

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140849.12 + phase: 0
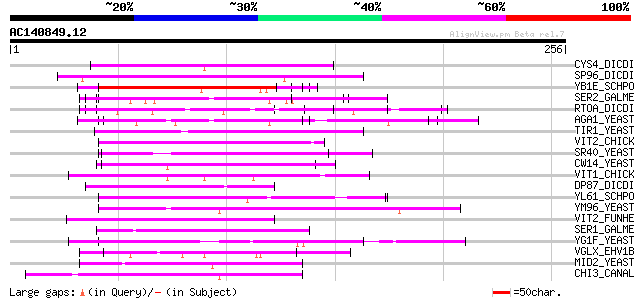
(256 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CYS4_DICDI (P54639) Cysteine proteinase 4 precursor (EC 3.4.22.-) 57 3e-08
SP96_DICDI (P14328) Spore coat protein SP96 56 1e-07
YB1E_SCHPO (P87179) Serine-rich protein C30B4.01c precursor 53 6e-07
SER2_GALME (O96615) Sericin-2 (Silk gum protein 2) (Fragment) 51 2e-06
RTOA_DICDI (P54681) Protein rtoA (Ratio-A) 50 5e-06
AGA1_YEAST (P32323) A-agglutinin attachment subunit precursor 50 5e-06
TIR1_YEAST (P10863) Cold shock induced protein TIR1 precursor (S... 50 7e-06
VIT2_CHICK (P02845) Vitellogenin II precursor (Major vitellogeni... 47 3e-05
SR40_YEAST (P32583) Suppressor protein SRP40 46 8e-05
CW14_YEAST (O13547) Covalently-linked cell wall protein 14 precu... 46 1e-04
VIT1_CHICK (P87498) Vitellogenin I precursor (Minor vitellogenin... 45 1e-04
DP87_DICDI (Q04503) Prespore protein DP87 precursor 45 2e-04
YL61_SCHPO (Q8TFG9) Hypothetical serine/threonine-rich protein P... 45 2e-04
YM96_YEAST (Q04893) Hypothetical 113.1 kDa protein in PRE5-FET4 ... 44 3e-04
VIT2_FUNHE (Q98893) Vitellogenin II precursor (VTG II) [Contains... 44 3e-04
SER1_GALME (O96614) Sericin-1 (Silk gum protein 1) (Fragment) 44 3e-04
YG1F_YEAST (P53214) Hypothetical 57.5 kDa protein in VMA7-RPS25A... 44 4e-04
VGLX_EHV1B (P28968) Glycoprotein X precursor 44 4e-04
MID2_YEAST (P36027) Mating process protein MID2 (Serine-rich pro... 44 4e-04
CHI3_CANAL (P40954) Chitinase 3 precursor (EC 3.2.1.14) 44 4e-04
>CYS4_DICDI (P54639) Cysteine proteinase 4 precursor (EC 3.4.22.-)
Length = 442
Score = 57.4 bits (137), Expect = 3e-08
Identities = 39/113 (34%), Positives = 60/113 (52%), Gaps = 1/113 (0%)
Query: 38 LGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGS-DMASELTT 96
+G+GS SSSS+ ++ G S S ++ G +S S S + SS SG S+S S +S ++
Sbjct: 282 VGYGSGSSSSSGSSSGKSSSSSSTGGKTSSSSSSGKASSSSSGKASSSSSSGKTSSAASS 341
Query: 97 TSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSV 149
TS S S+ S SG T +Q+ QTS +++ + GS S G +V
Sbjct: 342 TSGSQSGSQSGSQSGQSTGSQSGQTSASGQASASGSGSGSGSGSGSGSGSGAV 394
Score = 38.1 bits (87), Expect = 0.021
Identities = 40/150 (26%), Positives = 62/150 (40%), Gaps = 8/150 (5%)
Query: 33 SQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMAS 92
S + G + SSSS+ A SS +G +S S S + SS S T S SGS S
Sbjct: 300 SSSSSTGGKTSSSSSSGKASSSS------SGKASSSSSSGKTSSAASSTSGSQSGSQSGS 353
Query: 93 ELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWN 152
+ + + GS S +SASG +++ + S + + G+ S + +NS +
Sbjct: 354 Q--SGQSTGSQSGQTSASGQASASGSGSGSGSGSGSGSGSGAVEASSGNYWIVKNSWGTS 411
Query: 153 GARTTETLGITQRKNDDFSLSTGDFPTLGS 182
R N+ + FPT S
Sbjct: 412 WGMDGYIFMSKDRNNNCGIATMASFPTASS 441
Score = 32.3 bits (72), Expect = 1.1
Identities = 23/93 (24%), Positives = 39/93 (41%)
Query: 9 TSSTRRGGMTVLGKVAVPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPS 68
+SS + + GK + SQ G+ S+ S + +S S A+A S
Sbjct: 321 SSSGKASSSSSSGKTSSAASSTSGSQSGSQSGSQSGQSTGSQSGQTSASGQASASGSGSG 380
Query: 69 HLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S S GSG ++SG+ + + ++WG
Sbjct: 381 SGSGSGSGSGSGAVEASSGNYWIVKNSWGTSWG 413
>SP96_DICDI (P14328) Spore coat protein SP96
Length = 600
Score = 55.8 bits (133), Expect = 1e-07
Identities = 47/143 (32%), Positives = 66/143 (45%), Gaps = 2/143 (1%)
Query: 23 VAVPKPINLP-SQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGT 81
V++P+P S + GS S SSAS + S S + ++ ASS SA SS S
Sbjct: 414 VSLPRPTTTTGSTSDSSALGSTSESSASGSSAVSSSASGSSAASSSPSSSAASSSPSSSA 473
Query: 82 RPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRP-RSAETRPGSSHLSRF 140
S+ S AS ++SA SSS SSAS + ++ +S P SA + SS +
Sbjct: 474 ASSSPSSSAASSSPSSSASSSSSPSSSASSSSAPSSSASSSSAPSSSASSSSASSSSASS 533
Query: 141 AEHVGENSVAWNGARTTETLGIT 163
A ++A A TT T T
Sbjct: 534 AATTAATTIATTAATTTATTTAT 556
Score = 42.0 bits (97), Expect = 0.001
Identities = 32/122 (26%), Positives = 61/122 (49%), Gaps = 5/122 (4%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S SSSA+++ SS + ++++ +SS S SA SS S + PS+S S ++ ++ S+
Sbjct: 476 SSPSSSAASSSPSSSASSSSSPSSSASSSSAPSSSASSSSAPSSSASSSSASSSSASSAA 535
Query: 102 SSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTETLG 161
+++ + A+ T+ T+ + +A T ++ A + + A A TT T
Sbjct: 536 TTAATTIATTAATTTATTTATTATTTATTTATTT-----AATIATTTAATTTATTTATTA 590
Query: 162 IT 163
T
Sbjct: 591 TT 592
Score = 33.5 bits (75), Expect = 0.52
Identities = 33/148 (22%), Positives = 59/148 (39%), Gaps = 14/148 (9%)
Query: 54 SSLSPNANAGASSPSHLSARPSS--------------GGSGTRPSTSGSDMASELTTTSA 99
S+++ + GA+SP ++ P+ + T STS S + +SA
Sbjct: 381 STIASTGSTGATSPCSVAQCPTGYVCVAQNNVAVSLPRPTTTTGSTSDSSALGSTSESSA 440
Query: 100 WGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTET 159
GSS+ SSASG ++ + +S S + SS S A +S A + + + +
Sbjct: 441 SGSSAVSSSASGSSAASSSPSSSAASSSPSSSAASSSPSSSAASSSPSSSASSSSSPSSS 500
Query: 160 LGITQRKNDDFSLSTGDFPTLGSEKDKS 187
+ + S S+ + S S
Sbjct: 501 ASSSSAPSSSASSSSAPSSSASSSSASS 528
Score = 31.6 bits (70), Expect = 2.0
Identities = 26/115 (22%), Positives = 55/115 (47%), Gaps = 8/115 (6%)
Query: 24 AVPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRP 83
A P + S + + SSS+ S++ SS +P+++A +SS S SA ++ + T
Sbjct: 483 ASSSPSSSASSSSSPSSSASSSSAPSSSASSSSAPSSSASSSSASSSSASSAATTAATTI 542
Query: 84 STSGSDM--------ASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAET 130
+T+ + A+ TT+A +++ ++ + T+ T+ T+ + T
Sbjct: 543 ATTAATTTATTTATTATTTATTTATTTAATIATTTAATTTATTTATTATTTATTT 597
>YB1E_SCHPO (P87179) Serine-rich protein C30B4.01c precursor
Length = 374
Score = 53.1 bits (126), Expect = 6e-07
Identities = 35/99 (35%), Positives = 59/99 (59%), Gaps = 5/99 (5%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S SSS +S++ ++ SP++++ +SS S S+ SS S + S+S S +S +++S+
Sbjct: 139 SSSSSPSSSSTTTTTSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 198
Query: 102 SSSRPSSASGPPT-----SNQTSQTSLRPRSAETRPGSS 135
SSS SS+S P T S+ +S +S S+ +RP SS
Sbjct: 199 SSSSSSSSSVPITSSTSSSHSSSSSSSSSSSSSSRPSSS 237
Score = 48.9 bits (115), Expect = 1e-05
Identities = 35/99 (35%), Positives = 56/99 (56%), Gaps = 3/99 (3%)
Query: 32 PSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMA 91
PS T S SSSS+S+ SS S ++++ +SS S S+ SS S + S+S S +
Sbjct: 144 PSSSSTTTTTSPSSSSSSS---SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 200
Query: 92 SELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAET 130
S +++S +SS SS S +S+ +S +S RP S+ +
Sbjct: 201 SSSSSSSVPITSSTSSSHSSSSSSSSSSSSSSRPSSSSS 239
Score = 47.0 bits (110), Expect = 5e-05
