

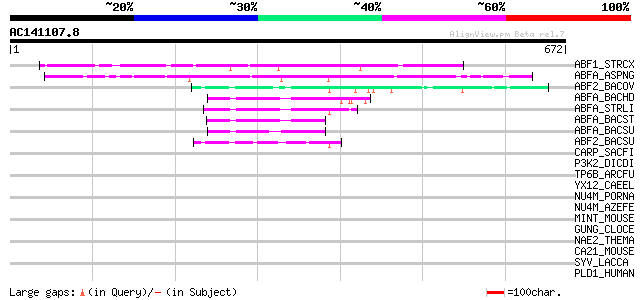
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141107.8 + phase: 0
(672 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ABF1_STRCX (P82593) Alpha-N-arabinofuranosidase I precursor (EC ... 290 8e-78
ABFA_ASPNG (P42254) Alpha-N-arabinofuranosidase A precursor (EC ... 252 2e-66
ABF2_BACOV (Q59219) Alpha-N-arabinofuranosidase 2 (EC 3.2.1.55) ... 84 2e-15
ABFA_BACHD (Q9KBR4) Alpha-N-arabinofuranosidase (EC 3.2.1.55) (A... 68 7e-11
ABFA_STRLI (P53627) Alpha-N-arabinofuranosidase (EC 3.2.1.55) (A... 68 9e-11
ABFA_BACST (Q9XBQ3) Alpha-N-arabinofuranosidase (EC 3.2.1.55) (A... 67 2e-10
ABFA_BACSU (P94531) Alpha-N-arabinofuranosidase 1 (EC 3.2.1.55) ... 60 1e-08
ABF2_BACSU (P94552) Alpha-N-arabinofuranosidase 2 (EC 3.2.1.55) ... 60 1e-08
CARP_SACFI (P22929) Acid protease precursor (EC 3.4.23.-) 36 0.29
P3K2_DICDI (P54674) Phosphatidylinositol 3-kinase 2 (EC 2.7.1.13... 36 0.38
TP6B_ARCFU (O29605) Type II DNA topoisomerase VI subunit B (EC 5... 33 2.4
YX12_CAEEL (Q11123) Hypothetical protein C03F11.2 in chromosome X 32 4.2
NU4M_PORNA (O03772) NADH-ubiquinone oxidoreductase chain 4 (EC 1... 32 4.2
NU4M_AZEFE (P92494) NADH-ubiquinone oxidoreductase chain 4 (EC 1... 32 4.2
MINT_MOUSE (Q62504) Msx2-interacting protein (SMART/HDAC1 associ... 32 4.2
GUNG_CLOCE (P37700) Endoglucanase G precursor (EC 3.2.1.4) (Endo... 32 4.2
NAE2_THEMA (Q9X0Y0) Probable glutamine-dependent NAD(+) syntheta... 32 5.4
CA21_MOUSE (Q01149) Collagen alpha 2(I) chain precursor 32 5.4
SYV_LACCA (P36420) Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--t... 32 7.1
PLD1_HUMAN (Q13393) Phospholipase D1 (EC 3.1.4.4) (PLD 1) (Choli... 32 7.1
>ABF1_STRCX (P82593) Alpha-N-arabinofuranosidase I precursor (EC
3.2.1.55) (Arabinosidase I) (Alpha-N-AFase I)
Length = 825
Score = 290 bits (742), Expect = 8e-78
Identities = 191/543 (35%), Positives = 269/543 (49%), Gaps = 56/543 (10%)
Query: 37 TLVVDASQASGRRIPETLFGIFFEEINHAGAGGLWAELVSNRGFEAGGPNIPSN--IDPW 94
++ VD + A G I +T++G+FFE+IN A GGL+AELV NR FE + S + W
Sbjct: 36 SITVDPA-AKGAAIDDTMYGVFFEDINRAADGGLYAELVQNRSFEYSTDDNRSYTPLTSW 94
Query: 95 SIIGNATYINVETDRTSCFERNKVALRLEVLCDGTCPTDGVGVYNPGF-WGMNIEQGKKY 153
+ G +N D ERN+ L L G V N G+ G+ +EQGK+Y
Sbjct: 95 IVDGTGEVVN---DAGRLNERNRNYLSLGA---------GSSVTNAGYNTGIRVEQGKRY 142
