

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141110.2 - phase: 0
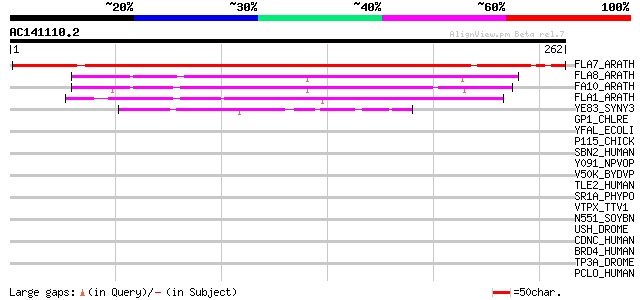
(262 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
FLA7_ARATH (Q9SJ81) Fasciclin-like arabinogalactan protein 7 pre... 286 3e-77
FLA8_ARATH (O22126) Fasciclin-like arabinogalactan protein 8 pre... 107 3e-23
FA10_ARATH (Q9LZX4) Fasciclin-like arabinogalactan protein 10 pr... 97 4e-20
FLA1_ARATH (Q9FM65) Fasciclin-like arabinogalactan protein 1 pre... 80 7e-15
YE83_SYNY3 (P74615) Hypothetical protein sll1483 precursor 44 5e-04
GP1_CHLRE (Q9FPQ6) Vegetative cell wall protein gp1 precursor (H... 38 0.022
YFAL_ECOLI (P45508) Hypothetical protein yfaL precursor 37 0.063
P115_CHICK (Q98917) 115 kDa melanosomal matrix protein precursor 37 0.063
SBN2_HUMAN (Q8WWQ8) Stabilin 2 precursor (FEEL-2 protein) (Fasci... 36 0.083
Y091_NPVOP (O10341) Hypothetical 29.3 kDa protein (ORF92) 36 0.11
V50K_BYDVP (P09516) 50 kDa protein (ORF 4) 36 0.11
TLE2_HUMAN (Q04725) Transducin-like enhancer protein 2 (ESG2) 36 0.11
SR1A_PHYPO (P09350) Spherulin 1A precursor 36 0.11
VTPX_TTV1 (P19274) Viral protein TPX 35 0.14
N551_SOYBN (Q05544) Early nodulin 55-1 (N-55-1) (Fragment) 35 0.14
USH_DROME (Q9VPQ6) Zinc-finger protein ush (U-shaped protein) 35 0.18
CDNC_HUMAN (P49918) Cyclin-dependent kinase inhibitor 1C (Cyclin... 35 0.18
BRD4_HUMAN (O60885) Bromodomain-containing protein 4 (HUNK1 prot... 35 0.18
TP3A_DROME (Q9NG98) DNA topoisomerase III alpha (EC 5.99.1.2) 35 0.24
PCLO_HUMAN (Q9Y6V0) Piccolo protein (Aczonin) 35 0.24
>FLA7_ARATH (Q9SJ81) Fasciclin-like arabinogalactan protein 7
precursor
Length = 254
Score = 286 bits (733), Expect = 3e-77
Identities = 148/261 (56%), Positives = 196/261 (74%), Gaps = 9/261 (3%)
Query: 2 KVSMIFLVSTLLLLSSSAFAKTVSPPSPPAESPTPAPAPAPTPDFVNLTELLTVAGPFHT 61
K+ + ++ + L+ SA AKT SPP+P P PAPAP P+ VNLTELL+VAGPFHT
Sbjct: 3 KMQLSIFIAVVALIVCSASAKTASPPAPVLP---PTPAPAPAPENVNLTELLSVAGPFHT 59
Query: 62 FLQYLQSTKVLDTFQNQANNTEEGITIFVPKDSSFASLKKPSLSKLKDDEIKQVILFHAL 121
FL YL ST V++TFQNQANNTEEGITIFVPKD +F + K P LS L D++KQ++LFHAL
Sbjct: 60 FLDYLLSTGVIETFQNQANNTEEGITIFVPKDDAFKAQKNPPLSNLTKDQLKQLVLFHAL 119
Query: 122 PHFYSLADFKNLSQTASTPTFAGGDYTLNFTDNSGTVKINSGWSITKVTSAVHATDPVAI 181
PH+YSL++FKNLSQ+ TFAGG Y+L FTD SGTV+I+S W+ TKV+S+V +TDPVA+
Sbjct: 120 PHYYSLSEFKNLSQSGPVSTFAGGQYSLKFTDVSGTVRIDSLWTRTKVSSSVFSTDPVAV 179
Query: 182 YQVDKVLLPEAIFGTDIPPVLAPAPTPEIAPAADSPTEQSADSKSSSPSSSPDRSSSYKI 241
YQV++VLLPEAIFGTD+PP+ APAP P ++ +DSP+ ADS+ +S S ++S K+
Sbjct: 180 YQVNRVLLPEAIFGTDVPPMPAPAPAPIVSAPSDSPS--VADSEGASSPKSSHKNSGQKL 237
Query: 242 VSYGIWGNLVLATFGLVVVIL 262
+ I ++V++ GLV + L
Sbjct: 238 LLAPI--SMVIS--GLVALFL 254
>FLA8_ARATH (O22126) Fasciclin-like arabinogalactan protein 8
precursor (AtAGP8)
Length = 420
Score = 107 bits (267), Expect = 3e-23
Identities = 73/223 (32%), Positives = 108/223 (47%), Gaps = 16/223 (7%)
Query: 30 PAESPTPAPAPAPTPDFVNLTELLTVAGPFHTFLQYLQSTKVLDTFQNQANNTEEGITIF 89
P +P APAP+ N+T LL AG TF L S+ VL T+++ E+G+T+F
Sbjct: 171 PIIAPGVLTAPAPSASLSNITGLLEKAG-CKTFANLLVSSGVLKTYESAV---EKGLTVF 226
Query: 90 VPKDSSFASLKKPSLSKLKDDEIKQVILFHALPHFYSLADFKNLSQTAST-PTFAGGDYT 148