Identities = 38/103 (36%), Positives = 57/103 (54%), Gaps = 5/103 (4%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSG--SDMASELTTTSA 99
S SSSS+S++ SS S ++++ +SS S S+ SS S + P TS S +S +++S+
Sbjct: 168 SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSVPITSSTSSSHSSSSSSSSS 227
Query: 100 WGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAE 142
SSSRPSS+S T T +S + P SS S +E
Sbjct: 228 SSSSSRPSSSSSFIT---TMSSSTFISTVTVTPSSSSSSTSSE 267
Score = 45.1 bits (105), Expect = 2e-04
Identities = 34/102 (33%), Positives = 53/102 (51%), Gaps = 5/102 (4%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S SS+ S++ SS S ++ +SPS S+ SS S + S+S S +S +++S+
Sbjct: 130 SSVSSTTSSSSSSSPSSSSTTTTTSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 189
Query: 102 SSSRPSSASGPPTSNQ-----TSQTSLRPRSAETRPGSSHLS 138
SSS SS+S +S+ TS TS S+ + SS S
Sbjct: 190 SSSSSSSSSSSSSSSSSSVPITSSTSSSHSSSSSSSSSSSSS 231
Score = 44.3 bits (103), Expect = 3e-04
Identities = 29/82 (35%), Positives = 50/82 (60%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S SSSS+S++ SS S ++++ +SS S S+ SS S + S+S +S ++ S+
Sbjct: 163 SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSVPITSSTSSSHSSSS 222
Query: 102 SSSRPSSASGPPTSNQTSQTSL 123
SSS SS+S P+S+ + T++
Sbjct: 223 SSSSSSSSSSRPSSSSSFITTM 244
Score = 43.9 bits (102), Expect = 4e-04
Identities = 33/97 (34%), Positives = 52/97 (53%), Gaps = 2/97 (2%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S SSSS+S++ SS P ++ +SS S S+ SS S +RPS+S S + + ++T
Sbjct: 193 SSSSSSSSSSSSSSSVPITSSTSSSHSSSSSSSSSSSSSSRPSSSSSFITTMSSSTFIST 252
Query: 102 SSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLS 138
+ PSS+S +S S T+ +A S+H S
Sbjct: 253 VTVTPSSSSSSTSSEVPSSTAALALNAS--KASNHTS 287
Score = 42.4 bits (98), Expect = 0.001
Identities = 31/97 (31%), Positives = 53/97 (53%), Gaps = 4/97 (4%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S SS S++ + SS SP++++ ++ S S+ SS S S+S S +S +++S+
Sbjct: 128 SSSSVSSTTSSSSSSSPSSSSTTTTTSPSSSSSSSSSS----SSSSSSSSSSSSSSSSSS 183
Query: 102 SSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLS 138
SSS SS+S +S+ +S +S + SSH S
Sbjct: 184 SSSSSSSSSSSSSSSSSSSSSSSSVPITSSTSSSHSS 220
Score = 37.7 bits (86), Expect = 0.027
Identities = 33/136 (24%), Positives = 58/136 (42%), Gaps = 4/136 (2%)
Query: 75 SSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGS 134
SS S T S+S S +S TTT+ SSS SS+S +S+ +S +S S+ + S
Sbjct: 129 SSSVSSTTSSSSSSSPSSSSTTTTTSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 188
Query: 135 SHLSRFAEHVGENSVAWNGARTTETLGITQRKNDDFSLSTGDFPTLGSEKDKSVHDVELQ 194
S S + +S + + T + + + S S+ S S +
Sbjct: 189 SSSSSSSSSSSSSSSSSSSVPITSSTSSSHSSSSSSSSSSSS----SSRPSSSSSFITTM 244
Query: 195 GSLLCITIISIDSSAA 210
S I+ +++ S++
Sbjct: 245 SSSTFISTVTVTPSSS 260
Score = 34.7 bits (78), Expect = 0.23
Identities = 23/87 (26%), Positives = 42/87 (47%)
Query: 52 WGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASG 111
W L+ N + S + +S S + PS+S + + +++S+ SSS SS+S
Sbjct: 113 WSVYLTGNGVLQTTVSSSSVSSTTSSSSSSSPSSSSTTTTTSPSSSSSSSSSSSSSSSSS 172
Query: 112 PPTSNQTSQTSLRPRSAETRPGSSHLS 138
+S+ +S +S S+ + SS S
Sbjct: 173 SSSSSSSSSSSSSSSSSSSSSSSSSSS 199
>SER2_GALME (O96615) Sericin-2 (Silk gum protein 2) (Fragment)
Length = 220
Score = 51.2 bits (121), Expect = 2e-06
Identities = 39/134 (29%), Positives = 70/134 (52%), Gaps = 2/134 (1%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S SSS +N+ GSS + NA+ G+SS ++ S + SG+ S + S +S ++ +
Sbjct: 16 SSGSSSTNNSSGSSSTNNAS-GSSSSNNTSGSSRNNSSGSSSSNNSSSGSSAGNSSGSSS 74
Query: 102 SSSRPSSASGPPTSNQTSQTSLRPR-SAETRPGSSHLSRFAEHVGENSVAWNGARTTETL 160
S+S SS G + N +S +S + S+ +R S+ S + ++ A N A ++ET+
Sbjct: 75 SNSSGSSGIGSLSRNSSSSSSSQAAGSSSSRRVSADGSSSSSSASNSAAAQNSASSSETI 134
Query: 161 GITQRKNDDFSLST 174
+N+ S S+
Sbjct: 135 NADGSENESSSSSS 148
Score = 47.0 bits (110), Expect = 5e-05
Identities = 42/129 (32%), Positives = 60/129 (45%), Gaps = 13/129 (10%)
Query: 41 GSKSSSSASNAWGS----SLSPNAN-------AGASSPSHLSARPSSGGSGTRPSTSGSD 89
G+ S SS+SN+ GS SLS N++ AG+SS +SA SS S S + +
Sbjct: 67 GNSSGSSSSNSSGSSGIGSLSRNSSSSSSSQAAGSSSSRRVSADGSSSSSSASNSAAAQN 126
Query: 90 MASELTTTSAWGSSSRPSSASGPPTSNQT--SQTSLRPRSAETRPGSSHLSRFAEHVGEN 147
AS T +A GS + SS+S N SQ S + SS S A +
Sbjct: 127 SASSSETINADGSENESSSSSSSAAQNSATRSQVINADGSQSSSSSSSSASNQASATSSS 186
Query: 148 SVAWNGART 156
SV+ +G+ +
Sbjct: 187 SVSADGSES 195
Score = 43.5 bits (101), Expect = 5e-04
Identities = 33/97 (34%), Positives = 53/97 (54%), Gaps = 4/97 (4%)
Query: 36 GTLGWGSKSSSSASNAWGSSLSPNANAGAS--SPSHLSARPSSGGSGTRPSTSGSDMASE 93
G+ + S SS +N+ GSS S N+++G+S + S S+ SSG SG + S +S
Sbjct: 36 GSSSSNNTSGSSRNNSSGSSSSNNSSSGSSAGNSSGSSSSNSSGSSGIGSLSRNSSSSS- 94
Query: 94 LTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAET 130
++ +A SSSR SA G +S+ S ++ SA +
Sbjct: 95 -SSQAAGSSSSRRVSADGSSSSSSASNSAAAQNSASS 130
Score = 43.1 bits (100), Expect = 7e-04
Identities = 42/126 (33%), Positives = 56/126 (44%), Gaps = 12/126 (9%)
Query: 33 SQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMAS 92
S R GS SSSSASN S A ASS ++A S S + S++ + A+
Sbjct: 103 SSRRVSADGSSSSSSASN------SAAAQNSASSSETINADGSENESSSSSSSAAQNSAT 156
Query: 93 ELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAE----TRPGSSHLSRFAEHVGENS 148
+A GS S SS+S SNQ S TS SA+ SS S + +S
Sbjct: 157 RSQVINADGSQS--SSSSSSSASNQASATSSSSVSADGSESESSSSSSSSSSSSSESSSS 214
Query: 149 VAWNGA 154
+W+ A
Sbjct: 215 SSWSSA 220
>RTOA_DICDI (P54681) Protein rtoA (Ratio-A)
Length = 400
Score = 50.1 bits (118), Expect = 5e-06
Identities = 39/144 (27%), Positives = 70/144 (48%), Gaps = 5/144 (3%)
Query: 36 GTLGWGSKSSSSASNAWGSSLSPNANAGA-----SSPSHLSARPSSGGSGTRPSTSGSDM 90
G+ GS+SS+ +SN+ + ++N+G+ SS S + S SG++ ST S+
Sbjct: 126 GSSNSGSQSSTDSSNSGSQGSTGSSNSGSQSSTDSSNSGSQSSTDSSNSGSQGSTGSSNS 185
Query: 91 ASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVA 150
SE + +S GS S SS SG +S+ +S + S + GS S + E+S
Sbjct: 186 GSESSGSSNSGSESSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSSG 245
Query: 151 WNGARTTETLGITQRKNDDFSLST 174
+ + + + G + ++ S S+
Sbjct: 246 SSNSGSESSSGSSNSGSESSSGSS 269
Score = 47.8 bits (112), Expect = 3e-05
Identities = 33/101 (32%), Positives = 52/101 (50%)
Query: 36 GTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELT 95
G+ GS+SS S+++ SS S N+ + +SS S S SS GS S S S ++ +
Sbjct: 181 GSSNSGSESSGSSNSGSESSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSSGSSNSGS 240
Query: 96 TTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSH 136
+S+ S+S S+SG S S + +E+ GSS+
Sbjct: 241 ESSSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSSGSSN 281
Score = 46.6 bits (109), Expect = 6e-05
Identities = 37/162 (22%), Positives = 73/162 (44%), Gaps = 3/162 (1%)