Query: 154 KVVFYARSTGPLNLKVSLTGSNGVGSLASTVITGSASDFSNWTKVETVLEAKATNPNSRL 213
+AR+ L V+L + G + A V W K A T+ RL
Sbjct: 143 DFSVWARAGSASTLTVALKDAAGTLATARQVAVEGG-----WAKYRATFTATRTSNRGRL 197
Query: 214 QLTTTTKGVIWLDQVSAMPLDTYKG--HGFRSDLLQMLVDLKPSFIRFPGGCFVE----- 266
+ LD VS P DTY+ +G R DL + + L P F+RFPGGC V
Sbjct: 198 AVAANDAAA--LDMVSLFPRDTYRNQQNGLRKDLAEKIAALHPGFVRFPGGCLVNTGSME 255
Query: 267 ------GDYLRNAFRWKAAVGPWEERPGHFGDVWKYWTDDGLGYYEFLQLSEDLGALPIW 320
G + +++WK VGP EER + + W Y GLGYYE+ + SED+GA+P+
Sbjct: 256 DYSAASGWQRKRSYQWKDTVGPVEERATN-ANFWGYNQSYGLGYYEYFRFSEDIGAMPLP 314
Query: 321 VFN---NGVSHNDEVDTSAVLP-FVQEALDGLEFARGDPTSKWGSMRAAMGHPEPFNLKY 376
V G N VD A+L +Q+ LD +EFA G TSKWG +RA MGHP PF L +
Sbjct: 315 VVPALVTGCGQNKAVDDEALLKRHIQDTLDLIEFANGPATSKWGKVRAEMGHPRPFRLTH 374
Query: 377 VAVGNEDCGKKNYRGNYLRFYDAIRRAYPDIQIISNC--DGSSRPLDHP--------ADM 426
+ VGNE+ + + +F AI YPDI ++SN D + D +M
Sbjct: 375 LEVGNEENLPDEFFDRFKQFRAAIEAEYPDITVVSNSGPDDAGTTFDTAWKLNREANVEM 434
Query: 427 YDYHIYTNANDMFSRSTTFNRVTRSGPKAFVSEYAVTGNDAGQGSLLAALAEAGFLIGLE 486
D H Y + N + ++ R GPK F+ EYA GN G L+EA F+ GLE
Sbjct: 435 VDEHYYNSPNWFLQNNDRYDSYDRGGPKVFLGEYASQGNAWKNG-----LSEAAFMTGLE 489
Query: 487 KNSDIVHMASYAPLFVNANDRRWNPDAIVFNSFQLYGTPSYWMQLFFSESNGATLLNSSL 546
+N+D+V +ASYAPL N + +W PD + FN+ + + +Y +Q F + G ++ S
Sbjct: 490 RNADVVKLASYAPLLANEDYVQWRPDLVWFNNRASWNSANYEVQKLFMNNVGDRVVPSKA 549
Query: 547 QTT 549
TT
Sbjct: 550 TTT 552
>ABFA_ASPNG (P42254) Alpha-N-arabinofuranosidase A precursor (EC
3.2.1.55) (Arabinosidase A) (ABF A)
Length = 628
Score = 252 bits (644), Expect = 2e-66
Identities = 187/609 (30%), Positives = 304/609 (49%), Gaps = 49/609 (8%)
Query: 43 SQASGRRIPETLFGIFFEEINHAGAGGLWAELVSNRGFEAGGPNIPSNIDPWSIIGNATY 102
S G L+G FE+INH+G GG++ +++ N G + PN+ + W+ +G+AT
Sbjct: 31 STQGGNSSSPILYGFMFEDINHSGDGGIYGQMLQNPGLQGTAPNLTA----WAAVGDATI 86
Query: 103 -INVETDRTSCFERNKVALRLEVLCDGTCPTDGVGVYNPGFWGMNIEQGKKYKVVFYARS 161
I+ ++ TS ++L + D T VG+ N G+WG+ ++ G ++ F+ +
Sbjct: 87 AIDGDSPLTSAIPST---IKLNIADDAT---GAVGLTNEGYWGIPVD-GSEFHSSFWIKG 139
Query: 162 TGPLNLKVSLTGSNGVGSLASTVITGSASDFSNWTKVETVLEA-KATNPNSRLQLT---T 217
++ V L G+ ST IT + S N+T+ KA + N +LT +
Sbjct: 140 DYSGDITVRLVGNYTGTEYGSTTITHT-STADNFTQASVKFPTTKAPDGNVLYELTVDGS 198
Query: 218 TTKGVIWLDQVSAMPLDTYKG--HGFRSDLLQMLVDLKPSFIRFPGGCFVEGDYLRNAFR 275
G + +TYK +G + L +L D+K SF+RFPGG +EG+ N ++
Sbjct: 199 VAAGSSLNFGYLTLFGETYKSRENGLKPQLANVLDDMKGSFLRFPGGNNLEGNSAENRWK 258
