

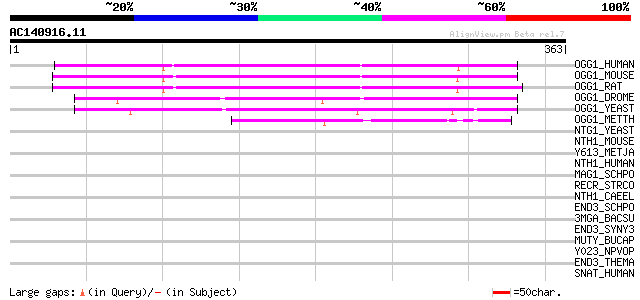
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140916.11 - phase: 0
(363 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
OGG1_HUMAN (O15527) N-glycosylase/DNA lyase [Includes: 8-oxoguan... 198 2e-50
OGG1_MOUSE (O08760) N-glycosylase/DNA lyase [Includes: 8-oxoguan... 197 3e-50
OGG1_RAT (O70249) N-glycosylase/DNA lyase [Includes: 8-oxoguanin... 196 6e-50
OGG1_DROME (Q9V3I8) N-glycosylase/DNA lyase (dOgg1) [Includes: 8... 191 3e-48
OGG1_YEAST (P53397) N-glycosylase/DNA lyase [Includes: 8-oxoguan... 153 6e-37
OGG1_METTH (O27397) Probable N-glycosylase/DNA lyase [Includes: ... 82 2e-15
NTG1_YEAST (P31378) DNA base excision repair N-glycosylase 1, mi... 43 0.001
NTH1_MOUSE (O35980) Endonuclease III-like protein 1 (EC 4.2.99.18) 43 0.001
Y613_METJA (Q58030) Putative endonuclease MJ0613 39 0.027
NTH1_HUMAN (P78549) Endonuclease III-like protein 1 (EC 4.2.99.18) 38 0.035
MAG1_SCHPO (Q92383) DNA-3-methyladenine glycosylase 1 (EC 3.2.2.... 38 0.035
RECR_STRCO (Q9XAI4) Recombination protein recR 36 0.17
NTH1_CAEEL (P54137) Probable endonuclease III homolog (EC 4.2.99... 36 0.17
END3_SCHPO (Q09907) Endonuclease III homolog (EC 4.2.99.18) (DNA... 35 0.23
3MGA_BACSU (P37878) DNA-3-methyladenine glycosylase (EC 3.2.2.21... 35 0.23
END3_SYNY3 (P73715) Endonuclease III (EC 4.2.99.18) (DNA-(apurin... 35 0.30
MUTY_BUCAP (Q8K926) A/G-specific adenine glycosylase (EC 3.2.2.-) 35 0.39
Y023_NPVOP (O10282) Hypothetical 73.1 kDa protein precursor (ORF21) 34 0.51
END3_THEMA (Q9WYK0) Endonuclease III (EC 4.2.99.18) (DNA-(apurin... 34 0.51
SNAT_HUMAN (Q16613) Serotonin N-acetyltransferase (EC 2.3.1.87) ... 34 0.66
>OGG1_HUMAN (O15527) N-glycosylase/DNA lyase [Includes: 8-oxoguanine
DNA glycosylase (EC 3.2.2.-); DNA-(apurinic or
apyrimidinic site) lyase (EC 4.2.99.18) (AP lyase)]
Length = 345
Score = 198 bits (503), Expect = 2e-50
Identities = 117/313 (37%), Positives = 172/313 (54%), Gaps = 12/313 (3%)
Query: 30 HSTNLSKSRAWIPLNITRQELSLPLTFPTGQTFRWKNTAPSQYTGVVGSHLISLKHLHNG 89
H T S W + R EL L L P+GQ+FRW+ +P+ ++GV+ + +L
Sbjct: 13 HRTLASTPALWASIPCPRSELRLDLVLPSGQSFRWREQSPAHWSGVLADQVWTLTQTEEQ 72
Query: 90 DVCYTLHSQS-----PSNDDCKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSGAR 144
