

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141107.7 - phase: 0
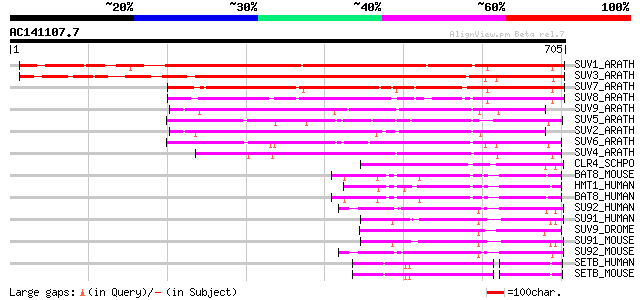
(705 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SUV1_ARATH (Q9FF80) Histone-lysine N-methyltransferase, H3 lysin... 637 0.0
SUV3_ARATH (Q9C5P4) Histone-lysine N-methyltransferase, H3 lysin... 617 e-176
SUV7_ARATH (Q9C5P1) Histone-lysine N-methyltransferase, H3 lysin... 412 e-114
SUV8_ARATH (Q9C5P0) Histone-lysine N-methyltransferase, H3 lysin... 375 e-103
SUV9_ARATH (Q9T0G7) Probable histone-lysine N-methyltransferase,... 362 2e-99
SUV5_ARATH (O82175) Histone-lysine N-methyltransferase, H3 lysin... 362 2e-99
SUV2_ARATH (O22781) Probable histone-lysine N-methyltransferase,... 333 7e-91
SUV6_ARATH (Q8VZ17) Histone-lysine N-methyltransferase, H3 lysin... 330 8e-90
SUV4_ARATH (Q8GZB6) Histone-lysine N-methyltransferase, H3 lysin... 301 3e-81
CLR4_SCHPO (O60016) Histone-lysine N-methyltransferase, H3 lysin... 157 9e-38
BAT8_MOUSE (Q9Z148) Histone-lysine N-methyltransferase, H3 lysin... 139 3e-32
HMT1_HUMAN (Q9H9B1) Histone-lysine N-methyltransferase, H3 lysin... 138 4e-32
BAT8_HUMAN (Q96KQ7) Histone-lysine N-methyltransferase, H3 lysin... 138 6e-32
SU92_HUMAN (Q9H5I1) Histone-lysine N-methyltransferase, H3 lysin... 120 1e-26
SU91_HUMAN (O43463) Histone-lysine N-methyltransferase, H3 lysin... 114 7e-25
SUV9_DROME (P45975) Histone-lysine N-methyltransferase, H3 lysin... 113 2e-24
SU91_MOUSE (O54864) Histone-lysine N-methyltransferase, H3 lysin... 113 2e-24
SU92_MOUSE (Q9EQQ0) Histone-lysine N-methyltransferase, H3 lysin... 111 6e-24
SETB_HUMAN (Q15047) Histone-lysine N-methyltransferase, H3 lysin... 99 3e-20
SETB_MOUSE (O88974) Histone-lysine N-methyltransferase, H3 lysin... 99 5e-20
>SUV1_ARATH (Q9FF80) Histone-lysine N-methyltransferase, H3 lysine-9
specific 1 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of
variegation 3-9 homolog 1) (Su(var)3-9 homolog 1)
Length = 670
Score = 637 bits (1642), Expect = 0.0
Identities = 342/712 (48%), Positives = 444/712 (62%), Gaps = 72/712 (10%)
Query: 13 DKSRVLNVKPLRTLVPVFPSPSNPSSSSNPQGGAPFVAVSPAGPFPAGVAPFYPFFVSPE 72
DK+RVL++KPLRTL PVFPS + PFV P GPFP G + FYPF S
Sbjct: 10 DKTRVLDIKPLRTLRPVFPSGNQ---------APPFVCAPPFGPFPPGFSSFYPFSSSQA 60
Query: 73 SQRLSEQHAPNPTPQRATPISAAVPINSFKTPTAATNGDVGSSRRKSRTRRGQLTEEEGY 132
+Q + + PQ P + P+ + P A+ + + R R+
Sbjct: 61 NQHTPDLNQAQYPPQHQQPQNPP-PVYQQQPPQHASEPSLVTPLRSFRS----------- 108
Query: 133 DNTEVIDVDAETGGGSSKR---KKRAKGRRASGAATDGSGVAAVDVDLDAVAHDILQSIN 189
+V + +AE G + KR KKR R + G VA
Sbjct: 109 --PDVSNGNAELEGSTVKRRIPKKRPISRPENMNFESGINVA------------------ 148
Query: 190 PMVFDVINHPDGSRDSVTYTLMIYEVLRRKLGQIEESTKDLHTGAKRPDLKAGNVMMTKG 249
+ +G+R+ V LM ++ LRR+ Q+E++ + + KRPDLK+G+ M +G
Sbjct: 149 -------DRENGNRELVLSVLMRFDALRRRFAQLEDAKEAVSGIIKRPDLKSGSTCMGRG 201
Query: 250 VRSNSKKRIGIVPGVEIGDIFFFRFEMCLVGLHSPSMAGIDYLTSKASQEEEPLAVSIVS 309
VR+N+KKR GIVPGVEIGD+FFFRFEMCLVGLHSPSMAGIDYL K EEEP+A SIVS
Sbjct: 202 VRTNTKKRPGIVPGVEIGDVFFFRFEMCLVGLHSPSMAGIDYLVVKGETEEEPIATSIVS 261
Query: 310 SGGYEDDTGDGDVLIYSGQGG-VNREKGASDQKLERGNLALEKSMHRGNDVRVIRGLKDV 368
SG Y++D G+ DVLIY+GQGG +++K +SDQKLERGNLALEKS+ R + VRVIRGLK+
Sbjct: 262 SGYYDNDEGNPDVLIYTGQGGNADKDKQSSDQKLERGNLALEKSLRRDSAVRVIRGLKEA 321
Query: 369 MHPSGKVYVYDGIYKIQDSWVEKAKSGFNVFKYKLARVRGQPEAYTIWKSIQQWTDKAAP 428
H + K+Y+YDG+Y+I++SWVEK KSG N FKYKL R GQP A+ W +IQ+W
Sbjct: 322 SH-NAKIYIYDGLYEIKESWVEKGKSGHNTFKYKLVRAPGQPPAFASWTAIQKWKTGVPS 380
Query: 429 RTGVILPDLTSGAEKVPVCLVNDVDNEKGPAYFTYIPTLKNLRGVAPVESSFGCSCIGGC 488
R G+ILPD+TSG E +PV LVN+VD + GPAYFTY T+K ++ SFGC C C
Sbjct: 381 RQGLILPDMTSGVESIPVSLVNEVDTDNGPAYFTYSTTVKYSESFKLMQPSFGCDCANLC 440
Query: 489 QPGNRNCPCIQKNGGYLPYTAAGLVADLKSVIHECGPSCQCPPTCRNRISQAGLKFRLEV 548
+PGN +C CI+KNGG PYT G++ K +I+EC PSC C TC+N+++Q G+K RLEV
Sbjct: 441 KPGNLDCHCIRKNGGDFPYTGNGILVSRKPMIYECSPSCPC-STCKNKVTQMGVKVRLEV 499
Query: 549 FRTSNKGWGLRSWDAIRAGTFICEYAGEVIDNARAEMLGAENEDEYIFDSTRIYQQL--- 605
F+T+N+GWGLRSWDAIRAG+FIC Y GE D ++ + A D+Y FD+T +Y
Sbjct: 500 FKTANRGWGLRSWDAIRAGSFICIYVGEAKDKSKVQQTMA--NDDYTFDTTNVYNPFKWN 557
Query: 606 --------EVFPANIEAPKIPSPLYITAKNEGNVARFMNHSCSPNVLWRPIVRENKNEPD 657
+ E +IP PL I+AKN GNVARFMNHSCSPNV W+P+ EN ++
Sbjct: 558 YEPGLADEDACEEMSEESEIPLPLIISAKNVGNVARFMNHSCSPNVFWQPVSYENNSQLF 617
Query: 658 LHIAFFAIRHIPPMMELTYDYGINLP--LQAGQ---RKKNCLCGSVKCRGYF 704
+H+AFFAI HIPPM ELTYDYG++ P Q G K+ C CGS CRG F
Sbjct: 618 VHVAFFAISHIPPMTELTYDYGVSRPSGTQNGNPLYGKRKCFCGSAYCRGSF 669
>SUV3_ARATH (Q9C5P4) Histone-lysine N-methyltransferase, H3 lysine-9
specific 3 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 3) (H3-K9-HMTase 3) (Suppressor of