Query: 276 WKAAVGPWEERPGHFGDVWKYWTDDGLGYYEFLQLSEDLGALPIWVFNNGVS----HNDE 331
W +G +RPG G W Y+ DGLG +E+ EDLG +P+ +G + N
Sbjct: 259 WNETIGDLCDRPGREG-TWTYYNTDGLGLHEYFYWCEDLGLVPVLGVWDGFALESGGNTP 317
Query: 332 VDTSAVLPFVQEALDGLEFARGDPTSKWGSMRAAMGHPEPFNLKYVAVGNEDC---GKKN 388
+ A+ P++ + L+ LE+ GD ++ +G+ RAA G EP+NL V +GNED G ++
Sbjct: 318 LTGDALTPYIDDVLNELEYILGDTSTTYGAWRAANGQEEPWNLTMVEIGNEDMLGGGCES 377
Query: 389 YRGNYLRFYDAIRRAYPD-IQIISNCDGSSRPLDHPADMY-DYHIYTNANDMFSRSTTFN 446
Y + FYDAI AYPD I I S + P P + DYH Y+ + + + F+
Sbjct: 378 YAERFTAFYDAIHAAYPDLILIASTSEADCLPESMPEGSWVDYHDYSTPDGLVGQFNYFD 437
Query: 447 RVTRSGPKAFVSEYAVTGNDAGQGSLLAALAEAGFLIGLEKNSDIVHMASYAPLFVNAND 506
+ RS P F+ EY+ D ++ ++AEA F+IG E+NSD+V MA+YAPL N
Sbjct: 438 NLNRSVP-YFIGEYSRWEID--WPNMKGSVAEAVFMIGFERNSDVVKMAAYAPLLQLINS 494
Query: 507 RRWNPDAIVFNSF--QLYGTPSYWMQLFFSESNGATLLNSSLQTTASNSLVASAITWQNS 564
+W PD I + ++ + SY++Q FS + G T+ + T+ + + W S
Sbjct: 495 TQWTPDLIGYTQSPGDIFLSTSYYVQEMFSRNRGDTI----KEVTSDSDF--GPLYWVAS 548
Query: 565 VDKKNYIRIKAVNFGTSAVNLKISFNGLDPNSLQSSGSTKTVLTSTNLMDENSFSQPKKV 624
+Y +K N+G+ +L +S G L TVL ++ NS +Q V
Sbjct: 549 SAGDSYY-VKLANYGSETQDLTVSIPGTSTGKL-------TVLADSDPDAYNSDTQ-TLV 599
Query: 625 IPIQSLLQS 633
P +S +Q+
Sbjct: 600 TPSESTVQA 608
>ABF2_BACOV (Q59219) Alpha-N-arabinofuranosidase 2 (EC 3.2.1.55)
(Arabinosidase 2)
Length = 514
Score = 83.6 bits (205), Expect = 2e-15
Identities = 113/491 (23%), Positives = 184/491 (37%), Gaps = 91/491 (18%)
Query: 221 GVIWLDQVSAMPLDTYKGHGFRSDLLQMLVDLKPSFIRFPGGCFVEGDYLRNAFRWKAAV 280
G +W+ + S +P G+R+D+ L DL +R+PGGCF + + W +
Sbjct: 55 GGLWVGENSDIP----NIKGYRTDVFNALKDLSVPVLRWPGGCFAD------EYHWMDGI 104
Query: 281 GPWEERPGHFGDVWKYWTDD-GLGYYEFLQLSEDLGALPIWVFNNGVSHNDEVDTSAVLP 339
GP E RP + W +D G +EFL L E LG P VS N T
Sbjct: 105 GPKENRPKMVNNNWGGTIEDNSFGTHEFLNLCEMLGCEPY------VSGNVGSGT----- 153
Query: 340 FVQEALDGLEFARGDPTSKWGSMRAAMGHPEPFNLKYVAVGNEDCG-----KKNYRGNYL 394
V+E +E+ D S ++R G + + LKY+ VGNE G + Y +
Sbjct: 154 -VEELAKWVEYMTSDGDSPMANLRRKNGRDKAWKLKYLGVGNESWGCGGSMRPEYYADLY 212
Query: 395 RFYDAIRRAYPDIQIISNCDGSS-----------RPLDHPADMYDYHIYT---------- 433
R Y R Y ++ G+S + H D H YT
Sbjct: 213 RRYSTYCRNYDGNRLFKIASGASDYDYKWTDVLMNRVGHRMDGLSLHYYTVTGWSGSKGS 272
Query: 434 ----NANDMF--------------SRSTTFNRVTRSGPKAFVSEY----------AVTGN 465
N +D + T ++ + A + + + G+