Query: 41 GSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAW 100
GS+SS+ +SN+ + ++N+G+ S ++ S GS S S S ++ + +S+
Sbjct: 164 GSQSSTDSSNSGSQGSTGSSNSGSESSGSSNSGSESSGSSNSGSESSSGSSNSGSESSSG 223
Query: 101 GSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTETL 160
S+S S+SG S S + +E+ GSS+ + NS + + + ++ +
Sbjct: 224 SSNSGSESSSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSSGSSNSG 283
Query: 161 GITQRKNDDFSLSTGDFPTLGSEKDKSVHDVELQGSLLCITI 202
+ ++ S S+ D GS D + +L T+
Sbjct: 284 SESSGSSNSGSESSSD---SGSSSDGKTTCISFHDTLSINTV 322
Score = 45.8 bits (107), Expect = 1e-04
Identities = 40/120 (33%), Positives = 64/120 (53%), Gaps = 9/120 (7%)
Query: 33 SQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMAS 92
S G+ GS+SSS +SN+ S S ++N+G+ S S S S SG+ S SGS+ +S
Sbjct: 209 SSSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSSGS--SNSGSESSS 266
Query: 93 ELTTT---SAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSV 149
+ + S+ GSS+ S +SG +SN S++S S + G + F + + N+V
Sbjct: 267 GSSNSGSESSSGSSNSGSESSG--SSNSGSESS--SDSGSSSDGKTTCISFHDTLSINTV 322
Score = 45.4 bits (106), Expect = 1e-04
Identities = 30/87 (34%), Positives = 45/87 (51%)
Query: 36 GTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELT 95
G+ GS+SSS +SN+ S S ++N+G+ S S S S SG+ S S S S +
Sbjct: 201 GSSNSGSESSSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSSGSSNS 260
Query: 96 TTSAWGSSSRPSSASGPPTSNQTSQTS 122
+ + SS S S +SN S++S
Sbjct: 261 GSESSSGSSNSGSESSSGSSNSGSESS 287
Score = 43.5 bits (101), Expect = 5e-04
Identities = 45/170 (26%), Positives = 74/170 (43%), Gaps = 10/170 (5%)
Query: 36 GTLGWGSKSSSSA-SNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASEL 94
G+ GS+SS S+ S + SS S N+ + +SS S S SS GS S S S ++
Sbjct: 191 GSSNSGSESSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSSGSSNSG 250
Query: 95 TTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGA 154
+ +S+ S+S S+SG S S + +E+ S+ S + G +S
Sbjct: 251 SESSSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSGSSNSGSESSSDSGSSS----DG 306
Query: 155 RTT-----ETLGITQRKNDDFSLSTGDFPTLGSEKDKSVHDVELQGSLLC 199
+TT +TL I +D+ + S+ + + GS+ C
Sbjct: 307 KTTCISFHDTLSINTVDDDEIECTGKGETRCISDNNYKCATKQRHGSIEC 356
Score = 42.4 bits (98), Expect = 0.001
Identities = 47/161 (29%), Positives = 77/161 (47%), Gaps = 15/161 (9%)
Query: 36 GTLGWGSKSSS-SASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASEL 94
G+ GS+S+S S S A GSS S + ++ SS S S SG++ ST S+ S+
Sbjct: 108 GSSNSGSQSTSNSGSEASGSSNSGSQSSTDSSNSGSQGSTGSSNSGSQSSTDSSNSGSQS 167
Query: 95 TTTS-------AWGSSSRPSSASGPPTSNQTSQTSLRPRS-AETRPGSSHLSRFAEHVGE 146
+T S + GSS+ S +SG +SN S++S S +E+ GSS+ +
Sbjct: 168 STDSSNSGSQGSTGSSNSGSESSG--SSNSGSESSGSSNSGSESSSGSSNSGSESSSGSS 225
Query: 147 NSVAWNGARTTETLGITQRKNDDFSLSTGDFPTLGSEKDKS 187
NS G+ ++ + ++ S ++G + GS S
Sbjct: 226 NS----GSESSSGSSNSGSESSSGSSNSGSESSSGSSNSGS 262
Score = 42.0 bits (97), Expect = 0.001
Identities = 41/153 (26%), Positives = 76/153 (48%), Gaps = 5/153 (3%)
Query: 41 GSKSSSSASNAWGSSLSPNANAGA-SSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSA 99
GS+ S+ +SN+ GS S ++N+G+ SS S S SS GS S S S ++ + +S+
Sbjct: 175 GSQGSTGSSNS-GSESSGSSNSGSESSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSS 233
Query: 100 WGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTET 159
S+S S+SG +SN S++S ++ + S + +E +S + + + +
Sbjct: 234 GSSNSGSESSSG--SSNSGSESSSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSGSSN 291
Query: 160 LGITQRKNDDFSLSTGDFPTLGSEKDKSVHDVE 192
G ++ +D S S G + S++ V+
Sbjct: 292 SG-SESSSDSGSSSDGKTTCISFHDTLSINTVD 323
Score = 41.6 bits (96), Expect = 0.002
Identities = 44/173 (25%), Positives = 75/173 (42%), Gaps = 22/173 (12%)
Query: 33 SQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPS------------SGGSG 80
S G++ S S S +++ GS S ++N+G+ S S+ + S S SG
Sbjct: 83 SSEGSVSSSSNSGSQSTSNSGSEASGSSNSGSQSTSNSGSEASGSSNSGSQSSTDSSNSG 142
Query: 81 TRPSTSGSDMASELTTTS--AWGSSSRPSSASGPP----TSNQTSQTSLRPRSAETRPGS 134
++ ST S+ S+ +T S + SS SS SG +SN S++S S GS
Sbjct: 143 SQGSTGSSNSGSQSSTDSSNSGSQSSTDSSNSGSQGSTGSSNSGSESSGSSNSGSESSGS 202
Query: 135 SHLSRFAEHVGENSVAWNGARTTETLGITQRKNDDFSLSTGDFPTLGSEKDKS 187
S+ + NS G+ ++ + ++ S ++G + GS S
Sbjct: 203 SNSGSESSSGSSNS----GSESSSGSSNSGSESSSGSSNSGSESSSGSSNSGS 251
Score = 38.5 bits (88), Expect = 0.016
Identities = 30/106 (28%), Positives = 53/106 (49%), Gaps = 4/106 (3%)
Query: 31 LPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDM 90
+ + G++G S+S SS ++ S S +N+ ASS +S SS SG++ +++
Sbjct: 50 IKKKNGSIGSSSQSLSSEVDSSDIS-SSGSNSTASSEGSVS---SSSNSGSQSTSNSGSE 105
Query: 91 ASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSH 136
AS + + + +S+ S ASG S S T ++ GSS+
Sbjct: 106 ASGSSNSGSQSTSNSGSEASGSSNSGSQSSTDSSNSGSQGSTGSSN 151
>AGA1_YEAST (P32323) A-agglutinin attachment subunit precursor
Length = 725
Score = 50.1 bits (118), Expect = 5e-06
Identities = 49/178 (27%), Positives = 76/178 (42%), Gaps = 13/178 (7%)
Query: 44 SSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSS 103
SS+++SN +SLS + +SPS S PSS T S+S + +S T+TS+ +S
Sbjct: 174 SSTTSSNPTTTSLSSTS----TSPSSTSTSPSS----TSTSSSSTSTSSSSTSTSSSSTS 225
Query: 104 SRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTETLGIT 163
+ PSS S + TS +S + T SS S +S + + + ++++ +
Sbjct: 226 TSPSSTSTSSSLTSTSSSSTSTSQSSTSTSSSSTSTSPSSTSTSSSSTSTSPSSKSTSAS 285
Query: 164 QRKNDDFSLS-----TGDFPTLGSEKDKSVHDVELQGSLLCITIISIDSSAAFFCLYS 216
+S S T PTL S S SI SS+ LYS
Sbjct: 286 STSTSSYSTSTSPSLTSSSPTLASTSPSSTSISSTFTDSTSSLGSSIASSSTSVSLYS 343
Score = 48.5 bits (114), Expect = 2e-05
Identities = 47/155 (30%), Positives = 72/155 (46%), Gaps = 8/155 (5%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S SS+S S + S+ S + + +SS S S+ S+ S T S+S + +S T+TS
Sbjct: 192 SPSSTSTSPSSTSTSSSSTSTSSSSTSTSSSSTSTSPSSTSTSSSLTSTSSSSTSTSQSS 251
Query: 102 SSSRPSSASGPPTSNQT--SQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTET 159
+S+ SS S P+S T S TS P S T S+ S ++ S + + ++ T
Sbjct: 252 TSTSSSSTSTSPSSTSTSSSSTSTSPSSKSTSASSTSTSSYS-----TSTSPSLTSSSPT 306
Query: 160 LGITQRKNDDFSLS-TGDFPTLGSEKDKSVHDVEL 193
L T + S + T +LGS S V L
Sbjct: 307 LASTSPSSTSISSTFTDSTSSLGSSIASSSTSVSL 341
Score = 47.8 bits (112), Expect = 3e-05
Identities = 34/107 (31%), Positives = 53/107 (48%), Gaps = 3/107 (2%)
Query: 32 PSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPS---SGGSGTRPSTSGS 88
PS T + +SSS+++ SS S ++++ ++SPS S S + S T S S +
Sbjct: 193 PSSTSTSPSSTSTSSSSTSTSSSSTSTSSSSTSTSPSSTSTSSSLTSTSSSSTSTSQSST 252
Query: 89 DMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSS 135
+S T+TS +S+ SS S P+S TS +S S T S
Sbjct: 253 STSSSSTSTSPSSTSTSSSSTSTSPSSKSTSASSTSTSSYSTSTSPS 299