P D +F + P L+KL E+ ++ +HAL + K ST T G +
Sbjct: 227 APSDEAFKAEGVPDLTKLTQAEVVSLLEYHALAEYKPKGSLKTNKNNISTLATNGAGKFD 286
Query: 149 LNFTDNSGTVKINSGWSITKVTSAVHATDPVAIYQVDKVLLPEAIFGTDIPPVLAPAPTP 208
L + + V +++G + +++ V PV I+ VD VLLP +FG P APAP P
Sbjct: 287 LTTSTSGDEVILHTGVAPSRLADTVLDATPVVIFTVDNVLLPAELFGKSKSPSPAPAPEP 346
Query: 209 EIAP-----------AADSPTEQSADSKSSSPSSSPDRSSSYK 240
AP AA P + +S S+PS SP S++ K
Sbjct: 347 VTAPTPSPADAPSPTAASPPAPPTDESPESAPSDSPTGSANSK 389
Score = 33.9 bits (76), Expect = 0.41
Identities = 43/176 (24%), Positives = 76/176 (42%), Gaps = 12/176 (6%)
Query: 41 APTPDFVNLTELLTVAGPFHTFLQYLQSTKVLDTFQNQANNTEEGITIFVPKDSSFASLK 100
A T N+T++L + + +F YL TK+ D N+ IT+ V + + ++L
Sbjct: 20 ASTVSSHNITQILADSPDYSSFNSYLSQTKLADEI-----NSRTTITVLVLNNGAMSALA 74
Query: 101 -KPSLSKLKDDEIKQVILFHALPH-FYSLADFKNLSQT--ASTPTFAGGDYTLNFTD-NS 155
K LS +K V+L + P + ++ LS T +T G +N TD
Sbjct: 75 GKHPLSVIKSALSLLVLLDYYDPQKLHKISKGTTLSTTLYQTTGNAPGNLGFVNITDLKG 134
Query: 156 GTVKINSGWSITKVTSAVHATDPVAIYQVDKVLLPEAIFGTDIPPVLAPAPTPEIA 211
G V S S +K+ S+ + Y + + + I + + APAP+ ++
Sbjct: 135 GKVGFGSAASGSKLDSSYTKSVKQIPYNISILEIDAPIIAPGV--LTAPAPSASLS 188
Score = 33.9 bits (76), Expect = 0.41
Identities = 20/44 (45%), Positives = 28/44 (63%), Gaps = 5/44 (11%)
Query: 5 MIFLVSTLLLLSSSAFAKTVSP---PSP-PAESPTPAPAPAPTP 44
+IF V +LL + F K+ SP P+P P +PTP+PA AP+P
Sbjct: 318 VIFTVDNVLL-PAELFGKSKSPSPAPAPEPVTAPTPSPADAPSP 360
>FA10_ARATH (Q9LZX4) Fasciclin-like arabinogalactan protein 10
precursor
Length = 422
Score = 97.1 bits (240), Expect = 4e-20
Identities = 71/215 (33%), Positives = 104/215 (48%), Gaps = 13/215 (6%)
Query: 30 PAESPTPAPAPAPTPDFV-NLTELLTVAGPFHTFLQYLQSTKVLDTFQNQANNTEEGITI 88
P +P APAP+ V N+T LL AG TF L S+ V+ TF++ E+G+T+
Sbjct: 171 PIIAPGILTAPAPSSAGVSNITGLLEKAG-CKTFANLLVSSGVIKTFES---TVEKGLTV 226
Query: 89 FVPKDSSFASLKKPSLSKLKDDEIKQVILFHALPHFYSLADFKNLSQTAST-PTFAGGDY 147
F P D +F + P L+ L E+ ++ +HAL + K ST T G Y
Sbjct: 227 FAPSDEAFKARGVPDLTNLTQAEVVSLLEYHALAEYKPKGSLKTNKDAISTLATNGAGKY 286
Query: 148 TLNFTDNSGTVKINSGWSITKVTSAVHATDPVAIYQVDKVLLPEAIFGTDIPPVLAPAPT 207
L + + V +++G +++ V PV I+ VD VLLP +FG P APAP
Sbjct: 287 DLTTSTSGDEVILHTGVGPSRLADTVVDETPVVIFTVDNVLLPAELFGKSSSP--APAPE 344
Query: 208 PEIAPA-----ADSPTEQSADSKSSSPSSSPDRSS 237
P AP + SP E + + +S P+ D SS
Sbjct: 345 PVSAPTPTPAKSPSPVEAPSPTAASPPAPPVDESS 379
Score = 33.5 bits (75), Expect = 0.54
Identities = 19/42 (45%), Positives = 27/42 (64%), Gaps = 3/42 (7%)
Query: 5 MIFLVSTLLLLSSSAFAKTVSP-PSP-PAESPTPAPAPAPTP 44
+IF V +LL + F K+ SP P+P P +PTP PA +P+P
Sbjct: 319 VIFTVDNVLL-PAELFGKSSSPAPAPEPVSAPTPTPAKSPSP 359
Score = 30.4 bits (67), Expect = 4.5
Identities = 42/165 (25%), Positives = 72/165 (43%), Gaps = 12/165 (7%)
Query: 48 NLTELLTVAGPFHTFLQYLQSTKVLDTFQNQANNTEEGITIFVPKDSSFASLK-KPSLSK 106
N+T++L+ + +F YL TK+ D N+ IT+ V + + +SL K LS
Sbjct: 27 NITQILSDTPEYSSFNNYLSQTKLADEI-----NSRTTITVLVLNNGAMSSLAGKHPLSV 81
Query: 107 LKDDEIKQVILFHALP-HFYSLADFKNLSQT--ASTPTFAGGDYTLNFTD-NSGTVKINS 162
+K+ V+L + P + L+ L+ T +T G +N TD G V S
Sbjct: 82 VKNALSLLVLLDYYDPLKLHQLSKGTTLTTTLYQTTGHALGNLGFVNVTDLKGGKVGFGS 141
Query: 163 GWSITKVTSAVHATDPVAIYQVDKVLLPEAIFGTDIPPVLAPAPT 207
