

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140913.6 - phase: 0
(831 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
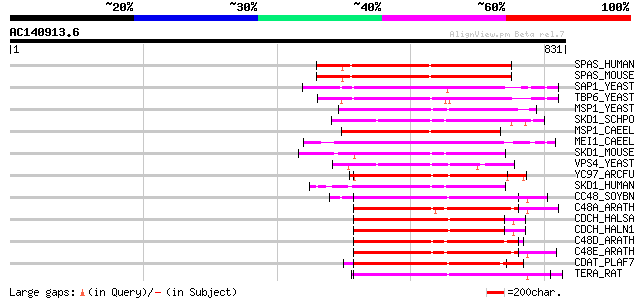
Score E
Sequences producing significant alignments: (bits) Value
SPAS_HUMAN (Q9UBP0) Spastin 236 2e-61
SPAS_MOUSE (Q9QYY8) Spastin 236 2e-61
SAP1_YEAST (P39955) SAP1 protein 226 1e-58
TBP6_YEAST (P40328) Probable 26S protease subunit YTA6 (TAT-bind... 218 4e-56
MSP1_YEAST (P28737) MSP1 protein (TAT-binding homolog 4) 217 9e-56
SKD1_SCHPO (Q09803) Suppressor protein of bem1/bed5 double mutants 209 3e-53
MSP1_CAEEL (P54815) MSP1 protein homolog 207 9e-53
MEI1_CAEEL (P34808) Meiotic spindle formation protein mei-1 201 9e-51
SKD1_MOUSE (P46467) SKD1 protein (Vacuolar sorting protein 4b) 194 1e-48
VPS4_YEAST (P52917) Vacuolar protein sorting-associated protein ... 192 2e-48
YC97_ARCFU (O28972) Cell division cycle protein 48 homolog AF1297 191 5e-48
SKD1_HUMAN (O75351) SKD1 protein (Vacuolar sorting protein 4b) 186 3e-46
CC48_SOYBN (P54774) Cell division cycle protein 48 homolog (Valo... 186 3e-46
C48A_ARATH (P54609) Cell division control protein 48 homolog A (... 183 1e-45
CDCH_HALSA (P46464) CdcH protein 183 2e-45
CDCH_HALN1 (Q9HPF0) CdcH protein 183 2e-45
C48D_ARATH (Q9SCN8) Putative cell division control protein 48 ho... 182 3e-45
C48E_ARATH (Q9LZF6) Cell division control protein 48 homolog E (... 182 4e-45
CDAT_PLAF7 (P46468) Putative cell division cycle ATPase 181 5e-45
TERA_RAT (P46462) Transitional endoplasmic reticulum ATPase (TER... 181 7e-45
>SPAS_HUMAN (Q9UBP0) Spastin
Length = 616
Score = 236 bits (602), Expect = 2e-61
Identities = 134/300 (44%), Positives = 188/300 (62%), Gaps = 11/300 (3%)
Query: 460 GKTESPAPAVKTEAEIPTSVRKTDGENSVPASKAEVP-------DNEFEKRIRPEVIPAN 512
G + PA T P + R A++ + D+ I E++ N
Sbjct: 277 GVKQGSGPAPTTHKGTPKTNRTNKPSTPTTATRKKKDLKNFRNVDSNLANLIMNEIVD-N 335
Query: 513 EIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKA 572
V F DI D K +LQE+V+LP RP+LF G L P RG+LLFGPPG GKTMLAKA
Sbjct: 336 GTAVKFDDIAGQDLAKQALQEIVILPSLRPELFTG-LRAPARGLLLFGPPGNGKTMLAKA 394
Query: 573 IANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVG 632
+A E+ A+F N+S +++TSK+ GE EK VRALF +A ++ P+IIF+DEVDS+L +R R G
Sbjct: 395 VAAESNATFFNISAASLTSKYVGEGEKLVRALFAVARELQPSIIFIDEVDSLLCER-REG 453
Query: 633 EHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAEN 692
EH+A R++K EF+ +DG+ S +DR+LV+ ATNRP +LDEA++RRF +R+ V LP+ E
Sbjct: 454 EHDASRRLKTEFLIEFDGVQSAGDDRVLVMGATNRPQELDEAVLRRFIKRVYVSLPNEET 513
Query: 693 RENILRTLLAKE-KVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDS 751
R +L+ LL K+ + +LA MT+GYSGSDL L AA P+REL ++++ S
Sbjct: 514 RLLLLKNLLCKQGSPLTQKELAQLARMTDGYSGSDLTALAKDAALGPIRELKPEQVKNMS 573
>SPAS_MOUSE (Q9QYY8) Spastin
Length = 614
Score = 236 bits (601), Expect = 2e-61
Identities = 134/300 (44%), Positives = 186/300 (61%), Gaps = 11/300 (3%)
Query: 460 GKTESPAPAVKTEAEIPTSVRKTDGENSVPASKAEVP-------DNEFEKRIRPEVIPAN 512
G P PA T P R A + + D+ I E++ N
Sbjct: 275 GARPGPGPAATTHKGTPKPNRTNKPSTPTTAVRKKKDLKNFRNVDSNLANLIMNEIVD-N 333
Query: 513 EIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKA 572
V F DI + K +LQE+V+LP RP+LF G L P RG+LLFGPPG GKTMLAKA
Sbjct: 334 GTAVKFDDIAGQELAKQALQEIVILPSLRPELFTG-LRAPARGLLLFGPPGNGKTMLAKA 392
Query: 573 IANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVG 632
+A E+ A+F N+S +++TSK+ GE EK VRALF +A ++ P+IIF+DEVDS+L +R R G
Sbjct: 393 VAAESNATFFNISAASLTSKYVGEGEKLVRALFAVARELQPSIIFIDEVDSLLCER-REG 451
Query: 633 EHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAEN 692
EH+A R++K EF+ +DG+ S +DR+LV+ ATNRP +LDEA++RRF +R+ V LP+ E
Sbjct: 452 EHDASRRLKTEFLIEFDGVQSAGDDRVLVMGATNRPQELDEAVLRRFIKRVYVSLPNEET 511
Query: 693 RENILRTLLAKE-KVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDS 751
R +L+ LL K+ + +LA MT+GYSGSDL L AA P+REL ++++ S
Sbjct: 512 RLLLLKNLLCKQGSPLTQKELAQLARMTDGYSGSDLTALAKDAALGPIRELKPEQVKNMS 571
>SAP1_YEAST (P39955) SAP1 protein
Length = 897
Score = 226 bits (577), Expect = 1e-58
Identities = 149/402 (37%), Positives = 216/402 (53%), Gaps = 50/402 (12%)
Query: 439 KLEAQAVTSEKKEEGAAVKPEGKTESPAPAVKTEAEIPTSVRKTDGENSVPASKAEVPDN 498
K+ AQ K E + + + + E EI V + E+ + S V D
Sbjct: 527 KIPAQKKIGSPKIEDVGTEDATEHATSLNEQREEPEIDKKVLREILEDEIIDSLQGV-DR 585
Query: 499 EFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILL 558
+ K+I E++ + V + DI L+ K SL+E V+ P RPDLF G L +P RG+LL
Sbjct: 586 QAAKQIFAEIVVHGD-EVHWDDIAGLESAKYSLKEAVVYPFLRPDLFRG-LREPVRGMLL 643
