

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141111.7 - phase: 0 /pseudo
(1730 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
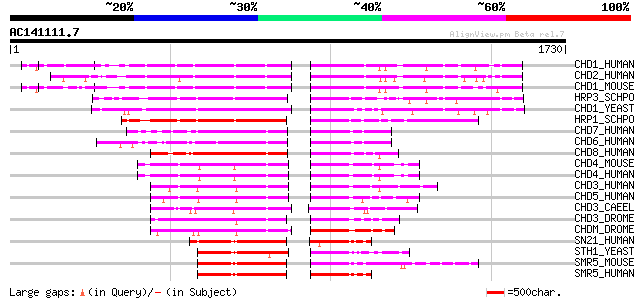
Score E
Sequences producing significant alignments: (bits) Value
CHD1_HUMAN (O14646) Chromodomain-helicase-DNA-binding protein 1 ... 419 e-116
CHD2_HUMAN (O14647) Chromodomain-helicase-DNA-binding protein 2 ... 416 e-115
CHD1_MOUSE (P40201) Chromodomain-helicase-DNA-binding protein 1 ... 414 e-114
HRP3_SCHPO (O14139) Chromodomain helicase hrp3 392 e-108
CHD1_YEAST (P32657) Chromo domain protein 1 379 e-104
HRP1_SCHPO (Q9US25) Chromodomain helicase hrp1 358 5e-98
CHD7_HUMAN (Q9P2D1) Chromodomain-helicase-DNA-binding protein 7 ... 310 2e-83
CHD6_HUMAN (Q8TD26) Chromodomain-helicase-DNA-binding protein 6 ... 305 5e-82
CHD8_HUMAN (Q9HCK8) Chromodomain-helicase-DNA-binding protein 8 ... 302 6e-81
CHD4_MOUSE (Q6PDQ2) Chromodomain helicase-DNA-binding protein 4 ... 296 4e-79
CHD4_HUMAN (Q14839) Chromodomain helicase-DNA-binding protein 4 ... 296 4e-79
CHD3_HUMAN (Q12873) Chromodomain helicase-DNA-binding protein 3 ... 290 2e-77
CHD5_HUMAN (Q8TDI0) Chromodomain helicase-DNA-binding protein 5 ... 290 3e-77
CHD3_CAEEL (Q22516) Chromodomain helicase-DNA-binding protein 3 ... 286 3e-76
CHD3_DROME (O16102) Chromodomain helicase-DNA-binding protein 3 273 2e-72
CHDM_DROME (O97159) Chromodomain helicase-DNA-binding protein Mi... 273 3e-72
SN21_HUMAN (P28370) Possible global transcription activator SNF2... 250 3e-65
STH1_YEAST (P32597) Nuclear protein STH1/NPS1 (Chromatin structu... 244 1e-63
SMR5_MOUSE (Q91ZW3) SWI/SNF related matrix associated actin depe... 244 1e-63
SMR5_HUMAN (O60264) SWI/SNF related matrix associated actin depe... 243 3e-63
>CHD1_HUMAN (O14646) Chromodomain-helicase-DNA-binding protein 1
(CHD-1)
Length = 1709
Score = 419 bits (1077), Expect = e-116
Identities = 274/798 (34%), Positives = 420/798 (52%), Gaps = 67/798 (8%)
Query: 90 SPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVPAE 149
S Q+ +SG S SGS + +SS+G + GS D D +SG +SD E
Sbjct: 17 SSQSDDDSGSASGSGSGSSS-----GSSSDGSSSQSGSSDSDSGSESGSQSESESDTSRE 71
Query: 150 EMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRILDDAEDDD- 208
+ DG E +S S + ++ L+ ++ H+ S ED
Sbjct: 72 NKVQAKPPKVDGAEFWKSSPSI---LAVQRSAILKKQQQQQQQQQHQASSNSGSEEDSSS 128
Query: 209 -DDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDEDIDVSDNDDLY 267
+D+D E D D++ + SG + D E E+ E DE + +
Sbjct: 129 SEDSDDSSSEVKRKKHKDEDWQMSGSGSPSQSGSDSESEEEREKSSCDETESDYEPKNKV 188
Query: 268 FDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENSTAEDSDSESDEDFK 327
+K + R + K G + ++R++ SS ED+DE ED D++
Sbjct: 189 KSRKPQNRSKSKNGKKI----------LGQKKRQIDSSEEDDDE----EDYDND------ 228
Query: 328 SLKKRGVRVRKNNGRSSAATSFSRPSNEVRSSSRTIRKVSYVESDESEGADEGTKKSQKE 387
K + R A + S +E + D E E + ++E
Sbjct: 229 ----------KRSSRRQATVNVSYKEDEEMKTD---------SDDLLEVCGEDVPQPEEE 269
Query: 388 EIEVDDGDSVEKVLWHQPKGMAAEAQRNNQSMEPVLMSHLFDSEPDWNNMEFLIKWKGQS 447
E E + ++ G +P + F+ + +++LIKWKG S
Sbjct: 270 EFETIERFMDCRIGRKGATGATTTIYAVEADGDP---NAGFEKNKEPGEIQYLIKWKGWS 326
Query: 448 HLHCQWKSFVDL--QNLSGFKKVLNYTKRVTEEIRNRMGISREEIEVNDVSKEMDIDIIK 505
H+H W++ L QN+ G KK+ NY K+ E R S E++E + +E+ D+ K
Sbjct: 327 HIHNTWETEETLKQQNVRGMKKLDNYKKKDQETKRWLKNASPEDVEYYNCQQELTDDLHK 386
Query: 506 QNSQVERIIADRISNDNSGNVFPEYLVKWQGLSYAEVTWEKDIDIAFA-QHTIDEYKTRE 564
Q V RIIA SN S +P+Y KWQGL Y+E +WE I+ Q IDEY +R
Sbjct: 387 QYQIVGRIIAH--SNQKSAAGYPDYYCKWQGLPYSECSWEDGALISKKFQACIDEYFSRN 444
Query: 565 AAMSVQGKMVDFQRRQSKGSLRKLDEQPEWL---KGGKLRDYQLEGLNFLVNSWKNDTNV 621
+ + K D + + + L +QP ++ +G +LRDYQL GLN+L +SW +
Sbjct: 445 QSKTTPFK--DCKVLKQRPRFVALKKQPSYIGGHEGLELRDYQLNGLNWLAHSWCKGNSC 502
Query: 622 VLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYV 681
+LADEMGLGKT+Q++S L +L + Q++GPFL+VVPLSTL++W +E + W +N +VY+
Sbjct: 503 ILADEMGLGKTIQTISFLNYLFHEHQLYGPFLLVVPLSTLTSWQREIQTWASQMNAVVYL 562
Query: 682 GTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLK 741
G +SR + + +E+ + + K++KFN LLTTYE++LKDKA L + W ++ VDEAHRLK
Sbjct: 563 GDINSRNMIRTHEWTHHQT--KRLKFNILLTTYEILLKDKAFLGGLNWAFIGVDEAHRLK 620
Query: 742 NSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHFLDSDKFKSKDEFAQNYKNLSS 801
N ++ LY L +F + ++LLITGTPLQNS++ELW+LLHF+ +KF S ++F + +
Sbjct: 621 NDDSLLYKTLIDFKSNHRLLITGTPLQNSLKELWSLLHFIMPEKFSSWEDFEEEH---GK 677
Query: 802 FNENELSNLHMELRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYKWILERNFRDL 861
E ++LH EL P +LRRV KDVEKSLP K+E+ILR++MS LQKQYYKWIL RN++ L
Sbjct: 678 GREYGYASLHKELEPFLLRRVKKDVEKSLPAKVEQILRMEMSALQKQYYKWILTRNYKAL 737
Query: 862 NKGVRGNQSKYCSRIEEV 879
+KG +G+ S + + + E+
Sbjct: 738 SKGSKGSTSGFLNIMMEL 755
Score = 345 bits (885), Expect = 6e-94
Identities = 253/717 (35%), Positives = 358/717 (49%), Gaps = 95/717 (13%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DILA+Y+ R F FQRLDGS K ELR+QA+DHFNA GS+DFCFLLSTRAGGLGINLA+AD
Sbjct: 818 DILAEYLKYRQFPFQRLDGSIKGELRKQALDHFNAEGSEDFCFLLSTRAGGLGINLASAD 877
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TV+IFDSDWNPQNDLQA +RAHRIGQ++ VNIYR VT SVEEDILERAKKKMVLDHLVI
Sbjct: 878 TVVIFDSDWNPQNDLQAQARAHRIGQKKQVNIYRLVTKGSVEEDILERAKKKMVLDHLVI 937
Query: 1058 QKLNAEGK--LEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDE 1115
Q+++ GK L A + F+K ELSAIL+FGAEELFKE +E+ + MDIDE
Sbjct: 938 QRMDTTGKTVLHTGSAPSSSTPFNKEELSAILKFGAEELFKEPEGEEQEPQ---EMDIDE 994
Query: 1116 ILERAEKVEEKENGGEQAHELLSAFKVANFCN-DEDD--------GSFWSRWIKADSVAQ 1166
IL+RAE E + ELLS FKVANF N DEDD W I D +
Sbjct: 995 ILKRAETHENEPGPLTVGDELLSQFKVANFSNMDEDDIELEPERNSKNWEEIIPEDQRRR 1054
Query: 1167 AENA----------LAPRAARNIKSYA-EADQSERSKKRKKKENEPTE----RIPKRRKA 1211
E + PR K + + RS+ R+ ++ + PK+R
Sbjct: 1055 LEEEERQKELEEIYMLPRMRNCAKQISFNGSEGRRSRSRRYSGSDSDSISEGKRPKKR-- 1112
Query: 1212 DYSAHVISMIDGASAQVRSWSYGNLSKRDALRFSRSVMKFGNESQINLIVAEVGGAIEAA 1271
G + + S + RF +S KFG + +A ++ +
Sbjct: 1113 -----------GRPRTIPRENIKGFSDAEIRRFIKSYKKFGGPLERLDAIARDAELVDKS 1161
Query: 1272 PLKAQVELFNALIDGCREAVEVGSL----------DLKGPLLDFYGVPMKANELLIRVQE 1321
+ L + +GC +A++ S +KGP GV + A ++ +E
Sbjct: 1162 ETDLR-RLGELVHNGCIKALKDSSSGTERTGGRLGKVKGPTFRISGVQVNAKLVISHEEE 1220
Query: 1322 LQLLAKRISRYEDPIAQFRVLTYLKPSNWSKGCGWNQIDDARLLLGVHYHGYGNWEVIRL 1381
L L K I + Q+ + + K +++ W + DD+ LL+G++ +GYG+WE+I++
Sbjct: 1221 LIPLHKSIPSDPEERKQYTIPCHTKAAHFD--IDWGKEDDSNLLIGIYEYGYGSWEMIKM 1278
Query: 1382 DERLGLTKKIAPVELQHHETFLPRAPNLRDRANALLEQELAVLGVKNASSKVGRKTSKKE 1441
D L LT KI P + P+A L+ RA+ L++ L K A S G +K
Sbjct: 1279 DPDLSLTHKILPDDPDKK----PQAKQLQTRADYLIKLLSRDLAKKEALSGAGSSKRRKA 1334
Query: 1442 REEREHLV--------------DISLSRGQEKKKNIGSSKVNVQMRKDR---LQKPLNV- 1483
R ++ + + + E + SK + + R + P+++
Sbjct: 1335 RAKKNKAMKSIKVKEEIKSDSSPLPSEKSDEDDDKLSESKSDGRERSKKSSVSDAPVHIT 1394
Query: 1484 ---EPI-VKEEGEMSDDDDVYEQFKEGKWKEWCQDLMVEEMKTLKRLHRLQTTSASLPKE 1539
EP+ + EE E D K C++ M LK+L R L +
Sbjct: 1395 ASGEPVPISEESEELDQ----------KTFSICKERMRPVKAALKQLDR---PEKGLSER 1441
Query: 1540 KVLSKIRNYLQLLGRRIDQIVSEQEDEPHKQDRMTTRLWKYVSTFSHLSGERLHQIY 1596
+ L R L +G I + + E + P + + LW +VS F+ +LH++Y
Sbjct: 1442 EQLEHTRQCLIKIGDHITECLKEYTN-PEQIKQWRKNLWIFVSKFTEFDARKLHKLY 1497
Score = 61.6 bits (148), Expect = 2e-08
Identities = 60/238 (25%), Positives = 103/238 (43%), Gaps = 36/238 (15%)
Query: 38 SDAEPDDASGKQNEAAAVDRLSTRESNVETTSRNPSASERWGSS----------YLKDCQ 87
SD++ SG Q+E+ + ++RE+ V+ +E W SS LK Q
Sbjct: 50 SDSDSGSESGSQSESESD---TSRENKVQAKPPKVDGAEFWKSSPSILAVQRSAILKKQQ 106
Query: 88 PMSPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQ-KDSGKGQRGDSDV 146
Q ++ +S S D + ++ +D+SSE + +K +DED Q SG + SD
Sbjct: 107 QQQQQQQHQASSNSGSEEDSSSSEDSDDSSSEVKRKK--HKDEDWQMSGSGSPSQSGSDS 164
Query: 147 PAEEMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRILDDAED 206
+EE S + + + +P S S N + + +K R +D +E+
Sbjct: 165 ESEEEREKSSCDETESDYEPKNKVKSRKPQNRSK------SKNGKKILGQKKRQIDSSEE 218
Query: 207 DDDDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDEDIDVSDND 264
DDD+ DY+ D+ +S R A ++ ED + DSD+ ++V D
Sbjct: 219 DDDEEDYDNDK-------------RSSRRQATVNVSYK-EDEEMKTDSDDLLEVCGED 262
>CHD2_HUMAN (O14647) Chromodomain-helicase-DNA-binding protein 2
(CHD-2)
Length = 1739
Score = 416 bits (1070), Expect = e-115
Identities = 280/810 (34%), Positives = 426/810 (52%), Gaps = 112/810 (13%)
Query: 127 SEDEDGQKDSGKGQRGDSDVPAEEMLSDDSYGQDGEEQG---------------ESVHSR 171
++D+ ++DS S +EE DS Q EQG ES S+
Sbjct: 4 NKDKSQEEDSSLHSNASSHSASEEASGSDSGSQSESEQGSDPGSGHGSESNSSSESSESQ 63
Query: 172 GFRPSTGSNSCLQPTSTNVNRR-VHRKSRILDDA---EDDDDDADYEEDEPDEDDPDDAD 227
S + S QP + +K RI D E+ D +P +
Sbjct: 64 SESESESAGSKSQPVLPEAKEKPASKKERIADVKKMWEEYPDVYGVRRSNRSRQEPSRFN 123
Query: 228 F-EPATSG-----------RGANKYKDWEGEDSDEVDDSDEDIDVSDNDDLYFDKKAKGR 275
E A+SG R K + W+ E S+ D+ ++ + KK K R
Sbjct: 124 IKEEASSGSESGSPKRRGQRQLKKQEKWKQEPSE--DEQEQGTSAESEPE---QKKVKAR 178
Query: 276 QRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENSTAEDSDSESDEDFKSLKKRGVR 335
+ P R T + RVK + + +DS E D+D ++ K++ R
Sbjct: 179 R-----PVPRRTVP---------KPRVKKQPKTQRGKRKKQDSSDEDDDDDEAPKRQTRR 224
Query: 336 VRKNNGRSSAATSFSRPSNEVRSSSRTIRKVSYVESDESEGADEGTKKSQKEEIEV--DD 393
R + VSY E D+ E + + E ++ D+
Sbjct: 225 -------------------------RAAKNVSYKEDDDFETDSDDLIEMTGEGVDEQQDN 259
Query: 394 GDSVEKVLWHQ-----PKGMAAEAQRNNQSMEPVLMSHLFDSEPDWNNMEFLIKWKGQSH 448
+++EKVL + G + + +P S FD+E D +++LIKWKG S+
Sbjct: 260 SETIEKVLDSRLGKKGATGASTTVYAIEANGDP---SGDFDTEKDEGEIQYLIKWKGWSY 316
Query: 449 LHCQWKSFVDLQN--LSGFKKVLNYTKRVTEEIRNRMG-ISREEIEVNDVSKEMDIDIIK 505
+H W+S LQ + G KK+ N+ K+ +EI+ +G +S E++E + +E+ ++ K
Sbjct: 317 IHSTWESEESLQQQKVKGLKKLENFKKK-EDEIKQWLGKVSPEDVEYFNCQQELASELNK 375
Query: 506 QNSQVERIIADRISNDNSGNVF-------------PEYLVKWQGLSYAEVTWEKDIDIAF 552
Q VER+IA + S G PEYL KW GL Y+E +WE + I
Sbjct: 376 QYQIVERVIAVKTSKSTLGQTDFPAHSRKPAPSNEPEYLCKWMGLPYSECSWEDEALIGK 435
Query: 553 A-QHTIDEYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEWLKGG--KLRDYQLEGLN 609
Q+ ID + +R + ++ + + + + + L +QP +L G +LRDYQLEGLN
Sbjct: 436 KFQNCIDSFHSRNNSKTIPTR--ECKALKQRPRFVALKKQPAYLGGENLELRDYQLEGLN 493
Query: 610 FLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEFR 669
+L +SW + +V+LADEMGLGKT+Q++S L +L + Q++GPFL+VVPLSTL++W +EF
Sbjct: 494 WLAHSWCKNNSVILADEMGLGKTIQTISFLSYLFHQHQLYGPFLIVVPLSTLTSWQREFE 553