variegation 3-9 homolog 3) (Su(var)3-9 homolog 3)
Length = 669
Score = 617 bits (1590), Expect = e-176
Identities = 336/710 (47%), Positives = 430/710 (60%), Gaps = 76/710 (10%)
Query: 13 DKSRVLNVKPLRTLVPVFPSPSNPSSSSNPQGGAPFVAVSPAGPFPAGVAPFYPFFVSPE 72
DKS VL++KPLR+L PVFP N G PFV P GP + + F+PF
Sbjct: 17 DKSIVLDIKPLRSLKPVFP---------NGNQGPPFVGCPPFGPSSSEYSSFFPFGAQQP 67
Query: 73 SQRLSEQHAPNPTPQRATPISAAVP-INSFKTPTAATNGDVGSSRRKSRTRRGQLTEEEG 131
+ P+ + TPI + VP + S++TPT TNG SS K R +
Sbjct: 68 TH-----DTPDLNQTQNTPIPSFVPPLRSYRTPTK-TNGPSSSSGTKRGVGRPK------ 115
Query: 132 YDNTEVIDVDAETGGGSSKRKKRAKGRRASGAATDGSGVAAVDVDLDAVAHDILQSINPM 191
G++ KK+ K A+ D V D D+
Sbjct: 116 ---------------GTTSVKKKEKKTVANEPNLDVQVVKKFSSDFDSG----------- 149
Query: 192 VFDVINHPDGSRDSVTYTLMIYEVLRRKLGQIEESTKDLHTGAKRPDLKAGNVMMTKGVR 251
DG+ V+ LM ++ +RR+L Q+E + K KA +M+ GVR
Sbjct: 150 -ISAAEREDGNAYLVSSVLMRFDAVRRRLSQVEFT--------KSATSKAAGTLMSNGVR 200
Query: 252 SNSKKRIGIVPGVEIGDIFFFRFEMCLVGLHSPSMAGIDYLTSKASQEEEPLAVSIVSSG 311
+N KKR+G VPG+E+GDIFF R EMCLVGLH +MAGIDY+ SKA +EE LA SIVSSG
Sbjct: 201 TNMKKRVGTVPGIEVGDIFFSRIEMCLVGLHMQTMAGIDYIISKAGSDEESLATSIVSSG 260
Query: 312 GYEDDTGDGDVLIYSGQGG-VNREKGASDQKLERGNLALEKSMHRGNDVRVIRGLKDVMH 370
YE + D + LIYSGQGG ++ + ASDQKLERGNLALE S+ +GN VRV+RG +D
Sbjct: 261 RYEGEAQDPESLIYSGQGGNADKNRQASDQKLERGNLALENSLRKGNGVRVVRGEEDAAS 320
Query: 371 PSGKVYVYDGIYKIQDSWVEKAKSGFNVFKYKLARVRGQPEAYTIWKSIQQWTDKAAPRT 430
+GK+Y+YDG+Y I +SWVEK KSG N FKYKL R GQP A+ WKS+Q+W + R
Sbjct: 321 KTGKIYIYDGLYSISESWVEKGKSGCNTFKYKLVRQPGQPPAFGFWKSVQKWKEGLTTRP 380
Query: 431 GVILPDLTSGAEKVPVCLVNDVDNEKGPAYFTYIPTLKNLRGVAPVESSFGCSCIGGCQP 490
G+ILPDLTSGAE PV LVNDVD +KGPAYFTY +LK + GCSC G C P
Sbjct: 381 GLILPDLTSGAESKPVSLVNDVDEDKGPAYFTYTSSLKYSETFKLTQPVIGCSCSGSCSP 440
Query: 491 GNRNCPCIQKNGGYLPYTAAGLVADLKSVIHECGPSCQCPPTCRNRISQAGLKFRLEVFR 550
GN NC CI+KN G LPY ++ + VI+ECGP+C C +C+NR+ Q GLK RLEVF+
Sbjct: 441 GNHNCSCIRKNDGDLPYLNGVILVSRRPVIYECGPTCPCHASCKNRVIQTGLKSRLEVFK 500
Query: 551 TSNKGWGLRSWDAIRAGTFICEYAGEVIDNARAEMLGAENEDEYIFDSTRIYQ------- 603
T N+GWGLRSWD++RAG+FICEYAGEV DN + G + ED Y+FD++R++
Sbjct: 501 TRNRGWGLRSWDSLRAGSFICEYAGEVKDN--GNLRGNQEEDAYVFDTSRVFNSFKWNYE 558
Query: 604 -QLEVFPANIEAPK---IPSPLYITAKNEGNVARFMNHSCSPNVLWRPIVRENKNEPDLH 659
+L + E P+ +PSPL I+AK GNVARFMNHSCSPNV W+P++RE E +H
Sbjct: 559 PELVDEDPSTEVPEEFNLPSPLLISAKKFGNVARFMNHSCSPNVFWQPVIREGNGESVIH 618
Query: 660 IAFFAIRHIPPMMELTYDYGINLPLQAGQR-----KKNCLCGSVKCRGYF 704
IAFFA+RHIPPM ELTYDYGI+ +A ++ CLCGS +CRG F
Sbjct: 619 IAFFAMRHIPPMAELTYDYGISPTSEARDESLLHGQRTCLCGSEQCRGSF 668
>SUV7_ARATH (Q9C5P1) Histone-lysine N-methyltransferase, H3 lysine-9
specific 7 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 7) (H3-K9-HMTase 7) (Suppressor of
variegation 3-9 homolog 7) (Su(var)3-9 homolog 7)
Length = 693
Score = 412 bits (1060), Expect = e-114
Identities = 236/535 (44%), Positives = 322/535 (60%), Gaps = 50/535 (9%)
Query: 201 GSRDSVTYTLMIYEVLRRKLGQIEESTKDLHTGAKRPDLKAGNVMMTKGVRSNSKKRIGI 260
G+++ V +M ++ +RR+L QI L T + GN GV++N+++RIG
Sbjct: 177 GNQEIVDSVMMRFDAVRRRLCQINHPEDILTTAS-------GNCTKM-GVKTNTRRRIGA 228
Query: 261 VPGVEIGDIFFFRFEMCLVGLHSPSMAGIDYLTSKASQEEEPLAVSIVSSGGYEDDTGDG 320
VPG+ +GDIF++ EMCLVGLH + GID+ T+ S E A+ +V++G Y+ +T
Sbjct: 229 VPGIHVGDIFYYWGEMCLVGLHKSNYGGIDFFTAAESAVEGHAAMCVVTAGQYDGETEGL 288
Query: 321 DVLIYSGQGGVNREKGASDQKLERGNLALEKSMHRGNDVRVIRGLKDVMHP---SGKVYV 377
D LIYSGQGG + A DQ+++ GNLALE S+ +GNDVRV+RG V+HP + K+Y+
Sbjct: 289 DTLIYSGQGGTDVYGNARDQEMKGGNLALEASVSKGNDVRVVRG---VIHPHENNQKIYI 345
Query: 378 YDGIYKIQDSWVEKAKSGFNVFKYKLARVRGQPEAYTIWKSIQQWT--DKAAPRTGVILP 435
YDG+Y + W KSGF F++KL R QP AY IWK+++ D R G IL
Sbjct: 346 YDGMYLVSKFWTVTGKSGFKEFRFKLVRKPNQPPAYAIWKTVENLRNHDLIDSRQGFILE 405
Query: 436 DLTSGAEKVPVCLVNDVDNEKG--PAYFTYIPTLKNLRGVAPVESSFGCSCIGGCQP--- 490
DL+ GAE + V LVN+VD + P F YIP+ + G+ E F + GCQ
Sbjct: 406 DLSFGAELLRVPLVNEVDEDDKTIPEDFDYIPSQCH-SGMMTHEFHFDRQSL-GCQNCRH 463
Query: 491 ---GNRNCPCIQKNGGYLPYTAAGLVADLKSVIHECGPSCQCPPTCRNRISQAGLKFRLE 547
++NC C+Q+NG LPY LV K +I+ECG SC CP C R+ Q GLK LE
Sbjct: 464 QPCMHQNCTCVQRNGDLLPYHNNILVC-RKPLIYECGGSCPCPDHCPTRLVQTGLKLHLE 522
Query: 548 VFRTSNKGWGLRSWDAIRAGTFICEYAGEVIDNARAEMLGAENEDEYIFDSTRIYQQL-- 605
VF+T N GWGLRSWD IRAGTFICE+AG R E +D+Y+FD+++IYQ+
Sbjct: 523 VFKTRNCGWGLRSWDPIRAGTFICEFAG-----LRKTKEEVEEDDDYLFDTSKIYQRFRW 577
Query: 606 ---------EVFPANIEAPKIPSPLYITAKNEGNVARFMNHSCSPNVLWRPIVRENKNEP 656
+ + E +P+ + I+AK +GNV RFMNHSCSPNV W+PI EN+ +
Sbjct: 578 NYEPELLLEDSWEQVSEFINLPTQVLISAKEKGNVGRFMNHSCSPNVFWQPIEYENRGDV 637
Query: 657 DLHIAFFAIRHIPPMMELTYDYGINLPLQAGQ-------RKKNCLCGSVKCRGYF 704
L I FA++HIPPM ELTYDYG++ ++ + KK CLCGSVKCRG F
Sbjct: 638 YLLIGLFAMKHIPPMTELTYDYGVSCVERSEEDEVLLYKGKKTCLCGSVKCRGSF 692