C P+ D+ + A+ + DV+LA + + D F E+AQ G R
Sbjct: 73 LHCTVYRGDKSQASRPTPDELE-AVRKYFQLDVTLAQLYHHWGSVDSHFQEVAQKFQGVR 131
Query: 145 VLRQDPFECLIQFMCSSNNHISRITKMVDYV-SSLGTYLGCVEGFDFHAFPTLNQLSLVS 203
+LRQDP ECL F+CSSNN+I+RIT MV+ + + G L ++ +H FP+L L+
Sbjct: 132 LLRQDPIECLFSFICSSNNNIARITGMVERLCQAFGPRLIQLDDVTYHGFPSLQALAGPE 191
Query: 204 -EQQLRDAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPKV 262
E LR G GYRA+Y++ + + + +GG WL LR+ E+ L LPGVG KV
Sbjct: 192 VEAHLRKLGLGYRARYVSASARAILEE-QGGLAWLQQLRESSYEEAHKALCILPGVGTKV 250
Query: 263 AACIALYSLDQHHAIPVDVHIWRIAQKYIL--PELAGSK-LTQKLHSRVAEAFVTKFGKY 319
A CI L +LD+ A+PVDVH+W IAQ+ P + +K + + + + F + +G Y
Sbjct: 251 ADCICLMALDKPQAVPVDVHMWHIAQRDYSWHPTTSQAKGPSPQTNKELGNFFRSLWGPY 310
Query: 320 AGWAQAVLFIAEL 332
AGWAQAVLF A+L
Sbjct: 311 AGWAQAVLFSADL 323
>OGG1_MOUSE (O08760) N-glycosylase/DNA lyase [Includes: 8-oxoguanine
DNA glycosylase (EC 3.2.2.-); DNA-(apurinic or
apyrimidinic site) lyase (EC 4.2.99.18) (AP lyase)]
Length = 345
Score = 197 bits (502), Expect = 3e-50
Identities = 118/314 (37%), Positives = 171/314 (53%), Gaps = 12/314 (3%)
Query: 29 RHSTNLSKSRAWIPLNITRQELSLPLTFPTGQTFRWKNTAPSQYTGVVGSHLISLKHLHN 88
RH T S W + R EL L L +GQ+FRWK +P+ ++GV+ + +L +
Sbjct: 12 RHRTLSSSPALWASIPCPRSELRLDLVLASGQSFRWKEQSPAHWSGVLADQVWTLTQTED 71
Query: 89 GDVCYTLHSQS-----PSNDDCKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSGA 143
C P+ ++ +T L + DVSLA + ++ D F +AQ G
Sbjct: 72 QLYCTVYRGDDSQVSRPTLEELET-LHKYFQLDVSLAQLYSHWASVDSHFQRVAQKFQGV 130
Query: 144 RVLRQDPFECLIQFMCSSNNHISRITKMVDYV-SSLGTYLGCVEGFDFHAFPTLNQLS-L 201
R+LRQDP ECL F+CSSNN+I+RIT MV+ + + G L ++ +H FP L+ L+
Sbjct: 131 RLLRQDPTECLFSFICSSNNNIARITGMVERLCQAFGPRLIQLDDVTYHGFPNLHALAGP 190
Query: 202 VSEQQLRDAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPK 261
+E LR G GYRA+Y+ + + + +GG WL LR E+ L LPGVG K
Sbjct: 191 EAETHLRKLGLGYRARYVRASAKAILEE-QGGPAWLQQLRVAPYEEAHKALCTLPGVGAK 249
Query: 262 VAACIALYSLDQHHAIPVDVHIWRIAQKYI--LPELAGSKLTQKL-HSRVAEAFVTKFGK 318
VA CI L +LD+ A+PVDVH+W+IA + P+ + +K L + + F +G
Sbjct: 250 VADCICLMALDKPQAVPVDVHVWQIAHRDYGWHPKTSQAKGPSPLANKELGNFFRNLWGP 309
Query: 319 YAGWAQAVLFIAEL 332
YAGWAQAVLF A+L
Sbjct: 310 YAGWAQAVLFSADL 323