+K+ S+ + Y + + + I I + APAP+
Sbjct: 142 AAPGSKLDSSYTKSVKQIPYNISVLEINAPIIAPGI--LTAPAPS 184
>FLA1_ARATH (Q9FM65) Fasciclin-like arabinogalactan protein 1
precursor
Length = 424
Score = 79.7 bits (195), Expect = 7e-15
Identities = 59/208 (28%), Positives = 94/208 (44%), Gaps = 11/208 (5%)
Query: 27 PSPPAESPTPAPAPAPTPDFVNLTELLTVAGPFHTFLQYLQSTKVLDTFQNQANNTEEGI 86
PS A +PTPAPA +NLT +++ G L + T+Q + E G+
Sbjct: 171 PSETAAAPTPAPAE------MNLTGIMSAHGCKVFAETLLTNPGASKTYQE---SLEGGM 221
Query: 87 TIFVPKDSSFASLKKPSLSKLKDDEIKQVILFHALPHFYSLADFKNLSQTASTPTFAGGD 146
T+F P D + P L + + + F A+P +YS+A K+ + +T G +
Sbjct: 222 TVFCPGDDAMKGFL-PKYKNLTAPKKEAFLDFLAVPTYYSMAMLKSNNGPMNTLATDGAN 280
Query: 147 -YTLNFTDNSGTVKINSGWSITKVTSAVHATDPVAIYQVDKVLLPEAIFGTDIPPVLAPA 205
+ L ++ V + + + K+ + P+AIY DKVLLP+ +F APA
Sbjct: 281 KFELTVQNDGEKVTLKTRINTVKIVDTLIDEQPLAIYATDKVLLPKELFKASAVEAPAPA 340
Query: 206 PTPEIAPAADSPTEQSADSKSSSPSSSP 233
P PE ADSP +K ++P
Sbjct: 341 PAPEDGDVADSPKAAKGKAKGKKKKAAP 368
>YE83_SYNY3 (P74615) Hypothetical protein sll1483 precursor
Length = 180
Score = 43.5 bits (101), Expect = 5e-04
Identities = 37/143 (25%), Positives = 69/143 (47%), Gaps = 16/143 (11%)
Query: 52 LLTVAGPFHTFLQYLQSTKVLDTFQNQANNTEEGITIFVPKDSSFASLKKPSLSKL---- 107
++ VA TF + + K D + A + E T+F P + +FA+L ++ L
Sbjct: 48 IVEVAAGNETFSTLVAAVKAADLVE--ALSAEGPFTVFAPTNDAFAALPAGTVESLLLPE 105
Query: 108 KDDEIKQVILFHALPHFYSLADFKNLSQTASTPTFAGGDYTLNFTDNSGTVKINSGWSIT 167
D++ +++ +H +P + A Q+ + AG L F G VK+N T
Sbjct: 106 NKDKLVKILTYHVVPGKITAAQV----QSGEVASLAG--EALTFKVKDGKVKVNKA---T 156
Query: 168 KVTSAVHATDPVAIYQVDKVLLP 190
+++ V A++ V I+ +D+V+LP
Sbjct: 157 VISADVDASNGV-IHVIDQVILP 178
>GP1_CHLRE (Q9FPQ6) Vegetative cell wall protein gp1 precursor
(Hydroxyproline-rich glycoprotein 1)
Length = 555
Score = 38.1 bits (87), Expect = 0.022
Identities = 20/49 (40%), Positives = 25/49 (50%)
Query: 190 PEAIFGTDIPPVLAPAPTPEIAPAADSPTEQSADSKSSSPSSSPDRSSS 238
P + + PP AP+P P AP SP+ + S S SPS SP S S
Sbjct: 318 PAPVPPSPAPPSPAPSPPPSPAPPTPSPSPSPSPSPSPSPSPSPSPSPS 366
Score = 35.0 bits (79), Expect = 0.18
Identities = 14/24 (58%), Positives = 16/24 (66%)
Query: 21 AKTVSPPSPPAESPTPAPAPAPTP 44
A T PPSPP+ P+PAP PTP
Sbjct: 282 ANTPMPPSPPSPPPSPAPPTPPTP 305
Score = 32.3 bits (72), Expect = 1.2
Identities = 10/18 (55%), Positives = 16/18 (88%)
Query: 27 PSPPAESPTPAPAPAPTP 44
P+PP SP+P+P+P+P+P
Sbjct: 338 PAPPTPSPSPSPSPSPSP 355
Score = 31.6 bits (70), Expect = 2.0
Identities = 10/19 (52%), Positives = 16/19 (83%)
Query: 26 PPSPPAESPTPAPAPAPTP 44
PPSP +P+P+P+P+P+P
Sbjct: 335 PPSPAPPTPSPSPSPSPSP 353
Score = 31.6 bits (70), Expect = 2.0
Identities = 19/56 (33%), Positives = 30/56 (52%), Gaps = 2/56 (3%)
Query: 199 PPVLAPAPTPEIAPA-ADSPTEQSADSKSSSPSSSPDRSSSYKIVSYGIWGNLVLA 253
PP +P+P+P +P+ + SP+ + S S PS SP S S V +W + +A
Sbjct: 340 PPTPSPSPSPSPSPSPSPSPSPSPSPSPSPIPSPSPKPSPSPVAVKL-VWADDAIA 394
Score = 31.2 bits (69), Expect = 2.7
Identities = 11/21 (52%), Positives = 19/21 (90%), Gaps = 1/21 (4%)
Query: 25 SPPSP-PAESPTPAPAPAPTP 44
+PP+P P+ SP+P+P+P+P+P
Sbjct: 339 APPTPSPSPSPSPSPSPSPSP 359
Score = 31.2 bits (69), Expect = 2.7