Sbjct: 273 ATQFNKDDYYWTMGKCLEVEDVLKKHCTIMDKYDKDKKIALLLDEWGTWWDEEPGTIKGH 332
Query: 466 DAGQGSLLAALAEAGFLIGLEKNSDIVHMASYAPLFVNANDRRWNPDAIVFNSFQLYGTP 525
Q +L A + L K +D + MA+ A + VN I+ ++ TP
Sbjct: 333 LYQQNTLRDAFVASLSLDVFHKYTDRLKMANIAQI-VNVLQ-----SMILTKDKEMVLTP 386
Query: 526 SYWMQLFFSESNGATLLNSSL---QTTASNSLVASAITWQNSVDKKNYIRIKAVNFGTSA 582
+Y++ + AT L L + + ++ ++ S +K I I N
Sbjct: 387 TYYVFKMYKVHQDATYLPIDLTCEKMSVRDNRTVPMVSATASKNKDGVIHISLSNVDADE 446
Query: 583 VNLKISFNGLDPNSLQSSGSTKTVLTSTNLMDENSFSQPKKVIPIQSLLQSVGK-DMNVI 641
+I+ N D + ++ G +LT++ L D NSF +P V P + K M V
Sbjct: 447 AQ-EITINLGDTKAKKAIGE---ILTASKLTDYNSFEKPNIVKPAPFKEVKINKGTMKVK 502
Query: 642 VPPHSFTSFDL 652
+P S + +L
Sbjct: 503 LPAKSIVTLEL 513
>ABFA_BACHD (Q9KBR4) Alpha-N-arabinofuranosidase (EC 3.2.1.55)
(Arabinosidase)
Length = 500
Score = 68.2 bits (165), Expect = 7e-11
Identities = 56/219 (25%), Positives = 89/219 (40%), Gaps = 39/219 (17%)
Query: 240 GFRSDLLQMLVDLKPSFIRFPGGCFVEGDYLRNAFRWKAAVGPWEERPGHFGDVWKYWTD 299
GFR D+++++ +L+ +R+PGG FV G + W+ VGP ERP W+
Sbjct: 50 GFRKDVIRLVQELQVPLVRYPGGNFVSG------YNWEDGVGPVSERPKRLDLAWRTTET 103
Query: 300 DGLGYYEFLQLSEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGLEFARGDPTSKW 359
+ +G EF+ ++ +GA N G V A + +E+ S W
Sbjct: 104 NEIGTNEFVDWAKKVGAEVNMAVNLGSRG------------VDAARNLVEYCNHPSGSYW 151
Query: 360 GSMRAAMGHPEPFNLKYVAVGNEDCGKKNYRGNYLRFYDAI--------RRAYPDIQII- 410
+R + G+ +P N+K +GNE G Y + + P I+++
Sbjct: 152 SDLRISHGYKDPHNIKTWCLGNEMDGPWQIGQKTAEEYGRVAAEAGKVMKLVDPSIELVA 211
Query: 411 ---SN------CDGSSRPLDHPADMYDY---HIYTNAND 437
SN D + LDH D DY H Y D
Sbjct: 212 CGSSNSKMATFADWEATVLDHTYDYVDYISLHTYYGNRD 250
>ABFA_STRLI (P53627) Alpha-N-arabinofuranosidase (EC 3.2.1.55)
(Arabinosidase) (ABF) (Alpha-L-AF)
Length = 662
Score = 67.8 bits (164), Expect = 9e-11
Identities = 50/197 (25%), Positives = 89/197 (44%), Gaps = 29/197 (14%)
Query: 235 TYKGHGFRSDLLQMLVDLKPSFIRFPGGCFVEGDYLRNAFRWKAAVGPWEERPGHFGDVW 294
T G R D+L+++ +L + +R+PGG FV G ++W+ +VGP E+RP W
Sbjct: 44 TADAEGLRQDVLELVRELGVTAVRYPGGNFVSG------YKWEDSVGPVEDRPRRLDLAW 97
Query: 295 KYWTDDGLGYYEFLQLSEDLG--ALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGLEFAR 352
+ + G E++ + +G A P+ N G V EAL+ E+A
Sbjct: 98 RSTETNRFGLSEYIAFLKKIGPQAEPMMAVNLGTRG------------VAEALELQEYAN 145
Query: 353 GDPTSKWGSMRAAMGHPEPFNLKYVAVGNEDCG--------KKNYRGNYLRFYDAIRRAY 404