Query: 670 KWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVLKDKAVLSKIKW 729
W P++NV+VY+G SR ++YE+ + + K++KFNAL+TTYE++LKDK VL I W
Sbjct: 554 IWAPEINVVVYIGDLMSRNTIREYEWIHSQT--KRLKFNALITTYEILLKDKTVLGSINW 611
Query: 730 NYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHFLDSDKFKSK 789
+L VDEAHRLKN ++ LY L +F + ++LLITGTPLQNS++ELW+LLHF+ +KF+
Sbjct: 612 AFLGVDEAHRLKNDDSLLYKTLIDFKSNHRLLITGTPLQNSLKELWSLLHFIMPEKFEFW 671
Query: 790 DEFAQNYKNLSSFNENELSNLHMELRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQY 849
++F +++ EN +LH L P +LRRV KDVEKSLP K+E+ILRV+MS LQKQY
Sbjct: 672 EDFEEDH---GKGRENGYQSLHKVLEPFLLRRVKKDVEKSLPAKVEQILRVEMSALQKQY 728
Query: 850 YKWILERNFRDLNKGVRGNQSKYCSRIEEV 879
YKWIL RN++ L KG RG+ S + + + E+
Sbjct: 729 YKWILTRNYKALAKGTRGSTSGFLNIVMEL 758
Score = 347 bits (891), Expect = 1e-94
Identities = 263/767 (34%), Positives = 377/767 (48%), Gaps = 143/767 (18%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DILA+Y++++ + FQRLDGS K E+R+QA+DHFNA GS+DFCFLLSTRAGGLGINLA+AD
Sbjct: 821 DILAEYLTIKHYPFQRLDGSIKGEIRKQALDHFNADGSEDFCFLLSTRAGGLGINLASAD 880
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TV+IFDSDWNPQNDLQA +RAHRIGQ++ VNIYR VT +VEE+I+ERAKKKMVLDHLVI
Sbjct: 881 TVVIFDSDWNPQNDLQAQARAHRIGQKKQVNIYRLVTKGTVEEEIIERAKKKMVLDHLVI 940
Query: 1058 QKLNAEGK--LEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDE 1115
Q+++ G+ LE + + F+K EL+AIL+FGAE+LFKE +E + MDIDE
Sbjct: 941 QRMDTTGRTILENNSGRSNSNPFNKEELTAILKFGAEDLFKELEGEESEPQ---EMDIDE 997
Query: 1116 ILERAEKVEEKENGGEQAHELLSAFKVANFCNDEDDGSF-------WSRWIKADSVAQAE 1168
IL AE E E ELLS FKVANF ED+ W I + + E
Sbjct: 998 ILRLAE-TRENEVSTSATDELLSQFKVANFATMEDEEELEERPHKDWDEIIPEEQRKKVE 1056
Query: 1169 NA----------LAPRAARNIKSYA--EADQSERSKKRKKK--------ENEPTERIPKR 1208
+ PR + K ++D SK++ ++ E+ ++ PKR
Sbjct: 1057 EEERQKELEEIYMLPRIRSSTKKAQTNDSDSDTESKRQAQRSSASESETEDSDDDKKPKR 1116
Query: 1209 RKADYSAHVISMIDGASAQVRSWSYGNLSKRDALRFSRSVMKFGNESQINLIVAEVGGAI 1268
R G VR + + RF ++ KFG + +A +
Sbjct: 1117 R-------------GRPRSVRKDLVEGFTDAEIRRFIKAYKKFGLPLERLECLARDAELV 1163
Query: 1269 E--AAPLKAQVELF-NALIDGCREAVE---------VGSLDLKGPLLDFYGVPMKANELL 1316
+ A LK EL N+ + +E E G +GP + GV + ++
Sbjct: 1164 DKSVADLKRLGELIHNSCVSAMQEYEEQLKENASEGKGPGKRRGPTIKISGVQVNVKSII 1223
Query: 1317 IRVQELQLLAKRISRYEDPIAQFRVLTYLKPSNWSKGCGWNQIDDARLLLGVHYHGYGNW 1376
+E ++L K I + ++ + +K +++ W DD+RLLLG++ HGYGNW
Sbjct: 1224 QHEEEFEMLHKSIPVDPEEKKKYCLTCRVKAAHFD--VEWGVEDDSRLLLGIYEHGYGNW 1281
Query: 1377 EVIRLDERLGLTKKIAPVELQHHETFLPRAPNLRDRANAL-------LEQELAVLGVKNA 1429
E+I+ D L LT KI PVE P+ L+ RA+ L LE++ AV G + A
Sbjct: 1282 ELIKTDPELKLTDKILPVETDKK----PQGKQLQTRADYLLKLLRKGLEKKGAVTGGEEA 1337
Query: 1430 SSKVGRKTSKKE----REEREHLVDIS-----------------------LSRGQEKKKN 1462
K + KKE R + EH +++S + + Q+KK+N
Sbjct: 1338 KLKKRKPRVKKENKVPRLKEEHGIELSSPRHSDNPSEEGEVKDDGLEKSPMKKKQKKKEN 1397
Query: 1463 IGSSKVNVQMRKD-----------------------------RLQKPLNV----EPIVKE 1489
+ + + RKD R Q P+++ EP+
Sbjct: 1398 KENKEKQMSSRKDKEGDKERKKSKDKKEKPKSGDAKSSSKSKRSQGPVHITAGSEPV--P 1455
Query: 1490 EGEMSDDDDVYEQFKEGKWKEWCQDLMVEEMKTLKRLHRLQTTSASLPKEKVLSKIRNYL 1549
GE DDD E F C++ M K LK+L + L ++ L RN L
Sbjct: 1456 IGEDEDDDLDQETF------SICKERMRPVKKALKQLDK---PDKGLNVQEQLEHTRNCL 1506
Query: 1550 QLLGRRIDQIVSEQEDEPHKQDRMTTRLWKYVSTFSHLSGERLHQIY 1596
+G RI + + D+ H + LW +VS F+ +LH++Y
Sbjct: 1507 LKIGDRIAECLKAYSDQEHIK-LWRRNLWIFVSKFTEFDARKLHKLY 1552
>CHD1_MOUSE (P40201) Chromodomain-helicase-DNA-binding protein 1
(CHD-1)
Length = 1711
Score = 414 bits (1063), Expect = e-114
Identities = 282/804 (35%), Positives = 425/804 (52%), Gaps = 81/804 (10%)
Query: 90 SPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVPAE 149
S Q+G + G S SGS + +SS+G + GS D D DSG +SD E
Sbjct: 17 SSQSGDDCGSASGSGSGSSS-----GSSSDGSSSQSGSSDSDSGSDSGSQSESESDTSRE 71
Query: 150 EMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRILDDAEDDDD 209
+ DG E +S S S L + + R+ ++++ A +
Sbjct: 72 NKVQAKPPKVDGAEFWKS-----------SPSILAVQRSAMLRKQPQQAQQQRPASSNSG 120
Query: 210 DADYEEDEPDEDDPDDADFEPATSGRGANKY--KDWEGEDSDEVD------DSDEDIDVS 261
EED +D DD+ +SG K+ +DW+ S +S+E+ D S
Sbjct: 121 S---EEDSSSSEDSDDS-----SSGAKRKKHNDEDWQMSGSGSPSQLGSDSESEEERDKS 172
Query: 262 DNDDLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENSTAEDSDSE 321
D D + K + R + P RS ++R++ SS EDED+ ED D++
Sbjct: 173 SCDGTESDYEPKNKVRSR-KPQNRSKSKNGKKILGQKKRQIDSS-EDEDD----EDYDND 226
Query: 322 SDEDFKSLKKRGVRVRKNNGRSSAATSFSRPSNEVRSSSRTIRKVSYVESDESEGADEGT 381
K + R A + S +E + D E E
Sbjct: 227 ----------------KRSSRRQATVNVSYKEDEEMKTD---------SDDLLEVCGEDV 261
Query: 382 KKSQKEEIEVDDGDSVEKVLWHQPKGMAAEAQRNNQSMEPVLMSHLFDSEPDWNNMEFLI 441
+ + EE E + +V G +P + F+ + ++++LI
Sbjct: 262 PQPEDEEFETIERVMDCRVGRKGATGATTTIYAVEADGDP---NAGFERNKEPGDIQYLI 318
Query: 442 KWKGQSHLHCQWKSFVDL--QNLSGFKKVLNYTKRVTEEIRNRMGISREEIEVNDVSKEM 499
KWKG SH+H W++ L QN+ G KK+ NY K+ E R S E++E + +E+
Sbjct: 319 KWKGWSHIHNTWETEETLKQQNVRGMKKLDNYKKKDQETKRWLKNASPEDVEYYNCQQEL 378
Query: 500 DIDIIKQNSQVERIIADRISNDNSGNVFPEYLVKWQGLSYAEVTWEKDIDIAFAQHT-ID 558
D+ KQ VERIIA SN S P+Y KWQGL Y+E +WE I+ T ID
Sbjct: 379 TDDLHKQYQIVERIIAH--SNQKSAAGLPDYYCKWQGLPYSECSWEDGALISKKFQTCID 436
Query: 559 EYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEWL---KGGKLRDYQLEGLNFLVNSW 615
EY +R + + K D + + + L +QP ++ +G +LRDYQL GLN+L +SW
Sbjct: 437 EYFSRNQSKTTPFK--DCKVLKQRPRFVALKKQPSYIGGHEGLELRDYQLNGLNWLAHSW 494
Query: 616 KNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEFRKWLPDL 675
+ +LADEMGLGKT+Q++S L +L + Q++GPFL+VVPLSTL++W +E + W +
Sbjct: 495 CKGNSCILADEMGLGKTIQTISFLNYLFHEHQLYGPFLLVVPLSTLTSWQREIQTWASQM 554
Query: 676 NVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVLKDKAVLSKIKWNYLMVD 735
N +VY+G +SR + + +E+ + + K++KFN LLTTYE++LKDKA L + W ++ VD
Sbjct: 555 NAVVYLGDINSRNMIRTHEWMHPQT--KRLKFNILLTTYEILLKDKAFLGGLNWAFIGVD 612
Query: 736 EAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHFLDSDKFKSKDEFAQN 795
EAHRLKN ++ LY L +F + ++LLITGTPLQNS++ELW+LLHF+ +KF S ++F +
Sbjct: 613 EAHRLKNDDSLLYKTLIDFKSNHRLLITGTPLQNSLKELWSLLHFIMPEKFSSWEDFEEE 672
Query: 796 YKNLSSFNENELSNLHMELRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYKWILE 855
+ E ++LH EL P +LRRV KDVEKSLP K+E+ILR++MS LQKQYYKWIL
Sbjct: 673 H---GKGREYGYASLHKELEPFLLRRVKKDVEKSLPAKVEQILRMEMSALQKQYYKWILT 729
Query: 856 RNFRDLNKGVRGNQSKYCSRIEEV 879
RN++ L+KG +G+ S + + + E+
Sbjct: 730 RNYKALSKGSKGSTSGFLNIMMEL 753
Score = 343 bits (881), Expect = 2e-93
Identities = 249/720 (34%), Positives = 354/720 (48%), Gaps = 99/720 (13%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DILA+Y+ R F FQRLDGS K ELR+QA+DHFNA GS+DFCFLLSTRAGGLGINLA+AD
Sbjct: 816 DILAEYLKYRQFPFQRLDGSIKGELRKQALDHFNAEGSEDFCFLLSTRAGGLGINLASAD 875
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TV+IFDSDWNPQNDLQA +RAHRIGQ++ VNIYR VT SVEEDILERAKKKMVLDHLVI
Sbjct: 876 TVVIFDSDWNPQNDLQAQARAHRIGQKKQVNIYRLVTKGSVEEDILERAKKKMVLDHLVI 935
Query: 1058 QKLNAEGK--LEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDE 1115
Q+++ GK L A + F+K ELSAIL+FGAEELFKE +E+ + MDIDE
Sbjct: 936 QRMDTTGKTVLHTGSAPSSSTPFNKEELSAILKFGAEELFKEPEGEEQEPQ---EMDIDE 992
Query: 1116 ILERAEKVEEKENGGEQAHELLSAFKVANFCN-DEDD--------GSFWSRWIKADSVAQ 1166
IL+RAE E + ELLS FKVANF N DEDD W I + +
Sbjct: 993 ILKRAETHENEPGPLSVGDELLSQFKVANFSNMDEDDIELEPERNSKNWEEIIPEEQRRR 1052
Query: 1167 AENA----------LAPRAARNIKSYAEADQSERSKKRKKKENEPTERIPKRRKADYSAH 1216
E + PR K + R + ++ ++ I +R++
Sbjct: 1053 LEEEERQKELEEIYMLPRMRNCAKQISFNGSEGRRSRSRRYSGSDSDSISERKRPKKR-- 1110
Query: 1217 VISMIDGASAQVRSWSYGNLSKRDALRFSRSVMKFGNESQINLIVAEVGGAIEAAPLKAQ 1276
G + + S + RF +S KFG + +A ++ + +
Sbjct: 1111 ------GRPRTIPRENIKGFSDAEIRRFIKSYKKFGGPLERLDAIARDAELVDKSETDLR 1164
Query: 1277 VELFNALIDGCREAVEVGSL----------DLKGPLLDFYGVPMKANELLIRVQELQLLA 1326
L + +GC +A++ S +KGP GV + A ++ EL L
Sbjct: 1165 -RLGELVHNGCVKALKDSSSGTERAGGRLGKVKGPTFRISGVQVNAKLVIAHEDELIPLH 1223
Query: 1327 KRISRYEDPIAQFRVLTYLKPSNWSKGCGWNQIDDARLLLGVHYHGYGNWEVIRLDERLG 1386
K I + Q+ + + K +++ W + DD+ LL+G++ +GYG+WE+I++D L
Sbjct: 1224 KSIPSDPEERKQYTIPCHTKAAHFD--IDWGKEDDSNLLIGIYEYGYGSWEMIKMDPDLS 1281
Query: 1387 LTKKIAPVELQHHETFLPRAPNLRDRANALLEQELAVLGVKNASSKVGRKTSKKEREERE 1446
LT KI P + P+A L+ RA+ L++ L + A G SK+ +
Sbjct: 1282 LTHKILPDDPDKK----PQAKQLQTRADYLIKLLSRDLAKREAQRLCGAGGSKRRKTR-- 1335
Query: 1447 HLVDISLSRGQEKKKNIGSSKVNVQMRKDRLQKPLNVEPIVKEEGEMSDD--DDVYEQFK 1504
+K K + S KV +++ D P+ E+ + DD +D + K
Sbjct: 1336 ----------AKKSKAMKSIKVKEEIKSDS-------SPLPSEKSDEDDDKLNDSKPESK 1378
Query: 1505 EGKWKEWCQDLMV---------------------------EEMKTLK-RLHRLQTTSASL 1536
+ K D V E M+ +K L +L L
Sbjct: 1379 DRSKKSVVSDAPVHITASGEPVPIAEESEELDQKTFSICKERMRPVKAALKQLDRPEKGL 1438
Query: 1537 PKEKVLSKIRNYLQLLGRRIDQIVSEQEDEPHKQDRMTTRLWKYVSTFSHLSGERLHQIY 1596
+ + L R L +G I + + E + P + + LW +VS F+ +LH++Y
Sbjct: 1439 SEREQLEHTRQCLIKIGDHITECLKEYSN-PEQIKQWRKNLWIFVSKFTEFDARKLHKLY 1497
Score = 52.8 bits (125), Expect = 8e-06
Identities = 58/237 (24%), Positives = 97/237 (40%), Gaps = 36/237 (15%)
Query: 38 SDAEPDDASGKQNEAAAVDRLSTRESNVETTSRNPSASERWGSS---------YLKDCQP 88
SD++ SG Q+E+ + ++RE+ V+ +E W SS + QP
Sbjct: 50 SDSDSGSDSGSQSESESD---TSRENKVQAKPPKVDGAEFWKSSPSILAVQRSAMLRKQP 106
Query: 89 MSPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQKDSGKGQRGDSDVPA 148
Q + +S S D + ++ +D+SS + +K ED Q G
Sbjct: 107 QQAQQQRPASSNSGSEEDSSSSEDSDDSSSGAKRKKHNDEDWQMSGSGSPSQLGSDSESE 166
Query: 149 EEMLSDDSYGQDGE-EQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRILDDAEDD 207
EE G + + E V SR +P S S N + + +K R +D +ED+
Sbjct: 167 EERDKSSCDGTESDYEPKNKVRSR--KPQNRSK------SKNGKKILGQKKRQIDSSEDE 218
Query: 208 DDDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDEDIDVSDND 264
DD+ DY+ D+ +S R A ++ ED + DSD+ ++V D
Sbjct: 219 DDE-DYDNDK-------------RSSRRQATVNVSYK-EDEEMKTDSDDLLEVCGED 260
>HRP3_SCHPO (O14139) Chromodomain helicase hrp3
Length = 1388
Score = 392 bits (1007), Expect = e-108
Identities = 241/617 (39%), Positives = 356/617 (57%), Gaps = 64/617 (10%)
Query: 258 IDVSDNDDLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENSTAED 317
+D D +D++ K KG + G ++ SR R ++ +DE E+ T+E+
Sbjct: 75 VDTDDQEDVFPSKHRKGTRNG---------------SSFSRHRTIRD-LDDEAESVTSEE 118
Query: 318 SDSESDEDFKSLKKRGVRVRKNNGRSSAATSFSRPSNEVRSSSRTIRKVSY-----VESD 372