Identities = 11/22 (50%), Positives = 17/22 (77%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T SP P+ SP+P+P+P+P+P
Sbjct: 342 TPSPSPSPSPSPSPSPSPSPSP 363
Score = 31.2 bits (69), Expect = 2.7
Identities = 11/19 (57%), Positives = 13/19 (67%)
Query: 26 PPSPPAESPTPAPAPAPTP 44
PPSP SP P+P P+P P
Sbjct: 322 PPSPAPPSPAPSPPPSPAP 340
Score = 30.0 bits (66), Expect = 5.9
Identities = 10/20 (50%), Positives = 16/20 (80%)
Query: 25 SPPSPPAESPTPAPAPAPTP 44
SP P+ SP+P+P+P+P+P
Sbjct: 346 SPSPSPSPSPSPSPSPSPSP 365
Score = 30.0 bits (66), Expect = 5.9
Identities = 10/20 (50%), Positives = 16/20 (80%)
Query: 25 SPPSPPAESPTPAPAPAPTP 44
SP P+ SP+P+P+P+P+P
Sbjct: 348 SPSPSPSPSPSPSPSPSPSP 367
Score = 30.0 bits (66), Expect = 5.9
Identities = 10/20 (50%), Positives = 16/20 (80%)
Query: 25 SPPSPPAESPTPAPAPAPTP 44
SP P+ SP+P+P+P+P+P
Sbjct: 350 SPSPSPSPSPSPSPSPSPSP 369
Score = 29.6 bits (65), Expect = 7.7
Identities = 11/20 (55%), Positives = 14/20 (70%)
Query: 25 SPPSPPAESPTPAPAPAPTP 44
SPPSP SP+P P+P+P
Sbjct: 120 SPPSPAPPSPSPPAPPSPSP 139
Score = 29.6 bits (65), Expect = 7.7
Identities = 12/20 (60%), Positives = 16/20 (80%), Gaps = 1/20 (5%)
Query: 25 SPPSPPAESPTPAPAPAPTP 44
SPP PA PTP+P+P+P+P
Sbjct: 333 SPPPSPAP-PTPSPSPSPSP 351
>YFAL_ECOLI (P45508) Hypothetical protein yfaL precursor
Length = 1250
Score = 36.6 bits (83), Expect = 0.063
Identities = 14/25 (56%), Positives = 17/25 (68%)
Query: 21 AKTVSPPSPPAESPTPAPAPAPTPD 45
++ V+PPSPP PTP P P P PD
Sbjct: 915 SQEVTPPSPPDPDPTPDPDPTPDPD 939
>P115_CHICK (Q98917) 115 kDa melanosomal matrix protein precursor
Length = 762
Score = 36.6 bits (83), Expect = 0.063
Identities = 25/87 (28%), Positives = 38/87 (42%), Gaps = 6/87 (6%)
Query: 147 YTLNFTDNSGTVKINSGWSITKVTSAVHATDPVAIYQVDKVLLPEAIFGTDIPPVLAPA- 205
Y+ +F D SGT+ S VT A V + +P + GT PPV+ P
Sbjct: 264 YSWDFGDQSGTLISRS----PTVTHTYLQAGSFAARLVLQAAIPLSSCGTSAPPVVDPTT 319
Query: 206 -PTPEIAPAADSPTEQSADSKSSSPSS 231
P P + P A P + +++PS+
Sbjct: 320 GPVPSLGPTATQPVGPTGSGTATAPSN 346
>SBN2_HUMAN (Q8WWQ8) Stabilin 2 precursor (FEEL-2 protein) (Fasciclin
EGF-like laminin-type EGF-like and link domain-containing
scavenger receptor-1) (FAS1 EGF-like and X-link domain
containing adhesion molecule-2) (Hyaluronan receptor for
endocytosis) [C
Length = 2551
Score = 36.2 bits (82), Expect = 0.083
Identities = 37/146 (25%), Positives = 66/146 (44%), Gaps = 11/146 (7%)
Query: 63 LQYLQSTKVLDTFQNQAN-----NTEEGITIFVPKDSSFASLKKPSLSK-LKDDEIKQVI 116
L +L + + + N A+ + +T+ VP + + + S L I +I
Sbjct: 1001 LSFLSEAAIFNRWINNASLQPTLSATSNLTVLVPSQQATEDMDQDEKSFWLSQSNIPALI 1060
Query: 117 LFHALPHFYSLADFKNLSQTASTPTFAGGDYTLNFTDNSGTVKINSGWSITKVTSAVHAT 176
+H L Y +AD + LS + T G++ L+ G + I G SI +A AT
Sbjct: 1061 KYHMLLGTYRVADLQTLSSSDMLATSLQGNF-LHLAKVDGNITI-EGASIVDGDNA--AT 1116
Query: 177 DPVAIYQVDKVLLPEAIFGTDIPPVL 202
+ V I+ ++KVL+P+ +P +L
Sbjct: 1117 NGV-IHIINKVLVPQRRLTGSLPNLL 1141
>Y091_NPVOP (O10341) Hypothetical 29.3 kDa protein (ORF92)
Length = 279
Score = 35.8 bits (81), Expect = 0.11
Identities = 18/41 (43%), Positives = 24/41 (57%), Gaps = 1/41 (2%)
Query: 5 MIFLVSTLLLLSSSAFAKTVSPPSP-PAESPTPAPAPAPTP 44
+I ++ TL LL+S PP P P +PTP+P P PTP