>SUV8_ARATH (Q9C5P0) Histone-lysine N-methyltransferase, H3 lysine-9
specific 8 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 8) (H3-K9-HMTase 8) (Suppressor of
variegation 3-9 homolog 8) (Su(var)3-9 homolog 8)
Length = 755
Score = 375 bits (962), Expect = e-103
Identities = 231/529 (43%), Positives = 308/529 (57%), Gaps = 59/529 (11%)
Query: 201 GSRDSVTYTLMIYEVLRRKLGQIEESTKDLHTGAKRPDLKAGNVMMTKGVRSNSKKRIGI 260
G+++ V LM ++ +RR+L Q+ + L A M GVR+N +RIG
Sbjct: 260 GNQEIVDSILMRFDAVRRRLCQLNYRKDKI--------LTASTNCMNLGVRTNMTRRIGP 311
Query: 261 VPGVEIGDIFFFRFEMCLVGLHSPSMAGIDYLTSKASQEEEPLAVSIVSSGGYEDDTGDG 320
+PGV++GDIF++ EMCLVGLH + GID L +K S + P A S+V+SG Y+++T D
Sbjct: 312 IPGVQVGDIFYYWCEMCLVGLHRNTAGGIDSLLAKESGVDGPAATSVVTSGKYDNETEDL 371
Query: 321 DVLIYSGQGGVNREKGASDQKLERGNLALEKSMHRGNDVRVIRGLKDVMHPSGKVYVYDG 380
+ LIYSG GG DQ L+RGN ALE S+ R N+VRVIRG ++ + KVY+YDG
Sbjct: 372 ETLIYSGHGG-----KPCDQVLQRGNRALEASVRRRNEVRVIRG---ELYNNEKVYIYDG 423
Query: 381 IYKIQDSWVEKAKSGFNVFKYKLARVRGQPEAYTIWKSIQQWTDKAA--PRTGVILPDLT 438
+Y + D W KSGF +++KL R GQP Y IWK ++ + PR G IL DL+
Sbjct: 424 LYLVSDCWQVTGKSGFKEYRFKLLRKPGQPPGYAIWKLVENLRNHELIDPRQGFILGDLS 483
Query: 439 SGAEKVPVCLVNDVDNEKG--PAYFTYIPT--LKNLRGVAPVES-SFGCSCIGGCQPGNR 493
G E + V LVN+VD E P F YI + + V+S S S I ++
Sbjct: 484 FGEEGLRVPLVNEVDEEDKTIPDDFDYIRSQCYSGMTNDVNVDSQSLVQSYI------HQ 537
Query: 494 NCPCIQKNGGYLPYTAAGLVADLKSVIHECGPSCQCPPTCRNRISQAGLKFRLEVFRTSN 553
NC CI KN G LPY LV K +I+ECG SC PT R+ + GLK LEVF+TSN
Sbjct: 538 NCTCILKNCGQLPYHDNILVC-RKPLIYECGGSC---PT---RMVETGLKLHLEVFKTSN 590
Query: 554 KGWGLRSWDAIRAGTFICEYAGEVIDNARAEMLGAENEDEYIFDSTRIYQQL-------- 605
GWGLRSWD IRAGTFICE+ G + + E+ E +D+Y+FD++RIY
Sbjct: 591 CGWGLRSWDPIRAGTFICEFTG--VSKTKEEV---EEDDDYLFDTSRIYHSFRWNYEPEL 645
Query: 606 ---EVFPANIEAPKIPSPLYITAKNEGNVARFMNHSCSPNVLWRPI-VRENKNEPDLHIA 661
+ E +P+ + I+AK +GNV RFMNH+C PNV W+PI +N + I
Sbjct: 646 LCEDACEQVSEDANLPTQVLISAKEKGNVGRFMNHNCWPNVFWQPIEYDDNNGHIYVRIG 705
Query: 662 FFAIRHIPPMMELTYDYGINLPLQAGQ------RKKNCLCGSVKCRGYF 704
FA++HIPPM ELTYDYGI+ + G+ KK CLCGSVKCRG F
Sbjct: 706 LFAMKHIPPMTELTYDYGISCVEKTGEDEVIYKGKKICLCGSVKCRGSF 754
>SUV9_ARATH (Q9T0G7) Probable histone-lysine N-methyltransferase, H3
lysine-9 specific 9 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 9) (H3-K9-HMTase 9) (Suppressor of
variegation 3-9 homolog 9) (Su(var)3-9 homolog 9)
Length = 650
Score = 362 bits (929), Expect = 2e-99
Identities = 210/501 (41%), Positives = 294/501 (57%), Gaps = 30/501 (5%)
Query: 203 RDSVTYTLMIYEVLRRKLGQIEESTKDLHTGAKRPDL-----KAGNVMMTKGVRSNSKKR 257
R+ V T MIY+ LR L + E K G +R KAG++M + N KR
Sbjct: 145 REHVRKTRMIYDSLRMFL--MMEEAKRNGVGGRRARADGKAGKAGSMMRDCMLWMNRDKR 202
Query: 258 I-GIVPGVEIGDIFFFRFEMCLVGLHSPSMAGIDYLTSKASQEEEPLAVSIVSSGGYEDD 316
I G +PGV++GDIFFFRFE+C++GLH +GID+LT S EP+A S++ SGGYEDD
Sbjct: 203 IVGSIPGVQVGDIFFFRFELCVMGLHGHPQSGIDFLTGSLSSNGEPIATSVIVSGGYEDD 262
Query: 317 TGDGDVLIYSGQGGVNR-EKGASDQKLERGNLALEKSMHRGNDVRVIRGLKDVMHPSGKV 375
GDV++Y+GQGG +R + A Q+LE GNLA+E+SM+ G +VRVIRGLK S +V
Sbjct: 263 DDQGDVIMYTGQGGQDRLGRQAEHQRLEGGNLAMERSMYYGIEVRVIRGLKYENEVSSRV 322
Query: 376 YVYDGIYKIQDSWVEKAKSGFNVFKYKLARVRGQPE----AYTIWKSIQQWTDKAAPRTG 431
YVYDG+++I DSW + KSGF VFKY+L R+ GQ E ++++ PR G
Sbjct: 323 YVYDGLFRIVDSWFDVGKSGFGVFKYRLERIEGQAEMGSSVLKFARTLKTNPLSVRPR-G 381
Query: 432 VILPDLTSGAEKVPVCLVNDVDNEKGPAYFTYIPTLKNLRGVAPVES--SFGCSCIGGCQ 489
I D+++G E VPV L ND+D+++ P Y+ Y+ G+ +S + GC C+ GC
Sbjct: 382 YINFDISNGKENVPVYLFNDIDSDQEPLYYEYLAQTSFPPGLFVQQSGNASGCDCVNGCG 441
Query: 490 PGNRNCPCIQKNGGYLPYTAAGLVADLKSVIHECGPSCQCPPTCRNRISQAGLKFRLEVF 549
G C C KN G + Y G + K +IHECG +CQCPP+CRNR++Q GL+ RLEVF
Sbjct: 442 SG---CLCEAKNSGEIAYDYNGTLIRQKPLIHECGSACQCPPSCRNRVTQKGLRNRLEVF 498
Query: 550 RTSNKGWGLRSWDAIRAGTFICEYAGEVIDNARAEMLGAENEDEYI----FDSTR--IYQ 603
R+ GWG+RS D + AG FICEYAG + +A +L N D + F S R +
Sbjct: 499 RSLETGWGVRSLDVLHAGAFICEYAGVALTREQANIL-TMNGDTLVYPARFSSARWEDWG 557
Query: 604 QLEVFPANIEAPKIPS----PLYITAKNEGNVARFMNHSCSPNVLWRPIVRENKNEPDLH 659
L A+ E P P + NVA +++HS PNV+ + ++ ++ +
Sbjct: 558 DLSQVLADFERPSYPDIPPVDFAMDVSKMRNVACYISHSTDPNVIVQFVLHDHNSLMFPR 617
Query: 660 IAFFAIRHIPPMMELTYDYGI 680
+ FA +IPPM EL+ DYG+
Sbjct: 618 VMLFAAENIPPMTELSLDYGV 638
>SUV5_ARATH (O82175) Histone-lysine N-methyltransferase, H3 lysine-9
specific 5 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 5) (H3-K9-HMTase 5) (Suppressor of
variegation 3-9 homolog 5) (Su(var)3-9 homolog 5)
Length = 794
Score = 362 bits (929), Expect = 2e-99
Identities = 206/516 (39%), Positives = 303/516 (57%), Gaps = 41/516 (7%)
Query: 200 DGSRDSVTYTLMIYEVLRRKLGQIEESTKDLHTGAK-RPDLKAGNVMMTKGVRSNSKKRI 258