+ +RA G +PF ++ +GNE G + Y A+R+
Sbjct: 146 HPSGTALSDLRAEHGDKDPFGIRLWCLGNEMDGPWQTGHKTAEEYGRVAAETARAMRQID 205
Query: 405 PDIQIISNCDGSSRPLD 421
PD+++++ C S + ++
Sbjct: 206 PDVELVA-CGSSGQSME 221
>ABFA_BACST (Q9XBQ3) Alpha-N-arabinofuranosidase (EC 3.2.1.55)
(Arabinosidase)
Length = 501
Score = 66.6 bits (161), Expect = 2e-10
Identities = 41/144 (28%), Positives = 68/144 (46%), Gaps = 18/144 (12%)
Query: 239 HGFRSDLLQMLVDLKPSFIRFPGGCFVEGDYLRNAFRWKAAVGPWEERPGHFGDVWKYWT 298
+GFR D+++++ +L+ IR+PGG FV G + W+ VGP E+RP WK
Sbjct: 49 NGFRQDVIELVKELQVPIIRYPGGNFVSG------YNWEDGVGPKEQRPRRLDLAWKSVE 102
Query: 299 DDGLGYYEFLQLSEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGLEFARGDPTSK 358
+ +G EF+ ++ +GA N G + A + +E+ S
Sbjct: 103 TNEIGLNEFMDWAKMVGAEVNMAVNLGTRG------------IDAARNLVEYCNHPSGSY 150
Query: 359 WGSMRAAMGHPEPFNLKYVAVGNE 382
+ +R A G+ EP +K +GNE
Sbjct: 151 YSDLRIAHGYKEPHKIKTWCLGNE 174
>ABFA_BACSU (P94531) Alpha-N-arabinofuranosidase 1 (EC 3.2.1.55)
(Arabinosidase 1)
Length = 500
Score = 60.5 bits (145), Expect = 1e-08
Identities = 39/145 (26%), Positives = 66/145 (44%), Gaps = 22/145 (15%)
Query: 240 GFRSDLLQMLVDLKPSFIRFPGGCFVEGDYLRNAFRWKAAVGPWEERPGHFGDVWKYWTD 299
GFR D+ ++ +L+ IR+PGG F+ G + W+ VGP E RP W+
Sbjct: 49 GFRKDVQSLIKELQVPIIRYPGGNFLSG------YNWEDGVGPVENRPRRLDLAWQTTET 102
Query: 300 DGLGYYEFLQLSEDLGALPIWVFNNGVSHNDEVDTSAVLPF--VQEALDGLEFARGDPTS 357
+ +G EFL + + N EV+ + L + A + +E+ S
Sbjct: 103 NEVGTNEFLSWPKKV--------------NTEVNMAVNLGTRGIDAARNLVEYCNHPKGS 148
Query: 358 KWGSMRAAMGHPEPFNLKYVAVGNE 382
W +R + G+ +P+ +K +GNE
Sbjct: 149 YWSDLRRSHGYEQPYGIKTWCLGNE 173
>ABF2_BACSU (P94552) Alpha-N-arabinofuranosidase 2 (EC 3.2.1.55)
(Arabinosidase 2)
Length = 495
Score = 60.5 bits (145), Expect = 1e-08
Identities = 54/186 (29%), Positives = 78/186 (41%), Gaps = 32/186 (17%)
Query: 223 IWLDQVSAMPLDTYKGHGFRSDLLQMLVDLKPSFIRFPGGCFVEGDYLRNAFRWKAAVGP 282
IW+ S +P +G R D+L+ L L +R+PGGCF + + W VG
Sbjct: 37 IWVGTDSDIP----NINGIRKDVLEALKQLHIPVLRWPGGCFAD------EYHWANGVGD 86
Query: 283 WEERPG-HFGDVWKYWTDDGLGYYEFLQLSEDLGALPIWVFNNGVSHNDEVDTSAVLPFV 341
+ H+G + + G +EF+ L E L P N G S + +
Sbjct: 87 RKTMLNTHWGGTIE---SNEFGTHEFMMLCELLECEPYICGNVG---------SGTVQEM 134
Query: 342 QEALDGLEFARGDPTSKWGSMRAAMGHPEPFNLKYVAVGNEDCG------KKNYRGNYLR 395
E ++ + F G P S W R G EP+ LKY VGNE+ G + Y Y R
Sbjct: 135 SEWIEYMTFEEGTPMSDW---RKQNGREEPWKLKYFGVGNENWGCGGNMHPEYYADLYRR 191