>OGG1_RAT (O70249) N-glycosylase/DNA lyase [Includes: 8-oxoguanine
DNA glycosylase (EC 3.2.2.-); DNA-(apurinic or
apyrimidinic site) lyase (EC 4.2.99.18) (AP lyase)]
Length = 345
Score = 196 bits (499), Expect = 6e-50
Identities = 116/317 (36%), Positives = 170/317 (53%), Gaps = 12/317 (3%)
Query: 29 RHSTNLSKSRAWIPLNITRQELSLPLTFPTGQTFRWKNTAPSQYTGVVGSHLISLKHLHN 88
RH T S W + R EL L L +GQ+FRW+ +P+ ++GV+ + +L +
Sbjct: 12 RHRTLTSSPALWASIPCPRSELRLDLVLASGQSFRWREQSPAHWSGVLADQVWTLTQTED 71
Query: 89 GDVCYTLHSQS-----PSNDDCKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSGA 143
C P+ ++ +T L + DVSL + ++ D F +AQ G
Sbjct: 72 QLYCTVYRGDKGQVGRPTLEELET-LHKYFQLDVSLTQLYSHWASVDSHFQSVAQKFQGV 130
Query: 144 RVLRQDPFECLIQFMCSSNNHISRITKMVDYV-SSLGTYLGCVEGFDFHAFPTLNQLSLV 202
R+LRQDP ECL F+CSSNN+I+RIT MV+ + + G L ++ +H FP L+ L+
Sbjct: 131 RLLRQDPTECLFSFICSSNNNIARITGMVERLCQAFGPRLVQLDDVTYHGFPNLHALAGP 190
Query: 203 S-EQQLRDAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPK 261
E LR G GYRA+Y+ + + + +GG WL LR E+ L LPGVG K
Sbjct: 191 EVETHLRKLGLGYRARYVCASAKAILEE-QGGPAWLQQLRVASYEEAHKALCTLPGVGTK 249
Query: 262 VAACIALYSLDQHHAIPVDVHIWRIAQKYI--LPELAGSKLTQKL-HSRVAEAFVTKFGK 318
VA CI L +LD+ A+PVD+H+W+IA + P+ + +K L + + F +G
Sbjct: 250 VADCICLMALDKPQAVPVDIHVWQIAHRDYGWQPKTSQTKGPSPLANKELGNFFRNLWGP 309
Query: 319 YAGWAQAVLFIAELPSQ 335
YAGWAQAVLF A+L Q
Sbjct: 310 YAGWAQAVLFSADLRQQ 326
>OGG1_DROME (Q9V3I8) N-glycosylase/DNA lyase (dOgg1) [Includes:
8-oxoguanine DNA glycosylase (EC 3.2.2.-); DNA-(apurinic
or apyrimidinic site) lyase (EC 4.2.99.18) (AP lyase)]
Length = 343
Score = 191 bits (484), Expect = 3e-48
Identities = 112/300 (37%), Positives = 168/300 (55%), Gaps = 14/300 (4%)
Query: 43 LNITRQELSLPLTFPTGQTFRWKNTAP---SQYTGVVGSHLISLKHLHNGDVCYTLHSQS 99
+ ++ +E L T GQ+FRW++ ++Y GVV + L+ + + S
Sbjct: 27 IGLSLEECDLERTLLGGQSFRWRSICDGNRTKYGGVVFNTYWVLQQEESFITYEAYGTSS 86
Query: 100 P-SNDDCKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSGARVLRQDPFECLIQFM 158
P + D + + D+L D L K + D+ F + R+L Q+PFE + F+
Sbjct: 87 PLATKDYSSLISDYLRVDFDLKVNQKDWLSKDDNFVKFLS--KPVRLLSQEPFENIFSFL 144
Query: 159 CSSNNHISRITKMVD-YVSSLGTYLGCVEGFDFHAFPTLNQLSLVS----EQQLRDAGFG 213
CS NN+I RI+ M++ + ++ GT +G G D + FPT+N+ + QLR A FG