D +R V T+ ++ +K+ Q EE+ G + +A ++ +KG S +I
Sbjct: 304 DSARYKVKETMRLFHETCKKIMQEEEARPRKRDGGNFKVVCEASKILKSKGKNLYSGTQI 363
Query: 259 -GIVPGVEIGDIFFFRFEMCLVGLHSPSMAGIDYLTSKASQEEEPLAVSIVSSGGYEDDT 317
G VPGVE+GD F +R E+ L+G+H PS +GIDY+ E +A SIVSSGGY D
Sbjct: 364 IGTVPGVEVGDEFQYRMELNLLGIHRPSQSGIDYMKDDGG---ELVATSIVSSGGYNDVL 420
Query: 318 GDGDVLIYSGQGG-VNREKG---ASDQKLERGNLALEKSMHRGNDVRVIRGLKDVMHPSG 373
+ DVLIY+GQGG V ++K DQ+L GNLAL+ S+++ N VRVIRG+K+ S
Sbjct: 421 DNSDVLIYTGQGGNVGKKKNNEPPKDQQLVTGNLALKNSINKKNPVRVIRGIKNTTLQSS 480
Query: 374 KV---YVYDGIYKIQDSWVEKAKSGFNVFKYKLARVRGQPEAYTIWKSIQQWTDKAAPRT 430
V YVYDG+Y +++ W E G VFK+KL R+ GQPE WK + + + K+ R
Sbjct: 481 VVAKNYVYDGLYLVEEYWEETGSHGKLVFKFKLRRIPGQPELP--WKEVAK-SKKSEFRD 537
Query: 431 GVILPDLTSGAEKVPVCLVNDVDNEKGPAYFTYIPTLKNLRGVAPVESSFGCSCIGGCQP 490
G+ D+T G E +P+C VN++D+EK P F Y + P+ C C GC
Sbjct: 538 GLCNVDITEGKETLPICAVNNLDDEKPPP-FIYTAKMIYPDWCRPIPPK-SCGCTNGCSK 595
Query: 491 GNRNCPCIQKNGGYLPYTAAGLVADLKSVIHECGPSCQCPPTCRNRISQAGLKFRLEVFR 550
++NC CI KNGG +PY G + ++K +++ECGP C+CPP+C R+SQ G+K +LE+F+
Sbjct: 596 -SKNCACIVKNGGKIPYYD-GAIVEIKPLVYECGPHCKCPPSCNMRVSQHGIKIKLEIFK 653
Query: 551 TSNKGWGLRSWDAIRAGTFICEYAGEVIDNARAEMLGAENEDEYIFDSTRIYQQLEVFPA 610
T ++GWG+RS ++I G+FICEYAGE++++ +AE L +DEY+FD
Sbjct: 654 TESRGWGVRSLESIPIGSFICEYAGELLEDKQAESL--TGKDEYLFD------------- 698
Query: 611 NIEAPKIPSPLYITAKNEGNVARFMNHSCSPNVLWRPIVRENKNEPDLHIAFFAIRHIPP 670
P I A +GN+ RF+NHSCSPN+ + ++ +++ HI FFA+ +IPP
Sbjct: 699 ---LGDEDDPFTINAAQKGNIGRFINHSCSPNLYAQDVLYDHEEIRIPHIMFFALDNIPP 755
Query: 671 MMELTYDYGINL----PLQAGQRKKNCLCGSVKCRG 702
+ EL+YDY + +KK C CGS +C G
Sbjct: 756 LQELSYDYNYKIDQVYDSNGNIKKKFCYCGSAECSG 791
>SUV2_ARATH (O22781) Probable histone-lysine N-methyltransferase, H3
lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of
variegation 3-9 homolog 2) (Su(var)3-9 homolog 2)
Length = 651
Score = 333 bits (855), Expect = 7e-91
Identities = 205/501 (40%), Positives = 287/501 (56%), Gaps = 33/501 (6%)
Query: 203 RDSVTYTLMIYEVLRRKLGQIEESTKDLHTGA---KRPDLKAGNVMMTKGVRSNSKKRI- 258
R + T M YE LR L + ES K+ G +R D+ A +M +G+ N K I
Sbjct: 149 RQVMKRTRMTYESLRIHL--MAESMKNHVLGQGRRRRSDMAAAYIMRDRGLWLNYDKHIV 206
Query: 259 GIVPGVEIGDIFFFRFEMCLVGLHSPSMAGIDYLTSKASQEEEPLAVSIVSSGGYEDDTG 318
G V GVE+GDIFF+R E+C++GLH + AGID LT++ S EP+A SIV SGGYEDD
Sbjct: 207 GPVTGVEVGDIFFYRMELCVLGLHGQTQAGIDCLTAERSATGEPIATSIVVSGGYEDDED 266
Query: 319 DGDVLIYSGQGGVNRE-KGASDQKLERGNLALEKSMHRGNDVRVIRGLKDVMHPSGKVYV 377
GDVL+Y+G GG + + K +Q+L GNL +E+SMH G +VRVIRG+K S KVYV
Sbjct: 267 TGDVLVYTGHGGQDHQHKQCDNQRLVGGNLGMERSMHYGIEVRVIRGIKYENSISSKVYV 326
Query: 378 YDGIYKIQDSWVEKAKSGFNVFKYKLARVRGQPE-AYTIWKSIQQWTDKAAP--RTGVIL 434
YDG+YKI D W KSGF VFK++L R+ GQP + + Q +K + TG +
Sbjct: 327 YDGLYKIVDWWFAVGKSGFGVFKFRLVRIEGQPMMGSAVMRFAQTLRNKPSMVRPTGYVS 386
Query: 435 PDLTSGAEKVPVCLVNDVDNEKGPAYFTYI------PTLKNLRGVAPVESSFGCSCIGGC 488
DL++ E VPV L NDVD ++ P ++ YI P + G+ S GC C C
Sbjct: 387 FDLSNKKENVPVFLYNDVDGDQEPRHYEYIAKAVFPPGIFGQGGI----SRTGCECKLSC 442
Query: 489 QPGNRNCPCIQKNGGYLPYTAAGLVADLKSVIHECGPSCQCPPTCRNRISQAGLKFRLEV 548
+C C +KNGG Y G + K V+ ECG C C P+C++R++Q GL+ RLEV
Sbjct: 443 TD---DCLCARKNGGEFAYDDNGHLLKGKHVVFECGEFCTCGPSCKSRVTQKGLRNRLEV 499
Query: 549 FRTSNKGWGLRSWDAIRAGTFICEYAGEVIDNARAEMLGAENEDEYIF-----DSTRIYQ 603
FR+ GWG+R+ D I AG FICEYAG V+ +AE+L + N D ++ D R +
Sbjct: 500 FRSKETGWGVRTLDLIEAGAFICEYAGVVVTRLQAEIL-SMNGDVMVYPGRFTDQWRNWG 558
Query: 604 QLEVFPANIEAPKIPS--PL--YITAKNEGNVARFMNHSCSPNVLWRPIVRENKNEPDLH 659
L + P PS PL + NVA +++HS PNV+ + ++ ++ +
Sbjct: 559 DLSQVYPDFVRPNYPSLPPLDFSMDVSRMRNVACYISHSKEPNVMVQFVLHDHNHLMFPR 618
Query: 660 IAFFAIRHIPPMMELTYDYGI 680
+ FA+ +I P+ EL+ DYG+
Sbjct: 619 VMLFALENISPLAELSLDYGL 639
>SUV6_ARATH (Q8VZ17) Histone-lysine N-methyltransferase, H3 lysine-9
specific 6 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 6) (H3-K9-HMTase 6) (Suppressor of
variegation 3-9 homolog 6) (Su(var)3-9 homolog 6)
Length = 790
Score = 330 bits (846), Expect = 8e-90
Identities = 202/532 (37%), Positives = 294/532 (54%), Gaps = 42/532 (7%)
Query: 200 DGSRDSVTYTLMIYEVLRRKLGQIEEST-KDLHTGAK--RPDLKAGNVMMTKGVRSNSKK 256
D SR+ V TL ++ + RK+ Q +E+ +D K R D +A ++ G NS
Sbjct: 267 DSSRNKVKETLRLFHGVCRKILQEDEAKPEDQRRKGKGLRIDFEASTILKRNGKFLNSGV 326
Query: 257 RI-GIVPGVEIGDIFFFRFEMCLVGLHSPSMAGIDYLTSKASQEEEPLAVSIVSSGGYED 315
I G VPGVE+GD F +R E+ ++G+H PS AGIDY+ ++ +A SIV+SGGY+D
Sbjct: 327 HILGEVPGVEVGDEFQYRMELNILGIHKPSQAGIDYMKYGKAK----VATSIVASGGYDD 382
Query: 316 DTGDGDVLIYSGQGG----VNRE----KGASDQKLERGNLALEKSMHRGNDVRVIRGLKD 367
+ DVL Y+GQGG V ++ K DQKL GNLAL S+ + VRVIRG
Sbjct: 383 HLDNSDVLTYTGQGGNVMQVKKKGEELKEPEDQKLITGNLALATSIEKQTPVRVIRGKHK 442