Query: 396 FYDAIR 401
F +R
Sbjct: 192 FQTYVR 197
>CARP_SACFI (P22929) Acid protease precursor (EC 3.4.23.-)
Length = 390
Score = 36.2 bits (82), Expect = 0.29
Identities = 47/227 (20%), Positives = 80/227 (34%), Gaps = 14/227 (6%)
Query: 308 LQLSEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGLEFARGDPTSK---WGSMRA 364
LQ+ D G+ +WV G S ++ G + GD +S W
Sbjct: 88 LQVDVDTGSSDLWVPGQGTSSLYGTYDHTKSTSYKKDRSGFSISYGDGSSARGDWAQETV 147
Query: 365 AMGHPEPFNLKYVAVGNEDCGKK----NYRGNYLRFYDAIRRAYPDIQIISNCDGSSRPL 420
++G L++ ++D G+ +GN + Y ++ + G
Sbjct: 148 SIGGASITGLEFGDATSQDVGQGLLGIGLKGNEASAQSSNSFTYDNLPLKLKDQGL---- 203
Query: 421 DHPADMYDYHIYTNANDMFSRSTTFNRVTRSGPKAFVSEYAVTGNDAGQGSLLAALAEAG 480
D Y +Y N+ D S S F S ++ + D S A+A
Sbjct: 204 ---IDKAAYSLYLNSEDATSGSILFGGSDSSKYSGSLATLDLVNIDDEGDSTSGAVAFFV 260
Query: 481 FLIGLEKNSDIVHMASYAPLFVNANDRRWNPDAIVFNSFQLYGTPSY 527
L G+E S + +Y L + + P +I + + YGT SY
Sbjct: 261 ELEGIEAGSSSITKTTYPALLDSGTTLIYAPSSIASSIGREYGTYSY 307
>P3K2_DICDI (P54674) Phosphatidylinositol 3-kinase 2 (EC 2.7.1.137)
(PI3-kinase) (PtdIns-3-kinase) (PI3K)
Length = 1858
Score = 35.8 bits (81), Expect = 0.38
Identities = 26/92 (28%), Positives = 38/92 (41%), Gaps = 1/92 (1%)
Query: 580 TSAVNLKISFNGLDPNSLQSSGSTKTVLTSTNLMDENSFSQPKKVIPIQSLLQSVGKDMN 639
+S+ N+ +S + P+S SS ST TV+ L NS + P +P L
Sbjct: 361 SSSSNITLSLSSSSPSS-SSSSSTSTVVPIVQLSSSNSTNSPSTSLPTTPRLSQPTTSYT 419
Query: 640 VIVPPHSFTSFDLLKESSNLKMLESDSSSWSS 671
++P SSN + SSS SS
Sbjct: 420 QLIPSQQQQQPTESNSSSNTNTTTTSSSSSSS 451
Score = 32.3 bits (72), Expect = 4.2
Identities = 23/67 (34%), Positives = 38/67 (56%), Gaps = 3/67 (4%)
Query: 534 SESNGATLLNSSLQTTASNSLVASAITWQNSVDKKNYIRIKAVNFGTSAVNLKISFNGLD 593
+ESN ++ N++ T++S+S +S++T + N IRI A FG S+ IS NG+
Sbjct: 431 TESNSSSNTNTTT-TSSSSSSSSSSLTISSPQPSNNSIRISA--FGRSSTQFTISSNGIP 487
Query: 594 PNSLQSS 600
+ Q S
Sbjct: 488 SSPGQVS 494
>TP6B_ARCFU (O29605) Type II DNA topoisomerase VI subunit B (EC
5.99.1.3) (TopoVI-B)
Length = 602
Score = 33.1 bits (74), Expect = 2.4
Identities = 16/59 (27%), Positives = 30/59 (50%), Gaps = 4/59 (6%)
Query: 103 INVETDRTSCFERNKVALRLEVLCDGTCPTDGVGVYNPGF----WGMNIEQGKKYKVVF 157
+ V T R S F R+K +++L +C G DG V G+ W + ++ ++ +V +
Sbjct: 514 VKVVTIRVSNFTRSKKSIKLYEMCSGNVEADGAKVSGSGYSTVTWSLEVKPDEEVEVSY 572
>YX12_CAEEL (Q11123) Hypothetical protein C03F11.2 in chromosome X