Sbjct: 145 CSQNNNIKRISSMIEWFCATFGTKIGHFNGADAYTFPTINRFHDIPCEDLNAQLRAAKFG 204
Query: 214 YRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPKVAACIALYSLDQ 273
YRAK+I T+ +Q K GG+ W SL+ + E EL LPG+G KVA CI L S+
Sbjct: 205 YRAKFIAQTLQEIQKK--GGQNWFISLKSMPFEKAREELTLLPGIGYKVADCICLMSMGH 262
Query: 274 HHAIPVDVHIWRIAQKYILPELAGSK-LTQKLHSRVAEAFVTKFGKYAGWAQAVLFIAEL 332
++PVD+HI+RIAQ Y LP L G K +T+K++ V++ F GKYAGWAQA+LF A+L
Sbjct: 263 LESVPVDIHIYRIAQNYYLPHLTGQKNVTKKIYEEVSKHFQKLHGKYAGWAQAILFSADL 322
>OGG1_YEAST (P53397) N-glycosylase/DNA lyase [Includes: 8-oxoguanine
DNA glycosylase (EC 3.2.2.-); DNA-(apurinic or
apyrimidinic site) lyase (EC 4.2.99.18) (AP lyase)]
Length = 376
Score = 153 bits (387), Expect = 6e-37
Identities = 113/324 (34%), Positives = 166/324 (50%), Gaps = 37/324 (11%)
Query: 43 LNITRQELSLPLTFPTGQTFRWK-NTAPSQYTGVVG-------SHLISLKHLHNGDVCYT 94
L I + EL L GQ+FRW + +QY+ + S +I + N + +
Sbjct: 8 LAINKSELCLANVLQAGQSFRWIWDEKLNQYSTTMKIGQQEKYSVVILRQDEENEILEFV 67
Query: 95 LHSQSPSNDDCKTALLDFLNADVSLADTW-KVFSDSDERFAELAQHLSGARVLRQDPFEC 153
+ D KT L+ + DVSL + V+ SD+ FA+L+ G R+L Q+P+E
Sbjct: 68 AVGDCGNQDALKTHLMKYFRLDVSLKHLFDNVWIPSDKAFAKLSPQ--GIRILAQEPWET 125
Query: 154 LIQFMCSSNNHISRITKMVDYV-SSLGTYLGCVEGFDFHAFPTLNQL-SLVSEQQLRDAG 211
LI F+CSSNN+ISRIT+M + + S+ G + ++G +H+FPT +L S +E +LR+ G
Sbjct: 126 LISFICSSNNNISRITRMCNSLCSNFGNLITTIDGVAYHSFPTSEELTSRATEAKLRELG 185
Query: 212 FGYRAKYITGTVNVL-----QSKPEGGEEWLYSLRK-LELEDVISELIKLPGVGPKVAAC 265
FGYRAKYI T L ++ +L S+ K + EDV L+ GVGPKVA C
Sbjct: 186 FGYRAKYIIETARKLVNDKAEANITSDTTYLQSICKDAQYEDVREHLMSYNGVGPKVADC 245
Query: 266 IALYSLDQHHAIPVDVHIWRIAQ-----------------KYILPELAGSKLTQKLHSRV 308
+ L L +PVDVH+ RIA+ KY ++ K+ +L +
Sbjct: 246 VCLMGLHMDGIVPVDVHVSRIAKRDYQISANKNHLKELRTKYNALPISRKKINLEL-DHI 304
Query: 309 AEAFVTKFGKYAGWAQAVLFIAEL 332
K+G YAGWAQ VLF E+
Sbjct: 305 RLMLFKKWGSYAGWAQGVLFSKEI 328
>OGG1_METTH (O27397) Probable N-glycosylase/DNA lyase [Includes:
8-oxoguanine DNA glycosylase (EC 3.2.2.-); DNA-(apurinic
or apyrimidinic site) lyase (EC 4.2.99.18) (AP lyase)]
Length = 312
Score = 82.0 bits (201), Expect = 2e-15
Identities = 62/202 (30%), Positives = 93/202 (45%), Gaps = 31/202 (15%)
Query: 146 LRQDPFECLIQFMCSSNNHISRITKMVDYVSSLGTYLGCVEGFDFHAFPTLNQLSLVSE- 204