Sbjct: 11 IIIILLTLWLLTSLKQNIETKPPLPSPTPTPTPSPTPTPTP 51
Score = 33.1 bits (74), Expect = 0.70
Identities = 15/30 (50%), Positives = 15/30 (50%)
Query: 15 LSSSAFAKTVSPPSPPAESPTPAPAPAPTP 44
L S T SP P SPTP P P PTP
Sbjct: 34 LPSPTPTPTPSPTPTPTPSPTPTPTPTPTP 63
Score = 32.7 bits (73), Expect = 0.91
Identities = 12/22 (54%), Positives = 16/22 (72%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T SP P+ +PTP+P P+PTP
Sbjct: 124 TPSPTPTPSPTPTPSPTPSPTP 145
Score = 32.3 bits (72), Expect = 1.2
Identities = 12/22 (54%), Positives = 15/22 (67%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T +P P SPTP+P P+PTP
Sbjct: 128 TPTPSPTPTPSPTPSPTPSPTP 149
Score = 32.0 bits (71), Expect = 1.6
Identities = 12/28 (42%), Positives = 17/28 (59%)
Query: 17 SSAFAKTVSPPSPPAESPTPAPAPAPTP 44
S + T SP P+ +P+P P+P PTP
Sbjct: 84 SPTLSPTPSPTPTPSPTPSPTPSPTPTP 111
Score = 31.6 bits (70), Expect = 2.0
Identities = 11/22 (50%), Positives = 16/22 (72%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T +P P+ +PTP+P P+PTP
Sbjct: 148 TPTPSPTPSPTPTPSPTPSPTP 169
Score = 31.6 bits (70), Expect = 2.0
Identities = 15/24 (62%), Positives = 19/24 (78%), Gaps = 2/24 (8%)
Query: 23 TVSP-PSP-PAESPTPAPAPAPTP 44
T SP PSP P+ +PTP+P P+PTP
Sbjct: 96 TPSPTPSPTPSPTPTPSPTPSPTP 119
Score = 31.6 bits (70), Expect = 2.0
Identities = 11/22 (50%), Positives = 16/22 (72%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T +P P+ +PTP+P P+PTP
Sbjct: 108 TPTPSPTPSPTPTPSPTPSPTP 129
Score = 31.6 bits (70), Expect = 2.0
Identities = 12/22 (54%), Positives = 14/22 (63%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T SP P SPTP P+P P+P
Sbjct: 136 TPSPTPSPTPSPTPTPSPTPSP 157
Score = 31.6 bits (70), Expect = 2.0
Identities = 11/22 (50%), Positives = 16/22 (72%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T +P P+ +PTP+P P+PTP
Sbjct: 158 TPTPSPTPSPTPTPSPTPSPTP 179
Score = 31.2 bits (69), Expect = 2.7
Identities = 12/22 (54%), Positives = 14/22 (63%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T SP P SPTP P+P P+P
Sbjct: 82 TPSPTLSPTPSPTPTPSPTPSP 103
Score = 30.8 bits (68), Expect = 3.5
Identities = 11/22 (50%), Positives = 15/22 (68%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T SP P+ +P+P P P+PTP
Sbjct: 154 TPSPTPTPSPTPSPTPTPSPTP 175
Score = 30.8 bits (68), Expect = 3.5
Identities = 11/22 (50%), Positives = 15/22 (68%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T SP P+ +P+P P+P PTP
Sbjct: 130 TPSPTPTPSPTPSPTPSPTPTP 151
Score = 30.8 bits (68), Expect = 3.5
Identities = 11/20 (55%), Positives = 13/20 (65%)
Query: 25 SPPSPPAESPTPAPAPAPTP 44
+P P SPTP P+P PTP
Sbjct: 118 TPTPSPTPSPTPTPSPTPTP 137
Score = 30.8 bits (68), Expect = 3.5
Identities = 11/22 (50%), Positives = 14/22 (63%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T +P P +PTP P+P PTP
Sbjct: 54 TPTPTPTPTPTPTPTPSPTPTP 75
Score = 30.8 bits (68), Expect = 3.5
Identities = 11/22 (50%), Positives = 15/22 (68%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T SP P+ +P+P P P+PTP
Sbjct: 114 TPSPTPTPSPTPSPTPTPSPTP 135
Score = 30.8 bits (68), Expect = 3.5
Identities = 11/22 (50%), Positives = 15/22 (68%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T SP P+ +P+P P P+PTP