Query: 368 VMHPSGKV--YVYDGIYKIQDSWVEKAKSGFNVFKYKLARVRGQPEAYTIWKSIQQWTDK 425
H K YVYDG+Y ++ W + G NVFK++L R+ GQPE W +++ K
Sbjct: 443 STHDKSKGGNYVYDGLYLVEKYWQQVGSHGMNVFKFQLRRIPGQPELS--WVEVKK--SK 498
Query: 426 AAPRTGVILPDLTSGAEKVPVCLVNDVDNEKGPAYFTYIPTLKNLRGVAPVESSFGCSCI 485
+ R G+ D++ G E+ P+ VN++D+EK P FTY L PV C C
Sbjct: 499 SKYREGLCKLDISEGKEQSPISAVNEIDDEK-PPLFTYTVKLIYPDWCRPVPPK-SCCCT 556
Query: 486 GGCQPGN-RNCPCIQKNGGYLPYTAAGLVADLKSVIHECGPSCQCPPTCRNRISQAGLKF 544
C R C C++KNGG +PY G + K I+ECGP C+CP +C R++Q G+K
Sbjct: 557 TRCTEAEARVCACVEKNGGEIPYNFDGAIVGAKPTIYECGPLCKCPSSCYLRVTQHGIKL 616
Query: 545 RLEVFRTSNKGWGLRSWDAIRAGTFICEYAGEVIDNARAEMLGAENEDEYIFDSTRIY-- 602
LE+F+T ++GWG+R +I G+FICEY GE+++++ AE DEY+FD Y
Sbjct: 617 PLEIFKTKSRGWGVRCLKSIPIGSFICEYVGELLEDSEAER--RIGNDEYLFDIGNRYDN 674
Query: 603 ---QQLEVFPANIEAPKI------PSPLYITAKNEGNVARFMNHSCSPNVLWRPIVRENK 653
Q + +A + S I A ++GNV RF+NHSCSPN+ + ++ +++
Sbjct: 675 SLAQGMSELMLGTQAGRSMAEGDESSGFTIDAASKGNVGRFINHSCSPNLYAQNVLYDHE 734
Query: 654 NEPDLHIAFFAIRHIPPMMELTYDYGINL----PLQAGQRKKNCLCGSVKCR 701
+ H+ FFA +IPP+ EL YDY L + ++K C CG+ CR
Sbjct: 735 DSRIPHVMFFAQDNIPPLQELCYDYNYALDQVRDSKGNIKQKPCFCGAAVCR 786
>SUV4_ARATH (Q8GZB6) Histone-lysine N-methyltransferase, H3 lysine-9
specific 4 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 4) (H3-K9-HMTase 4) (Suppressor of
variegation 3-9 homolog 4) (Su(var)3-9 homolog 4)
(KRYPTONITE protein)
Length = 624
Score = 301 bits (772), Expect = 3e-81
Identities = 187/499 (37%), Positives = 266/499 (52%), Gaps = 37/499 (7%)
Query: 236 RPDLKAGNVMMTKGVRSNSKKRIGIVPGVEIGDIFFFRFEMCLVGLHSPSMAGIDYLTSK 295
RPDLK M+ +K IG +PG+++G FF R EMC VG H+ + GIDY++ +
Sbjct: 126 RPDLKGVTEMIKAKAILYPRKIIGDLPGIDVGHRFFSRAEMCAVGFHNHWLNGIDYMSME 185
Query: 296 ASQEEE----PLAVSIVSSGGYEDDTGDGDVLIYSGQGGVN---REKGASDQKLERGNLA 348
+E PLAVSIV SG YEDD + D + Y+GQGG N ++ DQ LERGNLA
Sbjct: 186 YEKEYSNYKLPLAVSIVMSGQYEDDLDNADTVTYTGQGGHNLTGNKRQIKDQLLERGNLA 245
Query: 349 LEKSMHRGNDVRVIRGLKDVMHPSGKVYVYDGIYKIQDSWVEKAKSGFNVFKYKLARVRG 408
L+ VRV RG + +VY YDG+YK++ W +K SGF V+KY+L R+ G
Sbjct: 246 LKHCCEYNVPVRVTRGHNCKSSYTKRVYTYDGLYKVEKFWAQKGVSGFTVYKYRLKRLEG 305
Query: 409 QPEAYTIWKSIQ--QWTDKAAPRTGVILPDLTSGAEKVPVCLVNDVDNE--KGPAYFTYI 464
QPE T + + + G++ D++ G E + N VD+ + FTYI
Sbjct: 306 QPELTTDQVNFVAGRIPTSTSEIEGLVCEDISGGLEFKGIPATNRVDDSPVSPTSGFTYI 365
Query: 465 PTLKNLRGVAPVESSFGCSCIGGCQPGNRNCPCIQKNGGYLPYTAA--GLVADLKSVIHE 522
+L V +SS GC+C G C ++ C C + NGG PY G + + + V+ E
Sbjct: 366 KSLIIEPNVIIPKSSTGCNCRGSCTD-SKKCACAKLNGGNFPYVDLNDGRLIESRDVVFE 424
Query: 523 CGPSCQCPPTCRNRISQAGLKFRLEVFRTSNKGWGLRSWDAIRAGTFICEYAGEVIDNAR 582
CGP C C P C NR SQ L+F LEVFR++ KGW +RSW+ I AG+ +CEY G V A
Sbjct: 425 CGPHCGCGPKCVNRTSQKRLRFNLEVFRSAKKGWAVRSWEYIPAGSPVCEYIGVVRRTAD 484
Query: 583 AEMLGAENEDEYIF--DSTRIYQQLEVFPANIEAPKIP-------------SPLY-ITAK 626
+ + +++EYIF D + Q L + +P +P + I A
Sbjct: 485 VDTI---SDNEYIFEIDCQQTMQGLGGRQRRLRDVAVPMNNGVSQSSEDENAPEFCIDAG 541
Query: 627 NEGNVARFMNHSCSPNVLWRPIVRENKNEPDLHIAFFAIRHIPPMMELTYDYGINLPLQA 686
+ GN ARF+NHSC PN+ + ++ +++ + FA +I PM ELTYDYG L
Sbjct: 542 STGNFARFINHSCEPNLFVQCVLSSHQDIRLARVVLFAADNISPMQELTYDYGYALDSVH 601
Query: 687 GQ----RKKNCLCGSVKCR 701
G ++ C CG++ CR
Sbjct: 602 GPDGKVKQLACYCGALNCR 620
>CLR4_SCHPO (O60016) Histone-lysine N-methyltransferase, H3 lysine-9
specific (EC 2.1.1.43) (Histone H3-K9 methyltransferase)
(H3-K9-HMTase) (Cryptic loci regulator 4)
Length = 490
Score = 157 bits (397), Expect = 9e-38
Identities = 105/278 (37%), Positives = 145/278 (51%), Gaps = 31/278 (11%)
Query: 446 VCLVNDVDNEKGPAY-FTYIPTLKNLRGVAPVESSF--GCSC--IGGCQPGNRN-CPCIQ 499
V LVN+VD+E P+ F +I + +GV P + +F GC+C +GGC N + C C+
Sbjct: 221 VTLVNEVDDEPCPSLDFQFISQYRLTQGVIPPDPNFQSGCNCSSLGGCDLNNPSRCECLD 280
Query: 500 K--NGGYLPYTAAGLV-ADLKSVIHECGPSCQCPPTCRNRISQAGLKFRLEVFRTSNKGW 556
+ Y A G V AD +VI+EC C C C NR+ Q G LE+F+T KGW
Sbjct: 281 DLDEPTHFAYDAQGRVRADTGAVIYECNSFCSCSMECPNRVVQRGRTLPLEIFKTKEKGW 340
Query: 557 GLRSWDAIRAGTFICEYAGEVIDNARAEMLGAENEDEYIFDSTRIYQQLEVFPANIEAPK 616
G+RS AGTFI Y GEVI +A A A+ + Y D L++F
Sbjct: 341 GVRSLRFAPAGTFITCYLGEVITSAEA----AKRDKNYDDDGITYLFDLDMFDD------ 390
Query: 617 IPSPLYITAKNEGNVARFMNHSCSPNVLWRPIVRENKNEPDLHIAFFAIRHIPPMMELTY 676
S + A+N G+V+RF NHSCSPN+ VR + +AFFAI+ I P+ ELT+
Sbjct: 391 -ASEYTVDAQNYGDVSRFFNHSCSPNIAIYSAVRNHGFRTIYDLAFFAIKDIQPLEELTF 449
Query: 677 DYG---INLPLQAGQRKKN--------CLCGSVKCRGY 703
DY P+Q+ + ++N C CGS CRG+
Sbjct: 450 DYAGAKDFSPVQSQKSQQNRISKLRRQCKCGSANCRGW 487
>BAT8_MOUSE (Q9Z148) Histone-lysine N-methyltransferase, H3 lysine-9
specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase
3) (H3-K9-HMTase 3) (HLA-B associated transcript 8) (G9a)
(NG36)
Length = 1263