S+S+ D + K+ R +K+N + +E+R SSR + V+Y ES
Sbjct: 119 SESD-DSSYGGTPKKRSRQKKSN---------TYVQDEIRFSSRNSKGVNYNEDAYFESF 168
Query: 373 ESEGADEGTKKSQKEEIEVDDGDSVEKVLWHQPKGMAAEAQRNNQSMEPVLMSHLFDSEP 432
E E +E + + + E +D +++ VL H+ ++ H D
Sbjct: 169 EEEEEEEMYEYATEVSEEPEDTRAIDVVLDHR-----------------LIEGH--DGST 209
Query: 433 DWNNMEFLIKWKGQSHLHCQWKSFVDLQNLSGFKKVLNYTKRVT---EEIRNRMGISREE 489
+ EFLIKW SHLHC W+ + ++ + G KKV N+ K+V EIR +RE+
Sbjct: 210 PSEDYEFLIKWVNFSHLHCTWEPYNNISMIRGSKKVDNHIKQVILLDREIREDPTTTRED 269
Query: 490 IEVNDVSKEMDIDIIKQNSQVERIIADRISNDNSGNVFPEYLVKWQGLSYAEVTWEKDID 549
IE D+ KE + ++ QV+RI+A +++D S EYLVKW+ L Y TWE
Sbjct: 270 IEAMDIEKERKRENYEEYKQVDRIVAKHLNSDGS----VEYLVKWKQLLYDFCTWEASSI 325
Query: 550 IA-FAQHTIDEYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEWLKGGKLRDYQLEGL 608
I A I ++ RE + + ++ + K RKL++QP ++ GG+LRD+QL G+
Sbjct: 326 IEPIAATEIQAFQEREESALSPSRGTNYGNSRPK--YRKLEQQPSYITGGELRDFQLTGV 383
Query: 609 NFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEF 668
N++ W + N +LADEMGLGKTVQ+V+ L +L ++ + HGPFLVVVPLST+ W +
Sbjct: 384 NWMAYLWHKNENGILADEMGLGKTVQTVAFLSYLAHSLRQHGPFLVVVPLSTVPAWQETL 443
Query: 669 RKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVLKDKAVLSKIK 728
W D+N I Y+G +SR+V + YEF + ++IKFN LLTTYE VLKD++VLS IK
Sbjct: 444 ALWASDMNCISYLGNTTSRQVIRDYEFYVD--GTQKIKFNLLLTTYEYVLKDRSVLSNIK 501
Query: 729 WNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHFLDSDKFKS 788
W Y+ +DEAHRLKNSE+ LY ALS+F N+LLITGTPLQN++ EL AL+ FL KF+
Sbjct: 502 WQYMAIDEAHRLKNSESSLYEALSQFKNSNRLLITGTPLQNNIRELAALVDFLMPGKFEI 561
Query: 789 KDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQ 848
++E N + E + +L L+P++LRR+ KDVEKSLP K ERILRV++S LQ
Sbjct: 562 REEI--NLEAPDEEQEAYIRSLQEHLQPYILRRLKKDVEKSLPSKSERILRVELSDLQMY 619
Query: 849 YYKWILERNFRDLNKGV 865
+YK IL RN+R L + +
Sbjct: 620 WYKNILTRNYRVLTQSI 636
Score = 255 bits (651), Expect = 8e-67
Identities = 222/723 (30%), Positives = 348/723 (47%), Gaps = 114/723 (15%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DIL Y+SLRG+ QRLDG+ + +R+ ++DHFNAP S DF FLLSTRAGGLGINL TAD
Sbjct: 720 DILGDYLSLRGYPHQRLDGTVPAAVRRTSIDHFNAPNSPDFVFLLSTRAGGLGINLMTAD 779
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVIIFDSDWNPQ DLQAM+RAHRIGQ+ V +YR ++ ++EED+LERA++KM+L++ +I
Sbjct: 780 TVIIFDSDWNPQADLQAMARAHRIGQKNHVMVYRLLSKDTIEEDVLERARRKMILEYAII 839
Query: 1058 QKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDEIL 1117
+ G +K++ K F ELSAIL+FGA +FK E N +K+L M++DEIL
Sbjct: 840 ----SLGVTDKQKNSKNDK-FSAEELSAILKFGASNMFKAENN----QKKLEDMNLDEIL 890
Query: 1118 ERAEKVEEKENGGEQA---HELLSAFKVANFCNDEDDGSFWSRWIKADSVAQAENALAPR 1174
E AE + + G + E L F+V ++ KAD + + + P
Sbjct: 891 EHAEDHDTSNDVGGASMGGEEFLKQFEVTDY--------------KAD---VSWDDIIPL 933
Query: 1175 AARNIKSYAEADQSERSKKRKKKENEPTERIPKRRKADYSAHVISMIDGASAQVRSWSYG 1234
R + + E D+ ++ K+E E + R R Y S+ V S SY
Sbjct: 934 TER--EKFEEEDRLREEEEALKQEIELSSR---RGNRPY----------PSSAVESPSYS 978
Query: 1235 NLSKRDALR---------------FSRSVMKFGN-ESQINLIVAEVGGAIEAAPLKAQVE 1278
S+R + + R+++++G+ E + N IV + A + ++
Sbjct: 979 GTSERKSKKQMLKDEVLLEKEIRLLYRAMIRYGSLEHRYNDIVKYADLTTQDAHVIKKIA 1038
Query: 1279 LFNALIDGCREAVEVGSLDL-----------KGPLLDFYGVP-MKANELLIRVQELQLL- 1325
L+ R+AV DL K L+ F GV + A L+ R+ +L +L
Sbjct: 1039 A--DLVTASRKAVSAAEKDLSNDQSNNKSSRKALLITFKGVKNINAETLVQRLNDLDILY 1096
Query: 1326 -AKRISRYEDPIAQFRVLTYLKP-SNWSKGCGWNQIDDARLLLGVHYHGYGNWEVIRLDE 1383
A S Y + F++ +++ WS C W +D+ LL G+ HG+G W IR D
Sbjct: 1097 DAMPTSGY----SNFQIPMHVRSVHGWS--CQWGPREDSMLLSGICKHGFGAWLEIRDDP 1150
Query: 1384 RLGLTKKI-----------APVELQHHETFLPRAPNLRDRANAL---LEQELAVLGVKN- 1428
L + KI P + ++ E +P A +L R L L + G+K+
Sbjct: 1151 ELKMKDKIFLEDTKQTDNSVPKDKENKEKKVPSAVHLVRRGEYLLSALREHHQNFGIKSS 1210
Query: 1429 -ASSKVGRKTSKKEREEREHLVDISLSRGQE-KKKNIGSSKVNVQMRKDRLQKPLNVEPI 1486
A S G+ KK+ R ++ Q+ + S + RK K +E
Sbjct: 1211 PAISTNGKTQPKKQTANRRQSGKPNVKSAQKIESATRTPSPAISESRKKPSSKDTKIETP 1270
Query: 1487 VKEEGEMSDDDDVYEQFKEG------KWKEWCQDLMVEEMKTLKRLHRLQTTSASLPKEK 1540
+E+ V + +G + K C++LM K +KRL + S+ L + +
Sbjct: 1271 SREQSRSQTASPVKSEKDDGNVSLNAEQKARCKELMYPVRKHMKRLRK---DSSGLGRAE 1327
Query: 1541 VLSKIRNYLQLLGRRIDQIVSEQEDEPHKQDRMTTRLWKYVSTF---SHLSGERLHQIYS 1597
++ + L +G+ I++ V++ E ++ + LW + F + L +Y
Sbjct: 1328 LVKLLTECLTTIGKHIEKTVNDTPSE--EKATVRKNLWMFACYFWPKEEVKYTSLISMYE 1385
Query: 1598 KLK 1600
K+K
Sbjct: 1386 KMK 1388
>CHD1_YEAST (P32657) Chromo domain protein 1
Length = 1468
Score = 379 bits (972), Expect = e-104
Identities = 233/665 (35%), Positives = 368/665 (55%), Gaps = 54/665 (8%)
Query: 255 DEDIDVSDNDDLYF---DKKAKGRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDE 311
D +V N +LY +A Q+ F S D + SR++R+ + +DEDE
Sbjct: 5 DISTEVLQNPELYGLRRSHRAAAHQQNYFNDS--DDEDDEDNIKQSRRKRMTTIEDDEDE 62
Query: 312 NSTAE-DSDSESDEDFKSLKKR----GVRVRKNNGRSSAAT---SFSRPSNE-------- 355
E + DS DED + ++ G +++N + + T S S+P ++
Sbjct: 63 FEDEEGEEDSGEDEDEEDFEEDDDYYGSPIKQNRSKPKSRTKSKSKSKPKSQSEKQSTVK 122
Query: 356 --VRSSSRTIRKVSY---------VESDESEGADEGTKKSQKEEIEV----DDGDSVEKV 400
R S+R + V+Y +ES++ G++E + E +D ++ V
Sbjct: 123 IPTRFSNRQNKTVNYNIDYSDDDLLESEDDYGSEEALSEENVHEASANPQPEDFHGIDIV 182
Query: 401 LWHQPKGMAAEAQRNNQSMEPVLMSHLFDSEPDWNNMEFLIKWKGQSHLHCQWKSFVDLQ 460
+ H+ K E + VL + D N EFLIKW +SHLH W+++ +
Sbjct: 183 INHRLKTSLEEGK--------VLEKTVPDLNNCKENYEFLIKWTDESHLHNTWETYESIG 234
Query: 461 NLSGFKKVLNYTKRVT---EEIRNRMGISREEIEVNDVSKEMDIDIIKQNSQVERIIADR 517
+ G K++ NY K+ +++R ++ E+IE+ D+ +E +D ++ ERII +
Sbjct: 235 QVRGLKRLDNYCKQFIIEDQQVRLDPYVTAEDIEIMDMERERRLDEFEEFHVPERIIDSQ 294
Query: 518 ISNDNSGNVFPEYLVKWQGLSYAEVTWEKDIDIA-FAQHTIDEYKTREAAMSVQGKMVDF 576
++ G +YLVKW+ L+Y E TWE DI A + ++ RE + + ++
Sbjct: 295 RASLEDGTSQLQYLVKWRRLNYDEATWENATDIVKLAPEQVKHFQNRENSKILPQYSSNY 354
Query: 577 QRRQSKGSLRKLDEQPEWLKGGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSV 636
++ + KL QP ++KGG+LRD+QL G+N++ W N +LADEMGLGKTVQ+V
Sbjct: 355 TSQRPR--FEKLSVQPPFIKGGELRDFQLTGINWMAFLWSKGDNGILADEMGLGKTVQTV 412
Query: 637 SMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFC 696
+ + +L A++ +GP ++VVPLST+ W F KW PDLN I Y+G + SR+ ++YEF
Sbjct: 413 AFISWLIFARRQNGPHIIVVPLSTMPAWLDTFEKWAPDLNCICYMGNQKSRDTIREYEFY 472
Query: 697 NEKKA--GKQIKFNALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEF 754
+A K +KFN LLTTYE +LKD+A L IKW ++ VDEAHRLKN+E+ LY +L+ F
Sbjct: 473 TNPRAKGKKTMKFNVLLTTYEYILKDRAELGSIKWQFMAVDEAHRLKNAESSLYESLNSF 532
Query: 755 NTKNKLLITGTPLQNSVEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMEL 814
N++LITGTPLQN+++EL AL++FL +F E +++N E + +LH +
Sbjct: 533 KVANRMLITGTPLQNNIKELAALVNFLMPGRFTIDQEI--DFENQDEEQEEYIHDLHRRI 590
Query: 815 RPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYKWILERNFRDLNKGVRGNQSKYCS 874
+P +LRR+ KDVEKSLP K ERILRV++S +Q +YYK IL +N+ L G +G +
Sbjct: 591 QPFILRRLKKDVEKSLPSKTERILRVELSDVQTEYYKNILTKNYSALTAGAKGGHFSLLN 650
Query: 875 RIEEV 879
+ E+
Sbjct: 651 IMNEL 655
Score = 241 bits (615), Expect = 1e-62
Identities = 219/767 (28%), Positives = 339/767 (43%), Gaps = 126/767 (16%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DIL Y+S++G FQRLDG+ S R+ ++DHFN+P S+DF FLLSTRAGGLGINL TAD
Sbjct: 725 DILGDYLSIKGINFQRLDGTVPSAQRRISIDHFNSPDSNDFVFLLSTRAGGLGINLMTAD 784
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TV+IFDSDWNPQ DLQAM+RAHRIGQ+ V +YR V+ +VEE++LERA+KKM+L++ +I
Sbjct: 785 TVVIFDSDWNPQADLQAMARAHRIGQKNHVMVYRLVSKDTVEEEVLERARKKMILEYAII 844
Query: 1058 QKLNAEG-KLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDEI 1116
+G K KK G ELSAIL+FGA +F N +K+L +++D++
Sbjct: 845 SLGVTDGNKYTKKNEPNAG------ELSAILKFGAGNMFTATDN----QKKLEDLNLDDV 894
Query: 1117 LERAE------KVEEKENGGEQAHELLSAFKVANFCNDEDDGSFWSRWI----------- 1159
L AE + E GGE E L F+V ++ D D W I
Sbjct: 895 LNHAEDHVTTPDLGESHLGGE---EFLKQFEVTDYKADID----WDDIIPEEELKKLQDE 947
Query: 1160 ----KADSVAQAENALAPRAARNIKSYAEADQSERSKKRKKKENEPTERIPKRRKADYSA 1215
K + + + + R +K + + + +++ T R +RR
Sbjct: 948 EQKRKDEEYVKEQLEMMNRRDNALKKIKNSVNGDGTAANSDSDDDSTSRSSRRRARANDM 1007
Query: 1216 HVISMIDGASAQVRSWSYGNLSK------RDALRFSRSVMKFG----------------- 1252
I + + +GNL + D +S K+G
Sbjct: 1008 DSIGESEVRALYKAILKFGNLKEILDELIADGTLPVKSFEKYGETYDEMMEAAKDCVHEE 1067
Query: 1253 --NESQINLIVAEVGGAIEAAPLKAQVELFNALIDGCREAVEVGSLDLKGPLLDFYGV-P 1309
N +I + + A A +++ N D + + + K L +F GV
Sbjct: 1068 EKNRKEILEKLEKHATAYRAKLKSGEIKAENQPKDNPLTRLSLKKREKKAVLFNFKGVKS 1127
Query: 1310 MKANELLIRVQELQLLAKRI-SRYEDPIAQFRV--LTYLKPSNWSKGCGWNQIDDARLLL 1366
+ A LL RV++L+ L I S Y+D +F + T NWS W + +D +LL+
Sbjct: 1128 LNAESLLSRVEDLKYLKNLINSNYKDDPLKFSLGNNTPKPVQNWSS--NWTKEEDEKLLI 1185
Query: 1367 GVHYHGYGNWEVIRLDERLGLTKKIAPVELQH-------------------------HET 1401
GV +GYG+W IR D LG+T KI E+ +
Sbjct: 1186 GVFKYGYGSWTQIRDDPFLGITDKIFLNEVHNPVAKKSASSSDTTPTPSKKGKGITGSSK 1245
Query: 1402 FLPRAPNLRDRANALLEQELAVLGVKNASSKVGRK------TSKKEREEREH-------L 1448
+P A +L R + LL L K+ S+ +G K + K++R+ H +
Sbjct: 1246 KVPGAIHLGRRVDYLLSFLRGGLNTKSPSADIGSKKLPTGPSKKRQRKPANHSKSMTPEI 1305
Query: 1449 VDISLSRGQEKKKNIGSSKVNVQMRKDRLQKPLNVEPIVK---------EEGEMSDDDDV 1499
+ G K+ K + + P + P +K +
Sbjct: 1306 TSSEPANGPPSKRMKALPKGPAALINNTRLSPNSPTPPLKSKVSRDNGTRQSSNPSSGSA 1365
Query: 1500 YEQFKEGKWKEWCQDLMVEEMKTLKRLHRLQTTSASLPKEKVLSKIRNYLQLLGRRID-Q 1558
+E+ + +E C+ M +LKRL R SL +++ ++ L +G I+ Q
Sbjct: 1366 HEKEYDSMDEEDCRHTMSAIRTSLKRLRR---GGKSLDRKEWAKILKTELTTIGNHIESQ 1422
Query: 1559 IVSEQEDEPHKQDRMTTRLWKYVSTF--SHLSGERLHQIYSKLKLEQ 1603
S ++ P K + LW Y + F + + +L +Y K+ Q
Sbjct: 1423 KGSSRKASPEKYRK---HLWSYSANFWPADVKSTKLMAMYDKITESQ 1466
>HRP1_SCHPO (Q9US25) Chromodomain helicase hrp1
Length = 1373
Score = 358 bits (920), Expect = 5e-98
Identities = 210/518 (40%), Positives = 314/518 (60%), Gaps = 36/518 (6%)
Query: 350 SRPSNEVRSSSRTIRKVSYVESDESEGAD--EGTKKSQKEEIEVDDGDSVEKVLWHQPKG 407
S E+R+S R+ + S V +E E D E ++ +E++E + ++ VL H+ +
Sbjct: 161 SASGTEIRTSLRSSKGKS-VNYNEQEFYDDFEDEEEEVEEQVEEEYEPIIDFVLNHRKR- 218
Query: 408 MAAEAQRNNQSMEPVLMSHLFDSEPDWNNMEFLIKWKGQSHLHCQWKSFVDLQNLSGFKK 467
A+AQ D +P ++ ++LIKW+ SHLH W+ + L ++ G+KK
Sbjct: 219 --ADAQ---------------DDDPK-SSYQYLIKWQEVSHLHNTWEDYSTLSSVRGYKK 260
Query: 468 VLNYTKR---VTEEIRNRMGISREEIEVNDVSKEMDIDIIKQNSQVERIIADRISNDNSG 524
V NY K+ EIR + E+IE D+ +E + ++ VERI+A + +
Sbjct: 261 VDNYIKQNIIYDREIREDPTTTFEDIEALDIERERKNMLFEEYKIVERIVASETNEEGK- 319