Query: 559 FGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFV 618
FGPPGTGKTMLA+A+A E+ ++F ++S S++TSK+ GE EK VRALF +A K+SP+IIFV
Sbjct: 644 FGPPGTGKTMLARAVATESHSTFFSISASSLTSKYLGESEKLVRALFAIAKKLSPSIIFV 703
Query: 619 DEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTS------------------KSEDRIL 660
DE+DS++G R E+E+ R+IKNEF+ W L+S + + R+L
Sbjct: 704 DEIDSIMGSRNNENENESSRRIKNEFLVQWSSLSSAAAGSNKSNTNNSDTNGDEDDTRVL 763
Query: 661 VLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEK-VHGGLDFKELATMT 719
VLAATN P+ +DEA RRF RR + LP + R + LL+ +K DF EL +T
Sbjct: 764 VLAATNLPWSIDEAARRRFVRRQYIPLPEDQTRHVQFKKLLSHQKHTLTESDFDELVKIT 823
Query: 720 EGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERVIT 779
EGYSGSD+ +L AA P+R+L D E E+E +
Sbjct: 824 EGYSGSDITSLAKDAAMGPLRDL-----------------------GDKLLETEREMI-- 858
Query: 780 LRPLNMQDFKMAKSQVAASFAAEGAGMNELRQWNDLYGEGGS 821
RP+ + DFK + + S + + G+ + +W +G GS
Sbjct: 859 -RPIGLVDFKNSLVYIKPSVSQD--GLVKYEKWASQFGSSGS 897
>TBP6_YEAST (P40328) Probable 26S protease subunit YTA6 (TAT-binding
homolog 6)
Length = 754
Score = 218 bits (556), Expect = 4e-56
Identities = 140/377 (37%), Positives = 216/377 (57%), Gaps = 48/377 (12%)
Query: 462 TESPAPAVKTEAEIPTSVRKTDGENSVPASKAEV------PDNEFEKRIRPEVIPANEIG 515
T PA + ++ T +++ +S+ + K ++ D ++I E++ +E
Sbjct: 409 TAVPAESKPMRSKSGTPDKESSASSSLDSRKEDILKSVQGVDRNACEQILNEILVTDE-K 467
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
V + DI L K+SL+E V+ P RPDLF+G L +P RG+LLFGPPGTGKTM+AKA+A
Sbjct: 468 VYWEDIAGLRNAKNSLKEAVVYPFLRPDLFKG-LREPVRGMLLFGPPGTGKTMIAKAVAT 526
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHE 635
E+ ++F +VS S++ SK+ GE EK VRALF +A K+SP+IIF+DE+DSML R+ E+E
Sbjct: 527 ESNSTFFSVSASSLLSKYLGESEKLVRALFYMAKKLSPSIIFIDEIDSMLTARSD-NENE 585
Query: 636 AMRKIKNEFMSNWDGL---TSKSED-------RILVLAATNRPFDLDEAIIRRFERRIMV 685
+ R+IK E + W L T++SED R+LVL ATN P+ +D+A RRF R++ +
Sbjct: 586 SSRRIKTELLIQWSSLSSATAQSEDRNNTLDSRVLVLGATNLPWAIDDAARRRFSRKLYI 645
Query: 686 GLPSAENRENILRTLLAKEKVH-GGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQ 744
LP E R L+ L+AK+K LD++ + MTEG+SGSDL +L AA P+R+L
Sbjct: 646 PLPDYETRLYHLKRLMAKQKNSLQDLDYELITEMTEGFSGSDLTSLAKEAAMEPIRDLGD 705
Query: 745 QEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERVITLRPLNMQDFKMAKSQVAASFAAEGA 804
+ + D K +R + ++DF+ A + S ++E
Sbjct: 706 KLMFADFDK--------------------------IRGIEIKDFQNALLTIKKSVSSE-- 737
Query: 805 GMNELRQWNDLYGEGGS 821
+ + +W+ +G GS
Sbjct: 738 SLQKYEEWSSKFGSNGS 754
>MSP1_YEAST (P28737) MSP1 protein (TAT-binding homolog 4)
Length = 362
Score = 217 bits (553), Expect = 9e-56
Identities = 122/298 (40%), Positives = 177/298 (58%), Gaps = 20/298 (6%)
Query: 493 AEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGG-LLK 551
AEV + +E+ I ++ +EI +TF DIG LD L E V+ PL P+++ LL+
Sbjct: 65 AEVTLDAYERTILSSIVTPDEINITFQDIGGLDPLISDLHESVIYPLMMPEVYSNSPLLQ 124
Query: 552 PCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKV 611
G+LL+GPPG GKTMLAKA+A E+GA+FI++ MS+I KW+GE K V A+F+LA K+
Sbjct: 125 APSGVLLYGPPGCGKTMLAKALAKESGANFISIRMSSIMDKWYGESNKIVDAMFSLANKL 184
Query: 612 SPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDL 671
P IIF+DE+DS L +R+ +HE +K EFM+ WDGL + R++++ ATNR D+
Sbjct: 185 QPCIIFIDEIDSFLRERSST-DHEVTATLKAEFMTLWDGLLNNG--RVMIIGATNRINDI 241
Query: 672 DEAIIRRFERRIMVGLPSAENRENILRTLLAKEKV-HGGLDFKELATMTEGYSGSDLKNL 730
D+A +RR +R +V LP ++ R IL LL K+ D + +A T+G+SGSDLK L
Sbjct: 242 DDAFLRRLPKRFLVSLPGSDQRYKILSVLLKDTKLDEDEFDLQLIADNTKGFSGSDLKEL 301
Query: 731 CTTAAYRPVRELIQQEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERVITLRPLNMQDF 788
C AA +E I+Q+ Q D +S + +RPL +DF
Sbjct: 302 CREAALDAAKEYIKQKRQLIDSGTIDVNDTSS---------------LKIRPLKTKDF 344
>SKD1_SCHPO (Q09803) Suppressor protein of bem1/bed5 double mutants
Length = 432
Score = 209 bits (531), Expect = 3e-53
Identities = 133/327 (40%), Positives = 188/327 (56%), Gaps = 12/327 (3%)
Query: 483 DGENSVPASKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRP 542
+G NS A++A D + + I + V + DI L+ K++L+E V+LP++ P
Sbjct: 93 EGSNSPTANEALDSDAKKLRSALTSAILVEKPNVRWDDIAGLENAKEALKETVLLPIKLP 152
Query: 543 DLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVR 602
LF G KP GILL+GPPGTGK+ LAKA+A EAG++F ++S S + SKW GE E+ VR
Sbjct: 153 QLFSHGR-KPWSGILLYGPPGTGKSYLAKAVATEAGSTFFSISSSDLVSKWMGESERLVR 211
Query: 603 ALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVL 662
LF +A + P+IIF+DE+DS+ G R+ GE E+ R+IK EF+ +G+ K E +LVL
Sbjct: 212 QLFEMAREQKPSIIFIDEIDSLCGSRSE-GESESSRRIKTEFLVQMNGV-GKDESGVLVL 269