Score = 139 bits (349), Expect = 3e-32
Identities = 95/309 (30%), Positives = 149/309 (47%), Gaps = 41/309 (13%)
Query: 410 PEAYTIWKSIQQWT-------DKAAPRTGVILPDLTSGAEKVPVCLVNDVDNEKGPAYFT 462
PE +W ++Q ++A +I D+ G E VP+ VN VD E P +
Sbjct: 945 PERSDVWFALQLNRKLRLGVGNRAVRTEKIICRDVARGYENVPIPCVNGVDGEPCPEDYK 1004
Query: 463 YIP------TLKNLRGVAPVESSFGCSCIGGCQPGNRNCPCIQKNGGYLPYTAAGLVADL 516
YI T+ R + ++ C+C+ C + NC C Q + L+ +
Sbjct: 1005 YISENCETSTMNIDRNITHLQH---CTCVDDCS--SSNCLCGQLSIRCWYDKDGRLLQEF 1059
Query: 517 KSV----IHECGPSCQCPPTCRNRISQAGLKFRLEVFRTSNKGWGLRSWDAIRAGTFICE 572
+ I EC +C C +C+NR+ Q+G+K RL+++RT+ GWG+R+ I GTFICE
Sbjct: 1060 NKIEPPLIFECNQACSCWRSCKNRVVQSGIKVRLQLYRTAKMGWGVRALQTIPQGTFICE 1119
Query: 573 YAGEVIDNARAEMLGAENEDEYIFDSTRIYQQLEVFPANIEAPKIPSPLYITAKNEGNVA 632
Y GE+I +A A++ +D Y+FD + EV+ I A+ GN++
Sbjct: 1120 YVGELISDAEADV---REDDSYLFDLDN--KDGEVY-------------CIDARYYGNIS 1161
Query: 633 RFMNHSCSPNVLWRPIVRENKNEPDLHIAFFAIRHIPPMMELTYDYGINLPLQAGQRKKN 692
RF+NH C PN++ + +++ IAFF+ R I EL +DYG +
Sbjct: 1162 RFINHLCDPNIIPVRVFMLHQDLRFPRIAFFSSRDIRTGEELGFDYGDRF-WDIKSKYFT 1220
Query: 693 CLCGSVKCR 701
C CGS KC+
Sbjct: 1221 CQCGSEKCK 1229
>HMT1_HUMAN (Q9H9B1) Histone-lysine N-methyltransferase, H3 lysine-9
specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase
5) (H3-K9-HMTase 5) (Euchromatic histone
methyltransferase 1) (Eu-HMTase1) (G9a-like protein 1)
(GLP1)
Length = 1267
Score = 138 bits (348), Expect = 4e-32
Identities = 96/293 (32%), Positives = 147/293 (49%), Gaps = 44/293 (15%)
Query: 424 DKAAPRTGVILPDLTSGAEKVPVCLVNDVDNEKGPAYFTYIPT------LKNLRGVAPVE 477
D+ +P ++ D+ G E++P+ VN VD+E P+ + Y+ + R + ++
Sbjct: 970 DRPSPVERIVSRDIARGYERIPIPCVNAVDSEPCPSNYKYVSQNCVTSPMNIDRNITHLQ 1029
Query: 478 SSFGCSCIGGCQPGNRNCPCIQ--------KNGGYLP-YTAAGLVADLKSVIHECGPSCQ 528
C CI C + NC C Q K+G LP + A +I EC +C
Sbjct: 1030 Y---CVCIDDCS--SSNCMCGQLSMRCWYDKDGRLLPEFNMAE-----PPLIFECNHACS 1079
Query: 529 CPPTCRNRISQAGLKFRLEVFRTSNKGWGLRSWDAIRAGTFICEYAGEVIDNARAEMLGA 588
C CRNR+ Q GL+ RL+++RT + GWG+RS I GTF+CEY GE+I ++ A++
Sbjct: 1080 CWRNCRNRVVQNGLRARLQLYRTRDMGWGVRSLQDIPPGTFVCEYVGELISDSEADV--- 1136
Query: 589 ENEDEYIFDSTRIYQQLEVFPANIEAPKIPSPLYITAKNEGNVARFMNHSCSPNVLWRPI 648
ED Y+FD + EV+ I A+ GNV+RF+NH C PN++ +
Sbjct: 1137 REEDSYLFDLDN--KDGEVY-------------CIDARFYGNVSRFINHHCEPNLVPVRV 1181
Query: 649 VRENKNEPDLHIAFFAIRHIPPMMELTYDYGINLPLQAGQRKKNCLCGSVKCR 701
+++ IAFF+ R I +L +DYG G + +C CGS KCR
Sbjct: 1182 FMAHQDLRFPRIAFFSTRLIEAGEQLGFDYGERFWDIKG-KLFSCRCGSPKCR 1233
>BAT8_HUMAN (Q96KQ7) Histone-lysine N-methyltransferase, H3 lysine-9
specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase
3) (H3-K9-HMTase 3) (HLA-B associated transcript 8) (G9a)
(NG36)
Length = 1210
Score = 138 bits (347), Expect = 6e-32
Identities = 95/309 (30%), Positives = 148/309 (47%), Gaps = 41/309 (13%)
Query: 410 PEAYTIWKSIQQWT-------DKAAPRTGVILPDLTSGAEKVPVCLVNDVDNEKGPAYFT 462
PE +W ++Q ++A +I D+ G E VP+ VN VD E P +
Sbjct: 892 PERSDVWFALQLNRKLRLGVGNRAIRTEKIICRDVARGYENVPIPCVNGVDGEPCPEDYK 951
Query: 463 YIP------TLKNLRGVAPVESSFGCSCIGGCQPGNRNCPCIQKNGGYLPYTAAGLVADL 516
YI T+ R + ++ C+C+ C + NC C Q + L+ +
Sbjct: 952 YISENCETSTMNIDRNITHLQH---CTCVDDCS--SSNCLCGQLSIRCWYDKDGRLLQEF 1006
Query: 517 KSV----IHECGPSCQCPPTCRNRISQAGLKFRLEVFRTSNKGWGLRSWDAIRAGTFICE 572
+ I EC +C C C+NR+ Q+G+K RL+++RT+ GWG+R+ I GTFICE
Sbjct: 1007 NKIEPPLIFECNQACSCWRNCKNRVVQSGIKVRLQLYRTAKMGWGVRALQTIPQGTFICE 1066
Query: 573 YAGEVIDNARAEMLGAENEDEYIFDSTRIYQQLEVFPANIEAPKIPSPLYITAKNEGNVA 632
Y GE+I +A A++ +D Y+FD + EV+ I A+ GN++
Sbjct: 1067 YVGELISDAEADV---REDDSYLFDLDN--KDGEVY-------------CIDARYYGNIS 1108
Query: 633 RFMNHSCSPNVLWRPIVRENKNEPDLHIAFFAIRHIPPMMELTYDYGINLPLQAGQRKKN 692
RF+NH C PN++ + +++ IAFF+ R I EL +DYG +
Sbjct: 1109 RFINHLCDPNIIPVRVFMLHQDLRFPRIAFFSSRDIRTGEELGFDYGDRF-WDIKSKYFT 1167
Query: 693 CLCGSVKCR 701
C CGS KC+
Sbjct: 1168 CQCGSEKCK 1176
>SU92_HUMAN (Q9H5I1) Histone-lysine N-methyltransferase, H3 lysine-9
specific 2 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of
variegation 3-9 homolog 2) (Su(var)3-9 homolog 2)
Length = 410
Score = 120 bits (301), Expect = 1e-26
Identities = 97/303 (32%), Positives = 141/303 (46%), Gaps = 47/303 (15%)
Query: 418 SIQQWTDKAAPRTGVILPDLTSGAEKVPVCLVNDVDNEKGPAYFTYIPTLKNLRGVAPV- 476
++Q+W D+ R K + + N VD E P+ F YI K G++ V
Sbjct: 136 ALQRWQDELNRRKN----------HKGMIFVENTVDLEGPPSDFYYINEYKPAPGISLVN 185
Query: 477 ESSFGCSCIGGCQPGNRNCPCIQKNGGYLPYTAAGLVADLKSV-IHECGPSCQCPPTCRN 535
E++FGCSC + C C + G L Y + I+EC CQC P C N
Sbjct: 186 EATFGCSCTDCFF---QKC-CPAEAGVLLAYNKNQQIKIPPGTPIYECNSRCQCGPDCPN 241
Query: 536 RISQAGLKFRLEVFRTSN-KGWGLRSWDAIRAGTFICEYAGEVIDNARAEMLGAENEDE- 593