Query: 525 NVFPEYLVKWQGLSYAEVTWEK-DIDIAFAQHTIDEYKTREAAMSVQGKMVDFQRRQSKG 583
EY VKW+ L Y TWE D+ + A + + ++ RE + + K V + R
Sbjct: 320 ---TEYFVKWRQLPYDNCTWEDADVIYSMAPNEVYQFLQRENSPYLPYKGVFYNTRPP-- 374
Query: 584 SLRKLDEQPEWLKGGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQ 643
RKL++QP ++KGG++RD+QL G+N++ W + N +LADEMGLGKTVQ+V L +L
Sbjct: 375 -YRKLEKQPSYIKGGEIRDFQLTGINWMAYLWHRNENGILADEMGLGKTVQTVCFLSYLV 433
Query: 644 NAQQIHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGK 703
++ + HGPFL+VVPLST+ W + W PDLN I Y G SR ++YEF + +
Sbjct: 434 HSLKQHGPFLIVVPLSTVPAWQETLANWTPDLNSICYTGNTESRANIREYEFYLSTNS-R 492
Query: 704 QIKFNALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLIT 763
++KFN LLTTYE +LKDK L+ I+W YL +DEAHRLKNSE+ LY LS+F T N+LLIT
Sbjct: 493 KLKFNILLTTYEYILKDKQELNNIRWQYLAIDEAHRLKNSESSLYETLSQFRTANRLLIT 552
Query: 764 GTPLQNSVEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVI 823
GTPLQN+++EL +L++FL KF +DE N+ ++ E ++ +L L+P +LRR+
Sbjct: 553 GTPLQNNLKELASLVNFLMPGKFYIRDEL--NFDQPNAEQERDIRDLQERLQPFILRRLK 610
Query: 824 KDVEKSLPPKIERILRVDMSPLQKQYYKWILERNFRDL 861
KDVEKSLP K ERILRV++S +Q ++YK IL +N+R L
Sbjct: 611 KDVEKSLPSKSERILRVELSDMQTEWYKNILTKNYRAL 648
Score = 268 bits (686), Expect = 7e-71
Identities = 195/543 (35%), Positives = 297/543 (53%), Gaps = 51/543 (9%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
+IL +YMSLRG+ +QRLDG+ + +R+ ++DHFNAP S DF FLLSTRAGGLGINL TAD
Sbjct: 734 NILGEYMSLRGYNYQRLDGTIPASVRRVSIDHFNAPDSPDFVFLLSTRAGGLGINLNTAD 793
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVIIFDSDWNPQ DLQAM+RAHRIGQ+ VN+YRF++ +VEEDILERA++KM+L++ +I
Sbjct: 794 TVIIFDSDWNPQADLQAMARAHRIGQKNHVNVYRFLSKDTVEEDILERARRKMILEYAII 853
Query: 1058 QKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDEIL 1117
+ G EK + K +D ELSAIL+FGA +FK E++K+L +M++D+IL
Sbjct: 854 ----SLGVTEKSKNSKNDK-YDAQELSAILKFGASNMFKA----TENQKKLENMNLDDIL 904
Query: 1118 ERAEKVEEKENGGEQA---HELLSAFKVANFCNDEDDGSFWSRWIKADSVAQAENALAPR 1174
AE + + G + E L F+V ++ ++ + W I + + + E
Sbjct: 905 SHAEDRDSSNDVGGASMGGEEFLKQFEVTDYKAEDLN---WDDIIPEEEMERIEEEERML 961
Query: 1175 AARNIKSYAEADQSERSKKRKKKENEPTERIPKRRKADYSAHVISMIDGASAQVRSWSYG 1234
AA+ K E ++R+++E E E P R + + VR
Sbjct: 962 AAQRAK-------EEERERREEEERENDEDHPSRTYKRTTKSITKRQQRREEMVR----- 1009
Query: 1235 NLSKRDALRFSRSVMKFG-NESQINLIVAEVGGAIEAAPLKAQVELFNALIDGCREAVE- 1292
+++ R+++KFG + + + IV E ++A K L ++ C EAVE
Sbjct: 1010 ---EKEIRLLYRAMIKFGLVDERFDTIVKE--AELQATDPKRIYSLSADMVKACDEAVER 1064
Query: 1293 VGSLDLKGP------LLDFYGVP-MKANELLIRVQELQLLAKRISRYEDPIAQFRVLTYL 1345
+G+ D K L++F GV + A + +RV++L L R + DP+ Q ++ Y
Sbjct: 1065 LGADDTKNKQPRKAILIEFKGVKNINAETVTLRVKDLTHL-HRAYKGLDPLKQ--IIGYP 1121
Query: 1346 KPSNWSKGCGWNQIDDARLLLGVHYHGYGNWEVIRLDERLGLTKKIAPVELQH--HETFL 1403
S S C W +D+ LL G++ HG+G W+ I+ D LGL KI E ++ ++
Sbjct: 1122 IRSVHSWNCSWGIKEDSMLLAGINKHGFGCWQAIKNDPDLGLHDKIFLDEAKNDKESRYV 1181
Query: 1404 PRAPNLRDRANALLE--QELAVLGVKNASSKVGRKTSKK---EREEREHLVDISLSRGQE 1458
P A +L R LL +E L V RK ++K + R+ +D S+S ++
Sbjct: 1182 PSAVHLVRRGEYLLSVVREHPDLFVVKTDQPTKRKYNRKAPTKSSTRQTTLDGSISNTKK 1241
Query: 1459 KKK 1461
+
Sbjct: 1242 SSR 1244
>CHD7_HUMAN (Q9P2D1) Chromodomain-helicase-DNA-binding protein 7
(CHD-7) (Fragment)
Length = 1967
Score = 310 bits (795), Expect = 2e-83
Identities = 193/509 (37%), Positives = 299/509 (57%), Gaps = 51/509 (10%)
Query: 365 KVSYVESDESE--GADEGTKKSQKEEIEVDD--GDSVEKVLWHQPKGMAAEAQRNNQSME 420
K+S E+D+++ G D + SQ E+ E D G VEK++ + ++ +S E
Sbjct: 4 KISDEEADDADAAGRDSPSNTSQSEQQESVDAEGPVVEKIM------SSRSVKKQKESGE 57
Query: 421 PVLMSHLFDSEPDWNNMEFLIKWKGQSHLHCQWKSFVDLQNLSGFKKVLNYTKRVTEEIR 480
V + EF +K+K S+LHCQW S DL+ KR+ ++I+
Sbjct: 58 EVEIE------------EFYVKYKNFSYLHCQWASIEDLEK----------DKRIQQKIK 95
Query: 481 NRMGISREEIEVNDVSKEM-DIDIIKQNSQVERIIADRISNDNSGNVFPEYLVKWQGLSY 539
+ ++++ E+ + D + +V+RI+ S D+ G YLVKW L Y
Sbjct: 96 RFKAKQGQNKFLSEIEDELFNPDYV----EVDRIMDFARSTDDRGEPVTHYLVKWCSLPY 151
Query: 540 AEVTWEKDIDIAFAQHTIDEYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEWLKGGK 599
+ TWE+ DI A+ I+E+ E MS + + +R + +K + E+ K
Sbjct: 152 EDSTWERRQDIDQAK--IEEF---EKLMSREPETERVERPPAD-DWKKSESSREYKNNNK 205
Query: 600 LRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLS 659
LR+YQLEG+N+L+ +W N N +LADEMGLGKT+QS++ L + + IHGPFLV+ PLS
Sbjct: 206 LREYQLEGVNWLLFNWYNMRNCILADEMGLGKTIQSITFLYEIY-LKGIHGPFLVIAPLS 264
Query: 660 TLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIK----FNALLTTYE 715
T+ NW +EFR W +LNV+VY G+++SR Q YE + G+ IK F+A++TT+E
Sbjct: 265 TIPNWEREFRTWT-ELNVVVYHGSQASRRTIQLYEMYFKDPQGRVIKGSYKFHAIITTFE 323
Query: 716 VVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELW 775
++L D L I W +++DEAHRLKN +L L + ++K+L+TGTPLQN+VEEL+
Sbjct: 324 MILTDCPELRNIPWRCVVIDEAHRLKNRNCKLLEGLKMMDLEHKVLLTGTPLQNTVEELF 383
Query: 776 ALLHFLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVIKDVEKSLPPKIE 835
+LLHFL+ +F S+ F Q + +L + E ++ L L+P MLRR+ +DVEK+L PK E
Sbjct: 384 SLLHFLEPSRFPSETTFMQEFGDLKT--EEQVQKLQAILKPMMLRRLKEDVEKNLAPKEE 441
Query: 836 RILRVDMSPLQKQYYKWILERNFRDLNKG 864
I+ V+++ +QK+YY+ ILE+NF L+KG
Sbjct: 442 TIIEVELTNIQKKYYRAILEKNFTFLSKG 470
Score = 169 bits (429), Expect = 5e-41
Identities = 100/261 (38%), Positives = 142/261 (54%), Gaps = 17/261 (6%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DIL Y+ R + ++R+DG + LRQ A+D F+ P SD F FLL TRAGGLGINL AD
Sbjct: 558 DILEDYLIQRRYPYERIDGRVRGNLRQAAIDRFSKPDSDRFVFLLCTRAGGLGINLTAAD 617
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
T IIFDSDWNPQNDLQA +R HRIGQ + V IYR +T S E ++ ++A K+ LD V+
Sbjct: 618 TCIIFDSDWNPQNDLQAQARCHRIGQSKSVKIYRLITRNSYEREMFDKASLKLGLDKAVL 677
Query: 1058 QKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDEIL 1117
Q ++ ++ A G K E+ +LR GA +E E+ + DID+IL
Sbjct: 678 QSMSG-----RENATNGVQQLSKKEIEDLLRKGAYGALMDE---EDEGSKFCEEDIDQIL 729
Query: 1118 ERAEKVEEKENGGEQAHELLSAFKVANFCND--EDDGSFWSRWIK-----ADSVAQAENA 1170
R E+ G+ + ++F + D DD +FW +W K D++ N
Sbjct: 730 LRRTHTITIESEGKGSTFAKASFVASGNRTDISLDDPNFWQKWAKKAELDIDALNGRNNL 789
Query: 1171 L--APRAARNIKSYAEADQSE 1189
+ PR + + Y+ + E
Sbjct: 790 VIDTPRVRKQTRLYSAVKEDE 810
>CHD6_HUMAN (Q8TD26) Chromodomain-helicase-DNA-binding protein 6
(CHD-6)
Length = 2713
Score = 305 bits (782), Expect = 5e-82
Identities = 202/634 (31%), Positives = 330/634 (51%), Gaps = 84/634 (13%)
Query: 270 KKAKGRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENSTAEDSDSESDEDFKSL 329
KKAK + K + R + + +++ + K S D + + + E
Sbjct: 135 KKAKEHKEPKQKDGAKKARKPRE-ASGTKEAKEKRSCTDSAARTKSRKASKEQGPTPVEK 193
Query: 330 KKRGVRVRKNNGRS-------------SAATSFSRPSNEVRSSSRTIRKVSYVESDESEG 376
KK+G R + S S S ++ R S R +++ Y E + +
Sbjct: 194 KKKGKRKSETTVESLELDQGLTNPSLRSPEESTESTDSQKRRSGRQVKRRKYNEDLDFKV 253
Query: 377 ADE------------------GTKKSQKEEIEVDDGDSVEKVLWHQPKGMAAEAQRNNQS 418
D+ T Q EE DD + +EK+L A + Q
Sbjct: 254 VDDDGETIAVLGAGRTSALSASTLAWQAEEPPEDDANIIEKIL----------ASKTVQE 303
Query: 419 MEPVLMSHLFDSEPDWNNMEFLIKWKGQSHLHCQWKSFVDLQNLSGFKKVLNYTKRVTEE 478
+ P EP ++ F +K++ S+LHC+W + +L+ R+ ++
Sbjct: 304 VHP--------GEPPFDLELFYVKYRNFSYLHCKWATMEELEK----------DPRIAQK 345
Query: 479 IRNRMGISREEIEVNDVSKEMDIDIIKQNS-QVERIIADRISND-NSGNVFPEYLVKWQG 536
I+ ++ ++ + E D D+ + +V+RI+ + D +G YLVKW
Sbjct: 346 IKR---FRNKQAQMKHIFTEPDEDLFNPDYVEVDRILEVAHTKDAETGEEVTHYLVKWCS 402
Query: 537 LSYAEVTWEKDIDIAFAQHTIDEYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEWLK 596
L Y E TWE + D+ A+ + E+++ + ++ R + S +KL++ E+
Sbjct: 403 LPYEESTWELEEDVDPAK--VKEFESLQVLPEIK-----HVERPASDSWQKLEKSREYKN 455
Query: 597 GGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLG--FLQNAQQIHGPFLV 654
+LR+YQLEG+N+L+ +W N N +LADEMGLGKT+QS++ L FL+ IHGPFL+
Sbjct: 456 SNQLREYQLEGMNWLLFNWYNRKNCILADEMGLGKTIQSITFLSEIFLRG---IHGPFLI 512
Query: 655 VVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQI----KFNAL 710
+ PLST++NW +EFR W ++N IVY G++ SR++ QQYE G + KF+ +
Sbjct: 513 IAPLSTITNWEREFRTWT-EMNAIVYHGSQISRQMIQQYEMVYRDAQGNPLSGVFKFHVV 571
Query: 711 LTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNS 770
+TT+E++L D L KI W+ +++DEAHRLKN +L L ++K+L+TGTPLQNS
Sbjct: 572 ITTFEMILADCPELKKIHWSCVIIDEAHRLKNRNCKLLEGLKLMALEHKVLLTGTPLQNS 631
Query: 771 VEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVIKDVEKSL 830
VEEL++LL+FL+ +F S+ F + + +L + E ++ L L+P MLRR+ DVEK+L
Sbjct: 632 VEELFSLLNFLEPSQFPSETAFLEEFGDLKT--EEQVKKLQSILKPMMLRRLKDDVEKNL 689
Query: 831 PPKIERILRVDMSPLQKQYYKWILERNFRDLNKG 864
PK E I+ V+++ +QK+YY+ ILE+NF L KG
Sbjct: 690 APKQETIIEVELTNIQKKYYRAILEKNFSFLTKG 723
Score = 170 bits (430), Expect = 4e-41
Identities = 101/261 (38%), Positives = 141/261 (53%), Gaps = 18/261 (6%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DIL Y+ R + ++R+DG + LRQ A+D F P SD F FLL TRAGGLGINL AD
Sbjct: 811 DILEDYLIQRRYTYERIDGRVRGNLRQAAIDRFCKPDSDRFVFLLCTRAGGLGINLTAAD 870
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
T IIFDSDWNPQNDLQA +R HRIGQ + V +YR +T S E ++ ++A K+ LD V+
Sbjct: 871 TCIIFDSDWNPQNDLQAQARCHRIGQSKAVKVYRLITRNSYEREMFDKASLKLGLDKAVL 930
Query: 1058 QKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDEIL 1117
Q +N +G G K E+ +LR GA +E E+ + DID+IL
Sbjct: 931 QDINRKG------GTNGVQQLSKMEVEDLLRKGAYGALMDE---EDEGSKFCEEDIDQIL 981
Query: 1118 ERAEKVEEKENGGEQAHELLSAFKVANFCND--EDDGSFWSRW---IKADSVAQAENAL- 1171
+R ++ G+ + ++F + D DD +FW +W K D+ A+ E
Sbjct: 982 QRRTHTITIQSEGKGSTFAKASFVASGNRTDISLDDPNFWQKWAKIAKLDTEAKNEKESL 1041
Query: 1172 ---APRAARNIKSYAEADQSE 1189
PR + K Y ++ E
Sbjct: 1042 VIDRPRVRKQTKHYNSFEEDE 1062
>CHD8_HUMAN (Q9HCK8) Chromodomain-helicase-DNA-binding protein 8
(CHD-8) (Helicase with SNF2 domain 1) (Fragment)
Length = 2004
Score = 302 bits (773), Expect = 6e-81
Identities = 173/434 (39%), Positives = 266/434 (60%), Gaps = 32/434 (7%)
Query: 438 EFLIKWKGQSHLHCQWKSFVDLQNLSGFKKVLNYTKRVTEEIRNRMGISREEIEVNDVSK 497
EF +K+K S+LHC+W + L+ + L K ++R+ E + V
Sbjct: 90 EFFVKYKNYSYLHCEWATISQLEKDKRIHQKLKRFKTKMAQMRHFFHEDEEPFNPDYV-- 147
Query: 498 EMDIDIIKQNSQVERIIADRISND-NSGNVFPEYLVKWQGLSYAEVTWEKDIDIAFAQHT 556
+V+RI+ + S D ++G YLVKW L Y + TWE D+
Sbjct: 148 -----------EVDRILDESHSIDKDNGEPVIYYLVKWCSLPYEDSTWELKEDV------ 190
Query: 557 IDEYKTREAAMSVQGKMVDFQR--RQSKGSLRKLDEQPEWLKGGKLRDYQLEGLNFLVNS 614
DE K RE +Q + + +R R + +KL+ E+ +LR+YQLEG+N+L+ +
Sbjct: 191 -DEGKIREFKR-IQSRHPELKRVNRPQASAWKKLELSHEYKNRNQLREYQLEGVNWLLFN 248