Length = 362
Score = 32.3 bits (72), Expect = 4.2
Identities = 25/81 (30%), Positives = 39/81 (47%), Gaps = 4/81 (4%)
Query: 537 NGATLLNSSLQTTASNSLVASAITWQNSVDKKNYIRIKAVNFGTSAVNLKISFNGLDPNS 596
NGA T N V S+ +QNS+++ IR A +SA + +SFN + S
Sbjct: 186 NGADCFTKYETTLDDNKTVQSSPQFQNSIEQ--VIRKPAETLFSSANS--VSFNYIGSPS 241
Query: 597 LQSSGSTKTVLTSTNLMDENS 617
+S ST+ + T+T D +
Sbjct: 242 TSASSSTRAIETTTAYSDSTT 262
>NU4M_PORNA (O03772) NADH-ubiquinone oxidoreductase chain 4 (EC
1.6.5.3) (Fragment)
Length = 231
Score = 32.3 bits (72), Expect = 4.2
Identities = 16/51 (31%), Positives = 24/51 (46%)
Query: 509 WNPDAIVFNSFQLYGTPSYWMQLFFSESNGATLLNSSLQTTASNSLVASAI 559
W P I+ + T SY + +F S G TLLN+ + T S + A+
Sbjct: 165 WCPTTIILLGLSMLITASYSLHMFLSTQMGPTLLNNQTEPTHSREHLLMAL 215
>NU4M_AZEFE (P92494) NADH-ubiquinone oxidoreductase chain 4 (EC
1.6.5.3) (Fragment)
Length = 231
Score = 32.3 bits (72), Expect = 4.2
Identities = 16/51 (31%), Positives = 24/51 (46%)
Query: 509 WNPDAIVFNSFQLYGTPSYWMQLFFSESNGATLLNSSLQTTASNSLVASAI 559
W P I+ + T SY + +F S G TLLN+ + T S + A+
Sbjct: 165 WCPTTIILLGLSMLITASYSLHMFLSTQAGPTLLNNQTEPTHSREHLLMAL 215
>MINT_MOUSE (Q62504) Msx2-interacting protein (SMART/HDAC1 associated
repressor protein)
Length = 3644
Score = 32.3 bits (72), Expect = 4.2
Identities = 18/58 (31%), Positives = 28/58 (48%), Gaps = 6/58 (10%)
Query: 591 GLDPNSLQSSGSTKTVLTSTNLMDENSFSQPKKVIPIQSLLQSVGKDMNVIVPPHSFT 648
GL P S +T T L++ N+ IP+ + S+ + +VI+PPHS T
Sbjct: 3009 GLFPRPCHPSSTTSTALST------NATVMLAAGIPVPQFISSIHPEQSVIMPPHSIT 3060
>GUNG_CLOCE (P37700) Endoglucanase G precursor (EC 3.2.1.4)
(Endo-1,4-beta-glucanase G) (Cellulase G) (EGCCG)
Length = 725
Score = 32.3 bits (72), Expect = 4.2
Identities = 25/107 (23%), Positives = 47/107 (43%), Gaps = 9/107 (8%)
Query: 27 DVQVQADLNAT--------LVVDASQASGRRIPETLFGIFFEEINHAGAGGLWA-ELVSN 77
+V ++A LN+T VV R+ + + +F +++ A G+ LV++
Sbjct: 493 EVIIKAGLNSTGPNYTEIKAVVYNQTGWPARVTDKISFKYFMDLSEIVAAGIDPLSLVTS 552
Query: 78 RGFEAGGPNIPSNIDPWSIIGNATYINVETDRTSCFERNKVALRLEV 124
+ G S + PW + N Y+NV+ + + + A R EV
Sbjct: 553 SNYSEGKNTKVSGVLPWDVSNNVYYVNVDLTGENIYPGGQSACRREV 599
>NAE2_THEMA (Q9X0Y0) Probable glutamine-dependent NAD(+) synthetase
(EC 6.3.5.1) (NAD(+) synthase [glutamine-hydrolyzing])
Length = 576
Score = 32.0 bits (71), Expect = 5.4
Identities = 16/48 (33%), Positives = 27/48 (55%), Gaps = 1/48 (2%)