L +DPFEC+I + S+N + R T+ ++ + L G FH FP+ + L+ V+E
Sbjct: 106 LAKDPFECVISSIASANCSVVRWTRSIEDIRRLWGQANTFNGETFHTFPSPHVLTGVAEG 165
Query: 205 ------------------QQLRDAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLELE 246
LR G GYRA YI T +L + + + + ++ +
Sbjct: 166 SLEDLQRAEDNLPSDFSFNDLRSCGVGYRAPYIRETSRILAEEMD-----IRRIDGMDYD 220
Query: 247 DVISELIKLPGVGPKVAACIALYSLDQHHAIPVDVHIWRIAQKYILPELAGSKLTQKLHS 306
D L++L GVGPKVA CI LY + A PVDV I RI +I P G +
Sbjct: 221 DARELLLELSGVGPKVADCILLYGFRKTEAFPVDVWIRRI-MNHIHP---GRNFNDR--- 273
Query: 307 RVAEAFVTKFGKYAGWAQAVLF 328
+ E ++G+ A + Q LF
Sbjct: 274 SMVEFARREYGEMADYVQLYLF 295
>NTG1_YEAST (P31378) DNA base excision repair N-glycosylase 1,
mitochondrial precursor
Length = 399
Score = 43.1 bits (100), Expect = 0.001
Identities = 37/124 (29%), Positives = 55/124 (43%), Gaps = 22/124 (17%)
Query: 186 EGFDFHAFPTLNQLSLVSEQQLRDAGFGYR-AKYITGTVNVLQSKPEGGEEWLYSLRKLE 244
EG A +N+ L ++ + GF R AKYI T +LQ + +
Sbjct: 180 EGMTLEAVLQINETKL--DELIHSVGFHTRKAKYILSTCKILQDQFSS-----------D 226
Query: 245 LEDVISELIKLPGVGPKVAACIALYSLDQHHAIPVDVHI--------WRIAQKYILPELA 296
+ I+EL+ LPGVGPK+A + + I VDVH+ W AQK P+
Sbjct: 227 VPATINELLGLPGVGPKMAYLTLQKAWGKIEGICVDVHVDRLTKLWKWVDAQKCKTPDQT 286
Query: 297 GSKL 300
++L
Sbjct: 287 RTQL 290
>NTH1_MOUSE (O35980) Endonuclease III-like protein 1 (EC 4.2.99.18)
Length = 300
Score = 42.7 bits (99), Expect = 0.001
Identities = 25/75 (33%), Positives = 37/75 (49%), Gaps = 11/75 (14%)
Query: 215 RAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPKVAACIALYSLDQH 274
+ KYI T +LQ + EG ++ ++EL+ LPGVGPK+A +
Sbjct: 173 KVKYIKQTTAILQQRYEG-----------DIPASVAELVALPGVGPKMAHLAMAVAWGTI 221
Query: 275 HAIPVDVHIWRIAQK 289
I VD H+ RIA +
Sbjct: 222 SGIAVDTHVHRIANR 236
>Y613_METJA (Q58030) Putative endonuclease MJ0613
Length = 344
Score = 38.5 bits (88), Expect = 0.027
Identities = 39/167 (23%), Positives = 73/167 (43%), Gaps = 21/167 (12%)
Query: 190 FHAFPTLNQLSLVSEQQLRD----AGF-GYRAKYITGTVNVLQSKPEGGEEWLYSLRKLE 244
F ++ L + E++L D AGF +AK + +L+ G +
Sbjct: 52 FKEIKDVDDLLNIDEEKLADLIYPAGFYKNKAKNLKKLAKILKENYNG-----------K 100
Query: 245 LEDVISELIKLPGVGPKVAACIALYSLDQHHAIPVDVHIWRIAQKYILPELAGSKLTQKL 304
+ D + EL+KLPGVG K A + + ++ I VD H+ RI ++ E+ ++ ++