Score = 47.8 bits (112), Expect = 3e-05
Identities = 34/119 (28%), Positives = 56/119 (46%), Gaps = 12/119 (10%)
Query: 32 PSQRGTLGWGSKSSSSASNAWGSSLS------------PNANAGASSPSHLSARPSSGGS 79
PS T + +SSS+++ SS S + ++ ++S S S SS +
Sbjct: 200 PSSTSTSSSSTSTSSSSTSTSSSSTSTSPSSTSTSSSLTSTSSSSTSTSQSSTSTSSSST 259
Query: 80 GTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLS 138
T PS++ + +S T+ S+ +S+ +S S TS S TS P A T P S+ +S
Sbjct: 260 STSPSSTSTSSSSTSTSPSSKSTSASSTSTSSYSTSTSPSLTSSSPTLASTSPSSTSIS 318
Score = 47.8 bits (112), Expect = 3e-05
Identities = 35/104 (33%), Positives = 56/104 (53%), Gaps = 4/104 (3%)
Query: 32 PSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMA 91
PS T + +SSS+++ SS S ++++ ++SPS S SS + T PS+ + +
Sbjct: 228 PSSTSTSSSLTSTSSSSTSTSQSSTSTSSSSTSTSPSSTST--SSSSTSTSPSSKSTSAS 285
Query: 92 SELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSS 135
S T+TS++ +S+ PS S PT TS +S S T SS
Sbjct: 286 S--TSTSSYSTSTSPSLTSSSPTLASTSPSSTSISSTFTDSTSS 327
Score = 43.5 bits (101), Expect = 5e-04
Identities = 37/154 (24%), Positives = 62/154 (40%), Gaps = 7/154 (4%)
Query: 44 SSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSS 103
SSS+ + S +SP + +S+ S S + T PS++ T+ S+ +S
Sbjct: 154 SSSAIEPSSASIISPVTSTLSSTTSSNPTTTSLSSTSTSPSSTS-------TSPSSTSTS 206
Query: 104 SRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTETLGIT 163
S +S S TS +S TS P S T + S + ++S + + + T+ + T
Sbjct: 207 SSSTSTSSSSTSTSSSSTSTSPSSTSTSSSLTSTSSSSTSTSQSSTSTSSSSTSTSPSST 266
Query: 164 QRKNDDFSLSTGDFPTLGSEKDKSVHDVELQGSL 197
+ S S T S S + SL
Sbjct: 267 STSSSSTSTSPSSKSTSASSTSTSSYSTSTSPSL 300
Score = 42.4 bits (98), Expect = 0.001
Identities = 39/165 (23%), Positives = 67/165 (39%), Gaps = 3/165 (1%)
Query: 47 SASNAWGSSLSPNANAGA-SSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSR 105
S ++ + S + P + A SS S + S PS++ + S +T+T + +SS
Sbjct: 123 SVTSKFTSYICPTCHTTAISSLSEVGTTTVVSSSAIEPSSAS--IISPVTSTLSSTTSSN 180
Query: 106 PSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTETLGITQR 165
P++ S TS S TS P S T S+ S + +S + + + T+ + +T
Sbjct: 181 PTTTSLSSTSTSPSSTSTSPSSTSTSSSSTSTSSSSTSTSSSSTSTSPSSTSTSSSLTST 240
Query: 166 KNDDFSLSTGDFPTLGSEKDKSVHDVELQGSLLCITIISIDSSAA 210
+ S S T S S S + S +SA+
Sbjct: 241 SSSSTSTSQSSTSTSSSSTSTSPSSTSTSSSSTSTSPSSKSTSAS 285
Score = 39.3 bits (90), Expect = 0.009
Identities = 33/121 (27%), Positives = 62/121 (50%), Gaps = 12/121 (9%)
Query: 32 PSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMA 91
PS + T +SS++++++ +S SP+ +SSP+ S PSS + + S S +
Sbjct: 277 PSSKST-----SASSTSTSSYSTSTSPSLT--SSSPTLASTSPSSTSISSTFTDSTSSLG 329
Query: 92 SELTTTSAWGSSSRPSSA--SGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSV 149
S + ++S S PS+ S P TS+ + S+ + ET S S E++ ++S+
Sbjct: 330 SSIASSSTSVSLYSPSTPVYSVPSTSSNVATPSMTSSTVETTVSSQSSS---EYITKSSI 386
Query: 150 A 150
+
Sbjct: 387 S 387
Score = 29.6 bits (65), Expect = 7.4
Identities = 29/92 (31%), Positives = 46/92 (49%), Gaps = 1/92 (1%)
Query: 30 NLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSD 89
+L S TL S SS+S S+ + S S ++ ASS + +S S + PSTS S+
Sbjct: 299 SLTSSSPTLASTSPSSTSISSTFTDSTSSLGSSIASSSTSVSLYSPSTPVYSVPSTS-SN 357
Query: 90 MASELTTTSAWGSSSRPSSASGPPTSNQTSQT 121
+A+ T+S ++ S+S T + S T
Sbjct: 358 VATPSMTSSTVETTVSSQSSSEYITKSSISTT 389
>TIR1_YEAST (P10863) Cold shock induced protein TIR1 precursor
(Serine-rich protein 1)
Length = 254
Score = 49.7 bits (117), Expect = 7e-06
Identities = 35/124 (28%), Positives = 59/124 (47%), Gaps = 3/124 (2%)
Query: 40 WGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSA 99
W S A + S +A +S+ SA PSS + P++S + +SE ++SA
Sbjct: 97 WYSSRLEPALKSLNGDASSSAAPSSSAAPTSSAAPSSSAA---PTSSAASSSSEAKSSSA 153
Query: 100 WGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTET 159
SSS S+S P+S++ +S P S+E + S+ S + + + GA+T+
Sbjct: 154 APSSSEAKSSSAAPSSSEAKSSSAAPSSSEAKSSSAAPSSTEAKITSAAPSSTGAKTSAI 213
Query: 160 LGIT 163
IT
Sbjct: 214 SQIT 217
>VIT2_CHICK (P02845) Vitellogenin II precursor (Major vitellogenin)
[Contains: Lipovitellin I (LVI); Phosvitin (PV);
Lipovitellin II (LVII); YGP40]
Length = 1850
Score = 47.4 bits (111), Expect = 3e-05
Identities = 33/104 (31%), Positives = 56/104 (53%), Gaps = 1/104 (0%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S +SS S++ S+ S +++ +SS S S SS S + S S S + +++S
Sbjct: 1175 SSNSSKRSSSKSSNSSKRSSSSSSSSSSSSRSSSSSSSSSSNSKSSSSSSKSSSSSSRSR 1234
Query: 102 SSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVG 145
SSS+ SS+S +S+ +S++S S+ + SSH S + H G
Sbjct: 1235 SSSKSSSSSSSSSSSSSSKSSSSRSSSSSSKSSSHHSH-SHHSG 1277
Score = 42.4 bits (98), Expect = 0.001
Identities = 30/107 (28%), Positives = 56/107 (52%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
SK SSS+S++ SS ++++ +SS + S+ SS S + + S +S +++S+
Sbjct: 1190 SKRSSSSSSSSSSSSRSSSSSSSSSSNSKSSSSSSKSSSSSSRSRSSSKSSSSSSSSSSS 1249
Query: 102 SSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENS 148
SSS+ SS+ +S+++S + GSS S + V +S
Sbjct: 1250 SSSKSSSSRSSSSSSKSSSHHSHSHHSGHLNGSSSSSSSSRSVSHHS 1296
Score = 31.2 bits (69), Expect = 2.6
Identities = 31/136 (22%), Positives = 56/136 (40%), Gaps = 12/136 (8%)
Query: 64 ASSPSHLSARPSSGGS------GTRPSTSGSDMASELTTTSAWGSSSRPSSAS-----GP 112
A+ H RPS G+ GT P S +S ++T+ SSS SS +
Sbjct: 1093 ANKTRHPKNRPSKKGNTVLAEFGTEPDAKTSSSSSSASSTATSSSSSSASSPNRKKPMDE 1152
Query: 113 PTSNQTSQTSLRPRSAETRPG-SSHLSRFAEHVGENSVAWNGARTTETLGITQRKNDDFS 171
++Q Q + S+ +R SS+ S+ + NS + + ++ + ++ + S
Sbjct: 1153 EENDQVKQARNKDASSSSRSSKSSNSSKRSSSKSSNSSKRSSSSSSSSSSSSRSSSSSSS 1212
Query: 172 LSTGDFPTLGSEKDKS 187
S+ + S K S
Sbjct: 1213 SSSNSKSSSSSSKSSS 1228
>SR40_YEAST (P32583) Suppressor protein SRP40
Length = 406
Score = 46.2 bits (108), Expect = 8e-05
Identities = 37/126 (29%), Positives = 62/126 (48%), Gaps = 1/126 (0%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S SSSS S++ S S ++++ + S S + S SG+ S+S SD +S+ +TS+
Sbjct: 210 SDSSSSDSSSDSDSSSSSSSSSSDSDSDSDSSSDSDSSGSSDSSSSSDSSSD-ESTSSDS 268
Query: 102 SSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTETLG 161
S S S SG + +T + + AE P SS+ S + ++ N T+ +
Sbjct: 269 SDSDSDSDSGSSSELETKEATADESKAEETPASSNESTPSASSSSSANKLNIPAGTDEIK 328
Query: 162 ITQRKN 167
QRK+
Sbjct: 329 EGQRKH 334
Score = 45.1 bits (105), Expect = 2e-04
Identities = 35/105 (33%), Positives = 57/105 (53%), Gaps = 8/105 (7%)
Query: 43 KSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGS 102
KSSSS+S++ SS S ++++ +SS SSG S + S+S S +S+ + +S S
Sbjct: 24 KSSSSSSSSSSSSSSSSSSSSSSS--------SSGESSSSSSSSSSSSSSDSSDSSDSES 75
Query: 103 SSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGEN 147
SS SS+S +S+ S++S S+ + SS S E E+