Query: 663 AATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAK-EKVHGGLDFKELATMTEG 721
ATN P+ LD AI RRFE+RI + LP+A R + + K DFKELA MT+G
Sbjct: 270 GATNIPWTLDSAIRRRFEKRIYIPLPNAHARARMFELNVGKIPSELTSQDFKELAKMTDG 329
Query: 722 YSGSDLKNLCTTAAYRPVRELIQQEIQK---DSKKKKDAEGQNSQDAQDAKE----EVEQ 774
YSGSD+ + A PVR + K D+K + S DA E EV
Sbjct: 330 YSGSDISIVVRDAIMEPVRRIHTATHFKEVYDNKSNRTLVTPCSPGDPDAFESSWLEVNP 389
Query: 775 ERVITLRPLNMQDFKMAKSQVAASFAA 801
E ++ + L ++DF A +V + A
Sbjct: 390 EDIMEPK-LTVRDFYSAVRKVKPTLNA 415
>MSP1_CAEEL (P54815) MSP1 protein homolog
Length = 342
Score = 207 bits (527), Expect = 9e-53
Identities = 106/237 (44%), Positives = 153/237 (63%), Gaps = 2/237 (0%)
Query: 498 NEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGIL 557
+E E RI + + ++G + +IG +E L++ ++LPLR LL P RGIL
Sbjct: 62 SEHEIRIATQFVGGEDVGADWDEIGGCEELVAELKDRIILPLRFASQSGSHLLSPPRGIL 121
Query: 558 LFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIF 617
L+GPPG GKT+LAKA+A AG FIN+ +S +T KW+GE +K A+F++A K PTIIF
Sbjct: 122 LYGPPGCGKTLLAKAVARAAGCRFINLQVSNLTDKWYGESQKLAAAVFSVAQKFQPTIIF 181
Query: 618 VDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR 677
+DE+DS L R + +HE+ +K +FM+ WDG +S S D+I+V+ ATNRP D+D AI+R
Sbjct: 182 IDEIDSFLRDR-QSHDHESTAMMKAQFMTLWDGFSS-SGDQIIVMGATNRPRDVDAAILR 239
Query: 678 RFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTA 734
R R V +P+A+ R IL +L EK++ ++ E+A EG SGSDLK +C A
Sbjct: 240 RMTARFQVPVPNAKQRSQILNVILRNEKINNTVNLGEIAQAAEGLSGSDLKEVCRLA 296
>MEI1_CAEEL (P34808) Meiotic spindle formation protein mei-1
Length = 472
Score = 201 bits (510), Expect = 9e-51
Identities = 130/379 (34%), Positives = 200/379 (52%), Gaps = 41/379 (10%)
Query: 441 EAQAVTSEKKEEGAAVKPEGKTESPAPAVKTEAEIPTSVRKTDGENSVPAS-KAEVPDNE 499
E TS A VKP T+ P +NS S A D
Sbjct: 131 EISKSTSSMSTNPADVKPANPTQGILP-----------------QNSAGDSFDASAYDAY 173
Query: 500 FEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLF 559
+ +R + E ++ DI + + K L E V LPL P+ F+G L P + ++L
Sbjct: 174 IVQAVRGTMATNTENTMSLDDIIGMHDVKQVLHEAVTLPLLVPEFFQG-LRSPWKAMVLA 232
Query: 560 GPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVD 619
GPPGTGKT++A+AIA+E+ ++F VS + ++SKW G+ EK VR LF LA +P+IIF+D
Sbjct: 233 GPPGTGKTLIARAIASESSSTFFTVSSTDLSSKWRGDSEKIVRLLFELARFYAPSIIFID 292
Query: 620 EVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSED-RILVLAATNRPFDLDEAIIRR 678
E+D++ GQR GEHEA R++K+EF+ DG +K + R+ VLAATN P++LDEA+ RR
Sbjct: 293 EIDTLGGQRGNSGEHEASRRVKSEFLVQMDGSQNKFDSRRVFVLAATNIPWELDEALRRR 352
Query: 679 FERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRP 738
FE+RI + LP + R+ ++ + +++ +LA TEG+SG+D+ +LC TAA
Sbjct: 353 FEKRIFIPLPDIDARKKLIEKSMEGTPKSDEINYDDLAARTEGFSGADVVSLCRTAAINV 412
Query: 739 VRELIQQEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERVITLRPLNMQDFKMAKSQVAAS 798
+R + D + + A E ++ E V + DF+ A V+ S
Sbjct: 413 LR-------------RYDTKSLRGGELTAAMESLKAELVRNI------DFEAALQAVSPS 453
Query: 799 FAAEGAGMNELRQWNDLYG 817
+ M + ++W D +G
Sbjct: 454 AGPD--TMLKCKEWCDSFG 470
>SKD1_MOUSE (P46467) SKD1 protein (Vacuolar sorting protein 4b)
Length = 444
Score = 194 bits (492), Expect = 1e-48
Identities = 127/315 (40%), Positives = 180/315 (56%), Gaps = 14/315 (4%)
Query: 433 GDRDSLKLEAQAVTSEKKEEGAAVKPEGKTESPAPAVKTEAEIPTSVRKTDGENSVPASK 492
GD+ + A+ + E + K + P VK E P + D + +
Sbjct: 51 GDKAKQSIRAKCTEYLDRAEKLKEYLKKKEKKPQKPVKEEQSGPVDEKGNDSDG-----E 105
Query: 493 AEVPDNEFEKRIRPEVIPANEI---GVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGL 549
AE D E +K+++ ++ A I V +SD+ L+ K++L+E V+LP++ P LF G
Sbjct: 106 AESDDPE-KKKLQNQLQGAIVIERPNVKWSDVAGLEGAKEALKEAVILPIKFPHLFTGKR 164
Query: 550 LKPCRGILLFGPPGTGKTMLAKAIANEAGAS-FINVSMSTITSKWFGEDEKNVRALFTLA 608
P RGILLFGPPGTGK+ LAKA+A EA S F ++S S + SKW GE EK V+ LF LA
Sbjct: 165 T-PWRGILLFGPPGTGKSYLAKAVATEANNSTFFSISSSDLVSKWLGESEKLVKNLFQLA 223
Query: 609 AKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRP 668
+ P+IIF+DE+DS+ G R+ E EA R+IK EF+ G+ D ILVL ATN P
Sbjct: 224 RENKPSIIFIDEIDSLCGSRSE-NESEAARRIKTEFLVQMQGV-GVDNDGILVLGATNIP 281
Query: 669 FDLDEAIIRRFERRIMVGLPSAENRENILRTLL-AKEKVHGGLDFKELATMTEGYSGSDL 727
+ LD AI RRFE+RI + LP A R + R L + + DF+EL T+GYSG+D+
Sbjct: 282 WVLDSAIRRRFEKRIYIPLPEAHARAAMFRLHLGSTQNSLTEADFQELGRKTDGYSGADI 341
Query: 728 KNLCTTAAYRPVREL 742
+ A +PVR++
Sbjct: 342 SIIVRDALMQPVRKV 356
>VPS4_YEAST (P52917) Vacuolar protein sorting-associated protein
VPS4 (END13 protein)
Length = 437
Score = 192 bits (489), Expect = 2e-48