Sbjct: 101 VPDSLEELLKLPGVGRKTANLVITLAFNK-DGICVDTHVHRICNRW---EIVDTETPEET 156
Query: 305 HSRVAEAFVTKFGKYAGWAQAVLFIAELPSQKAILPSHLRATKQQSP 351
+ + K+ K V+F E+ S K+ + K++ P
Sbjct: 157 EFELRKKLPKKYWKVIN-NLLVVFGREICSSKSKCDKCFKEIKEKCP 202
>NTH1_HUMAN (P78549) Endonuclease III-like protein 1 (EC 4.2.99.18)
Length = 312
Score = 38.1 bits (87), Expect = 0.035
Identities = 24/75 (32%), Positives = 35/75 (46%), Gaps = 11/75 (14%)
Query: 215 RAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPKVAACIALYSLDQH 274
+ KYI T +LQ G ++ ++EL+ LPGVGPK+A +
Sbjct: 185 KVKYIKQTSAILQQHYGG-----------DIPASVAELVALPGVGPKMAHLAMAVAWGTV 233
Query: 275 HAIPVDVHIWRIAQK 289
I VD H+ RIA +
Sbjct: 234 SGIAVDTHVHRIANR 248
>MAG1_SCHPO (Q92383) DNA-3-methyladenine glycosylase 1 (EC 3.2.2.21)
(3-methyladenine DNA glycosidase 1) (3MEA DNA
glycosylase 1)
Length = 228
Score = 38.1 bits (87), Expect = 0.035
Identities = 42/199 (21%), Positives = 82/199 (41%), Gaps = 18/199 (9%)
Query: 127 SDSDERFAELAQHLSGARVLR----QDPFECLIQFMCSSNNHISRITKMVDYVSSLGTYL 182
S DE + L + + R R ++P+E LI+ + S H + + S+
Sbjct: 23 SGLDENWKRLVKLVGNYRPNRSMEKKEPYEELIRAVASQQLHSKAANAIFNRFKSISNN- 81
Query: 183 GCVEGFDFHAFPTLNQLSLVSEQQLRDAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRK 242
FPT ++ + + +R GF A+ I ++ ++ G +
Sbjct: 82 --------GQFPTPEEIRDMDFEIMRACGFS--ARKIDSLKSIAEATISGLIPTKEEAER 131
Query: 243 LELEDVISELIKLPGVGPKVAACIALYSLDQHHAIPVDVHIWRIAQKYI--LPELAGSKL 300
L E++I L ++ G+G + ++SL++ +P D R +Y+ LP++ +K+
Sbjct: 132 LSNEELIERLTQIKGIGRWTVEMLLIFSLNRDDVMPADDLSIRNGYRYLHRLPKIP-TKM 190
Query: 301 TQKLHSRVAEAFVTKFGKY 319
HS + F T Y
Sbjct: 191 YVLKHSEICAPFRTAAAWY 209
>RECR_STRCO (Q9XAI4) Recombination protein recR
Length = 199
Score = 35.8 bits (81), Expect = 0.17
Identities = 23/44 (52%), Positives = 29/44 (65%), Gaps = 5/44 (11%)
Query: 245 LEDVISELIKLPGVGPKVAACIALYSLDQHHAIPVDVHIWRIAQ 288
++D+I EL +LPGVGPK A IA + L A PVDV R+AQ
Sbjct: 6 VQDLIDELGRLPGVGPKSAQRIAFHIL---QAEPVDVR--RLAQ 44
>NTH1_CAEEL (P54137) Probable endonuclease III homolog (EC
4.2.99.18) (DNA-(Apurinic or apyrimidinic site) lyase)
Length = 259
Score = 35.8 bits (81), Expect = 0.17
Identities = 24/89 (26%), Positives = 41/89 (45%), Gaps = 11/89 (12%)