Sbjct: 76 SSSSSSSSSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSES 120
Score = 40.8 bits (94), Expect = 0.003
Identities = 29/79 (36%), Positives = 44/79 (54%), Gaps = 2/79 (2%)
Query: 44 SSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSS 103
SSSS S++ GSS S + +G+ S S S+ SS S S S SD S +++S+ SS
Sbjct: 151 SSSSESSSSGSSSSSESESGSESDSDSSS--SSSSSSDSESDSESDSQSSSSSSSSDSSS 208
Query: 104 SRPSSASGPPTSNQTSQTS 122
SS+S + + +S +S
Sbjct: 209 DSDSSSSDSSSDSDSSSSS 227
Score = 38.9 bits (89), Expect = 0.012
Identities = 43/167 (25%), Positives = 77/167 (45%), Gaps = 12/167 (7%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPST------SGSDMASEL- 94
S SSSS+S++ SS S ++++G+SS S S+ SS S + T S ++ A E
Sbjct: 83 SSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSESESEDETKKRARESDNEDAKETK 142
Query: 95 -TTTSAWGSSSRPSSASGPPTSNQT---SQTSLRPRSAETRPGSSHLSRFAEHVGENSVA 150
T SSS SS+SG +S+++ S++ S+ + S ++ +S +
Sbjct: 143 KAKTEPESSSSSESSSSGSSSSSESESGSESDSDSSSSSSSSSDSESDSESDSQSSSSSS 202
Query: 151 WNGARTTETLGITQRKNDDFSLSTGDFPTLGSEKDK-SVHDVELQGS 196
+ + + + +D S S+ + S+ D S D + GS
Sbjct: 203 SSDSSSDSDSSSSDSSSDSDSSSSSSSSSSDSDSDSDSSSDSDSSGS 249
Score = 35.4 bits (80), Expect = 0.14
Identities = 28/109 (25%), Positives = 49/109 (44%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S S SS+S+ S ++++ +SS S + S S+S S +S + +S+
Sbjct: 157 SSSGSSSSSESESGSESDSDSSSSSSSSSDSESDSESDSQSSSSSSSSDSSSDSDSSSSD 216
Query: 102 SSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVA 150
SSS S+S +S+ S + S GSS S ++ + S +
Sbjct: 217 SSSDSDSSSSSSSSSSDSDSDSDSSSDSDSSGSSDSSSSSDSSSDESTS 265
Score = 33.1 bits (74), Expect = 0.67
Identities = 39/167 (23%), Positives = 65/167 (38%), Gaps = 16/167 (9%)
Query: 33 SQRGTLGWGSKSSSSASNAWGSSLSPNA---------NAGASSPSHLSARPSSGGSGTRP 83
S + G S SSSS+ + S S + N A P S S +
Sbjct: 98 SDSSSSGSSSSSSSSSDESSSESESEDETKKRARESDNEDAKETKKAKTEPES-SSSSES 156
Query: 84 STSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEH 143
S+SGS +SE + S S S SS+S + + + S S+ + SS +
Sbjct: 157 SSSGSSSSSESESGSESDSDSSSSSSSSSDSESDSESDSQSSSSSSSSDSSS------DS 210
Query: 144 VGENSVAWNGARTTETLGITQRKNDDFSLSTGDFPTLGSEKDKSVHD 190
+S + + + ++ + + +D S S+ D + GS S D
Sbjct: 211 DSSSSDSSSDSDSSSSSSSSSSDSDSDSDSSSDSDSSGSSDSSSSSD 257
Score = 30.0 bits (66), Expect = 5.7
Identities = 24/110 (21%), Positives = 50/110 (44%)
Query: 83 PSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAE 142
P S + E ++S+ SSS SS+S +S+ +S S+ + SS S +
Sbjct: 12 PKLSVKEKEIEEKSSSSSSSSSSSSSSSSSSSSSSSSSGESSSSSSSSSSSSSSDSSDSS 71
Query: 143 HVGENSVAWNGARTTETLGITQRKNDDFSLSTGDFPTLGSEKDKSVHDVE 192
+S + + + ++ + ++ ++ S S+G + S D+S + E
Sbjct: 72 DSESSSSSSSSSSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSESE 121
>CW14_YEAST (O13547) Covalently-linked cell wall protein 14
precursor (Inner cell wall protein)
Length = 238
Score = 45.8 bits (107), Expect = 1e-04
Identities = 29/101 (28%), Positives = 50/101 (48%)
Query: 41 GSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAW 100
G SSS++S+A SS + ++ +SS + S + SS + + S S A + S+
Sbjct: 91 GDSSSSASSSASSSSKASSSTKASSSSASSSTKASSSSASSSTKASSSSAAPSSSKASST 150
Query: 101 GSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFA 141
SSS SS++ P+S ++S T + + +H S A
Sbjct: 151 ESSSSSSSSTKAPSSEESSSTYVSSSKQASSTSEAHSSSAA 191
Score = 44.7 bits (104), Expect = 2e-04
Identities = 34/112 (30%), Positives = 55/112 (48%), Gaps = 4/112 (3%)
Query: 43 KSSSSASNAWGSSLSPNANAGASSPSHLS----ARPSSGGSGTRPSTSGSDMASELTTTS 98
KSS S NA S +A++ ASS S S A SS S T+ S+S + +++ +++S
Sbjct: 80 KSSCSEQNASLGDSSSSASSSASSSSKASSSTKASSSSASSSTKASSSSASSSTKASSSS 139
Query: 99 AWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVA 150
A SSS+ SS +S+ +++ S+ T SS + +S A
Sbjct: 140 AAPSSSKASSTESSSSSSSSTKAPSSEESSSTYVSSSKQASSTSEAHSSSAA 191
Score = 38.1 bits (87), Expect = 0.021
Identities = 34/116 (29%), Positives = 60/116 (51%), Gaps = 3/116 (2%)
Query: 33 SQRGTLGWGSKSSSSASN--AWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDM 90
S + + + SSS++S+ A SS S + A +SS + S++ SS S + S+S
Sbjct: 104 SSKASSSTKASSSSASSSTKASSSSASSSTKASSSSAAPSSSKASSTESSSSSSSSTKAP 163
Query: 91 ASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSH-LSRFAEHVG 145
+SE ++++ SS + SS S +S+ S T + + P S+ +S F+E G
Sbjct: 164 SSEESSSTYVSSSKQASSTSEAHSSSAASSTVSQETVSSALPTSTAVISTFSEGSG 219
Score = 31.2 bits (69), Expect = 2.6
Identities = 27/111 (24%), Positives = 45/111 (40%)
Query: 9 TSSTRRGGMTVLGKVAVPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPS 68
+SST+ + S + + + SSS AS+ SS S ++ SS
Sbjct: 108 SSSTKASSSSASSSTKASSSSASSSTKASSSSAAPSSSKASSTESSSSSSSSTKAPSSEE 167
Query: 69 HLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTS 119
S SS + S + S A+ T + SS+ P+S + T ++ S
Sbjct: 168 SSSTYVSSSKQASSTSEAHSSSAASSTVSQETVSSALPTSTAVISTFSEGS 218
>VIT1_CHICK (P87498) Vitellogenin I precursor (Minor vitellogenin)
[Contains: Lipovitellin I (LVI); Phosvitin (PV);
Lipovitellin II (LVII); YGP42]
Length = 1912
Score = 45.4 bits (106), Expect = 1e-04
Identities = 45/162 (27%), Positives = 77/162 (46%), Gaps = 25/162 (15%)
Query: 28 PINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLS------ARPSSGGSGT 81
P S G+ S SSSS+S++ SS S +++ +SS S S ++ SS S +
Sbjct: 1146 PFGDSSSSGSSSSSSSSSSSSSDSSSSSRSSSSSDSSSSSSSSSSSSSSKSKSSSRSSKS 1205
Query: 82 RPSTSGS---DMASELTTTSAWGSSSRPSSASG--------------PPTSNQTSQTSLR 124
S+S S D +S + +++ GSSS S ASG P S + S+
Sbjct: 1206 NRSSSSSNSKDSSSSSSKSNSKGSSSSSSKASGTRQKAKKQSKTTSFPHASAAEGERSVH 1265
Query: 125 PRSAETRPGSSHLSRFAEHVGENSVAWNGARTTETLGITQRK 166
+ ET+ SS SR + + S + + + ++E+ G++ R+
Sbjct: 1266 EQKQETQSSSSSSSRASSN--SRSTSSSTSSSSESSGVSHRQ 1305
Score = 30.8 bits (68), Expect = 3.3
Identities = 36/118 (30%), Positives = 52/118 (43%), Gaps = 13/118 (11%)
Query: 40 WGSKSSSSASNAWGSSL---------SPNANAGASSPSHLSARPSSGGSGTRPSTSGSDM 90
+ S SS N W + L S + + + + S+ SS SG+ S S S
Sbjct: 1321 FNSHSSYDIPNEWETYLPKVYRLRFRSAHTHWHSGHRTSSSSSSSSSESGSSHSNSSSSD 1380
Query: 91 ASELTTTSAWGSSSRPSSASGPPT--SNQTSQTSLRPRSAETRPGSSHLSRFAEHVGE 146
+S + + SSS S G S++ S TS R SA RPGSS L+R +G+
Sbjct: 1381 SSSRRSHMSDSSSSSSSHRHGEKAAHSSRRSPTS-RAASAHHRPGSS-LTRERNFLGD 1436
Score = 29.3 bits (64), Expect = 9.7
Identities = 26/96 (27%), Positives = 45/96 (46%), Gaps = 2/96 (2%)
Query: 25 VPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPS 84
+PK L + W S +S+S++ SS S ++++ +SS S R S S + S
Sbjct: 1337 LPKVYRLRFRSAHTHWHSGHRTSSSSSSSSSESGSSHSNSSSSDSSSRR--SHMSDSSSS 1394
Query: 85 TSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQ 120
+S + +S +SR +SA P S+ T +
Sbjct: 1395 SSSHRHGEKAAHSSRRSPTSRAASAHHRPGSSLTRE 1430
>DP87_DICDI (Q04503) Prespore protein DP87 precursor