Identities = 115/283 (40%), Positives = 165/283 (57%), Gaps = 19/283 (6%)
Query: 484 GENSVPASKAEVPDNEFEKRIR---PEVIPANEIGVTFSDIGALDETKDSLQELVMLPLR 540
G + + E E K++R I + + V + D+ L+ K++L+E V+LP++
Sbjct: 95 GNKKISQEEGEDNGGEDNKKLRGALSSAILSEKPNVKWEDVAGLEGAKEALKEAVILPVK 154
Query: 541 RPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKN 600
P LF+G KP GILL+GPPGTGK+ LAKA+A EA ++F +VS S + SKW GE EK
Sbjct: 155 FPHLFKGNR-KPTSGILLYGPPGTGKSYLAKAVATEANSTFFSVSSSDLVSKWMGESEKL 213
Query: 601 VRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRIL 660
V+ LF +A + P+IIF+DEVD++ G R GE EA R+IK E + +G+ + S+ +L
Sbjct: 214 VKQLFAMARENKPSIIFIDEVDALTGTRGE-GESEASRRIKTELLVQMNGVGNDSQG-VL 271
Query: 661 VLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILR-------TLLAKEKVHGGLDFK 713
VL ATN P+ LD AI RRFERRI + LP R + +L KE D++
Sbjct: 272 VLGATNIPWQLDSAIRRRFERRIYIPLPDLAARTTMFEINVGDTPCVLTKE------DYR 325
Query: 714 ELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKD 756
L MTEGYSGSD+ + A +P+R++ KD + D
Sbjct: 326 TLGAMTEGYSGSDIAVVVKDALMQPIRKIQSATHFKDVSTEDD 368
>YC97_ARCFU (O28972) Cell division cycle protein 48 homolog AF1297
Length = 733
Score = 191 bits (486), Expect = 5e-48
Identities = 107/271 (39%), Positives = 165/271 (60%), Gaps = 14/271 (5%)
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
V + DIG L+ K L E V PL+ P++F +KP RGILLFGPPGTGKT+LAKA+AN
Sbjct: 452 VKWEDIGGLEHAKQELMEAVEWPLKYPEVFRAANIKPPRGILLFGPPGTGKTLLAKAVAN 511
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHE 635
E+ A+FI+V + SKW GE EK+VR +F A +V+P +IF DE+DS+ +R +G+
Sbjct: 512 ESNANFISVKGPELLSKWVGESEKHVREMFRKARQVAPCVIFFDEIDSLAPRRGGIGDSH 571
Query: 636 AMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAENR 693
++ ++ ++ DGL + ++V+AATNRP +D A++R R ER I + P + R
Sbjct: 572 VTERVVSQLLTELDGLEELKD--VVVIAATNRPDMIDPALLRPGRLERHIYIPPPDKKAR 629
Query: 694 ENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQ----QEIQK 749
I + L + + ++ +ELA TEGYSG+D++ +C A +RELI+ +E K
Sbjct: 630 VEIFKIHLRGKPLADDVNIEELAEKTEGYSGADIEAVCREAGMLAIRELIKPGMTREEAK 689
Query: 750 DSKKKKDAEGQNSQDAQD------AKEEVEQ 774
++ KK ++ ++A KE+VE+
Sbjct: 690 EAAKKLKITKKHFEEALKKVRPSLTKEDVEK 720
Score = 179 bits (453), Expect = 3e-44
Identities = 97/243 (39%), Positives = 155/243 (62%), Gaps = 10/243 (4%)
Query: 510 PANEI-----GVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGT 564
PA E+ VT+ DIG L ++E++ LPL+ P+LF+ ++P +G+LL+GPPGT
Sbjct: 168 PAEEVKRAVPDVTYEDIGGLKRELRLVREMIELPLKHPELFQRLGIEPPKGVLLYGPPGT 227
Query: 565 GKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSM 624
GKT++AKA+ANE A FI +S I SK++GE E+ +R +F A + +P+IIF+DE+DS+
Sbjct: 228 GKTLIAKAVANEVDAHFIPISGPEIMSKYYGESEQRLREIFEEAKENAPSIIFIDEIDSI 287
Query: 625 LGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERR 682
+R V E R++ + ++ DGL ++ + ++V+AATNRP +D A+ R RF+R
Sbjct: 288 APKREEV-TGEVERRVVAQLLALMDGLEARGD--VIVIAATNRPDAIDPALRRPGRFDRE 344
Query: 683 IMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVREL 742
I +G+P E R+ IL K + +D +ELA +T G+ G+DL+ LC AA +R +
Sbjct: 345 IEIGVPDKEGRKEILEIHTRKMPLAEDVDLEELAELTNGFVGADLEALCKEAAMHALRRV 404
Query: 743 IQQ 745
+ +
Sbjct: 405 LPE 407
>SKD1_HUMAN (O75351) SKD1 protein (Vacuolar sorting protein 4b)
Length = 444
Score = 186 bits (471), Expect = 3e-46
Identities = 122/296 (41%), Positives = 169/296 (56%), Gaps = 18/296 (6%)
Query: 449 KKEEGAAVKPEGKTESPAPAVKTEAEIPTSVRKTDGENSVPASKAEVPDNEFEKRIRPEV 508
K +E A KP K P+PA + + +DGE + + N+ + I E
Sbjct: 77 KNKEKKAQKPV-KEGQPSPADEKGND-------SDGEGESDDPEKKKLQNQLQGAIVIE- 127
Query: 509 IPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTM 568
V +SD+ L+ K++L+E V+LP++ P LF G P RGILLFGPPGTGK+
Sbjct: 128 ----RPNVKWSDVAGLEGAKEALKEAVILPIKFPHLFTGKRT-PWRGILLFGPPGTGKSY 182
Query: 569 LAKAIANEAGAS-FINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQ 627
LAKA+A EA S F ++S S + SKW GE EK V+ LF LA + P+IIF+DE+DS+ G
Sbjct: 183 LAKAVATEANNSTFFSISSSDLVSKWLGESEKLVKNLFQLARENKPSIIFIDEIDSLCGS 242
Query: 628 RTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGL 687
R+ E EA R+IK EF+ G+ D ILVL ATN P+ LD AI RRFE+RI + L
Sbjct: 243 RSE-NESEAARRIKTEFLVQMQGV-GVDNDGILVLGATNIPWVLDSAIRRRFEKRIYIPL 300
Query: 688 PSAENRENILRTLL-AKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVREL 742
P R + + L + DF+EL T+GYSG+D+ + A +PVR++
Sbjct: 301 PEPHARAAMFKLHLGTTQNSLTEADFRELGRKTDGYSGADISIIVRDALMQPVRKV 356
>CC48_SOYBN (P54774) Cell division cycle protein 48 homolog (Valosin
containing protein homolog) (VCP)
Length = 807
