.9)
(tRNA-intron endonuclease)
Length = 284
Score = 34.7 bits (78), Expect = 0.22
Identities = 26/97 (26%), Positives = 42/97 (42%), Gaps = 14/97 (14%)
Query: 111 LNDEELWHYFRSKKETFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGV 170
LND E+ K YK L + ++V+SG +YG + +Y+ + HS+Y V
Sbjct: 179 LNDYEIRFLNNDVKSNVDVIYK---DLIKRGFIVKSGFKYGSNFRIYK-NSMNEHSDYLV 234
Query: 171 LVLSHDKDGDLNGRLRVWSDVHCTTRLLGGVAKTLLV 207
+ HD +W + RL V K L++
Sbjct: 235 NYMDHD----------LWYVIARAVRLASNVRKRLII 261
>ENDA_HALVO (O07118) tRNA-splicing endonuclease (EC 3.1.27.9)
(tRNA-intron endonuclease)
Length = 339
Score = 33.5 bits (75), Expect = 0.49
Identities = 27/87 (31%), Positives = 39/87 (44%), Gaps = 10/87 (11%)
Query: 134 YSHLRMKNWVVRSGAQYGVDLIVYRHHPA---RVHSEYGVLVLSHDKDGDLNGRLRVWSD 190
Y+ LR V +SG ++G D VY + HSE+ V V++ D V D
Sbjct: 251 YAALREAKTVPKSGFKFGSDFRVYTEFESVDDLSHSEFLVRVVAPD-------HTFVPRD 303
Query: 191 VHCTTRLLGGVAKTLLVLYVNKNGNND 217
+ RL GGV K ++ + NG D
Sbjct: 304 LSLDVRLAGGVRKRMVFALTDDNGEID 330
>KU70_CHICK (O93257) ATP-dependent DNA helicase II, 70 kDa subunit
(Ku autoantigen protein p70 homolog) (Ku70)
Length = 632
Score = 32.0 bits (71), Expect = 1.4
Identities = 24/74 (32%), Positives = 35/74 (46%), Gaps = 10/74 (13%)
Query: 6 KGKDAKSKKEAEAEALKEPMSKIISQLQSSLVQSNTCG-----FLSDNCVHLTVQA---- 56
+GK AK K+ +A+A K P +I S VQ+ T G L D C H +++
Sbjct: 559 EGKAAKRKQAGDAQAEKRPKIEISEDSLRSYVQNGTLGKLTVSALKDTCRHYGLRSGGKK 618
Query: 57 -EQLDLLDKACFGR 69
E +D L + GR
Sbjct: 619 QELIDALTEYFSGR 632
>ENDA_THEAC (Q9HIY5) tRNA-splicing endonuclease (EC 3.1.27.9)
(tRNA-intron endonuclease)
Length = 289
Score = 32.0 bits (71), Expect = 1.4
Identities = 21/80 (26%), Positives = 38/80 (47%), Gaps = 10/80 (12%)
Query: 134 YSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGDLNGRLRVWSDVHC 193
YS L + +V++G +YG + VY ++ H+EY V V+ ++ W +
Sbjct: 206 YSDLVGRGCIVKTGFKYGANFRVYLGRDSQ-HAEYLVSVMPEEER---------WYSISR 255
Query: 194 TTRLLGGVAKTLLVLYVNKN 213
R+ V KT++ + KN
Sbjct: 256 GVRVASSVRKTMIYASIYKN 275
>ENDA_METMA (Q8PZI1) tRNA-splicing endonuclease (EC 3.1.27.9)
(tRNA-intron endonuclease)
Length = 353
Score = 31.6 bits (70), Expect = 1.8
Identities = 29/87 (33%), Positives = 43/87 (49%), Gaps = 8/87 (9%)
Query: 131 YKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPA---RVHSEYGVLVLSHDKDGDL---NGR 184
Y AY LR VV++G ++G VYR + HSEY V V+ D + L +G
Sbjct: 263 YSAYKALRDSGHVVKTGFKFGTHFRVYRKVESIEKIPHSEYLVNVIPEDYEFRLPVMSGA 322
Query: 185 LRVWSDVHCTTRLLGGVAKTLLVLYVN 211
+R+ + V R+L V K V Y++
Sbjct: 323 VRLANSVR--KRMLFAVEKGEEVEYLD 347
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.136 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,879,735
Number of Sequences: 164201
Number of extensions: 1227856
Number of successful extensions: 2728
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 2703
Number of HSP's gapped (non-prelim): 27
length of query: 246
length of database: 59,974,054
effective HSP length: 107
effective length of query: 139
effective length of database: 42,404,547
effective search space: 5894232033
effective search space used: 5894232033
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC141114.7