Length = 555
Score = 45.1 bits (105), Expect = 2e-04
Identities = 29/87 (33%), Positives = 51/87 (58%), Gaps = 1/87 (1%)
Query: 36 GTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELT 95
GT GS S SSA+++ SS + ++ + +++ S S+ PSS + + +S S A
Sbjct: 466 GTTTGGSTSDSSAASSADSSAASSSPSSSAASSAASSEPSSSAASSSAPSSASSSAPSSA 525
Query: 96 TTSAWGSSSRPSSASGPPTSNQTSQTS 122
++SA SSS SSA+ S+++S++S
Sbjct: 526 SSSA-PSSSASSSAASSAASSESSESS 551
Score = 41.2 bits (95), Expect = 0.002
Identities = 25/94 (26%), Positives = 48/94 (50%)
Query: 45 SSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSS 104
+++ G S S ++ A ++ S S+ PSS + + S+ S A+ + S+ SS+
Sbjct: 462 TTTGGTTTGGSTSDSSAASSADSSAASSSPSSSAASSAASSEPSSSAASSSAPSSASSSA 521
Query: 105 RPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLS 138
S++S P+S+ +S + S+E+ SS S
Sbjct: 522 PSSASSSAPSSSASSSAASSAASSESSESSSATS 555
Score = 40.4 bits (93), Expect = 0.004
Identities = 30/100 (30%), Positives = 47/100 (47%), Gaps = 3/100 (3%)
Query: 9 TSSTRRGGMTVLGKVAVPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPS 68
T+ T GG T G + + S + S SSSA+++ SS ++ A +S+PS
Sbjct: 459 TAGTTTGGTTTGGSTS---DSSAASSADSSAASSSPSSSAASSAASSEPSSSAASSSAPS 515
Query: 69 HLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSS 108
S+ S S + PS+S S A+ +S SS +S
Sbjct: 516 SASSSAPSSASSSAPSSSASSSAASSAASSESSESSSATS 555
Score = 40.0 bits (92), Expect = 0.006
Identities = 37/138 (26%), Positives = 63/138 (44%), Gaps = 5/138 (3%)
Query: 20 LGKVAVPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGS 79
+G + + P N + R S S+++ A+ A++ S +A ++GG+
Sbjct: 408 VGMICIQVPSNCTNTRFPCCPSHPICIHPSTTAASTIATTASTVATTTSATTAGTTTGGT 467
Query: 80 GTRPSTSGSDMASELTTTSAW---GSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSH 136
T STS S AS +++A SS+ S+AS P+S+ S ++ P SA + SS
Sbjct: 468 TTGGSTSDSSAASSADSSAASSSPSSSAASSAASSEPSSSAASSSA--PSSASSSAPSSA 525
Query: 137 LSRFAEHVGENSVAWNGA 154
S +S A + A
Sbjct: 526 SSSAPSSSASSSAASSAA 543
>YL61_SCHPO (Q8TFG9) Hypothetical serine/threonine-rich protein
PB15E9.01c in chromosome I precursor
Length = 943
Score = 44.7 bits (104), Expect = 2e-04
Identities = 38/145 (26%), Positives = 65/145 (44%), Gaps = 12/145 (8%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S SS ++S+A SSL+ +++ +SS + ++ + S T S + S +AS TT+S+
Sbjct: 71 SSSSLTSSSAASSSLTSSSSLASSSTNSTTSASPTSSSLTSSSATSSSLASSSTTSSSLA 130
Query: 102 SSSRPSS------------ASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSV 149
SSS SS AS TS+ + +S ++ T S+ S + NS
Sbjct: 131 SSSITSSSLASSSITSSSLASSSTTSSSLASSSTNSTTSATPTSSATSSSLSSTAASNSA 190
Query: 150 AWNGARTTETLGITQRKNDDFSLST 174
+ ++ T SLS+
Sbjct: 191 TSSSLASSSLNSTTSATATSSSLSS 215
Score = 43.9 bits (102), Expect = 4e-04
Identities = 39/132 (29%), Positives = 65/132 (48%), Gaps = 6/132 (4%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S S +S+S S+ S + + +++ S L++ + S S + S +AS TT+S+
Sbjct: 101 SASPTSSSLTSSSATSSSLASSSTTSSSLASSSITSSSLASSSITSSSLASSSTTSSSLA 160
Query: 102 SSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTETLG 161
SSS S+ S PTS+ TS +SL +A SS L+ + + ++ A T+ +L
Sbjct: 161 SSSTNSTTSATPTSSATS-SSLSSTAASNSATSSSLASSSLNSTTSATA-----TSSSLS 214
Query: 162 ITQRKNDDFSLS 173
T N S S
Sbjct: 215 STAASNSATSSS 226
Score = 39.3 bits (90), Expect = 0.009
Identities = 39/141 (27%), Positives = 69/141 (48%), Gaps = 3/141 (2%)
Query: 9 TSSTRRGGMTVLGKVAVPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPS 68
+SS+ T A P +L S T + SS+++S+ SS++ ++ A +S S
Sbjct: 87 SSSSLASSSTNSTTSASPTSSSLTSSSATSSSLASSSTTSSSLASSSITSSSLASSSITS 146
Query: 69 HLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPS-SASGPPTSNQTSQTSLRPRS 127
S+ SS + + ++S ++ + T TS+ SSS S +AS TS+ + +SL +
Sbjct: 147 --SSLASSSTTSSSLASSSTNSTTSATPTSSATSSSLSSTAASNSATSSSLASSSLNSTT 204
Query: 128 AETRPGSSHLSRFAEHVGENS 148
+ T SS S A + +S
Sbjct: 205 SATATSSSLSSTAASNSATSS 225
Score = 37.7 bits (86), Expect = 0.027
Identities = 37/142 (26%), Positives = 64/142 (45%), Gaps = 9/142 (6%)
Query: 28 PINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSG 87
P + PS S +++SAS+ +S++ ASS S SS S T STS
Sbjct: 283 PSSTPSSTPLSSANSTTATSASSTPLTSVNSTTTTSASSTPLSSV--SSANSTTATSTSS 340
Query: 88 SDMASELTTTSAWGSSSRPSSASGPPTSNQTSQT------SLRPRSAETRPGSSHLSRFA 141
+ ++S + +T+A +SS P ++ T+ S T S SA + P +S S +
Sbjct: 341 TPLSS-VNSTTATSASSTPLTSVNSTTATSASSTPLTSVNSTSATSASSTPLTSANSTTS 399
Query: 142 EHVGENSVAWNGARTTETLGIT 163
V + ++N + T ++
Sbjct: 400 TSVSSTAPSYNTSSVLPTSSVS 421
Score = 37.0 bits (84), Expect = 0.047
Identities = 50/200 (25%), Positives = 80/200 (40%), Gaps = 43/200 (21%)
Query: 44 SSSSASNAWGSSLSPNANAGASSP-SHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGS 102
S++S ++ SS +P+ N + P S +S+ P S + T +++ S + + +T+A +
Sbjct: 507 SANSTTSTSVSSTAPSYNTSSVLPTSSVSSTPLSSANSTTATSASSTPLTSVNSTTATSA 566
Query: 103 SSRP-------SSASG--------------------PPTSNQTSQTSLRPRSAETRPG-- 133
SS P SSASG PTS S +++ P S T G
Sbjct: 567 SSTPFGNSTITSSASGSTGEFTNTNSGNGDVSGSVTTPTSTPLSNSTVAPTSTFTSSGFN 626
Query: 134 ----------SSHLSRFAEHVGENSVAWNGARTTETLGI--TQRKNDDFSLSTGDFPTLG 181
S+ LS + + S ++ +T T I T+ +TG PT G
Sbjct: 627 TTSGLPTSSVSTPLSNSSAYPSSGSSTFSRLSSTLTSSIIPTETFGSTSGSATGTRPT-G 685
Query: 182 SEKDKSVHDVELQGSLLCIT 201
S SV GS + T
Sbjct: 686 SSSQGSVVPTTSTGSSVTST 705
Score = 36.2 bits (82), Expect = 0.079
Identities = 35/130 (26%), Positives = 61/130 (46%), Gaps = 3/130 (2%)
Query: 9 TSSTRRGGMTVLGKVAVPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPS 68
TSS+ T +A + ++ S +SSS +++ +S S N+ A+ S
Sbjct: 115 TSSSLASSSTTSSSLASSSITSSSLASSSITSSSLASSSTTSSSLASSSTNSTTSATPTS 174
Query: 69 HLSARPSSGGSGTRPSTSGSDMASEL--TTTSAWGSSSRPS-SASGPPTSNQTSQTSLRP 125
++ S + + +TS S +S L TT++ SSS S +AS TS+ + +SL
Sbjct: 175 SATSSSLSSTAASNSATSSSLASSSLNSTTSATATSSSLSSTAASNSATSSSLASSSLNS 234
Query: 126 RSAETRPGSS 135
++ T SS
Sbjct: 235 TTSATATSSS 244
Score = 35.4 bits (80), Expect = 0.14
Identities = 27/95 (28%), Positives = 49/95 (51%), Gaps = 1/95 (1%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S + ++SN+ ++ S +A + ++ + S PSS S T S++ S A+ ++T
Sbjct: 251 SSTPLTSSNSTTAATSASATSSSAQYNTSSLLPSSTPSSTPLSSANSTTATSASSTPLTS 310
Query: 102 -SSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSS 135
+S+ +SAS P S+ +S S S + P SS
Sbjct: 311 VNSTTTTSASSTPLSSVSSANSTTATSTSSTPLSS 345
Score = 35.4 bits (80), Expect = 0.14
Identities = 32/98 (32%), Positives = 49/98 (49%), Gaps = 4/98 (4%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHL-SARP-SSGGSGTRPSTSGSDMASELTTTSA 99
S S+++A++A +S S N + PS S+ P SS S T S S + + S +TT+
Sbjct: 258 SNSTTAATSASATSSSAQYNTSSLLPSSTPSSTPLSSANSTTATSASSTPLTSVNSTTTT 317
Query: 100 WGSSSRPSSASGP--PTSNQTSQTSLRPRSAETRPGSS 135