RI Q G ++ L +FRTSN +GWG+++ I+ +F+ EY GEVI + AE G +++
Sbjct: 242 RIVQKGTQYSLCIFRTSNGRGWGVKTLVKIKRMSFVMEYVGEVITSEEAERRGQFYDNKG 301
Query: 594 --YIFDSTRIYQQLEVFPANIEAPKIPSPLYITAKNEGNVARFMNHSCSPNVLWRPIVRE 651
Y+FD Y+ E + A GNV+ F+NHSC PN+ + +
Sbjct: 302 ITYLFDLD--YESDE--------------FTVDAARYGNVSHFVNHSCDPNLQVFNVFID 345
Query: 652 NKNEPDLHIAFFAIRHIPPMMELTYDYGI-------NLPLQAGQRKKN----CLCGSVKC 700
N + IA F+ R I ELT+DY + + + KK C CG+V C
Sbjct: 346 NLDTRLPRIALFSTRTINAGEELTFDYQMKGSGDISSDSIDHSPAKKRVRTVCKCGAVTC 405
Query: 701 RGY 703
RGY
Sbjct: 406 RGY 408
>SU91_HUMAN (O43463) Histone-lysine N-methyltransferase, H3 lysine-9
specific 1 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of
variegation 3-9 homolog 1) (Su(var)3-9 homolog 1)
Length = 412
Score = 114 bits (286), Expect = 7e-25
Identities = 87/289 (30%), Positives = 129/289 (44%), Gaps = 54/289 (18%)
Query: 446 VCLVNDVDNEKGPAYFTYIPTLKNLRGVAPVESSFGCSC-------IGGCQPGNRNCPCI 498
+ + N+VD + P F YI + G+ + + GC C GGC PG
Sbjct: 145 ITVENEVDLDGPPRAFVYINEYRVGEGITLNQVAVGCECQDCLWAPTGGCCPGASLHKFA 204
Query: 499 QKNGGYLPYTAAGLVADLKSVIHECGPSCQCPPTCRNRISQAGLKFRLEVFRTSN-KGWG 557
+ G + A GL I+EC C+C C NR+ Q G+++ L +FRT + +GWG
Sbjct: 205 YNDQGQVRLRA-GLP------IYECNSRCRCGYDCPNRVVQKGIRYDLCIFRTDDGRGWG 257
Query: 558 LRSWDAIRAGTFICEYAGEVIDNARAEMLGAENEDE---YIFDSTRIYQQLEVFPANIEA 614
+R+ + IR +F+ EY GE+I + AE G + + Y+FD + V
Sbjct: 258 VRTLEKIRKNSFVMEYVGEIITSEEAERRGQIYDRQGATYLFDLDYVEDVYTV------- 310
Query: 615 PKIPSPLYITAKNEGNVARFMNHSCSPNVLWRPIVRENKNEPDLHIAFFAIRHIPPMMEL 674
A GN++ F+NHSC PN+ + +N +E IAFFA R I EL
Sbjct: 311 ---------DAAYYGNISHFVNHSCDPNLQVYNVFIDNLDERLPRIAFFATRTIRAGEEL 361
Query: 675 TYDYGINL-PLQ---------------AGQRKK----NCLCGSVKCRGY 703
T+DY + + P+ G KK C CG+ CR Y
Sbjct: 362 TFDYNMQVDPVDMESTRMDSNFGLAGLPGSPKKRVRIECKCGTESCRKY 410
>SUV9_DROME (P45975) Histone-lysine N-methyltransferase, H3 lysine-9
specific (EC 2.1.1.43) (Histone H3-K9 methyltransferase)
(H3-K9-HMTase) (Suppressor of variegation protein 3-9)
Length = 635
Score = 113 bits (283), Expect = 2e-24
Identities = 83/271 (30%), Positives = 125/271 (45%), Gaps = 27/271 (9%)
Query: 445 PVCLVNDVDNEKGPAYFTYIPTLKNLRGVAPVESSF-GCSCIGGCQPGNRNCPCIQKNGG 503
P+ + N++D + + F YI + V E+ GC C + + C + G
Sbjct: 374 PIRVENNIDLDTIDSNFMYIHDNIIGKDVPKPEAGIVGCKCTEDTEECTASTKCCARFAG 433
Query: 504 YL-PYTAAGLVADLK--SVIHECGPSCQCPPTCRNRISQAGLKFRLEVFRTSN-KGWGLR 559
L Y + L+ S I+EC C C +C NR+ Q G + L +F+T+N GWG+R
Sbjct: 434 ELFAYERSTRRLRLRPGSAIYECNSRCSCDSSCSNRLVQHGRQVPLVLFKTANGSGWGVR 493
Query: 560 SWDAIRAGTFICEYAGEVIDNARAEMLGAENEDE---YIFDSTRIYQQLEVFPANIEAPK 616
+ A+R G F+CEY GE+I + A G +D Y+FD Q
Sbjct: 494 AATALRKGEFVCEYIGEIITSDEANERGKAYDDNGRTYLFDLDYNTAQ------------ 541
Query: 617 IPSPLYITAKNEGNVARFMNHSCSPNVLWRPIVRENKNEPDLHIAFFAIRHIPPMMELTY 676
S I A N GN++ F+NHSC PN+ P E+ N H+ FF +R I EL++
Sbjct: 542 -DSEYTIDAANYGNISHFINHSCDPNLAVFPCWIEHLNVALPHLVFFTLRPIKAGEELSF 600
Query: 677 DY----GINLPLQ--AGQRKKNCLCGSVKCR 701
DY ++P + + + C CG CR
Sbjct: 601 DYIRADNEDVPYENLSTAVRVECRCGRDNCR 631
>SU91_MOUSE (O54864) Histone-lysine N-methyltransferase, H3 lysine-9
specific 1 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of
variegation 3-9 homolog 1) (Su(var)3-9 homolog 1)
(Position-effect variegation 3-9 homolog)
Length = 412
Score = 113 bits (283), Expect = 2e-24
Identities = 85/289 (29%), Positives = 127/289 (43%), Gaps = 54/289 (18%)
Query: 446 VCLVNDVDNEKGPAYFTYIPTLKNLRGVAPVESSFGCSC-------IGGCQPGNRNCPCI 498
+ + N+VD + P F YI + G+ + + GC C GGC PG
Sbjct: 145 ITVENEVDLDGPPRSFVYINEYRVGEGITLNQVAVGCECQDCLLAPTGGCCPGASLHKFA 204
Query: 499 QKNGGYLPYTAAGLVADLKSVIHECGPSCQCPPTCRNRISQAGLKFRLEVFRTSN-KGWG 557
+ G + A I+EC C C C NR+ Q G+++ L +FRT++ +GWG
Sbjct: 205 YNDQGQVRLKAG-------QPIYECNSRCCCGYDCPNRVVQKGIRYDLCIFRTNDGRGWG 257
Query: 558 LRSWDAIRAGTFICEYAGEVIDNARAEMLGAENEDE---YIFDSTRIYQQLEVFPANIEA 614
+R+ + IR +F+ EY GE+I + AE G + + Y+FD + V
Sbjct: 258 VRTLEKIRKNSFVMEYVGEIITSEEAERRGQIYDRQGATYLFDLDYVEDVYTV------- 310
Query: 615 PKIPSPLYITAKNEGNVARFMNHSCSPNVLWRPIVRENKNEPDLHIAFFAIRHIPPMMEL 674
A GN++ F+NHSC PN+ + +N +E IAFFA R I EL
Sbjct: 311 ---------DAAYYGNISHFVNHSCDPNLQVYNVFIDNLDERLPRIAFFATRTIWAGEEL 361
Query: 675 TYDYGINL-PLQ---------------AGQRKK----NCLCGSVKCRGY 703
T+DY + + P+ G KK C CG+ CR Y
Sbjct: 362 TFDYNMQVDPVDMESTRMDSNFGLAGLPGSPKKRVRIECKCGTTACRKY 410
>SU92_MOUSE (Q9EQQ0) Histone-lysine N-methyltransferase, H3 lysine-9
specific 2 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of
variegation 3-9 homolog 2) (Su(var)3-9 homolog 2)
Length = 477
Score = 111 bits (278), Expect = 6e-24
Identities = 91/303 (30%), Positives = 137/303 (45%), Gaps = 47/303 (15%)