Sbjct: 144 TPSPTPTPSPTPSPTPTPSPTP 165
Score = 30.8 bits (68), Expect = 3.5
Identities = 11/22 (50%), Positives = 15/22 (68%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T SP P+ +P+P P P+PTP
Sbjct: 104 TPSPTPTPSPTPSPTPTPSPTP 125
Score = 30.4 bits (67), Expect = 4.5
Identities = 11/22 (50%), Positives = 14/22 (63%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T SP P +P+P P P+PTP
Sbjct: 120 TPSPTPSPTPTPSPTPTPSPTP 141
Score = 30.4 bits (67), Expect = 4.5
Identities = 11/22 (50%), Positives = 14/22 (63%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T SP P +P+P P+P PTP
Sbjct: 100 TPSPTPSPTPTPSPTPSPTPTP 121
Score = 30.4 bits (67), Expect = 4.5
Identities = 11/22 (50%), Positives = 14/22 (63%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T SP P +P+P P+P PTP
Sbjct: 110 TPSPTPSPTPTPSPTPSPTPTP 131
Score = 30.4 bits (67), Expect = 4.5
Identities = 11/22 (50%), Positives = 15/22 (68%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T SP P+ +P+P P P+PTP
Sbjct: 164 TPSPTPTPSPTPSPTPPPSPTP 185
Score = 30.4 bits (67), Expect = 4.5
Identities = 11/22 (50%), Positives = 13/22 (59%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T +P P +PTP P P PTP
Sbjct: 48 TPTPSPTPTPTPTPTPTPTPTP 69
Score = 30.4 bits (67), Expect = 4.5
Identities = 11/22 (50%), Positives = 13/22 (59%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T +P P +PTP P P PTP
Sbjct: 46 TPTPTPSPTPTPTPTPTPTPTP 67
Score = 30.4 bits (67), Expect = 4.5
Identities = 11/22 (50%), Positives = 14/22 (63%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T SP P +P+P P+P PTP
Sbjct: 150 TPSPTPSPTPTPSPTPSPTPTP 171
Score = 30.4 bits (67), Expect = 4.5
Identities = 11/22 (50%), Positives = 14/22 (63%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T SP P +P+P P+P PTP
Sbjct: 140 TPSPTPSPTPTPSPTPSPTPTP 161
Score = 30.0 bits (66), Expect = 5.9
Identities = 11/22 (50%), Positives = 13/22 (59%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T SP P +PTP P P P+P
Sbjct: 50 TPSPTPTPTPTPTPTPTPTPSP 71
Score = 29.6 bits (65), Expect = 7.7
Identities = 10/22 (45%), Positives = 15/22 (67%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T +P P+ +PTP+P P P+P
Sbjct: 118 TPTPSPTPSPTPTPSPTPTPSP 139
Score = 29.6 bits (65), Expect = 7.7
Identities = 10/22 (45%), Positives = 15/22 (67%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T +P P+ +P+P P P+PTP
Sbjct: 134 TPTPSPTPSPTPSPTPTPSPTP 155
Score = 29.6 bits (65), Expect = 7.7
Identities = 12/22 (54%), Positives = 14/22 (63%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP 44
T +P P SPTP PA +PTP
Sbjct: 60 TPTPTPTPTPSPTPTPALSPTP 81
>V50K_BYDVP (P09516) 50 kDa protein (ORF 4)
Length = 450
Score = 35.8 bits (81), Expect = 0.11
Identities = 24/87 (27%), Positives = 39/87 (44%), Gaps = 18/87 (20%)
Query: 23 TVSPPSPPAESPTPAPAPAPTP----DFVNL----------TELLTVAGPFHTFLQYLQS 68
T +P PA PTP P PAP P +++ T+ ++V+ +QY+++
Sbjct: 15 TPTPQPTPAPQPTPEPTPAPVPKRFFEYIGTPTGTISTRENTDSISVSKLGGQSMQYIEN 74
Query: 69 ----TKVLDTFQNQANNTEEGITIFVP 91
TKV+D+F + NN P
Sbjct: 75 EKCETKVIDSFWSTNNNVSAQAAFVYP 101
>TLE2_HUMAN (Q04725) Transducin-like enhancer protein 2 (ESG2)