Query: 615 WKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEFRKWLPD 674
W N N +LADEMGLGKT+QS++ L + N IHGPFLV+ PLST++NW +EF W +
Sbjct: 249 WYNRQNCILADEMGLGKTIQSIAFLQEVYNVG-IHGPFLVIAPLSTITNWEREFNTWT-E 306
Query: 675 LNVIVYVGTRSSREVCQQYEFCNEKKAGKQI----KFNALLTTYEVVLKDKAVLSKIKWN 730
+N IVY G+ +SR++ QQYE + G+ I KF+AL+TT+E++L D L +I+W
Sbjct: 307 MNTIVYHGSLASRQMIQQYEMYCKDSRGRLIPGAYKFDALITTFEMILSDCPELREIEWR 366
Query: 731 YLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHFLDSDKFKSKD 790
+++DEAHRLKN +L +L + ++K+L+TGTPLQN+VEEL++LLHFL+ +F S+
Sbjct: 367 CVIIDEAHRLKNRNCKLLDSLKHMDLEHKVLLTGTPLQNTVEELFSLLHFLEPSQFPSES 426
Query: 791 EFAQNYKNLSSFNENELSNLHMELRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYY 850
EF +++ +L + E ++ L L+P MLRR+ +DVEK+L PK E I+ V+++ +QK+YY
Sbjct: 427 EFLKDFGDLKT--EEQVQKLQAILKPMMLRRLKEDVEKNLAPKQETIIEVELTNIQKKYY 484
Query: 851 KWILERNFRDLNKG 864
+ ILE+NF L+KG
Sbjct: 485 RAILEKNFSFLSKG 498
Score = 170 bits (430), Expect = 4e-41
Identities = 115/292 (39%), Positives = 154/292 (52%), Gaps = 32/292 (10%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DIL Y+ R + ++R+DG + LRQ A+D F+ P SD F FLL TRAGGLGINL AD
Sbjct: 586 DILEDYLIQRRYLYERIDGRVRGNLRQAAIDRFSKPDSDRFVFLLCTRAGGLGINLTAAD 645
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
T IIFDSDWNPQNDLQA +R HRIGQ + V +YR +T S E ++ ++A K+ LD V+
Sbjct: 646 TCIIFDSDWNPQNDLQAQARCHRIGQSKAVKVYRLITRNSYEREMFDKASLKLGLDKAVL 705
Query: 1058 QKLNA-EGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDEI 1116
Q ++ +G + G F K E+ +LR GA EE +DE SK DID+I
Sbjct: 706 QSMSGRDGNI------TGIQQFSKKEIEDLLRKGAYAAIMEE-DDEGSK--FCEEDIDQI 756
Query: 1117 LERAEKVEEKENGGEQAHELLSAFKVANFCNDE-------DDGSFWSRWI-KAD---SVA 1165
L R E+ G+ S F A+F E DD +FW +W KAD +
Sbjct: 757 LLRRTTTITIESEGKG-----STFAKASFVASENRTDISLDDPNFWQKWAKKADLDMDLL 811
Query: 1166 QAENAL---APRA---ARNIKSYAEADQSERSKKRKKKENEPTERIPKRRKA 1211
++N L PR R+ + + D E S + + P R R A
Sbjct: 812 NSKNNLVIDTPRVRKQTRHFSTLKDDDLVEFSDLESEDDERPRSRRHDRHHA 863
>CHD4_MOUSE (Q6PDQ2) Chromodomain helicase-DNA-binding protein 4
(CHD-4)
Length = 1915
Score = 296 bits (757), Expect = 4e-79
Identities = 189/519 (36%), Positives = 286/519 (54%), Gaps = 61/519 (11%)
Query: 397 VEKVL---WHQPKGMAA-----EAQRNNQSMEPVLMSHLFDSEPDWNNMEFLIKWKGQSH 448
V+K+L W QP +A N S +P+ + P+ +F +KW+G S+
Sbjct: 495 VQKILIWKWGQPPSPTPVPRPPDADPNTPSPKPL------EGRPE---RQFFVKWQGMSY 545
Query: 449 LHCQWKSFVDLQNLSGFKKVLNYTKR--VTEEIRNRMG----ISREEIEVNDVSKEMDID 502
HC W S + L+ L NY ++ + E G SR+ + EM+
Sbjct: 546 WHCSWVSELQLE-LHCQVMFRNYQRKNDMDEPPSGDFGGDEEKSRKRKNKDPKFAEMEER 604
Query: 503 IIKQNSQVERIIADRISN---DNSGNVFPEYLVKWQGLSYAEVTWEKDIDIAFAQHTIDE 559
+ + E ++ RI N D G+V YL+KW+ L Y + +WE + D+ + + +
Sbjct: 605 FYRYGIKPEWMMIHRILNHSVDKKGHV--HYLIKWRDLPYDQASWESE-DVEIQDYDLFK 661
Query: 560 YKTREAAMSVQGKMVDFQRRQSKGSLRKLDE---------------QPEWLK--GGKLRD 602
++G+ ++ K LRKL+ QPE+L GG L
Sbjct: 662 QSYWNHRELMRGEEGRPGKKLKKVKLRKLERPPETPTVDPTVKYERQPEYLDATGGTLHP 721
Query: 603 YQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLS 662
YQ+EGLN+L SW T+ +LADEMGLGKTVQ+ L L GPFLV PLST+
Sbjct: 722 YQMEGLNWLRFSWAQGTDTILADEMGLGKTVQTAVFLYSLYKEGHSKGPFLVSAPLSTII 781
Query: 663 NWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNE-------KKAGK-----QIKFNAL 710
NW +EF W PD+ V+ YVG + SR + ++ EF E KKA + +KF+ L
Sbjct: 782 NWEREFEMWAPDMYVVTYVGDKDSRAIIRENEFSFEDNAIRGGKKASRMKKEASVKFHVL 841
Query: 711 LTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNS 770
LT+YE++ D A+L I W L+VDEAHRLKN++++ + L+ ++ ++KLL+TGTPLQN+
Sbjct: 842 LTSYELITIDMAILGSIDWACLIVDEAHRLKNNQSKFFRVLNGYSLQHKLLLTGTPLQNN 901
Query: 771 VEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVIKDVEKSL 830
+EEL+ LL+FL ++F + + F + + +++ E+++ LH L PHMLRR+ DV K++
Sbjct: 902 LEELFHLLNFLTPERFHNLEGFLEEFADIA--KEDQIKKLHDMLGPHMLRRLKADVFKNM 959
Query: 831 PPKIERILRVDMSPLQKQYYKWILERNFRDLNKGVRGNQ 869
P K E I+RV++SP+QK+YYK+IL RNF LN GNQ
Sbjct: 960 PSKTELIVRVELSPMQKKYYKYILTRNFEALNARGGGNQ 998
Score = 194 bits (492), Expect = 2e-48
Identities = 138/372 (37%), Positives = 191/372 (51%), Gaps = 65/372 (17%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
D+L ++ G++++R+DG +RQ+A+D FNAPG+ FCFLLSTRAGGLGINLATAD
Sbjct: 1073 DLLEDFLEHEGYKYERIDGGITGNMRQEAIDRFNAPGAQQFCFLLSTRAGGLGINLATAD 1132
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVII+DSDWNP ND+QA SRAHRIGQ + V IYRFVT SVEE I + AKKKM+L HLV+
Sbjct: 1133 TVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERITQVAKKKMMLTHLVV 1192
Query: 1058 QKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERND------EESKKRLLSM 1111
+ K GS K EL IL+FG EELFK+E D E ++
Sbjct: 1193 R---------PGLGSKTGS-MSKQELDDILKFGTEELFKDEATDGGGDNKEGEDSSVIHY 1242
Query: 1112 D---IDEILER-AEKVEEKENGGEQAHELLSAFKVANFCNDED----------------- 1150
D I+ +L+R ++ E+ E G +E LS+FKVA + E+
Sbjct: 1243 DDKAIERLLDRNQDETEDTELQG--MNEYLSSFKVAQYVVREEEMGEEEEVEREIIKQEE 1300
Query: 1151 --DGSFWSRWIKADSVAQ----AENALAPRAARNIKSYAEADQSERSKKRKKKENEPTER 1204
D +W + ++ Q A N + R +Y + Q +R + + +N+
Sbjct: 1301 SVDPDYWEKLLRHHYEQQQEDLARNLGKGKRIRKQVNYNDGSQEDRDWQDDQSDNQ---- 1356
Query: 1205 IPKRRKADYSAHVISMIDGASAQVRSWSYGNLSKRDALRFSRSVMKFGNESQINLIVAEV 1264
+DYS V S RS R SR ++ + + ++A V
Sbjct: 1357 ------SDYS--VASEEGDEDFDERS--------EAPRRPSRKGLRNDKDKPLPPLLARV 1400
Query: 1265 GGAIEAAPLKAQ 1276
GG IE A+
Sbjct: 1401 GGNIEVLGFNAR 1412
Score = 33.5 bits (75), Expect = 5.2
Identities = 34/167 (20%), Positives = 59/167 (34%), Gaps = 20/167 (11%)
Query: 95 SESGDDSKSGSDY---------------RNEDEFEDNSSEGRGEKLGS----EDEDGQKD 135
++ G D+K G D RN+DE ED +G E L S + +++
Sbjct: 1225 TDGGGDNKEGEDSSVIHYDDKAIERLLDRNQDETEDTELQGMNEYLSSFKVAQYVVREEE 1284
Query: 136 SGKGQRGDSDVPAEEMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVH 195
G+ + + ++ +E D Y + + G ++ N N
Sbjct: 1285 MGEEEEVEREIIKQEESVDPDYWEKLLRHHYEQQQEDLARNLGKGKRIRK-QVNYNDGSQ 1343
Query: 196 RKSRILDDAEDDDDDADYEEDEPDEDDPDDADFEPATSGRGANKYKD 242
DD D+ D +E DED + ++ S +G KD
Sbjct: 1344 EDRDWQDDQSDNQSDYSVASEEGDEDFDERSEAPRRPSRKGLRNDKD 1390
Score = 32.7 bits (73), Expect = 8.9
Identities = 21/68 (30%), Positives = 31/68 (44%), Gaps = 1/68 (1%)
Query: 1356 WNQIDDARLLLGVHYHGYGNWEVIRLDERLGLTKKIAPVELQHHETFLPRAPNLRDRANA 1415
W++ D LL G+ HGY W+ I+ D R + + E+ + FL R
Sbjct: 1732 WHRRHDYWLLAGIINHGYARWQDIQNDPRYAILNEPFKGEM-NRGNFLEIKNKFLARRFK 1790
Query: 1416 LLEQELAV 1423
LLEQ L +
Sbjct: 1791 LLEQALVI 1798
>CHD4_HUMAN (Q14839) Chromodomain helicase-DNA-binding protein 4
(CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta)
Length = 1912
Score = 296 bits (757), Expect = 4e-79
Identities = 189/519 (36%), Positives = 286/519 (54%), Gaps = 61/519 (11%)
Query: 397 VEKVL---WHQPKGMAA-----EAQRNNQSMEPVLMSHLFDSEPDWNNMEFLIKWKGQSH 448
V+K+L W QP +A N S +P+ + P+ +F +KW+G S+
Sbjct: 502 VQKILIWKWGQPPSPTPVPRPPDADPNTPSPKPL------EGRPE---RQFFVKWQGMSY 552
Query: 449 LHCQWKSFVDLQNLSGFKKVLNYTKR--VTEEIRNRMG----ISREEIEVNDVSKEMDID 502
HC W S + L+ L NY ++ + E G SR+ + EM+
Sbjct: 553 WHCSWVSELQLE-LHCQVMFRNYQRKNDMDEPPSGDFGGDEEKSRKRKNKDPKFAEMEER 611
Query: 503 IIKQNSQVERIIADRISN---DNSGNVFPEYLVKWQGLSYAEVTWEKDIDIAFAQHTIDE 559
+ + E ++ RI N D G+V YL+KW+ L Y + +WE + D+ + + +
Sbjct: 612 FYRYGIKPEWMMIHRILNHSVDKKGHV--HYLIKWRDLPYDQASWESE-DVEIQDYDLFK 668
Query: 560 YKTREAAMSVQGKMVDFQRRQSKGSLRKLDE---------------QPEWLK--GGKLRD 602
++G+ ++ K LRKL+ QPE+L GG L
Sbjct: 669 QSYWNHRELMRGEEGRPGKKLKKVKLRKLERPPETPTVDPTVKYERQPEYLDATGGTLHP 728
Query: 603 YQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLS 662
YQ+EGLN+L SW T+ +LADEMGLGKTVQ+ L L GPFLV PLST+
Sbjct: 729 YQMEGLNWLRFSWAQGTDTILADEMGLGKTVQTAVFLYSLYKEGHSKGPFLVSAPLSTII 788
Query: 663 NWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNE-------KKAGK-----QIKFNAL 710
NW +EF W PD+ V+ YVG + SR + ++ EF E KKA + +KF+ L
Sbjct: 789 NWEREFEMWAPDMYVVTYVGDKDSRAIIRENEFSFEDNAIRGGKKASRMKKEASVKFHVL 848
Query: 711 LTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNS 770
LT+YE++ D A+L I W L+VDEAHRLKN++++ + L+ ++ ++KLL+TGTPLQN+
Sbjct: 849 LTSYELITIDMAILGSIDWACLIVDEAHRLKNNQSKFFRVLNGYSLQHKLLLTGTPLQNN 908
Query: 771 VEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVIKDVEKSL 830
+EEL+ LL+FL ++F + + F + + +++ E+++ LH L PHMLRR+ DV K++
Sbjct: 909 LEELFHLLNFLTPERFHNLEGFLEEFADIA--KEDQIKKLHDMLGPHMLRRLKADVFKNM 966
Query: 831 PPKIERILRVDMSPLQKQYYKWILERNFRDLNKGVRGNQ 869
P K E I+RV++SP+QK+YYK+IL RNF LN GNQ
Sbjct: 967 PSKTELIVRVELSPMQKKYYKYILTRNFEALNARGGGNQ 1005
Score = 194 bits (492), Expect = 2e-48
Identities = 138/372 (37%), Positives = 191/372 (51%), Gaps = 65/372 (17%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
D+L ++ G++++R+DG +RQ+A+D FNAPG+ FCFLLSTRAGGLGINLATAD
Sbjct: 1080 DLLEDFLEHEGYKYERIDGGITGNMRQEAIDRFNAPGAQQFCFLLSTRAGGLGINLATAD 1139
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVII+DSDWNP ND+QA SRAHRIGQ + V IYRFVT SVEE I + AKKKM+L HLV+
Sbjct: 1140 TVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERITQVAKKKMMLTHLVV 1199
Query: 1058 QKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERND------EESKKRLLSM 1111
+ K GS K EL IL+FG EELFK+E D E ++
Sbjct: 1200 R---------PGLGSKTGS-MSKQELDDILKFGTEELFKDEATDGGGDNKEGEDSSVIHY 1249
Query: 1112 D---IDEILER-AEKVEEKENGGEQAHELLSAFKVANFCNDED----------------- 1150
D I+ +L+R ++ E+ E G +E LS+FKVA + E+
Sbjct: 1250 DDKAIERLLDRNQDETEDTELQG--MNEYLSSFKVAQYVVREEEMGEEEEVEREIIKQEE 1307
Query: 1151 --DGSFWSRWIKADSVAQ----AENALAPRAARNIKSYAEADQSERSKKRKKKENEPTER 1204
D +W + ++ Q A N + R +Y + Q +R + + +N+
Sbjct: 1308 SVDPDYWEKLLRHHYEQQQEDLARNLGKGKRIRKQVNYNDGSQEDRDWQDDQSDNQ---- 1363
Query: 1205 IPKRRKADYSAHVISMIDGASAQVRSWSYGNLSKRDALRFSRSVMKFGNESQINLIVAEV 1264
+DYS V S RS R SR ++ + + ++A V
Sbjct: 1364 ------SDYS--VASEEGDEDFDERS--------EAPRRPSRKGLRNDKDKPLPPLLARV 1407
Query: 1265 GGAIEAAPLKAQ 1276
GG IE A+
Sbjct: 1408 GGNIEVLGFNAR 1419
Score = 33.5 bits (75), Expect = 5.2
Identities = 39/154 (25%), Positives = 66/154 (42%), Gaps = 22/154 (14%)
Query: 109 NEDEFEDNSSEGRGEKLGSEDE-----DGQKDSGKGQRGDSDVPAEEMLSDDSYGQDGEE 163
NE++ E++ SE KL + + D + K Q+ + + ++ G + E
Sbjct: 35 NEEDPEEDLSETETPKLKKKKKPKKPRDPKIPKSKRQKKERMLLCRQLGDSSGEGPEFVE 94
Query: 164 QGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRILDDAEDDDDDADYEEDEPDEDDP 223
+ E V R S S P +K + L ++ + +E+E ++DD