Score = 186 bits (471), Expect = 3e-46
Identities = 101/297 (34%), Positives = 170/297 (57%), Gaps = 9/297 (3%)
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
V++ DIG L+ K LQE V P+ P+ FE + P +G+L +GPPG GKT+LAKAIAN
Sbjct: 478 VSWEDIGGLENVKRELQETVQYPVEHPEKFEKFGMSPSKGVLFYGPPGCGKTLLAKAIAN 537
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQR-TRVGE- 633
E A+FI+V + + WFGE E NVR +F A + +P ++F DE+DS+ QR + VG+
Sbjct: 538 ECQANFISVKGPELLTMWFGESEANVREIFDKARQSAPCVLFFDELDSIATQRGSSVGDA 597
Query: 634 HEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAE 691
A ++ N+ ++ DG+++K + ++ ATNRP +D A++R R ++ I + LP +
Sbjct: 598 GGAADRVLNQLLTEMDGMSAKK--TVFIIGATNRPDIIDPALLRPGRLDQLIYIPLPDED 655
Query: 692 NRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDS 751
+R I + L K + +D + LA T+G+SG+D+ +C A +RE I+++I+++
Sbjct: 656 SRHQIFKACLRKSPIAKNVDLRALARHTQGFSGADITEICQRACKYAIRENIEKDIERER 715
Query: 752 KKKKDAEGQNSQDAQDAKEEVEQ---ERVITLRPLNMQDFKMAKSQVAASFAAEGAG 805
K +++ E + D E++ E + ++ D + K Q A + G
Sbjct: 716 KSRENPEAMDEDTVDDEVAEIKAAHFEESMKFARRSVSDADIRKYQAFAQTLQQSRG 772
Score = 174 bits (441), Expect = 9e-43
Identities = 111/286 (38%), Positives = 165/286 (56%), Gaps = 10/286 (3%)
Query: 479 VRKTDGENSVPASKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLP 538
V GE V A E+ E E R + +E+G + D+G + + ++ELV LP
Sbjct: 171 VETDPGEYCVVAPDTEI-FCEGEPLKREDEERLDEVG--YDDVGGVRKQMAQIRELVELP 227
Query: 539 LRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDE 598
LR P LF+ +KP +GILL+GPPG+GKT++A+A+ANE GA F ++ I SK GE E
Sbjct: 228 LRHPQLFKSIGVKPPKGILLYGPPGSGKTLIARAVANETGAFFFCINGPEIMSKLAGESE 287
Query: 599 KNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDR 658
N+R F A K +P+IIF+DE+DS+ +R + E R+I ++ ++ DGL KS
Sbjct: 288 SNLRKAFEEAEKNAPSIIFIDEIDSIAPKREKT-HGEVERRIVSQLLTLMDGL--KSRAH 344
Query: 659 ILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELA 716
++V+ ATNRP +D A+ R RF+R I +G+P R +LR K+ +D + +A
Sbjct: 345 VIVIGATNRPNSIDPALRRFGRFDREIDIGVPDEVGRLEVLRIHTKNMKLSDDVDLERIA 404
Query: 717 TMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAEGQNS 762
T GY G+DL LCT AA + +RE + ++ + DAE NS
Sbjct: 405 KDTHGYVGADLAALCTEAALQCIRE--KMDVIDLEDETIDAEVLNS 448
>C48A_ARATH (P54609) Cell division control protein 48 homolog A
(AtCDC48a)
Length = 809
Score = 183 bits (465), Expect = 1e-45
Identities = 103/314 (32%), Positives = 174/314 (54%), Gaps = 11/314 (3%)
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
V+++DIG L+ K LQE V P+ P+ FE + P +G+L +GPPG GKT+LAKAIAN
Sbjct: 477 VSWNDIGGLENVKRELQETVQYPVEHPEKFEKFGMSPSKGVLFYGPPGCGKTLLAKAIAN 536
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHE 635
E A+FI+V + + WFGE E NVR +F A + +P ++F DE+DS+ QR +
Sbjct: 537 ECQANFISVKGPELLTMWFGESEANVREIFDKARQSAPCVLFFDELDSIATQRGGGSGGD 596
Query: 636 ---AMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSA 690
A ++ N+ ++ DG+ +K + ++ ATNRP +D A++R R ++ I + LP
Sbjct: 597 GGGAADRVLNQLLTEMDGMNAKK--TVFIIGATNRPDIIDSALLRPGRLDQLIYIPLPDE 654
Query: 691 ENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKD 750
++R NI + L K + +D LA T+G+SG+D+ +C A +RE I+++I+K+
Sbjct: 655 DSRLNIFKAALRKSPIAKDVDIGALAKYTQGFSGADITEICQRACKYAIRENIEKDIEKE 714
Query: 751 SKKKKDAEGQNSQDAQDAKEEVEQ---ERVITLRPLNMQDFKMAKSQVAASFAAEGAGMN 807
++ ++ E +D D E++ E + ++ D + K Q A + G
Sbjct: 715 KRRSENPEAM-EEDGVDEVSEIKAAHFEESMKYARRSVSDADIRKYQAFAQTLQQSRGFG 773
Query: 808 ELRQWNDLYGEGGS 821
++ + G G +
Sbjct: 774 SEFRFENSAGSGAT 787
Score = 171 bits (434), Expect = 6e-42
Identities = 102/250 (40%), Positives = 153/250 (60%), Gaps = 9/250 (3%)
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
V + D+G + + ++ELV LPLR P LF+ +KP +GILL+GPPG+GKT++A+A+AN
Sbjct: 204 VGYDDVGGVRKQMAQIRELVELPLRHPQLFKSIGVKPPKGILLYGPPGSGKTLIARAVAN 263
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHE 635
E GA F ++ I SK GE E N+R F A K +P+IIF+DE+DS+ +R + E
Sbjct: 264 ETGAFFFCINGPEIMSKLAGESESNLRKAFEEAEKNAPSIIFIDEIDSIAPKREKT-NGE 322
Query: 636 AMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAENR 693
R+I ++ ++ DGL KS ++V+ ATNRP +D A+ R RF+R I +G+P R
Sbjct: 323 VERRIVSQLLTLMDGL--KSRAHVIVMGATNRPNSIDPALRRFGRFDREIDIGVPDEIGR 380
Query: 694 ENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQ-EIQKDSK 752
+LR K+ +D + ++ T GY G+DL LCT AA + +RE + +++ DS
Sbjct: 381 LEVLRIHTKNMKLAEDVDLERISKDTHGYVGADLAALCTEAALQCIREKMDVIDLEDDS- 439
Query: 753 KKKDAEGQNS 762
DAE NS
Sbjct: 440 --IDAEILNS 447
>CDCH_HALSA (P46464) CdcH protein
Length = 742
Score = 183 bits (464), Expect = 2e-45