SS+ SS S T+ TS T L ++ T +S
Sbjct: 318 SASSTPLSSVSSANSTTATSTSSTPLSSVNSTTATSAS 355
Score = 34.7 bits (78), Expect = 0.23
Identities = 24/104 (23%), Positives = 52/104 (49%), Gaps = 10/104 (9%)
Query: 42 SKSSSSASN----------AWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMA 91
S +++SAS+ A +S +P ++ +++ + S+ P + + T +++ S
Sbjct: 430 STTATSASSTPLSSVNSTTATSASSTPLSSVNSTTATSASSTPLTSVNSTTATSASSTPL 489
Query: 92 SELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSS 135
+ + +TSA +SS P +++ TS S T+ ++ P SS
Sbjct: 490 TSVNSTSATSASSTPLTSANSTTSTSVSSTAPSYNTSSVLPTSS 533
Score = 33.9 bits (76), Expect = 0.39
Identities = 32/117 (27%), Positives = 49/117 (41%), Gaps = 22/117 (18%)
Query: 41 GSKSSSSASNAWGSSLSPNANAGASSPSHLSARP-------------SSGGSGTRPSTSG 87
GS + S S+ SS+ P G++S S RP S+G S T T
Sbjct: 650 GSSTFSRLSSTLTSSIIPTETFGSTSGSATGTRPTGSSSQGSVVPTTSTGSSVTSTGTGT 709
Query: 88 SDMASELTTTSAW------GSSSRPSSASGPPTSNQTSQTSLRP---RSAETRPGSS 135
+ +E+T TS + S+ P++ASG N T+ + P ET G++
Sbjct: 710 TTGVTEVTETSTFETTEIITSTIEPTTASGTGGGNPTAAPTNEPTVTTGTETTEGTA 766
Score = 33.1 bits (74), Expect = 0.67
Identities = 32/119 (26%), Positives = 57/119 (47%), Gaps = 3/119 (2%)
Query: 42 SKSSSSASNAWGSS--LSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSA 99
S SS++ASN+ SS S + N+ S+ + S+ S+ S + S+S + + TT++
Sbjct: 180 SLSSTAASNSATSSSLASSSLNSTTSATATSSSLSSTAASNSATSSSLASSSLNSTTSAT 239
Query: 100 WGSSSRPSS-ASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTT 157
SSS S+ +S P ++ S T+ SA + + S +S + A +T
Sbjct: 240 ATSSSISSTVSSSTPLTSSNSTTAATSASATSSSAQYNTSSLLPSSTPSSTPLSSANST 298
Score = 32.0 bits (71), Expect = 1.5
Identities = 28/95 (29%), Positives = 51/95 (53%), Gaps = 4/95 (4%)
Query: 44 SSSSASNAWGSSLSPNANAGASSPSHLSARP-SSGGSGTRPSTSGSDMASELTTTSAWGS 102
SS +++ A +S +P + +++ + S+ P +S S + S S + + S +TTS S
Sbjct: 344 SSVNSTTATSASSTPLTSVNSTTATSASSTPLTSVNSTSATSASSTPLTSANSTTSTSVS 403
Query: 103 SSRPS--SASGPPTSNQTSQTSLRPRSAETRPGSS 135
S+ PS ++S PTS+ S T L ++ T +S
Sbjct: 404 STAPSYNTSSVLPTSS-VSSTPLSSANSTTATSAS 437
Score = 32.0 bits (71), Expect = 1.5
Identities = 28/95 (29%), Positives = 51/95 (53%), Gaps = 4/95 (4%)
Query: 44 SSSSASNAWGSSLSPNANAGASSPSHLSARP-SSGGSGTRPSTSGSDMASELTTTSAWGS 102
SS +++ A +S +P + +++ + S+ P +S S + S S + + S +TTS S
Sbjct: 458 SSVNSTTATSASSTPLTSVNSTTATSASSTPLTSVNSTSATSASSTPLTSANSTTSTSVS 517
Query: 103 SSRPS--SASGPPTSNQTSQTSLRPRSAETRPGSS 135
S+ PS ++S PTS+ S T L ++ T +S
Sbjct: 518 STAPSYNTSSVLPTSS-VSSTPLSSANSTTATSAS 551
Score = 32.0 bits (71), Expect = 1.5
Identities = 36/110 (32%), Positives = 55/110 (49%), Gaps = 18/110 (16%)
Query: 37 TLGWGSKSSSSASNAWGSSLSPNANAGASSPSHL-SARPSSGGSGTRPSTSGSDMASELT 95
+L S +S++++ A SSLS A + +++ S L S+ +S S T S+S S S T
Sbjct: 194 SLASSSLNSTTSATATSSSLSSTAASNSATSSSLASSSLNSTTSATATSSSISSTVSSST 253
Query: 96 -------TTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLS 138
TT+A +S+ SSA Q + +SL P S P S+ LS
Sbjct: 254 PLTSSNSTTAATSASATSSSA-------QYNTSSLLPSST---PSSTPLS 293
Score = 30.0 bits (66), Expect = 5.7
Identities = 35/144 (24%), Positives = 59/144 (40%), Gaps = 22/144 (15%)
Query: 46 SSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSS- 104
++AS G + P A A + P+ + ++ G+ T + TTT G+++
Sbjct: 735 TTASGTGGGN--PTA-APTNEPTVTTGTETTEGTATYTEPTTFTSTFSFTTTIIGGTTTI 791
Query: 105 -------RPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVA-WNGART 156
SS S PPT TS P GS + S G N+ + WN + +
Sbjct: 792 IPVNPGNPSSSVSAPPT------TSFTPGPG----GSGYPSYSNTTQGMNTTSIWNSSNS 841
Query: 157 TETLGITQRKNDDFSLSTGDFPTL 180
T +T + +++TGD T+
Sbjct: 842 TIVSNVTATITGNVTITTGDLTTI 865
>YM96_YEAST (Q04893) Hypothetical 113.1 kDa protein in PRE5-FET4
intergenic region
Length = 1140
Score = 44.3 bits (103), Expect = 3e-04
Identities = 46/172 (26%), Positives = 82/172 (46%), Gaps = 7/172 (4%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELT-TTSAW 100
S++ SS S++ S +S ++ SS + L+ SS S PS++ S ++SE++ TTS+
Sbjct: 310 SEAPSSTSSSVSSEISSTTSSSVSSEAPLAT--SSVVSSEAPSSTSSSVSSEISSTTSSS 367
Query: 101 GSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTETL 160
SS P + S +S S TS S SS +S A +SV+ + T ++
Sbjct: 368 VSSEAPLATSSVVSSEAPSSTSSSVSSEAPSSTSSSVSSEAPSSTSSSVSSEISSTKSSV 427
Query: 161 GITQRKNDDFSLSTGDFP----TLGSEKDKSVHDVELQGSLLCITIISIDSS 208
++ + SL + + P +L S + S + + +L+ S+ SS
Sbjct: 428 MSSEVSSATSSLVSSEAPSAISSLASSRLFSSKNTSVTSTLVATEASSVTSS 479
Score = 41.2 bits (95), Expect = 0.002
Identities = 49/181 (27%), Positives = 76/181 (41%), Gaps = 8/181 (4%)
Query: 10 SSTRRGGMTVLGKVAVPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASS--P 67
S T T+L + P PS S SS AS++ SS+S A SS
Sbjct: 243 SETLPFSSTILSITSSPVSSEAPSATS-----SSVSSEASSSTSSSVSSEAPLATSSVVS 297
Query: 68 SHLSARPSSGGSGTRPSTSGSDMASELT-TTSAWGSSSRPSSASGPPTSNQTSQTSLRPR 126
S + SS S PS++ S ++SE++ TTS+ SS P + S +S S TS
Sbjct: 298 SEAPSSTSSVVSSEAPSSTSSSVSSEISSTTSSSVSSEAPLATSSVVSSEAPSSTSSSVS 357
Query: 127 SAETRPGSSHLSRFAEHVGENSVAWNGARTTETLGITQRKNDDFSLSTGDFPTLGSEKDK 186
S + SS +S A + V+ +T + ++ + S + + P+ S
Sbjct: 358 SEISSTTSSSVSSEAPLATSSVVSSEAPSSTSSSVSSEAPSSTSSSVSSEAPSSTSSSVS 417
Query: 187 S 187
S
Sbjct: 418 S 418
Score = 34.3 bits (77), Expect = 0.30
Identities = 43/161 (26%), Positives = 62/161 (37%), Gaps = 14/161 (8%)
Query: 33 SQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSP--SHLSARPSSGGSGTRPSTSGSDM 90
S L S SS A ++ SS+S A + SS S + SS S ST S M
Sbjct: 369 SSEAPLATSSVVSSEAPSSTSSSVSSEAPSSTSSSVSSEAPSSTSSSVSSEISSTKSSVM 428
Query: 91 ASELTT-TSAWGSSSRPSSASGPPTSNQTSQ-----------TSLRPRSAETRPGSSHLS 138
+SE+++ TS+ SS PS+ S +S S T ++ RP S L+
Sbjct: 429 SSEVSSATSSLVSSEAPSAISSLASSRLFSSKNTSVTSTLVATEASSVTSSLRPSSETLA 488
Query: 139 RFAEHVGENSVAWNGARTTETLGITQRKNDDFSLSTGDFPT 179
+ S +N +T T + S S T
Sbjct: 489 SNSIIESSLSTGYNSTVSTTTSAASSTLGSKVSSSNSRMAT 529
Score = 33.5 bits (75), Expect = 0.52
Identities = 41/125 (32%), Positives = 63/125 (49%), Gaps = 11/125 (8%)
Query: 42 SKSSSSASNAWGSSLSPNANAGA-SSPSHLSARPSSGG-SGTRPSTSG-SDMASELTTTS 98
+ SSS +S+ S LS + SS S+ S+ SS + STSG S+ S L++T+
Sbjct: 67 TSSSSLSSDTIASILSSESLVSIFSSLSYTSSDISSTSVNDVESSTSGPSNSYSALSSTN 126
Query: 99 AWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTE 158
A SSS + S S+ QTS S+ GSS +E +G++SV A +++
Sbjct: 127 AQLSSSTTETDS---ISSSAIQTSSPQTSSSNGGGSS-----SEPLGKSSVLETTASSSD 178
Query: 159 TLGIT 163
T +T
Sbjct: 179 TTAVT 183
Score = 33.1 bits (74), Expect = 0.67
Identities = 41/165 (24%), Positives = 63/165 (37%), Gaps = 17/165 (10%)
Query: 44 SSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSS 103
S+S SN++ + S NA +SS + + SS + P TS S+ GSS
Sbjct: 111 STSGPSNSYSALSSTNAQL-SSSTTETDSISSSAIQTSSPQTSSSN---------GGGSS 160