Length = 743
Score = 35.8 bits (81), Expect = 0.11
Identities = 41/149 (27%), Positives = 62/149 (41%), Gaps = 33/149 (22%)
Query: 15 LSSSAFAKTVSPPSPPAESPTPAPA-----------PAPTPDFVNLTELLTVAGPFHTFL 63
L +S A SPP ++ TP P+ PAP+ D V L LT++ PF T
Sbjct: 294 LPASTPASKSCDSSPPQDASTPGPSSASHLCQLAAKPAPSTDSVALRSPLTLSSPFTT-- 351
Query: 64 QYLQSTKVLDTFQNQANNTEEGITIFVPKDSSFASLK-KPSLSKLKDDEIKQVILFHALP 122
+F +++T G + VP SS+ SL P +S V+ F + P
Sbjct: 352 ----------SFSLGSHSTLNG-DLSVP--SSYVSLHLSPQVSSSVVYGRSPVMAFESHP 398
Query: 123 HFYSLADFKNLSQTASTPTFAGGDYTLNF 151
H + S ++S P+ GG +F
Sbjct: 399 H------LRGSSVSSSLPSIPGGKPAYSF 421
>SR1A_PHYPO (P09350) Spherulin 1A precursor
Length = 246
Score = 35.8 bits (81), Expect = 0.11
Identities = 20/53 (37%), Positives = 26/53 (48%), Gaps = 2/53 (3%)
Query: 1 MKVSMIFLVSTLLLLSSSAFAKTVSPPSPPAESPTPAPAPAPTPDFVNLTELL 53
MK + +F + L L +S A T P +PP PTPAP P P EL+
Sbjct: 1 MKSTFLFALFVLFLAASEA--ATDYPTNPPTTPPTPAPTSTPLPSSAASPELV 51
>VTPX_TTV1 (P19274) Viral protein TPX
Length = 360
Score = 35.4 bits (80), Expect = 0.14
Identities = 19/47 (40%), Positives = 27/47 (57%), Gaps = 1/47 (2%)
Query: 23 TVSPPSPPAESPTPAPAPAPTPDFVNLTELLTVAGPFHTFLQYLQST 69
TV+P S P+ +PTP P P PTP + ++T ++ P T L ST
Sbjct: 269 TVTPISSPSPTPTPTPTPTPTPTY-DITYVVFDVTPSPTPTPTLTST 314
Score = 30.8 bits (68), Expect = 3.5
Identities = 11/21 (52%), Positives = 13/21 (61%)
Query: 24 VSPPSPPAESPTPAPAPAPTP 44
V+P P +PTP P P PTP
Sbjct: 332 VTPSPTPTPTPTPTPTPTPTP 352
Score = 29.6 bits (65), Expect = 7.7
Identities = 12/24 (50%), Positives = 13/24 (54%)
Query: 20 FAKTVSPPSPPAESPTPAPAPAPT 43
F T SP P +PTP P P PT
Sbjct: 330 FDVTPSPTPTPTPTPTPTPTPTPT 353
>N551_SOYBN (Q05544) Early nodulin 55-1 (N-55-1) (Fragment)
Length = 137
Score = 35.4 bits (80), Expect = 0.14
Identities = 12/33 (36%), Positives = 25/33 (75%)
Query: 12 LLLLSSSAFAKTVSPPSPPAESPTPAPAPAPTP 44
++++S++ K + PSP + SP+P+P+P+P+P
Sbjct: 63 VVVMSNNTKKKLIHSPSPSSPSPSPSPSPSPSP 95
>USH_DROME (Q9VPQ6) Zinc-finger protein ush (U-shaped protein)
Length = 1191
Score = 35.0 bits (79), Expect = 0.18
Identities = 16/31 (51%), Positives = 20/31 (63%), Gaps = 2/31 (6%)
Query: 25 SPPSPPAESPTPAPAPAPTPDF--VNLTELL 53
+PPS P SP P+P+P PTP + V L LL
Sbjct: 131 TPPSEPEASPCPSPSPCPTPKYPKVRLNALL 161
>CDNC_HUMAN (P49918) Cyclin-dependent kinase inhibitor 1C
(Cyclin-dependent kinase inhibitor p57) (p57KIP2)
Length = 316
Score = 35.0 bits (79), Expect = 0.18
Identities = 14/25 (56%), Positives = 16/25 (64%)
Query: 21 AKTVSPPSPPAESPTPAPAPAPTPD 45
A +P PA +P PAPAPAP PD
Sbjct: 193 APVAAPAPAPAPAPAPAPAPAPAPD 217
Score = 33.1 bits (74), Expect = 0.70
Identities = 15/36 (41%), Positives = 20/36 (54%)
Query: 9 VSTLLLLSSSAFAKTVSPPSPPAESPTPAPAPAPTP 44
V+ +L + A A +P P +P PAPAPAP P
Sbjct: 173 VAVAVLAPAPAPAPAPAPAPAPVAAPAPAPAPAPAP 208
Score = 32.3 bits (72), Expect = 1.2
Identities = 13/24 (54%), Positives = 16/24 (66%)
Query: 21 AKTVSPPSPPAESPTPAPAPAPTP 44
A +P + PA +P PAPAPAP P
Sbjct: 189 APAPAPVAAPAPAPAPAPAPAPAP 212
Score = 30.4 bits (67), Expect = 4.5
Identities = 18/49 (36%), Positives = 25/49 (50%), Gaps = 1/49 (2%)
Query: 187 VLLPEAIFGTDIPPVLAPAPTPEIAP-AADSPTEQSADSKSSSPSSSPD 234