Sbjct: 95 EEEEVALR----SDSEGSDYTPGK--------KKKKKLGPKKEKKSKSKRKEEEEEDDDD 142
Query: 224 DDADFEPATSGRGANKYKDWEGEDSDEVDDSDED 257
DD+ EP +S A +DW ED D V S+ED
Sbjct: 143 DDSK-EPKSS---AQLLEDWGMEDIDHV-FSEED 171
Score = 33.5 bits (75), Expect = 5.2
Identities = 34/167 (20%), Positives = 59/167 (34%), Gaps = 20/167 (11%)
Query: 95 SESGDDSKSGSDY---------------RNEDEFEDNSSEGRGEKLGS----EDEDGQKD 135
++ G D+K G D RN+DE ED +G E L S + +++
Sbjct: 1232 TDGGGDNKEGEDSSVIHYDDKAIERLLDRNQDETEDTELQGMNEYLSSFKVAQYVVREEE 1291
Query: 136 SGKGQRGDSDVPAEEMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVH 195
G+ + + ++ +E D Y + + G ++ N N
Sbjct: 1292 MGEEEEVEREIIKQEESVDPDYWEKLLRHHYEQQQEDLARNLGKGKRIRK-QVNYNDGSQ 1350
Query: 196 RKSRILDDAEDDDDDADYEEDEPDEDDPDDADFEPATSGRGANKYKD 242
DD D+ D +E DED + ++ S +G KD
Sbjct: 1351 EDRDWQDDQSDNQSDYSVASEEGDEDFDERSEAPRRPSRKGLRNDKD 1397
Score = 32.7 bits (73), Expect = 8.9
Identities = 21/68 (30%), Positives = 31/68 (44%), Gaps = 1/68 (1%)
Query: 1356 WNQIDDARLLLGVHYHGYGNWEVIRLDERLGLTKKIAPVELQHHETFLPRAPNLRDRANA 1415
W++ D LL G+ HGY W+ I+ D R + + E+ + FL R
Sbjct: 1729 WHRRHDYWLLAGIINHGYARWQDIQNDPRYAILNEPFKGEM-NRGNFLEIKNKFLARRFK 1787
Query: 1416 LLEQELAV 1423
LLEQ L +
Sbjct: 1788 LLEQALVI 1795
>CHD3_HUMAN (Q12873) Chromodomain helicase-DNA-binding protein 3
(CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha)
Length = 1944
Score = 290 bits (742), Expect = 2e-77
Identities = 179/472 (37%), Positives = 267/472 (55%), Gaps = 45/472 (9%)
Query: 438 EFLIKWKGQSHLHCQWKSFVDLQNLSGFKKVLNYTKRVTEEIRNRMGI-SREEIEVNDVS 496
EF +KW G S+ HC W + L+ + NY ++ + + S E+ +D
Sbjct: 549 EFFVKWVGLSYWHCSWAKELQLE-IFHLVMYRNYQRKNDMDEPPPLDYGSGEDDGKSDKR 607
Query: 497 K-------EMDIDIIKQNSQVERIIADRISN---DNSGNVFPEYLVKWQGLSYAEVTWEK 546
K EM+ + + E + RI N D GN YLVKW+ L Y + TWE+
Sbjct: 608 KVKDPHYAEMEEKYYRFGIKPEWMTVHRIINHSVDKKGNY--HYLVKWRDLPYDQSTWEE 665
Query: 547 D-IDIA-FAQHTIDEYKTREAAMS---VQGKMVDFQRRQSKGS----------LRKLDEQ 591
D ++I + +H ++ RE M Q + ++++ +G K + Q
Sbjct: 666 DEMNIPEYEEHKQSYWRHRELIMGEDPAQPRKYKKKKKELQGDGPPSSPTNDPTVKYETQ 725
Query: 592 PEWLK--GGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIH 649
P ++ GG L YQLEGLN+L SW T+ +LADEMGLGKT+Q++ L L
Sbjct: 726 PRFITATGGTLHMYQLEGLNWLRFSWAQGTDTILADEMGLGKTIQTIVFLYSLYKEGHTK 785
Query: 650 GPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFC---NEKKAGK--- 703
GPFLV PLST+ NW +EF+ W P V+ Y G + SR + ++ EF N K GK
Sbjct: 786 GPFLVSAPLSTIINWEREFQMWAPKFYVVTYTGDKDSRAIIRENEFSFEDNAIKGGKKAF 845
Query: 704 ------QIKFNALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTK 757
Q+KF+ LLT+YE++ D+A L I+W L+VDEAHRLKN++++ + L+ +
Sbjct: 846 KMKREAQVKFHVLLTSYELITIDQAALGSIRWACLVVDEAHRLKNNQSKFFRVLNGYKID 905
Query: 758 NKLLITGTPLQNSVEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMELRPH 817
+KLL+TGTPLQN++EEL+ LL+FL ++F + + F + + ++S E+++ LH L PH
Sbjct: 906 HKLLLTGTPLQNNLEELFHLLNFLTPERFNNLEGFLEEFADIS--KEDQIKKLHDLLGPH 963
Query: 818 MLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYKWILERNFRDLNKGVRGNQ 869
MLRR+ DV K++P K E I+RV++SP+QK+YYK+IL RNF LN GNQ
Sbjct: 964 MLRRLKADVFKNMPAKTELIVRVELSPMQKKYYKYILTRNFEALNSRGGGNQ 1015
Score = 201 bits (510), Expect = 2e-50
Identities = 156/440 (35%), Positives = 223/440 (50%), Gaps = 67/440 (15%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
D+L ++ G++++R+DG LRQ+A+D FNAPG+ FCFLLSTRAGGLGINLATAD
Sbjct: 1090 DLLEDFLDYEGYKYERIDGGITGALRQEAIDRFNAPGAQQFCFLLSTRAGGLGINLATAD 1149
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVIIFDSDWNP ND+QA SRAHRIGQ V IYRFVT SVEE I + AK+KM+L HLV+
Sbjct: 1150 TVIIFDSDWNPHNDIQAFSRAHRIGQANKVMIYRFVTRASVEERITQVAKRKMMLTHLVV 1209
Query: 1058 QKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDE--ESKKRLLSMD--- 1112
+ K GS K EL IL+FG EELFK+E E E ++ D
Sbjct: 1210 R---------PGLGSKAGS-MSKQELDDILKFGTEELFKDENEGENKEEDSSVIHYDNEA 1259
Query: 1113 IDEILERAEKVEEKENGGEQAHELLSAFKVANFCNDED-----------------DGSFW 1155
I +L+R + E + + +E LS+FKVA + E+ D +W
Sbjct: 1260 IARLLDRNQDATE-DTDVQNMNEYLSSFKVAQYVVREEDKIEEIEREIIKQEENVDPDYW 1318
Query: 1156 SRWIKADSVAQ----AENALAPRAARNIKSYAEA------DQSERSKKRKKKENEPTERI 1205
+ ++ Q A N + R +Y +A +QSE S ++++ + ER
Sbjct: 1319 EKLLRHHYEQQQEDLARNLGKGKRVRKQVNYNDAAQEDQDNQSEYSVGSEEEDEDFDERP 1378
Query: 1206 PKRRKADYSAHVISMIDGASAQVRSWSYGNL-----SKRDALRFSRSVMKFGNESQ---- 1256
RR++ + + D + + GN+ + R F +VM++G Q
Sbjct: 1379 EGRRQS--KRQLRNEKDKPLPPLLARVGGNIEVLGFNTRQRKAFLNAVMRWGMPPQDAFT 1436
Query: 1257 INLIVAEVGGAIEAAPLKAQVELFNALIDGCREAVEVGSLDLKGPLLDFYGVP---MKAN 1313
+V ++ G E KA V LF R E G+ D D GVP +
Sbjct: 1437 TQWLVRDLRGKTE-KEFKAYVSLF------MRHLCEPGA-DGSETFAD--GVPREGLSRQ 1486
Query: 1314 ELLIRVQELQLLAKRISRYE 1333
++L R+ + L+ K++ +E
Sbjct: 1487 QVLTRIGVMSLVKKKVQEFE 1506
>CHD5_HUMAN (Q8TDI0) Chromodomain helicase-DNA-binding protein 5
(CHD-5)
Length = 1954
Score = 290 bits (741), Expect = 3e-77
Identities = 178/475 (37%), Positives = 262/475 (54%), Gaps = 48/475 (10%)
Query: 438 EFLIKWKGQSHLHCQWKSFVDLQNLSGFKKVLNYTKRVTEE--------IRNRMGISREE 489
EF +KW G S+ HC W + L+ L NY ++ + + G S +
Sbjct: 510 EFFVKWAGLSYWHCSWVKELQLE-LYHTVMYRNYQRKNDMDEPPPFDYGSGDEDGKSEKR 568
Query: 490 IEVNDVSKEMDIDIIKQNSQVERIIADRISN---DNSGNVFPEYLVKWQGLSYAEVTWE- 545
+ + +M+ + + E ++ RI N D G+V YL+KW+ L Y + TWE
Sbjct: 569 KNKDPLYAKMEERFYRYGIKPEWMMIHRILNHSFDKKGDV--HYLIKWKDLPYDQCTWEI 626
Query: 546 KDIDIAFAQHTIDEYKTREAAMSVQGKMVDFQRRQSKGSLR-----------------KL 588
DIDI + + Y M + + + + LR K
Sbjct: 627 DDIDIPYYDNLKQAYWGHRELMLGEDTRLPKRLLKKGKKLRDDKQEKPPDTPIVDPTVKF 686
Query: 589 DEQPEWLK--GGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQ 646
D+QP ++ GG L YQLEGLN+L SW T+ +LADEMGLGKTVQ++ L L
Sbjct: 687 DKQPWYIDSTGGTLHPYQLEGLNWLRFSWAQGTDTILADEMGLGKTVQTIVFLYSLYKEG 746
Query: 647 QIHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFC---NEKKAGK 703
GP+LV PLST+ NW +EF W PD V+ Y G + SR V ++ EF N ++GK
Sbjct: 747 HSKGPYLVSAPLSTIINWEREFEMWAPDFYVVTYTGDKESRSVIRENEFSFEDNAIRSGK 806
Query: 704 ---------QIKFNALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEF 754
QIKF+ LLT+YE++ D+A+L I+W L+VDEAHRLKN++++ + L+ +
Sbjct: 807 KVFRMKKEVQIKFHVLLTSYELITIDQAILGSIEWACLVVDEAHRLKNNQSKFFRVLNSY 866
Query: 755 NTKNKLLITGTPLQNSVEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMEL 814
KLL+TGTPLQN++EEL+ LL+FL ++F + + F + + ++S E+++ LH L
Sbjct: 867 KIDYKLLLTGTPLQNNLEELFHLLNFLTPERFNNLEGFLEEFADIS--KEDQIKKLHDLL 924
Query: 815 RPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYKWILERNFRDLNKGVRGNQ 869
PHMLRR+ DV K++P K E I+RV++S +QK+YYK+IL RNF LN GNQ
Sbjct: 925 GPHMLRRLKADVFKNMPAKTELIVRVELSQMQKKYYKFILTRNFEALNSKGGGNQ 979
Score = 184 bits (468), Expect = 1e-45
Identities = 134/393 (34%), Positives = 193/393 (49%), Gaps = 73/393 (18%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
D+L ++ G++++R+DG LRQ+A+D FNAPG+ FCFLLSTRAGGLGINLATAD
Sbjct: 1054 DLLEDFLEYEGYKYERIDGGITGGLRQEAIDRFNAPGAQQFCFLLSTRAGGLGINLATAD 1113
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVII+DSDWNP ND+QA SRAHRIGQ + V IYRFVT SVEE I + AK+KM+L HLV+
Sbjct: 1114 TVIIYDSDWNPHNDIQAFSRAHRIGQNKKVMIYRFVTRASVEERITQVAKRKMMLTHLVV 1173
Query: 1058 QKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEE------------------- 1098
+ K GS K EL IL+FG EELFK++
Sbjct: 1174 R---------PGLGSKSGS-MTKQELDDILKFGTEELFKDDVEGMMSQGQRPVTPIPDVQ 1223
Query: 1099 ---------------------RNDEESKKRLLSMD---IDEILERAEKVEEKENGGEQAH 1134
N + ++ D I ++L+R + + + + +
Sbjct: 1224 SSKGGNLAASAKKKHGSTPPGDNKDVEDSSVIHYDDAAISKLLDRNQDATD-DTELQNMN 1282
Query: 1135 ELLSAFKVANFCNDEDDGSFWSRWIKADSVAQAENALAPRAARNIKSYAEADQSERSKKR 1194
E LS+FKVA + E+DG ++ + + Q EN + ++ + E Q + ++
Sbjct: 1283 EYLSSFKVAQYVVREEDG---VEEVEREIIKQEENVDPDYWEKLLRHHYEQQQEDLARNL 1339
Query: 1195 KKKENEPTERIPKR-RKADYSAHVISMIDGASAQVRSWSYGNLSK----------RDALR 1243
K +RI K+ D S D S +S G+ + + R
Sbjct: 1340 GK-----GKRIRKQVNYNDASQEDQEWQDELSDNQSEYSIGSEDEDEDFEERPEGQSGRR 1394
Query: 1244 FSRSVMKFGNESQINLIVAEVGGAIEAAPLKAQ 1276
SR +K + + ++A VGG IE A+
Sbjct: 1395 QSRRQLKSDRDKPLPPLLARVGGNIEVLGFNAR 1427
Score = 33.5 bits (75), Expect = 5.2
Identities = 21/68 (30%), Positives = 31/68 (44%), Gaps = 1/68 (1%)
Query: 1356 WNQIDDARLLLGVHYHGYGNWEVIRLDERLGLTKKIAPVELQHHETFLPRAPNLRDRANA 1415
W++ D LL G+ HGY W+ I+ D R + + E+ H +L R
Sbjct: 1736 WHRRHDYWLLAGIVTHGYARWQDIQNDPRYMILNEPFKSEV-HKGNYLEMKNKFLARRFK 1794
Query: 1416 LLEQELAV 1423
LLEQ L +
Sbjct: 1795 LLEQALVI 1802
Score = 32.7 bits (73), Expect = 8.9
Identities = 25/104 (24%), Positives = 44/104 (42%), Gaps = 13/104 (12%)
Query: 294 TASSRQRRVKSSFEDEDENSTAEDSDSESDEDFKSL-------KKRGVRVRKNNGRSSAA 346
T R E+EDE S ED E+ +DF + KK+ ++++N +
Sbjct: 7 TEEELPRLFAEEMENEDEMSEEEDGGLEAFDDFFPVEPVSLPKKKKPKKLKENKCKG--- 63
Query: 347 TSFSRPSNEVRSSSRTIRKVSYVESDESEGADEGTKKSQKEEIE 390
R E + + + E ESEG+D K +K++++
Sbjct: 64 ---KRKKKEGSNDELSENEEDLEEKSESEGSDYSPNKKKKKKLK 104
>CHD3_CAEEL (Q22516) Chromodomain helicase-DNA-binding protein 3
homolog (CHD-3)
Length = 1787
Score = 286 bits (733), Expect = 3e-76
Identities = 185/488 (37%), Positives = 267/488 (53%), Gaps = 54/488 (11%)
Query: 438 EFLIKWKGQSHLHCQWKSFVDLQNLSGFKKVLNYTKRVTEE---IRNRMGISREEIEVND 494
EF +KWK ++ C+W S L ++ V Y ++V E I +SR + +
Sbjct: 426 EFFVKWKYLAYWQCEWLSET-LMDVYFTALVRMYWRKVDSENPPIFEESTLSRHHSDHDP 484
Query: 495 VS---KEMDIDIIKQNSQVERIIADRISNDNSGNVFPEYLVKWQGLSYAEVTWEKD-IDI 550
+ + + Q+ RII + +S S +YLVKW+ LSY TWE+D DI
Sbjct: 485 YKLRERFYQYGVKPEWMQIHRII-NHLSYAKSQQ---DYLVKWKELSYEHATWERDDTDI 540
Query: 551 AFAQHTIDEY-------KTREAAMSVQGKMVDFQR------------------RQSKGSL 585
A + I +Y E +VQ KM+ QR R+ L
Sbjct: 541 ANYEDAIIKYWHHRERMLNDEVPRNVQ-KMIAKQREAKGLGPKEDEVTSRRKKREKIDIL 599
Query: 586 RKLDEQPEWLK--GGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQ 643
+K + QP+++ GG L YQLEG+N+L + W N T+ +LADEMGLGKTVQS++ L L
Sbjct: 600 KKYEVQPDFISETGGNLHPYQLEGINWLRHCWSNGTDAILADEMGLGKTVQSLTFLYTLM 659
Query: 644 NAQQIHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEF-------- 695
GPFL+ PLST+ NW +E W PD V+ YVG R SR V +++EF
Sbjct: 660 KEGHTKGPFLIAAPLSTIINWEREAELWCPDFYVVTYVGDRESRMVIREHEFSFVDGAVR 719
Query: 696 ----CNEKKAGKQIKFNALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTAL 751
++ K + +KF+ LLT+YE + DKA+LS I W L+VDEAHRLKN+++ + L
Sbjct: 720 GGPKVSKIKTLENLKFHVLLTSYECINMDKAILSSIDWAALVVDEAHRLKNNQSTFFKNL 779