Identities = 95/228 (41%), Positives = 142/228 (61%), Gaps = 4/228 (1%)
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
+T+ D+G L E K++++E V PL +P+ F ++P G+LL+GPPGTGKT++AKA+AN
Sbjct: 459 ITWDDVGGLTEAKNNVKESVEWPLNQPEKFTRMGVEPPAGVLLYGPPGTGKTLMAKAVAN 518
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHE 635
E A+FI+V + SKW GE EK +R F A +V+PT+IF DE+DS+ R + G +
Sbjct: 519 ETNANFISVRGPQLLSKWVGESEKAIRQTFRKARQVAPTVIFFDELDSLAPGRGQTGGNN 578
Query: 636 AMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAENR 693
++ N+ ++ DGL E ++V+AATNRP +D A+IR RF+R + VG P E R
Sbjct: 579 VSERVVNQLLTELDGLEEMEE--VMVIAATNRPDIIDPALIRSGRFDRLVQVGQPGIEGR 636
Query: 694 ENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRE 741
E IL+ + + +ELA +GY GSDL N+ AA +R+
Sbjct: 637 EQILKIHTQDTPLAADVSLRELAERADGYVGSDLANIAREAAIEALRD 684
Score = 164 bits (416), Expect = 7e-40
Identities = 99/272 (36%), Positives = 159/272 (58%), Gaps = 18/272 (6%)
Query: 515 GVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIA 574
G+T+ DIG L+ ++E+V LP++ P +F+ ++P +G+LL GPPGTGKT+LAKA+A
Sbjct: 185 GITYEDIGGLENEIQRVREMVELPMKHPQIFQKLGIEPPQGVLLHGPPGTGKTLLAKAVA 244
Query: 575 NEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEH 634
NE ASF +++ I SK++GE E+ +R +F A SP+IIF+DE+DS+ +R V
Sbjct: 245 NETSASFFSIAGPEIISKYYGESEQQLREIFEDAKDDSPSIIFIDELDSIAPKREDV-TG 303
Query: 635 EAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAEN 692
E R++ + ++ DGL + + ++V+AATNR +D A+ R RF+R I +G+P
Sbjct: 304 EVERRVVAQLLTMMDGLEGRGQ--VIVIAATNRVDAVDPALRRPGRFDREIEIGVPDEIG 361
Query: 693 RENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSK 752
RE IL+ + ++ LA T G+ G+D+++L AA R +R + EI D +
Sbjct: 362 REEILKIHTRGMPLSDDVNLSTLADDTHGFVGADIESLSKEAAMRALRRYL-PEIDLDEE 420
Query: 753 ------------KKKDAEGQNSQDAQDAKEEV 772
K++D +G S+ A EV
Sbjct: 421 DIPPSLIDRMIVKREDFKGALSEVEPSAMREV 452
>CDCH_HALN1 (Q9HPF0) CdcH protein
Length = 742
Score = 183 bits (464), Expect = 2e-45
Identities = 95/228 (41%), Positives = 142/228 (61%), Gaps = 4/228 (1%)
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
+T+ D+G L E K++++E V PL +P+ F ++P G+LL+GPPGTGKT++AKA+AN
Sbjct: 459 ITWDDVGGLTEAKNNVKESVEWPLNQPEKFTRMGVEPPAGVLLYGPPGTGKTLMAKAVAN 518
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHE 635
E A+FI+V + SKW GE EK +R F A +V+PT+IF DE+DS+ R + G +
Sbjct: 519 ETNANFISVRGPQLLSKWVGESEKAIRQTFRKARQVAPTVIFFDELDSLAPGRGQTGGNN 578
Query: 636 AMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAENR 693
++ N+ ++ DGL E ++V+AATNRP +D A+IR RF+R + VG P E R
Sbjct: 579 VSERVVNQLLTELDGLEEMEE--VMVIAATNRPDIIDPALIRSGRFDRLVQVGQPGIEGR 636
Query: 694 ENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRE 741
E IL+ + + +ELA +GY GSDL N+ AA +R+
Sbjct: 637 EQILKIHTQDTPLAADVSLRELAERADGYVGSDLANIAREAAIEALRD 684
Score = 164 bits (416), Expect = 7e-40
Identities = 99/272 (36%), Positives = 159/272 (58%), Gaps = 18/272 (6%)
Query: 515 GVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIA 574
G+T+ DIG L+ ++E+V LP++ P +F+ ++P +G+LL GPPGTGKT+LAKA+A
Sbjct: 185 GITYEDIGGLENEIQRVREMVELPMKHPQIFQKLGIEPPQGVLLHGPPGTGKTLLAKAVA 244
Query: 575 NEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEH 634
NE ASF +++ I SK++GE E+ +R +F A SP+IIF+DE+DS+ +R V
Sbjct: 245 NETSASFFSIAGPEIISKYYGESEQQLREIFEDAKDDSPSIIFIDELDSIAPKREDV-TG 303
Query: 635 EAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAEN 692
E R++ + ++ DGL + + ++V+AATNR +D A+ R RF+R I +G+P
Sbjct: 304 EVERRVVAQLLTMMDGLEGRGQ--VIVIAATNRVDAVDPALRRPGRFDREIEIGVPDEIG 361
Query: 693 RENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSK 752
RE IL+ + ++ LA T G+ G+D+++L AA R +R + EI D +
Sbjct: 362 REEILKIHTRGMPLSDDVNLSTLADDTHGFVGADIESLSKEAAMRALRRYL-PEIDLDEE 420
Query: 753 ------------KKKDAEGQNSQDAQDAKEEV 772
K++D +G S+ A EV
Sbjct: 421 DIPPSLIDRMIVKREDFKGALSEVEPSAMREV 452
>C48D_ARATH (Q9SCN8) Putative cell division control protein 48
homolog D (AtCDC48d) (Transitional endoplasmic reticulum
ATPase D)
Length = 815
Score = 182 bits (462), Expect = 3e-45
Identities = 95/258 (36%), Positives = 156/258 (59%), Gaps = 6/258 (2%)
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
V++ DIG L+ K LQE V P+ P+ FE + P +G+L +GPPG GKT+LAKAIAN
Sbjct: 478 VSWEDIGGLENVKRELQETVQYPVEHPEKFEKFGMSPSKGVLFYGPPGCGKTLLAKAIAN 537
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQR-TRVGE- 633
E A+FI++ + + WFGE E NVR +F A + +P ++F DE+DS+ QR VG+
Sbjct: 538 ECQANFISIKGPELLTMWFGESEANVREIFDKARQSAPCVLFFDELDSIATQRGNSVGDA 597