Query: 215 RAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPKVAACIALYSLDQH 274
+A Y+ T +L+ G ++ D + L LPGVGPK+A + + +
Sbjct: 84 KAVYLQKTAKILKDDFSG-----------DIPDSLDGLCALPGVGPKMANLVMQIAWGEC 132
Query: 275 HAIPVDVHIWRIAQKYILPELAGSKLTQK 303
I VD H+ RI+ + + + + TQK
Sbjct: 133 VGIAVDTHVHRISNRLGWIKTSTPEKTQK 161
>END3_SCHPO (Q09907) Endonuclease III homolog (EC 4.2.99.18)
(DNA-(apurinic or apyrimidinic site) lyase)
Length = 355
Score = 35.4 bits (80), Expect = 0.23
Identities = 16/43 (37%), Positives = 26/43 (60%)
Query: 244 ELEDVISELIKLPGVGPKVAACIALYSLDQHHAIPVDVHIWRI 286
++ D + +L+ LPGVGPK+ + ++ I VDVH+ RI
Sbjct: 125 DIPDTVEDLMTLPGVGPKMGYLCMSIAWNKTVGIGVDVHVHRI 167
>3MGA_BACSU (P37878) DNA-3-methyladenine glycosylase (EC 3.2.2.21)
(3-methyladenine-DNA glycosidase)
Length = 303
Score = 35.4 bits (80), Expect = 0.23
Identities = 25/95 (26%), Positives = 43/95 (44%), Gaps = 5/95 (5%)
Query: 187 GFDFHAFPTLNQLSLVSEQQLRDAGFGYR-AKYITGTVNVLQSKPEGGEEWLYSLRKLEL 245
G + FP +++ ++ L D + ++YI G ++ S GE L K+
Sbjct: 167 GKKYWVFPPYERIARLTPTDLADIKMTVKKSEYIIGIARLMAS----GELSREKLMKMNF 222
Query: 246 EDVISELIKLPGVGPKVAACIALYSLDQHHAIPVD 280
+D LIK+ G+GP A + + L A P+D
Sbjct: 223 KDAEKNLIKIRGIGPWTANYVLMRCLRFPTAFPID 257
>END3_SYNY3 (P73715) Endonuclease III (EC 4.2.99.18) (DNA-(apurinic
or apyrimidinic site) lyase)
Length = 219
Score = 35.0 bits (79), Expect = 0.30
Identities = 25/105 (23%), Positives = 45/105 (42%), Gaps = 16/105 (15%)
Query: 190 FHAFPTLNQLSLVSEQQLRD-----AGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLE 244
F +P N L+ Q++ + F +AK I G + + +G E
Sbjct: 64 FQRYPDANALAYGDRQEIEELIHSTGFFRNKAKNIQGACRKIVEEFDG-----------E 112
Query: 245 LEDVISELIKLPGVGPKVAACIALYSLDQHHAIPVDVHIWRIAQK 289
+ + EL+ LPGV K A + ++ + VD H+ R++Q+
Sbjct: 113 VPQRMEELLTLPGVARKTANVVLAHAFGILAGVTVDTHVKRLSQR 157
>MUTY_BUCAP (Q8K926) A/G-specific adenine glycosylase (EC 3.2.2.-)
Length = 347
Score = 34.7 bits (78), Expect = 0.39
Identities = 37/121 (30%), Positives = 59/121 (48%), Gaps = 24/121 (19%)
Query: 193 FPTLNQLSLVSEQQLRD-----AGFGY--RAKYITGTVNVLQSKPEGGEEWLYSLRKLEL 245
FP + L+ + +L D +G GY RA+ I TV +++ EE+ + +
Sbjct: 59 FPNIQSLN---QSKLDDILCLWSGLGYYKRAENIYKTVKIIK------EEF-----QEKF 104
Query: 246 EDVISELIKLPGVGPKVAACIALYSLDQHHAIPVDVHIWRIAQKY--ILPELAGSKLTQK 303