Query: 752 SEFNTKNKLLITGTPLQNSVEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENELSNLH 811
E+N + ++L+TGTPLQN++EEL+ LL+FL D+F + F + +S E+++ LH
Sbjct: 780 REYNIQYRVLLTGTPLQNNLEELFHLLNFLAPDRFNQLESFTAEFSEIS--KEDQIEKLH 837
Query: 812 MELRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYKWILERNFRDLNKGVRGNQSK 871
L PHMLRR+ DV +P K E I+RV++S +QK+YYK IL RNF LN G Q
Sbjct: 838 NLLGPHMLRRLKADVLTGMPSKQELIVRVELSAMQKKYYKNILTRNFDALNVKNGGTQMS 897
Query: 872 YCSRIEEV 879
+ I E+
Sbjct: 898 LINIIMEL 905
Score = 196 bits (499), Expect = 4e-49
Identities = 139/363 (38%), Positives = 189/363 (51%), Gaps = 39/363 (10%)
Query: 931 SYSDFLSDILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLG 990
S + DIL + + G++++R+DGS + RQ A+D +NAPG+ F FLLSTRAGGLG
Sbjct: 963 SQMTMMLDILEDFCDVEGYKYERIDGSITGQQRQDAIDRYNAPGAKQFVFLLSTRAGGLG 1022
Query: 991 INLATADTVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKM 1050
INLATADTVII+DSDWNP ND+QA SRAHR+GQ+ V IYRFVT SVEE I AKKKM
Sbjct: 1023 INLATADTVIIYDSDWNPHNDIQAFSRAHRLGQKHKVMIYRFVTKGSVEERITSVAKKKM 1082
Query: 1051 VLDHLVIQK-LNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEE------ 1103
+L HLV++ L A K G K EL +LR+G EELFKEE E
Sbjct: 1083 LLTHLVVRAGLGA----------KDGKSMSKTELDDVLRWGTEELFKEEEAPVEGADGEG 1132
Query: 1104 -SKKRLLSMDI-------DEILERAEKVEEKENGGEQAH---ELLSAFKVANF----CND 1148
S K+ +I D +L+R K EE ++G ++ H E LS+FKVA + +D
Sbjct: 1133 TSSKKPNEQEIVWDDAAVDFLLDR-NKEEEGQDGEKKEHWTNEYLSSFKVATYNTKEADD 1191
Query: 1149 EDDGSFWSRWIKADSVAQAENALAPRAARNIKSYAEADQSERSKK--RKKKENEPTERIP 1206
DD + IK + Q N + +K + E DQ +K + K+
Sbjct: 1192 ADDDEDETEVIKEGTEEQDPNYW----EKLLKHHYEQDQETELQKLGKGKRVRRQVNYAS 1247
Query: 1207 KRRKADYSAHVISMIDGASAQVRSWSYGNLSKRDALRFSRSVMKFGNESQINLIVAEVGG 1266
+ D+SA + G L + D R + ++ ++A+V G
Sbjct: 1248 ENMGQDWSAQNNQQQEEDDGSEYGSDNGELLQTDEDYEERRRRREERSEKLPPLLAKVNG 1307
Query: 1267 AIE 1269
IE
Sbjct: 1308 QIE 1310
Score = 35.0 bits (79), Expect = 1.8
Identities = 40/191 (20%), Positives = 77/191 (39%), Gaps = 34/191 (17%)
Query: 100 DSKSGSDYRNEDEFEDNSSE-GRGEKLGSEDEDGQKDSGKGQRGDSDVPAEEMLSDDSYG 158
D KS S +D + E + E+ E DG+ S K + + ++ ++ D
Sbjct: 1097 DGKSMSKTELDDVLRWGTEELFKEEEAPVEGADGEGTSSK-KPNEQEIVWDDAAVDFLLD 1155
Query: 159 QDGEEQGESVHSRGFRPSTGSNSCLQPTSTNVNRRVHRKSRILDDAEDDDDDADYEEDEP 218
++ EE+G+ G + +N L ++ DDA+DD+D+ + ++
Sbjct: 1156 RNKEEEGQD----GEKKEHWTNEYLSSFKVATYN-----TKEADDADDDEDETEVIKEGT 1206
Query: 219 DEDDPD------------DADFEPATSGRG-----------ANKYKDWEGEDSDEVDDSD 255
+E DP+ D + E G+G N +DW +++ + ++ D
Sbjct: 1207 EEQDPNYWEKLLKHHYEQDQETELQKLGKGKRVRRQVNYASENMGQDWSAQNNQQQEEDD 1266
Query: 256 EDIDVSDNDDL 266
SDN +L
Sbjct: 1267 GSEYGSDNGEL 1277
>CHD3_DROME (O16102) Chromodomain helicase-DNA-binding protein 3
Length = 892
Score = 273 bits (699), Expect = 2e-72
Identities = 165/438 (37%), Positives = 253/438 (57%), Gaps = 26/438 (5%)
Query: 438 EFLIKWKGQSHLHCQWKSFVDLQNLSGFKKVLNYTKRVTEEIRNRMGISREEIE-VNDVS 496
E+ IKW G S+ HC+W + + +L++ V + R + +E ++D
Sbjct: 112 EYFIKWHGMSYWHCEW--------IPEGQMLLHHASMVAS-FQRRSDMEEPSLEELDDQD 162
Query: 497 KEMDIDIIKQNSQVERIIADRISNDNSG-NVFPEYLVKWQGLSYAEVTWEKDID-IAFAQ 554
+ + + E ++ R+ N + N YLVKW+ LSY + +WE++ D I
Sbjct: 163 GNLHERFYRYGIKPEWLLVQRVINHSEEPNGGTMYLVKWRELSYNDSSWERESDSIPGLN 222
Query: 555 HTIDEYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEWLK--GGKLRDYQLEGLNFLV 612
I YK + S +G+ D + + +K ++QP +LK G KL +Q+EG+++L
Sbjct: 223 QAIALYKKLRS--SNKGRQRD-RPAPTIDLNKKYEDQPVFLKEAGLKLHPFQIEGVSWLR 279
Query: 613 NSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEFRKWL 672
SW +LADEMGLGKT+Q+V L L GPFL+ VPLSTL+NW +E W
Sbjct: 280 YSWGQGIPTILADEMGLGKTIQTVVFLYSLFKEGHCRGPFLISVPLSTLTNWERELELWA 339
Query: 673 PDLNVIVYVGTRSSREVCQQYEFCNEKKAGK-------QIKFNALLTTYEVVLKDKAVLS 725
P+L + YVG +++R V +++E E+ K Q KFN +LT+YE + D A L
Sbjct: 340 PELYCVTYVGGKTARAVIRKHEISFEEVTTKTMRENQTQYKFNVMLTSYEFISVDAAFLG 399
Query: 726 KIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHFLDSDK 785
I W L+VDEAHRL++++++ + LS++ KLL+TGTPLQN++EEL+ LL+FL S K
Sbjct: 400 CIDWAALVVDEAHRLRSNQSKFFRILSKYRIGYKLLLTGTPLQNNLEELFHLLNFLSSGK 459
Query: 786 FKSKDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVIKDVEKSLPPKIERILRVDMSPL 845
F F + ++S E ++ LH L PHMLRR+ DV KS+PPK E I+RV++S +
Sbjct: 460 FNDLQTFQAEFTDVS--KEEQVKRLHEILEPHMLRRLKADVLKSMPPKSEFIVRVELSSM 517
Query: 846 QKQYYKWILERNFRDLNK 863
QK++YK IL +NF+ LN+
Sbjct: 518 QKKFYKHILTKNFKALNQ 535
Score = 195 bits (496), Expect = 8e-49
Identities = 125/288 (43%), Positives = 171/288 (58%), Gaps = 39/288 (13%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
++L ++ G+Q+ R+DGS K +LRQ+A+D FN P S+ F FLLSTRAGGLGINLATAD
Sbjct: 616 NVLEHFLEGEGYQYDRIDGSIKGDLRQKAIDRFNDPVSEHFVFLLSTRAGGLGINLATAD 675
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVIIFDSDWNP ND+QA SRAHR+GQ++ V IYRFVT SVEE I++ AK KM+L HLV+
Sbjct: 676 TVIIFDSDWNPHNDVQAFSRAHRMGQKKKVMIYRFVTHNSVEERIMQVAKHKMMLTHLVV 735
Query: 1058 QKLNAEGKLEKKEAKKGG--SFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMD--- 1112
+ GG + F K+EL ILRFG E+LFK + K + D
Sbjct: 736 R------------PGMGGMTTNFSKDELEDILRFGTEDLFK------DGKSEAIHYDDKA 777
Query: 1113 IDEILERAEK-VEEKENGGEQAHELLSAFKVANFCNDEDDGSFWSRWIKADSVAQAENAL 1171
+ ++L+R + +EEKE+ A+E LS+FKVA++ ED D AEN
Sbjct: 778 VADLLDRTNRGIEEKES---WANEYLSSFKVASYATKEDHEE------HDDYNNDAENT- 827
Query: 1172 APRAARNIKSYAEADQSERSKKRKKKENEPTERIPK-----RRKADYS 1214
P N+ ++ ++ KK+ ++ E I R++ DYS
Sbjct: 828 DPFYWENLMGKSQPKLPKKQKKQSQQSQVDVESIMGKGKRIRKEIDYS 875
>CHDM_DROME (O97159) Chromodomain helicase-DNA-binding protein Mi-2
homolog (dMi-2)
Length = 1982
Score = 273 bits (698), Expect = 3e-72
Identities = 176/495 (35%), Positives = 260/495 (51%), Gaps = 61/495 (12%)
Query: 438 EFLIKWKGQSHLHCQWKSFVDL-----------QNLSGFKKVLNYTKRVTEEIRNRMGIS 486
E+ IKW S+ HC+W V L Q ++ + + + E I
Sbjct: 522 EYFIKWHNMSYWHCEWVPEVQLDVHHPLMIRSFQRKYDMEEPPKFEESLDEADTRYKRIQ 581
Query: 487 REEIEV----NDVSKEMDIDIIKQNSQVERIIADRISNDNSG-NVFPEYLVKWQGLSYAE 541
R + +V ND ++ ++ K + E +I R+ N + + YLVKW+ L Y +
Sbjct: 582 RHKDKVGMKANDDAEVLEERFYKNGVKPEWLIVQRVINHRTARDGSTMYLVKWRELPYDK 641
Query: 542 VTWEKD-IDIAFAQHTIDEYKTREAAMSVQG----------------KMVDFQRRQSKGS 584
TWE++ DI + ID Y+ A + + K+ D + R K
Sbjct: 642 STWEEEGDDIQGLRQAIDYYQDLRAVCTSETTQSRSKKSKKGRKSKLKVEDDEDRPVKHY 701
Query: 585 L-----------RKLDEQPEWLKGG--KLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGK 631
+K ++QP +L+G +L YQ+EG+N+L SW + +LADEMGLGK
Sbjct: 702 TPPPEKPTTDLKKKYEDQPAFLEGTGMQLHPYQIEGINWLRYSWGQGIDTILADEMGLGK 761
Query: 632 TVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQ 691
T+Q+V+ L L GPFLV VPLSTL NW +EF W PD I Y+G + SR V +
Sbjct: 762 TIQTVTFLYSLYKEGHCRGPFLVAVPLSTLVNWEREFELWAPDFYCITYIGDKDSRAVIR 821
Query: 692 QYEFCNEKKAGK----------QIKFNALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLK 741
+ E E+ A + Q KFN LLT+YE++ D A L I W L+VDEAHRLK
Sbjct: 822 ENELSFEEGAIRGSKVSRLRTTQYKFNVLLTSYELISMDAACLGSIDWAVLVVDEAHRLK 881
Query: 742 NSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHFLDSDKFKSKDEFAQNYKNLSS 801
+++++ + L+ + KLL+TGTPLQN++EEL+ LL+FL DKF F + ++S
Sbjct: 882 SNQSKFFRILNSYTIAYKLLLTGTPLQNNLEELFHLLNFLSRDKFNDLQAFQGEFADVS- 940
Query: 802 FNENELSNLHMELRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYKWILERNFRDL 861
E ++ LH L PHMLRR+ DV K++P K E I+RV++S +QK++YK+IL +N+ L
Sbjct: 941 -KEEQVKRLHEMLGPHMLRRLKTDVLKNMPSKSEFIVRVELSAMQKKFYKFILTKNYEAL 999
Query: 862 NKGVRGNQSKYCSRI 876
N G CS I
Sbjct: 1000 NSKSGGGS---CSLI 1011
Score = 201 bits (511), Expect = 1e-50
Identities = 122/269 (45%), Positives = 167/269 (61%), Gaps = 26/269 (9%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DIL ++ ++++R+DG LRQ+A+D FNAPG+ F FLLSTRAGGLGINLATAD
Sbjct: 1082 DILEDFLEGEQYKYERIDGGITGTLRQEAIDRFNAPGAQQFVFLLSTRAGGLGINLATAD 1141
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVII+DSDWNP ND+QA SRAHRIGQ V IYRFVT SVEE + + AK+KM+L HLV+
Sbjct: 1142 TVIIYDSDWNPHNDIQAFSRAHRIGQANKVMIYRFVTRNSVEERVTQVAKRKMMLTHLVV 1201
Query: 1058 QKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDEIL 1117
+ GK G+ F K EL ILRFG E+LFKE+ D+E + E+L
Sbjct: 1202 RP-GMGGK---------GANFTKQELDDILRFGTEDLFKED--DKEEAIHYDDKAVAELL 1249
Query: 1118 ERAEK-VEEKENGGEQAHELLSAFKVANFCNDEDDGSFWSRWIKADSVAQAENALAPRAA 1176
+R + +EEKE+ A+E LS+FKVA++ E++ + IK D AEN+
Sbjct: 1250 DRTNRGIEEKES---WANEYLSSFKVASYATKEEEEEEETEIIKQD----AENSDPAYWV 1302
Query: 1177 RNIKSYAEADQSE------RSKKRKKKEN 1199
+ ++ + E Q + + K+ +K+ N
Sbjct: 1303 KLLRHHYEQHQEDVGRSLGKGKRVRKQVN 1331
Score = 36.2 bits (82), Expect = 0.81
Identities = 39/171 (22%), Positives = 71/171 (40%), Gaps = 31/171 (18%)
Query: 204 AEDDDDDADYEEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDSDEDIDV-SD 262
+E+++DD EE+E ED+ A+ +DSD + +D
Sbjct: 3 SEEENDDNFQEEEEAQEDNAPAAELS----------------------NDSDAPLKPNND 40
Query: 263 NDDLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQRRVKSSFEDEDENSTAEDSDSES 322
DD Y + ++ +++GK R TR K ++++ K + +ED + D + E
Sbjct: 41 EDDDYDPEDSRRKKKGK----KRKTR--KGEEKGRKKKKRKKNESEEDSDFVQHDEEVEY 94
Query: 323 DEDFKSLKKRGVRVRKNNGRSSAATSFSRPSNEVRSSSRTIRKVSYVESDE 373
K +KR + K + + S PS E S+ ++ V S+E
Sbjct: 95 PSTSKRGRKR--KEEKQAAKEKESASSGMPSVEDVCSAFSVCNVEIEYSEE 143
Score = 33.5 bits (75), Expect = 5.2
Identities = 21/70 (30%), Positives = 34/70 (48%), Gaps = 6/70 (8%)
Query: 1356 WNQIDDARLLLGVHYHGYGNWEVIRLDERLGLTKKIAPVE------LQHHETFLPRAPNL 1409
W++ D LL G+ HGYG W+ I+ D R + + ++ L+ FL R L
Sbjct: 1759 WHRRHDYWLLAGIVTHGYGRWQDIQNDIRFAIINEPFKMDVGKGNFLEIKNKFLARRFKL 1818
Query: 1410 RDRANALLEQ 1419
++A + EQ
Sbjct: 1819 LEQALVIEEQ 1828
>SN21_HUMAN (P28370) Possible global transcription activator SNF2L1
(SWI/SNF related matrix associated actin dependent
regulator of chromatin subfamily A member 1)
Length = 976
Score = 250 bits (638), Expect = 3e-65
Identities = 126/305 (41%), Positives = 201/305 (65%), Gaps = 9/305 (2%)
Query: 559 EYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEWLKGGKLRDYQLEGLNFLVNSWKND 618
+Y+ R +++ R+ S +R + P ++KGG LRDYQ+ GLN+L++ ++N
Sbjct: 65 DYRHRRTEQEEDEELLSESRKTSNVCIR-FEVSPSYVKGGPLRDYQIRGLNWLISLYENG 123
Query: 619 TNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEFRKWLPDLNVI 678
N +LADEMGLGKT+Q++++LG+L++ + I GP +V+VP STL NW EF++W+P L VI
Sbjct: 124 VNGILADEMGLGKTLQTIALLGYLKHYRNIPGPHMVLVPKSTLHNWMNEFKRWVPSLRVI 183