Query: 634 HEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAE 691
A ++ N+ ++ DG+ +K + ++ ATNRP +D A++R R ++ I + LP E
Sbjct: 598 GGAADRVLNQLLTEMDGMNAKK--TVFIIGATNRPDIIDPALLRPGRLDQLIYIPLPDEE 655
Query: 692 NRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDS 751
+R I ++ L K V +D + LA T+G+SG+D+ +C + +RE I+++I+K+
Sbjct: 656 SRYQIFKSCLRKSPVAKDVDLRALAKYTQGFSGADITEICQRSCKYAIRENIEKDIEKER 715
Query: 752 KKKKDAEGQNSQDAQDAK 769
K+ + E + + A+
Sbjct: 716 KRAESPEAMEEDEEEIAE 733
Score = 171 bits (433), Expect = 7e-42
Identities = 100/249 (40%), Positives = 152/249 (60%), Gaps = 7/249 (2%)
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
V + D+G + + ++ELV LPLR P LF+ +KP +GILL+GPPG+GKT++A+A+AN
Sbjct: 205 VGYDDVGGVRKQMAQIRELVELPLRHPQLFKSIGVKPPKGILLYGPPGSGKTLIARAVAN 264
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHE 635
E GA F ++ I SK GE E N+R F A K +P+IIF+DE+DS+ +R + E
Sbjct: 265 ETGAFFFCINGPEIMSKLAGESESNLRKAFEEAEKNAPSIIFIDEIDSIAPKREKT-HGE 323
Query: 636 AMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAENR 693
R+I ++ ++ DGL KS ++V+ ATNRP +D A+ R RF+R I +G+P R
Sbjct: 324 VERRIVSQLLTLMDGL--KSRAHVIVMGATNRPNSIDPALRRFGRFDREIDIGVPDEIGR 381
Query: 694 ENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKK 753
+LR K+ +D + ++ T GY G+DL LCT AA + +RE + ++ +
Sbjct: 382 LEVLRIHTKNMKLAEDVDLERVSKDTHGYVGADLAALCTEAALQCIRE--KMDVIDLDDE 439
Query: 754 KKDAEGQNS 762
+ DAE NS
Sbjct: 440 EIDAEILNS 448
>C48E_ARATH (Q9LZF6) Cell division control protein 48 homolog E
(AtCDC48e) (Transitional endoplasmic reticulum ATPase E)
Length = 810
Score = 182 bits (461), Expect = 4e-45
Identities = 103/311 (33%), Positives = 169/311 (54%), Gaps = 9/311 (2%)
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
V++ DIG L+ K LQE V P+ P+ FE + P +G+L +GPPG GKT+LAKAIAN
Sbjct: 477 VSWEDIGGLENVKRELQETVQYPVEHPEKFEKFGMSPSKGVLFYGPPGCGKTLLAKAIAN 536
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQR-TRVGE- 633
E A+FI+V + + WFGE E NVR +F A + +P ++F DE+DS+ QR G+
Sbjct: 537 ECQANFISVKGPELLTMWFGESEANVREIFDKARQSAPCVLFFDELDSIATQRGNSAGDA 596
Query: 634 HEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAE 691
A ++ N+ ++ DG+ +K + ++ ATNRP +D A++R R ++ I + LP +
Sbjct: 597 GGAADRVLNQLLTEMDGMNAKK--TVFIIGATNRPDIIDSALLRPGRLDQLIYIPLPDED 654
Query: 692 NRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDS 751
+R NI + L K V +D LA T+G+SG+D+ +C A +RE I+++I+ +
Sbjct: 655 SRLNIFKACLRKSPVAKDVDVTALAKYTQGFSGADITEICQRACKYAIRENIEKDIENER 714
Query: 752 KKKKDAEGQNSQDAQDAKEEVEQ---ERVITLRPLNMQDFKMAKSQVAASFAAEGAGMNE 808
++ ++ E D E+ E + ++ D + K Q A + G
Sbjct: 715 RRSQNPEAMEEDMVDDEVSEIRAAHFEESMKYARRSVSDADIRKYQAFAQTLQQSRGFGS 774
Query: 809 LRQWNDLYGEG 819
+++ G G
Sbjct: 775 EFRFDSTAGVG 785
Score = 171 bits (434), Expect = 6e-42
Identities = 102/250 (40%), Positives = 153/250 (60%), Gaps = 9/250 (3%)
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
V + D+G + + ++ELV LPLR P LF+ +KP +GILL+GPPG+GKT++A+A+AN
Sbjct: 204 VGYDDVGGVRKQMAQIRELVELPLRHPQLFKSIGVKPPKGILLYGPPGSGKTLIARAVAN 263
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHE 635
E GA F ++ I SK GE E N+R F A K +P+IIF+DE+DS+ +R + E
Sbjct: 264 ETGAFFFCINGPEIMSKLAGESESNLRKAFEEAEKNAPSIIFIDEIDSIAPKREKT-NGE 322
Query: 636 AMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAENR 693
R+I ++ ++ DGL KS ++V+ ATNRP +D A+ R RF+R I +G+P R
Sbjct: 323 VERRIVSQLLTLMDGL--KSRAHVIVMGATNRPNSIDPALRRFGRFDREIDIGVPDEIGR 380
Query: 694 ENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQ-EIQKDSK 752
+LR K+ +D + ++ T GY G+DL LCT AA + +RE + +++ DS
Sbjct: 381 LEVLRIHTKNMKLAEDVDLERISKDTHGYVGADLAALCTEAALQCIREKMDVIDLEDDS- 439
Query: 753 KKKDAEGQNS 762
DAE NS
Sbjct: 440 --IDAEILNS 447
>CDAT_PLAF7 (P46468) Putative cell division cycle ATPase
Length = 1229
Score = 181 bits (460), Expect = 5e-45
Identities = 100/256 (39%), Positives = 155/256 (60%), Gaps = 8/256 (3%)
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
VT++DIG ++E K+ L+E ++ PL L+ +GILL+GPPG GKT+LAKAIAN
Sbjct: 931 VTWNDIGGMNEVKEQLKETILYPLEYKHLYNKFNSNYNKGILLYGPPGCGKTLLAKAIAN 990
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHE 635
E A+FI+V + + WFGE E NVR LF A SP IIF DE+DS+ +R ++
Sbjct: 991 ECKANFISVKGPELLTMWFGESEANVRDLFDKARAASPCIIFFDEIDSLAKERNSNTNND 1050
Query: 636 AMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAENR 693
A ++ N+ ++ DG+ K I ++AATNRP LD+A+ R R ++ I + LP ++R
Sbjct: 1051 ASDRVINQILTEIDGINEKK--TIFIIAATNRPDILDKALTRPGRLDKLIYISLPDLKSR 1108
Query: 694 ENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKK 753