Query: 418 SIQQWTDKAAPRTGVILPDLTSGAEKVPVCLVNDVDNEKGPAYFTYIPTLKNLRGVA-PV 476
++Q+W D R K + + N VD E P F YI + G++
Sbjct: 203 ALQRWQDYLNRRKN----------HKGMIFVENTVDLEGPPLDFYYINEYRPAPGISINS 252
Query: 477 ESSFGCSCIGGCQPGNRNCPCIQKNGGYLPYTAAGLVADLKSV-IHECGPSCQCPPTCRN 535
E++FGCSC ++ CP + G L Y + I+EC C+C P C N
Sbjct: 253 EATFGCSCTDCFF--DKCCPA--EAGVVLAYNKKQQIKIQPGTPIYECNSRCRCGPECPN 308
Query: 536 RISQAGLKFRLEVFRTSNK-GWGLRSWDAIRAGTFICEYAGEVIDNARAEMLGAENEDE- 593
RI Q G ++ L +F+TSN GWG+++ I+ +F+ EY GEVI + AE G +++
Sbjct: 309 RIVQKGTQYSLCIFKTSNGCGWGVKTLVKIKRMSFVMEYVGEVITSEEAERRGQFYDNKG 368
Query: 594 --YIFDSTRIYQQLEVFPANIEAPKIPSPLYITAKNEGNVARFMNHSCSPNVLWRPIVRE 651
Y+FD Y+ E + A GNV+ F+NHSC PN+ + +
Sbjct: 369 ITYLFDLD--YESDE--------------FTVDAARYGNVSHFVNHSCDPNLQVFSVFID 412
Query: 652 NKNEPDLHIAFFAIRHIPPMMELTYDYGINLPLQAG-----------QRKKNCLCGSVKC 700
N + IA F+ R I ELT+DY + +A + + C CG+ C
Sbjct: 413 NLDTRLPRIALFSTRTINAGEELTFDYQMKGSGEASSDSIDHSPAKKRVRTQCKCGAETC 472
Query: 701 RGY 703
RGY
Sbjct: 473 RGY 475
>SETB_HUMAN (Q15047) Histone-lysine N-methyltransferase, H3 lysine-9
specific 4 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 4) (H3-K9-HMTase 4) (SET domain
bifurcated 1) (ERG-associated protein with SET domain)
(ESET)
Length = 1291
Score = 99.4 bits (246), Expect = 3e-20
Identities = 68/195 (34%), Positives = 97/195 (48%), Gaps = 19/195 (9%)
Query: 436 DLTSGAEKVPVCLVNDVDNEKGP--AYFTY-IPTLKNLRGVAPVESSFGCSCIGGCQPGN 492
D+T G E VP+ VN++D P AY IP P E GC C GC+ +
Sbjct: 681 DITYGKEDVPLSCVNEIDTTPPPQVAYSKERIPGKGVFINTGP-EFLVGCDCKDGCRDKS 739
Query: 493 RNCPCIQKN--------GGYLP----YTAAGLVADLKSVIHECGPSCQCPPT-CRNRISQ 539
+ C C Q GG + Y L L + ++EC C+C P C NR+ Q
Sbjct: 740 K-CACHQLTIQATACTPGGQINPNSGYQYKRLEECLPTGVYECNKRCKCDPNMCTNRLVQ 798
Query: 540 AGLKFRLEVFRTSNKGWGLRSWDAIRAGTFICEYAGEVIDNARAEMLGAENEDEYIFDST 599
GL+ RL++F+T NKGWG+R D I G+F+C YAG+++ + A+ G E DEY +
Sbjct: 799 HGLQVRLQLFKTQNKGWGIRCLDDIAKGSFVCIYAGKILTDDFADKEGLEMGDEYFANLD 858
Query: 600 RIYQQLEVFPANIEA 614
I + +E F E+
Sbjct: 859 HI-ESVENFKEGYES 872
Score = 61.6 bits (148), Expect = 7e-09
Identities = 31/80 (38%), Positives = 48/80 (59%), Gaps = 1/80 (1%)
Query: 623 ITAKNEGNVARFMNHSCSPNVLWRPIVRENKNEPDLHIAFFAIRHIPPMMELTYDYGINL 682
I AK EGN+ R++NHSCSPN+ + + + + +AFFA + I ELT+DY +
Sbjct: 1210 IDAKLEGNLGRYLNHSCSPNLFVQNVFVDTHDLRFPWVAFFASKRIRAGTELTWDYNYEV 1269
Query: 683 PLQAGQRKKNCLCGSVKCRG 702
G ++ C CG+++CRG
Sbjct: 1270 GSVEG-KELLCCCGAIECRG 1288
>SETB_MOUSE (O88974) Histone-lysine N-methyltransferase, H3 lysine-9
specific 4 (EC 2.1.1.43) (Histone H3-K9
methyltransferase 4) (H3-K9-HMTase 4) (SET domain
bifurcated 1) (ERG-associated protein with SET domain)
(ESET)
Length = 1307
Score = 98.6 bits (244), Expect = 5e-20
Identities = 68/195 (34%), Positives = 96/195 (48%), Gaps = 19/195 (9%)
Query: 436 DLTSGAEKVPVCLVNDVDNEKGP--AYFTY-IPTLKNLRGVAPVESSFGCSCIGGCQPGN 492
D+T G E VP+ VN++D P AY IP P E GC C GC+ +
Sbjct: 698 DITYGKEDVPLSCVNEIDTTPPPQVAYSKERIPGKGVFINTGP-EFLVGCDCKDGCRDKS 756
Query: 493 RNCPCIQKN--------GGYLP----YTAAGLVADLKSVIHECGPSCQCPPT-CRNRISQ 539
+ C C Q GG + Y L L + ++EC C C P C NR+ Q
Sbjct: 757 K-CACHQLTIQATACTPGGQVNPNSGYQYKRLEECLPTGVYECNKRCNCDPNMCTNRLVQ 815
Query: 540 AGLKFRLEVFRTSNKGWGLRSWDAIRAGTFICEYAGEVIDNARAEMLGAENEDEYIFDST 599
GL+ RL++F+T NKGWG+R D I G+F+C YAG+++ + A+ G E DEY +
Sbjct: 816 HGLQVRLQLFKTQNKGWGIRCLDDIAKGSFVCIYAGKILTDDFADKEGLEMGDEYFANLD 875
Query: 600 RIYQQLEVFPANIEA 614
I + +E F E+
Sbjct: 876 HI-ESVENFKEGYES 889
Score = 61.6 bits (148), Expect = 7e-09
Identities = 31/80 (38%), Positives = 48/80 (59%), Gaps = 1/80 (1%)
Query: 623 ITAKNEGNVARFMNHSCSPNVLWRPIVRENKNEPDLHIAFFAIRHIPPMMELTYDYGINL 682
I AK EGN+ R++NHSCSPN+ + + + + +AFFA + I ELT+DY +
Sbjct: 1226 IDAKLEGNLGRYLNHSCSPNLFVQNVFVDTHDLRFPWVAFFASKRIRAGTELTWDYNYEV 1285
Query: 683 PLQAGQRKKNCLCGSVKCRG 702
G ++ C CG+++CRG
Sbjct: 1286 GSVEG-KELLCCCGAIECRG 1304
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.136 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 90,867,110
Number of Sequences: 164201
Number of extensions: 4302398
Number of successful extensions: 12380
Number of sequences better than 10.0: 152
Number of HSP's better than 10.0 without gapping: 52
Number of HSP's successfully gapped in prelim test: 104
Number of HSP's that attempted gapping in prelim test: 11923
Number of HSP's gapped (non-prelim): 357
length of query: 705
length of database: 59,974,054
effective HSP length: 117
effective length of query: 588
effective length of database: 40,762,537
effective search space: 23968371756
effective search space used: 23968371756
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 69 (31.2 bits)
Medicago: description of AC141107.7