Query: 679 VYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVLKDKAVLSKIKWNYLMVDEAH 738
+VG + +R + +E G +++ +T+YE+V+K+K+V K W YL++DEAH
Sbjct: 184 CFVGDKDARAAFIR----DEMMPG---EWDVCVTSYEMVIKEKSVFKKFHWRYLVIDEAH 236
Query: 739 RLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHFLDSDKFKSKDEFAQNYKN 798
R+KN +++L + EF + N+LL+TGTPLQN++ ELWALL+FL D F S D+F +
Sbjct: 237 RIKNEKSKLSEIVREFKSTNRLLLTGTPLQNNLHELWALLNFLLPDVFNSADDFDSWFDT 296
Query: 799 LSSFNENEL-SNLHMELRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYKWILERN 857
+ + +L LH L+P +LRR+ DVEKSLPPK E + + +S +Q+++Y IL ++
Sbjct: 297 KNCLGDQKLVERLHAVLKPFLLRRIKTDVEKSLPPKKEIKIYLGLSKMQREWYTKILMKD 356
Query: 858 FRDLN 862
LN
Sbjct: 357 IDVLN 361
Score = 159 bits (402), Expect = 6e-38
Identities = 93/205 (45%), Positives = 126/205 (61%), Gaps = 26/205 (12%)
Query: 936 LSDILAQYMSLRGFQFQRLDGSTKSELRQ------------QAMDHFNAPGSDDFCFLLS 983
L DIL Y RG+++ RLDG T E R+ +A++ FNAP S F F+LS
Sbjct: 436 LLDILEDYCMWRGYEYCRLDGQTPHEEREDKFLEVEFLGQREAIEAFNAPNSSKFIFMLS 495
Query: 984 TRAGGLGINLATADTVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDIL 1043
TRAGGLGINLA+AD VI++DSDWNPQ DLQAM RAHRIGQ++ V ++R +T +VEE I+
Sbjct: 496 TRAGGLGINLASADVVILYDSDWNPQVDLQAMDRAHRIGQKKPVRVFRLITDNTVEERIV 555
Query: 1044 ERAKKKMVLDHLVIQKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEE 1103
ERA+ K+ LD +VIQ +G+L + + K K E+ ++R GA +F + ++
Sbjct: 556 ERAEIKLRLDSIVIQ----QGRLIDQRSNK----LAKEEMLQMIRHGATHVFASKESE-- 605
Query: 1104 SKKRLLSMDIDEILERAEKVEEKEN 1128
L DI ILER EK + N
Sbjct: 606 ----LTDEDITTILERGEKKTAEMN 626
>STH1_YEAST (P32597) Nuclear protein STH1/NPS1 (Chromatin structure
remodeling complex protein STH1) (SNF2 homolog)
Length = 1359
Score = 244 bits (623), Expect = 1e-63
Identities = 127/295 (43%), Positives = 194/295 (65%), Gaps = 22/295 (7%)
Query: 587 KLDEQPEWLKGGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQ 646
K+D+QP L GG L++YQL GL ++V+ + N N +LADEMGLGKT+QS+S++ +L +
Sbjct: 457 KIDKQPSILVGGTLKEYQLRGLEWMVSLYNNHLNGILADEMGLGKTIQSISLITYLYEVK 516
Query: 647 QIHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIK 706
+ GPFLV+VPLST++NW EF KW P LN I+Y GT + R Q ++ + G
Sbjct: 517 KDIGPFLVIVPLSTITNWTLEFEKWAPSLNTIIYKGTPNQRHSLQ-----HQIRVG---N 568
Query: 707 FNALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQL-YTALSEFNTKNKLLITGT 765
F+ LLTTYE ++KDK++LSK W ++++DE HR+KN++++L +T + T+N+L++TGT
Sbjct: 569 FDVLLTTYEYIIKDKSLLSKHDWAHMIIDEGHRMKNAQSKLSFTISHYYRTRNRLILTGT 628
Query: 766 PLQNSVEELWALLHFLDSDKFKS----KDEFAQNYKNLSSFNENELS---------NLHM 812
PLQN++ ELWALL+F+ F S +D F + N + + EL+ LH
Sbjct: 629 PLQNNLPELWALLNFVLPKIFNSAKTFEDWFNTPFANTGTQEKLELTEEETLLIIRRLHK 688
Query: 813 ELRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYKWILERNFRDLNKGVRG 867
LRP +LRR+ K+VEK LP K+E++++ +S LQ+Q Y+ +L+ N + G G
Sbjct: 689 VLRPFLLRRLKKEVEKDLPDKVEKVIKCKLSGLQQQLYQQMLKHNALFVGAGTEG 743
Score = 161 bits (407), Expect = 2e-38
Identities = 112/312 (35%), Positives = 168/312 (52%), Gaps = 48/312 (15%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DI+ ++ ++ ++ RLDGSTK+E R + ++ FNAP SD FCFLLSTRAGGLG+NL TAD
Sbjct: 821 DIMEDFLRMKDLKYMRLDGSTKTEERTEMLNAFNAPDSDYFCFLLSTRAGGLGLNLQTAD 880
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVIIFD+DWNP DLQA RAHRIGQ+ V I R +T+ SVEE ILERA +K+ +D VI
Sbjct: 881 TVIIFDTDWNPHQDLQAQDRAHRIGQKNEVRILRLITTDSVEEVILERAMQKLDIDGKVI 940
Query: 1058 QKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERN-DEESKKRLLSMDIDEI 1116
Q GK + K + E A LR L + E N D++ K L ++++
Sbjct: 941 QA----GKFDNKSTAE--------EQEAFLR----RLIESETNRDDDDKAELDDDELNDT 984
Query: 1117 LERAEKVEEKENGGEQAHELLSAFKVANFCNDEDDGSFWSRWIKADSVAQAENALAPRAA 1176
L R+ A E + K+ +++ +AD+ AQ PR
Sbjct: 985 LARS------------ADEKILFDKIDKERMNQE---------RADAKAQGLRVPPPRLI 1023
Query: 1177 RNIKSYAEADQSERSKKRKKKENEPTERIPKRRKADYSAHVISMIDGASAQ--VRSWSYG 1234
+ + + + + + KK+++EP RI ++++ Y DG + + + +
Sbjct: 1024 Q-LDELPKVFREDIEEHFKKEDSEPLGRIRQKKRVYYD-------DGLTEEQFLEAVEDD 1075
Query: 1235 NLSKRDALRFSR 1246
N+S DA++ R
Sbjct: 1076 NMSLEDAIKKRR 1087
>SMR5_MOUSE (Q91ZW3) SWI/SNF related matrix associated actin
dependent regulator of chromatin, subfamily A member 5
(EC 3.6.1.-) (Sucrose nonfermenting protein 2 homolog)
(mSnf2h)
Length = 1051
Score = 244 bits (623), Expect = 1e-63
Identities = 120/277 (43%), Positives = 187/277 (67%), Gaps = 8/277 (2%)
Query: 587 KLDEQPEWLKGGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQ 646
+ ++ P ++K GKLRDYQ+ GLN+L++ ++N N +LADEMGLGKT+Q++S+LG++++ +
Sbjct: 166 RFEDSPSYVKWGKLRDYQVRGLNWLISLYENGINGILADEMGLGKTLQTISLLGYMKHYR 225
Query: 647 QIHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIK 706
I GP +V+VP STL NW EF+KW+P L + +G + R F + +
Sbjct: 226 NIPGPHMVLVPKSTLHNWMSEFKKWVPTLRSVCLIGDKEQRAA-----FVRDVLLPGE-- 278
Query: 707 FNALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTP 766
++ +T+YE+++K+K+V K W YL++DEAHR+KN +++L + EF T N+LL+TGTP
Sbjct: 279 WDVCVTSYEMLIKEKSVFKKFNWRYLVIDEAHRIKNEKSKLSEIVREFKTTNRLLLTGTP 338
Query: 767 LQNSVEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENEL-SNLHMELRPHMLRRVIKD 825
LQN++ ELW+LL+FL D F S D+F + + + +L LHM LRP +LRR+ D
Sbjct: 339 LQNNLHELWSLLNFLLPDVFNSADDFDSWFDTNNCLGDQKLVERLHMVLRPFLLRRIKAD 398
Query: 826 VEKSLPPKIERILRVDMSPLQKQYYKWILERNFRDLN 862
VEKSLPPK E + V +S +Q+++Y IL ++ LN
Sbjct: 399 VEKSLPPKKEVKIYVGLSKMQREWYTRILMKDIDILN 435
Score = 164 bits (416), Expect = 2e-39
Identities = 158/576 (27%), Positives = 251/576 (43%), Gaps = 92/576 (15%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DIL Y R +++ RLDG T + RQ +++ +N P S F F+LSTRAGGLGINLATAD
Sbjct: 512 DILEDYCMWRNYEYCRLDGQTPHDERQDSINAYNEPNSTKFVFMLSTRAGGLGINLATAD 571
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
VI++DSDWNPQ DLQAM RAHRIGQ + V ++RF+T +VEE I+ERA+ K+ LD +VI
Sbjct: 572 VVILYDSDWNPQVDLQAMDRAHRIGQTKTVRVFRFITDNTVEERIVERAEMKLRLDSIVI 631
Query: 1058 QKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDEIL 1117
Q +G+L + K G K+E+ ++R GA +F + ++ + DID IL
Sbjct: 632 Q----QGRLVDQNLNKIG----KDEMLQMIRHGATHVFASKESE------ITDEDIDGIL 677
Query: 1118 ERAEK--VEEKENGGEQAHELLSAF------KVANFCND---EDDGSFWSRWIKADSVAQ 1166
ER K E E + L F V NF + E ++ WI+ +
Sbjct: 678 ERGAKKTAEMNEKLSKMGESSLRNFTMDTESSVYNFEGEDYREKQKIAFTEWIEPPKRER 737
Query: 1167 AENALAPRAARNIKSYAEADQSERSKKRKKKENEPTERIPKR------RKADYSAHVI-- 1218
N R +E + + K+ + + P R ++ Y I
Sbjct: 738 KANYAVDAYFREALRVSEPKAPKAPRPPKQPNVQDFQFFPPRLFELLEKEILYYRKTIGY 797
Query: 1219 ------SMIDGASAQ-----------------------VRSWSYGNLSKRDALRFSRSVM 1249
+ + A AQ + + + N +KRD +F ++
Sbjct: 798 KVPRSPDLPNAAQAQKEEQLKIDEAEPLNDEELEEKEKLLTQGFTNWNKRDFNQFIKANE 857
Query: 1250 KFGNESQINLIVAEVGGAIEAAPLKAQVELFNALIDGCREAVEVGSLDLKGPLLDFYGVP 1309
K+G + + + +E + +E + C E ++ + + +
Sbjct: 858 KWGRDD-----IENIAREVEGKTPEEVIEYSAVFWERCNELQDIEKIMAQ--------IE 904
Query: 1310 MKANELLIRVQELQLLAKRISRYEDPIAQFRVLTYLKPSNWSKGCGWNQIDDARLLLGVH 1369
+ R+ + L +I RY+ P Q R+ +KG + + +D L+ +H
Sbjct: 905 RGEARIQRRISIKKALDTKIGRYKAPFHQLRI-----SYGTNKGKNYTEEEDRFLICMLH 959
Query: 1370 YHGYGNWEVIRLDERLGLTKKIAPVELQHHETFLP--RAPNLRDRAN---ALLEQELAVL 1424
G+ V DE L + I + FL A L+ R N L+E+E L
Sbjct: 960 KLGFDKENV--YDE---LRQCIRNSPQFRFDWFLKSRTAMELQRRCNTLITLIERENMEL 1014
Query: 1425 GVKNASSKVGRKTSKKEREEREHLVDISLSRGQEKK 1460
K + K +K K ++ + RG++KK
Sbjct: 1015 EEKEKAEK--KKRGPKPSTQKRKMDGAPDGRGRKKK 1048
>SMR5_HUMAN (O60264) SWI/SNF related matrix associated actin
dependent regulator of chromatin subfamily A member 5
(EC 3.6.1.-) (SWI/SNF-related matrix-associated
actin-dependent regulator of chromatin A5) (Sucrose
nonfermenting protein 2 homolog) (hSNF2H
Length = 1052
Score = 243 bits (620), Expect = 3e-63
Identities = 119/277 (42%), Positives = 187/277 (66%), Gaps = 8/277 (2%)
Query: 587 KLDEQPEWLKGGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQ 646
+ ++ P ++K GKLRDYQ+ GLN+L++ ++N N +LADEMGLGKT+Q++S+LG++++ +
Sbjct: 167 RFEDSPSYVKWGKLRDYQVRGLNWLISLYENGINGILADEMGLGKTLQTISLLGYMKHYR 226
Query: 647 QIHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIK 706
I GP +V+VP STL NW EF++W+P L + +G + R F + +
Sbjct: 227 NIPGPHMVLVPKSTLHNWMSEFKRWVPTLRSVCLIGDKEQRAA-----FVRDVLLPGE-- 279
Query: 707 FNALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTP 766
++ +T+YE+++K+K+V K W YL++DEAHR+KN +++L + EF T N+LL+TGTP
Sbjct: 280 WDVCVTSYEMLIKEKSVFKKFNWRYLVIDEAHRIKNEKSKLSEIVREFKTTNRLLLTGTP 339
Query: 767 LQNSVEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENEL-SNLHMELRPHMLRRVIKD 825
LQN++ ELW+LL+FL D F S D+F + + + +L LHM LRP +LRR+ D
Sbjct: 340 LQNNLHELWSLLNFLLPDVFNSADDFDSWFDTNNCLGDQKLVERLHMVLRPFLLRRIKAD 399
Query: 826 VEKSLPPKIERILRVDMSPLQKQYYKWILERNFRDLN 862
VEKSLPPK E + V +S +Q+++Y IL ++ LN
Sbjct: 400 VEKSLPPKKEVKIYVGLSKMQREWYTRILMKDIDILN 436
Score = 164 bits (415), Expect = 2e-39
Identities = 90/191 (47%), Positives = 123/191 (64%), Gaps = 14/191 (7%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DIL Y R +++ RLDG T + RQ +++ +N P S F F+LSTRAGGLGINLATAD
Sbjct: 513 DILEDYCMWRNYEYCRLDGQTPHDERQDSINAYNEPNSTKFVFMLSTRAGGLGINLATAD 572
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
VI++DSDWNPQ DLQAM RAHRIGQ + V ++RF+T +VEE I+ERA+ K+ LD +VI
Sbjct: 573 VVILYDSDWNPQVDLQAMDRAHRIGQTKTVRVFRFITDNTVEERIVERAEMKLRLDSIVI 632
Query: 1058 QKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDEIL 1117
Q +G+L + K G K+E+ ++R GA +F + ++ + DID IL
Sbjct: 633 Q----QGRLVDQNLNKIG----KDEMLQMIRHGATHVFASKESE------ITDEDIDGIL 678
Query: 1118 ERAEKVEEKEN 1128
ER K + N
Sbjct: 679 ERGAKKTAEMN 689
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.133 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 206,359,959
Number of Sequences: 164201
Number of extensions: 9442037
Number of successful extensions: 58120
Number of sequences better than 10.0: 921
Number of HSP's better than 10.0 without gapping: 277
Number of HSP's successfully gapped in prelim test: 675
Number of HSP's that attempted gapping in prelim test: 40780
Number of HSP's gapped (non-prelim): 6739
length of query: 1730
length of database: 59,974,054
effective HSP length: 124
effective length of query: 1606
effective length of database: 39,613,130
effective search space: 63618686780
effective search space used: 63618686780
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 73 (32.7 bits)
Medicago: description of AC141111.7