+I + +L ++ +D ++A TEG+SG+D+ NLC +A V E I++ I + +
Sbjct: 1109 YSIFKAILKNTPLNEDVDIHDMAKRTEGFSGADITNLCQSA----VNEAIKETIHLLNIR 1164
Query: 754 KKDAEGQNSQDAQDAK 769
KK+ E Q ++ K
Sbjct: 1165 KKEQEEQRKKNKNSFK 1180
Score = 159 bits (402), Expect = 3e-38
Identities = 88/245 (35%), Positives = 149/245 (59%), Gaps = 5/245 (2%)
Query: 501 EKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFG 560
E+ ++ E N +T+ D+G + + + ++EL+ LPL+ P++F + +G+L+ G
Sbjct: 509 EEYLKREDYEENNDDITYEDLGGMKKQLNKIRELIELPLKYPEIFMSIGISAPKGVLMHG 568
Query: 561 PPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDE 620
PGTGKT +AKAIANE+ A ++ I SK GE E+ +R +F A++ +P IIF+DE
Sbjct: 569 IPGTGKTSIAKAIANESNAYCYIINGPEIMSKHIGESEQKLRKIFKKASEKTPCIIFIDE 628
Query: 621 VDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--R 678
+DS+ +R++ +E +++ ++ ++ DGL K + +LVLAATNRP +D A+ R R
Sbjct: 629 IDSIANKRSK-SNNELEKRVVSQLLTLMDGL--KKNNNVLVLAATNRPNSIDPALRRFGR 685
Query: 679 FERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRP 738
F+R I + +P + R IL T K K+ ++ +++A GY G+DL LC AA +
Sbjct: 686 FDREIEIPVPDEQGRYEILLTKTKKMKLDPDVNLRKIAKECHGYVGADLAQLCFEAAIQC 745
Query: 739 VRELI 743
++E I
Sbjct: 746 IKEHI 750
>TERA_RAT (P46462) Transitional endoplasmic reticulum ATPase (TER
ATPase) (15S Mg(2+)-ATPase p97 subunit)
(Valosin-containing protein) (VCP)
Length = 806
Score = 181 bits (459), Expect = 7e-45
Identities = 104/302 (34%), Positives = 171/302 (56%), Gaps = 10/302 (3%)
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
VT+ DIG L++ K LQELV P+ PD F + P +G+L +GPPG GKT+LAKAIAN
Sbjct: 474 VTWEDIGGLEDVKRELQELVQYPVEHPDKFLKFGMTPSKGVLFYGPPGCGKTLLAKAIAN 533
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQR-TRVGE- 633
E A+FI++ + + WFGE E NVR +F A + +P ++F DE+DS+ R +G+
Sbjct: 534 ECQANFISIKGPELLTMWFGESEANVREIFDKARQAAPCVLFFDELDSIAKARGGNIGDG 593
Query: 634 HEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAE 691
A ++ N+ ++ DG+++K + ++ ATNRP +D AI+R R ++ I + LP +
Sbjct: 594 GGAADRVINQILTEMDGMSTKK--NVFIIGATNRPDIIDPAILRPGRLDQLIYIPLPDEK 651
Query: 692 NRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDS 751
+R IL+ L K V +D + LA MT G+SG+DL +C A +RE I+ EI+++
Sbjct: 652 SRVAILKANLRKSPVAKDVDLEFLAKMTNGFSGADLTEICQRACKLAIRESIESEIRRER 711
Query: 752 KKKKDAEGQNSQDAQDAKEEVEQ---ERVITLRPLNMQDFKMAKSQVAASFAAEGAGMNE 808
+++ + ++ D E+ + E + ++ D + K ++ A + G
Sbjct: 712 ERQTNPSAMEVEE-DDPVPEIRRDHFEEAMRFARRSVSDNDIRKYEMFAQTLQQSRGFGS 770
Query: 809 LR 810
R
Sbjct: 771 FR 772
Score = 177 bits (448), Expect = 1e-43
Identities = 114/319 (35%), Positives = 172/319 (53%), Gaps = 28/319 (8%)
Query: 512 NEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAK 571
NE+G + DIG + ++E+V LPLR P LF+ +KP RGILL+GPPGTGKT++A+
Sbjct: 199 NEVG--YDDIGGCRKQLAQIKEMVELPLRHPALFKAIGVKPPRGILLYGPPGTGKTLIAR 256
Query: 572 AIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRV 631
A+ANE GA F ++ I SK GE E N+R F A K +P IIF+DE+D++ +R +
Sbjct: 257 AVANETGAFFFLINGPEIMSKLAGESESNLRKAFEEAEKNAPAIIFIDELDAIAPKREKT 316
Query: 632 GEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPS 689
E R+I ++ ++ DGL K ++V+AATNRP +D A+ R RF+R + +G+P
Sbjct: 317 -HGEVERRIVSQLLTLMDGL--KQRAHVIVMAATNRPNSIDPALRRFGRFDREVDIGIPD 373
Query: 690 AENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQK 749
A R IL+ K+ +D +++A T G+ G+DL LC+ AA + +R+
Sbjct: 374 ATGRLEILQIHTKNMKLADDVDLEQVANETHGHVGADLAALCSEAALQAIRK-------- 425
Query: 750 DSKKKKDAEGQNSQDAQDAKEEVEQERVITLRPLNMQDFKMAKSQVAASFAAEGAGMNEL 809
D D ++E V+ + M DF+ A SQ S E
Sbjct: 426 ------------KMDLIDLEDETIDAEVMNSLAVTMDDFRWALSQSNPSALRETVVEVPQ 473
Query: 810 RQWNDLYG-EGGSRKKEQL 827
W D+ G E R+ ++L
Sbjct: 474 VTWEDIGGLEDVKRELQEL 492
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.132 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 92,540,400
Number of Sequences: 164201
Number of extensions: 3918987
Number of successful extensions: 14641
Number of sequences better than 10.0: 840
Number of HSP's better than 10.0 without gapping: 531
Number of HSP's successfully gapped in prelim test: 309
Number of HSP's that attempted gapping in prelim test: 13009
Number of HSP's gapped (non-prelim): 1360
length of query: 831
length of database: 59,974,054
effective HSP length: 119
effective length of query: 712
effective length of database: 40,434,135
effective search space: 28789104120
effective search space used: 28789104120
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 70 (31.6 bits)
Medicago: description of AC140913.6