V P A+ P APAP P AP AA +P A + + +P+ +PD
Sbjct: 169 VAAPVAVAVLAPAPAPAPAPAPAPAPVAAPAPAPAPAPAPAPAPAPAPD 217
Score = 30.0 bits (66), Expect = 5.9
Identities = 15/38 (39%), Positives = 18/38 (46%)
Query: 21 AKTVSPPSPPAESPTPAPAPAPTPDFVNLTELLTVAGP 58
A +PP P +P PAPAPAP V + V P
Sbjct: 143 APASTPPPVPVLAPAPAPAPAPVAAPVAAPVAVAVLAP 180
Score = 29.6 bits (65), Expect = 7.7
Identities = 15/36 (41%), Positives = 19/36 (52%)
Query: 9 VSTLLLLSSSAFAKTVSPPSPPAESPTPAPAPAPTP 44
V+ L + A A +P A +P PAPAPAP P
Sbjct: 175 VAVLAPAPAPAPAPAPAPAPVAAPAPAPAPAPAPAP 210
>BRD4_HUMAN (O60885) Bromodomain-containing protein 4 (HUNK1
protein)
Length = 1362
Score = 35.0 bits (79), Expect = 0.18
Identities = 15/32 (46%), Positives = 19/32 (58%)
Query: 27 PSPPAESPTPAPAPAPTPDFVNLTELLTVAGP 58
P+PP TP P PA TPD + T ++TV P
Sbjct: 213 PNPPPVQATPHPFPAVTPDLIVQTPVMTVVPP 244
Score = 29.6 bits (65), Expect = 7.7
Identities = 11/17 (64%), Positives = 11/17 (64%)
Query: 26 PPSPPAESPTPAPAPAP 42
PP PP P PAPAP P
Sbjct: 251 PPVPPQPQPPPAPAPQP 267
>TP3A_DROME (Q9NG98) DNA topoisomerase III alpha (EC 5.99.1.2)
Length = 1250
Score = 34.7 bits (78), Expect = 0.24
Identities = 54/218 (24%), Positives = 79/218 (35%), Gaps = 43/218 (19%)
Query: 23 TVSPPSPPAESPTPAPAPAP-----TPDFVNLTELLTVAGPFHTFLQYLQSTKVLDTFQN 77
T PP+ P T P AP + ++ T A P ++ ++ LD F +
Sbjct: 849 TKKPPNEPKPKKTKEPKAAPNKKTSSKSSGSIRSFFTSAAPTNS------ASNGLDEFFD 902
Query: 78 QANNTEEGITIFVPKDSSFASLKKPSLSKLKDDEIKQVILFHALPHFYSLADFKNLSQTA 137
+ E+ + S + K S+ L DD F A A+F+ L
Sbjct: 903 SNDGFEDAMLAAAESVESSSQPKTISMVPLDDDI---AAAFAADDD----AEFEALVNGG 955
Query: 138 STPTFAGGDYTLNFTDNSGTVKINSGWSITKVTSAVHATDPVAIYQVDKVLLPEAIFGTD 197
+ PT + GD L+ K S W I + DK ++GT
Sbjct: 956 TMPTESNGDQQLD--------KSLSEW----------------IKEQDKADERPMLWGTR 991
Query: 198 IPPVLAPA-PTPEIAPAADSPTEQSADSKSSSPSSSPD 234
L A PTP PAA P S + S+ PSS P+
Sbjct: 992 ERASLGTAAPTPPPKPAAKRPRWDSVERDSTPPSSVPE 1029
>PCLO_HUMAN (Q9Y6V0) Piccolo protein (Aczonin)
Length = 5183
Score = 34.7 bits (78), Expect = 0.24
Identities = 24/72 (33%), Positives = 29/72 (39%), Gaps = 4/72 (5%)
Query: 7 FLVSTLLLLSSSAFAKTVSPPSPPAESPTPAPAPAPTPDFVNLTEL----LTVAGPFHTF 62
F S+ L +S+ PP PP P P P P P P T L LTVA P T
Sbjct: 2324 FFRSSSLDISAQPPPPPPPPPPPPPPPPPPPPPPLPPPTSPKPTILPKKKLTVASPVTTA 2383
Query: 63 LQYLQSTKVLDT 74
+ L+T
Sbjct: 2384 TPLFDAVTTLET 2395
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.313 0.130 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 31,410,913
Number of Sequences: 164201
Number of extensions: 1428996
Number of successful extensions: 16677
Number of sequences better than 10.0: 201
Number of HSP's better than 10.0 without gapping: 131
Number of HSP's successfully gapped in prelim test: 78
Number of HSP's that attempted gapping in prelim test: 13810
Number of HSP's gapped (non-prelim): 2010
length of query: 262
length of database: 59,974,054
effective HSP length: 108
effective length of query: 154
effective length of database: 42,240,346
effective search space: 6505013284
effective search space used: 6505013284
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 65 (29.6 bits)
Medicago: description of AC141110.2