S+LIKLPG+G A I SLD I ++ ++ RI +Y I+ + K+ QK
Sbjct: 105 PTGFSDLIKLPGIGRSTAGAILSLSLDYFFPI-LEGNVKRILMRYYGIIGYVTEKKIEQK 163
Query: 304 L 304
L
Sbjct: 164 L 164
>Y023_NPVOP (O10282) Hypothetical 73.1 kDa protein precursor (ORF21)
Length = 657
Score = 34.3 bits (77), Expect = 0.51
Identities = 34/130 (26%), Positives = 54/130 (41%), Gaps = 21/130 (16%)
Query: 69 PSQYTGVVGSHLISLKHLHNGDVCYTLHSQSPSNDDCKTALL-----DFLNADVSLADT- 122
P G + HL SL ++ V YTL+S P N++ T +L D D + AD
Sbjct: 91 PRATGGRISKHLASLTQINKEFVSYTLNSADPQNNEVFTDVLEVDYEDTRQQDAAFADAH 150
Query: 123 ----WKVFSDSDER----------FAELAQHLSGARVL-RQDPFECLIQFMCSSNNHISR 167
W V S +D + + H+S A V + +E I S++N
Sbjct: 151 NPPHWSVVSAADVKELLAVAPPRDRVRVLPHISTANVADKYLKYEACINEERSADNECLY 210
Query: 168 ITKMVDYVSS 177
+T+M ++S
Sbjct: 211 LTEMHGVMAS 220
>END3_THEMA (Q9WYK0) Endonuclease III (EC 4.2.99.18) (DNA-(apurinic
or apyrimidinic site) lyase)
Length = 213
Score = 34.3 bits (77), Expect = 0.51
Identities = 19/43 (44%), Positives = 27/43 (62%), Gaps = 1/43 (2%)
Query: 247 DVISELIKLPGVGPKVAACIALYSLDQHHAIPVDVHIWRIAQK 289
D + EL+KLPGVG K A I L+ + A+ VD H+ RI+ +
Sbjct: 99 DSLEELLKLPGVGRKTAN-IVLWVGFKKPALAVDTHVHRISNR 140
>SNAT_HUMAN (Q16613) Serotonin N-acetyltransferase (EC 2.3.1.87)
(Aralkylamine N-acetyltransferase) (AA-NAT) (Serotonin
acetylase)
Length = 207
Score = 33.9 bits (76), Expect = 0.66
Identities = 16/42 (38%), Positives = 21/42 (49%)
Query: 1 MSKRKRSPIKSLPPPLPPSTPPTPHTQTRHSTNLSKSRAWIP 42
MS + P+K P LPP P +P Q RH+ S+ R P
Sbjct: 1 MSTQSTHPLKPEAPRLPPGIPESPSCQRRHTLPASEFRCLTP 42
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.133 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 44,310,769
Number of Sequences: 164201
Number of extensions: 1881278
Number of successful extensions: 8634
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 26
Number of HSP's that attempted gapping in prelim test: 8525
Number of HSP's gapped (non-prelim): 93
length of query: 363
length of database: 59,974,054
effective HSP length: 111
effective length of query: 252
effective length of database: 41,747,743
effective search space: 10520431236
effective search space used: 10520431236
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC140916.11