Query: 591 GLDPNSLQSSG-STKTVLTSTNLMDENSFSQPKKVIPIQSLLQSVGKD 637
GLDP + S G KTVL T ++ +N + + + I ++ ++ GKD
Sbjct: 516 GLDPEEIASKGFDRKTVLDVTEMIRKNEYKRKQAAIGVKISTRAFGKD 563
>CA21_MOUSE (Q01149) Collagen alpha 2(I) chain precursor
Length = 1372
Score = 32.0 bits (71), Expect = 5.4
Identities = 26/76 (34%), Positives = 35/76 (45%), Gaps = 11/76 (14%)
Query: 146 NIEQGKKYKVVFYARSTGPLNLKVSLTGSNGVGSLA--------STVITGSASDFSNWTK 197
NI K + + TG LN V L GSN V +A S ++ G + + W K
Sbjct: 1273 NITYHCKNSIAYLDEETGSLNKAVLLQGSNDVELVAEGNSRFTYSVLVDGCSKKTNEWGK 1332
Query: 198 VETVLEAKATNPNSRL 213
T++E K TN SRL
Sbjct: 1333 --TIIEYK-TNKPSRL 1345
>SYV_LACCA (P36420) Valyl-tRNA synthetase (EC 6.1.1.9) (Valine--tRNA
ligase) (ValRS)
Length = 901
Score = 31.6 bits (70), Expect = 7.1
Identities = 28/108 (25%), Positives = 41/108 (37%), Gaps = 9/108 (8%)
Query: 295 KYWTDDGLGYYEFLQLSED--LGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGLEFAR 352
KY DG GY E + +G + V + D V T +LP + +E A
Sbjct: 226 KYPFADGSGYIEIATTRPETMMGDTAVAVHPGDERYKDMVGTELILPLANRKIPIIEDAY 285
Query: 353 GDPTSKWGSMRAAMGH-PEPF------NLKYVAVGNEDCGKKNYRGNY 393
DP G+++ H P F +LK + N+D G Y
Sbjct: 286 VDPEFGTGAVKITPAHDPNDFQVGNRHDLKRINTMNDDGTMNENAGKY 333
>PLD1_HUMAN (Q13393) Phospholipase D1 (EC 3.1.4.4) (PLD 1) (Choline
phosphatase 1) (Phosphatidylcholine-hydrolyzing
phospholipase D1) (hPLD1)
Length = 1074
Score = 31.6 bits (70), Expect = 7.1
Identities = 22/81 (27%), Positives = 37/81 (45%), Gaps = 10/81 (12%)
Query: 588 SFNGLDPNSLQSSGSTKTVLTSTNLMDENSFSQPKKVIPIQSLLQSVGKDMNVIVPPHSF 647
S L P +++S + S L D+N +P + +PIQ + V + I P F
Sbjct: 508 SLGSLPPAAMES-------MESLRLKDKN---EPVQNLPIQKSIDDVDSKLKGIGKPRKF 557
Query: 648 TSFDLLKESSNLKMLESDSSS 668
+ F L K+ + ++DS S
Sbjct: 558 SKFSLYKQLHRHHLHDADSIS 578
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 81,782,598
Number of Sequences: 164201
Number of extensions: 3612944
Number of successful extensions: 7601
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 7554
Number of HSP's gapped (non-prelim): 33
length of query: 672
length of database: 59,974,054
effective HSP length: 117
effective length of query: 555
effective length of database: 40,762,537
effective search space: 22623208035
effective search space used: 22623208035
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 69 (31.2 bits)
Medicago: description of AC141107.8