Query: 104 SRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTETLGIT 163
S P S + +S S T SS + + ++ +G+ T +
Sbjct: 161 SEPLGKSSVLETTASS-------SDTTAVTSSTFTTLTDVSSSPKISSSGSAVTSVGTTS 213
Query: 164 QRKNDDFSLSTGDFPTLGSEKDKSVHDVELQGSLLCITIISIDSS 208
+ FS ST D +L S + TI+SI SS
Sbjct: 214 DASKEVFSSSTSDVSSLLSSTSSPASSTISETLPFSSTILSITSS 258
Score = 29.3 bits (64), Expect = 9.7
Identities = 15/46 (32%), Positives = 27/46 (58%)
Query: 77 GGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTS 122
G SG++ +T+ + S TTTS+ S++ P++ S T++ TS
Sbjct: 2 GSSGSKSTTATTTSHSSTTTTSSTTSTTTPTTTSTTSTTSTKVTTS 47
>VIT2_FUNHE (Q98893) Vitellogenin II precursor (VTG II) [Contains:
Lipovitellin 1 (LV1); Phosvitin (PV); Lipovitellin 2
(LV2)]
Length = 1687
Score = 44.3 bits (103), Expect = 3e-04
Identities = 30/96 (31%), Positives = 49/96 (50%)
Query: 27 KPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTS 86
K I P + + S SS S+ ++ S S ++++ +SS S S+ S S R ++
Sbjct: 1074 KKILSPGLKNSTKASSSSSGSSRSSRSRSSSSSSSSSSSSSSRSSSSSSRSSSSLRRNSK 1133
Query: 87 GSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTS 122
D+A L TS SSS SS+S +S+ +S +S
Sbjct: 1134 MLDLADPLNITSKRSSSSSSSSSSSSSSSSSSSSSS 1169
Score = 30.8 bits (68), Expect = 3.3
Identities = 27/105 (25%), Positives = 50/105 (46%), Gaps = 18/105 (17%)
Query: 64 ASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSS------------------SR 105
ASS S S+R S S + S+S S +S +++S+ SS S+
Sbjct: 1087 ASSSSSGSSRSSRSRSSSSSSSSSSSSSSRSSSSSSRSSSSLRRNSKMLDLADPLNITSK 1146
Query: 106 PSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVA 150
SS+S +S+ +S +S S++T+ + +H+ ++SV+
Sbjct: 1147 RSSSSSSSSSSSSSSSSSSSSSSKTKWQLHERNFTKDHIHQHSVS 1191
>SER1_GALME (O96614) Sericin-1 (Silk gum protein 1) (Fragment)
Length = 115
Score = 44.3 bits (103), Expect = 3e-04
Identities = 33/98 (33%), Positives = 51/98 (51%), Gaps = 1/98 (1%)
Query: 41 GSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAW 100
GS SS +S + GSS S + ++G+S S S S GS +SGS +S + +S
Sbjct: 16 GSSGSSGSSGSSGSSGS-SGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGS 74
Query: 101 GSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLS 138
SS S +S +SN + +S +S+ ++ GSS S
Sbjct: 75 SGSSGSSGSSSSSSSNGSGSSSGSSQSSGSQGGSSSTS 112
Score = 42.4 bits (98), Expect = 0.001
Identities = 28/82 (34%), Positives = 45/82 (54%), Gaps = 1/82 (1%)
Query: 41 GSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAW 100
GS SS +S + GSS S ++ + S + SSG SG+ S SGS +S ++++
Sbjct: 34 GSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGS-SGSSGSSSSSSSNGS 92
Query: 101 GSSSRPSSASGPPTSNQTSQTS 122
GSSS S +SG + ++ +S
Sbjct: 93 GSSSGSSQSSGSQGGSSSTSSS 114
Score = 40.4 bits (93), Expect = 0.004
Identities = 26/95 (27%), Positives = 43/95 (44%)
Query: 41 GSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAW 100
GS SS +S + GSS S ++ + S + SSG SG+ S+ S + ++ +
Sbjct: 1 GSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSS 60
Query: 101 GSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSS 135
GSS S+ +S + + S+ GSS
Sbjct: 61 GSSGSSGSSGSSGSSGSSGSSGSSSSSSSNGSGSS 95
Score = 40.0 bits (92), Expect = 0.006
Identities = 34/98 (34%), Positives = 49/98 (49%), Gaps = 2/98 (2%)
Query: 41 GSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAW 100
GS SS +S + GSS S + ++G+S S S S GS +SGS +S + +S
Sbjct: 7 GSSGSSGSSGSSGSSGS-SGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGS 65
Query: 101 GSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLS 138
SS S +SG S+ +S +S S + GSS S
Sbjct: 66 SGSSGSSGSSGSSGSSGSSSSSSSNGSGSS-SGSSQSS 102
Score = 33.1 bits (74), Expect = 0.67
Identities = 20/63 (31%), Positives = 30/63 (46%)
Query: 36 GTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELT 95
G+ G S SS S+ S + ++G+S S S+ SG S +SGS S T
Sbjct: 52 GSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSSSSSSNGSGSSSGSSQSSGSQGGSSST 111
Query: 96 TTS 98
++S
Sbjct: 112 SSS 114
Score = 32.3 bits (72), Expect = 1.1
Identities = 19/49 (38%), Positives = 27/49 (54%)
Query: 41 GSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSD 89
GS SS +S + GSS S ++++ S S + SSG G STS S+
Sbjct: 67 GSSGSSGSSGSSGSSGSSSSSSSNGSGSSSGSSQSSGSQGGSSSTSSSN 115
>YG1F_YEAST (P53214) Hypothetical 57.5 kDa protein in VMA7-RPS25A
intergenic region
Length = 551
Score = 43.9 bits (102), Expect = 4e-04
Identities = 49/171 (28%), Positives = 81/171 (46%), Gaps = 12/171 (7%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S +S SA ++ SS ++A S+PS S +S + T S ++SEL+++S
Sbjct: 51 SITSLSAVESFTSSTDATSSASLSTPSIASVSFTSFPQSSSLLTLSSTLSSELSSSSMQV 110
Query: 102 SSSRPSSASGPPTSNQTSQTSLRPRSAETR--PGSSHLSRFAEHVGENSVAWNGARTTET 159
SSS SS+S TS+ +S +S+ P S+ + SS L F ++VA + T+ +
Sbjct: 111 SSSSTSSSSSEVTSS-SSSSSISPSSSSSTIISSSSSLPTFTVASTSSTVASSTLSTSSS 169
Query: 160 LGITQRKNDDFSLSTGDFPTLGSEKDKSVHDVELQGSLLCITIISIDSSAA 210
L I S S+ F T SE S+ + S+ ++ SS +
Sbjct: 170 LVI--------STSSSTF-TFSSESSSSLISSSISTSVSTSSVYVPSSSTS 211
Score = 43.5 bits (101), Expect = 5e-04
Identities = 42/138 (30%), Positives = 65/138 (46%), Gaps = 11/138 (7%)
Query: 28 PINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSG 87
P + S+ + + S SSSS ++ SS S ++++ +SS S S+ SS S STS
Sbjct: 213 PPSSSSELTSSSYSSSSSSSTLFSYSSSFSSSSSSSSSSSSSSSSSSSSSSSYFTLSTSS 272
Query: 88 SDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGS--SHLSRFAEHVG 145
S +S + SSS PS +S +SN TS + S+ P S S+L++ +
Sbjct: 273 S--------SSIYSSSSYPSFSSS-SSSNPTSSITSTSASSSITPASEYSNLAKTITSII 323
Query: 146 ENSVAWNGARTTETLGIT 163
E + TT T T
Sbjct: 324 EGQTILSNYYTTITYSPT 341
Score = 37.0 bits (84), Expect = 0.047
Identities = 31/94 (32%), Positives = 47/94 (49%), Gaps = 2/94 (2%)
Query: 44 SSSSASNAWGSSLSPNANAGAS--SPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
SS S+S+ SS+S + + + S S+ PSS T S S S +S L + S+
Sbjct: 182 SSESSSSLISSSISTSVSTSSVYVPSSSTSSPPSSSSELTSSSYSSSSSSSTLFSYSSSF 241
Query: 102 SSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSS 135
SSS SS+S +S+ +S +S + T SS
Sbjct: 242 SSSSSSSSSSSSSSSSSSSSSSSYFTLSTSSSSS 275
Score = 32.0 bits (71), Expect = 1.5
Identities = 35/142 (24%), Positives = 67/142 (46%), Gaps = 6/142 (4%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S SSS + S+ S A++ S+ S L SS +S S ++S ++T+ +
Sbjct: 142 SSSSSLPTFTVASTSSTVASSTLSTSSSLVISTSSSTFTFSSESSSSLISSSISTSVSTS 201
Query: 102 SSSRPSSA-SGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTETL 160
S PSS+ S PP+S+ +S S+ + S+ S F+ +S + + + ++ +
Sbjct: 202 SVYVPSSSTSSPPSSSSELTSSSYSSSSSSSTLFSYSSSFS-----SSSSSSSSSSSSSS 256
Query: 161 GITQRKNDDFSLSTGDFPTLGS 182
+ + F+LST ++ S
Sbjct: 257 SSSSSSSSYFTLSTSSSSSIYS 278
Score = 31.2 bits (69), Expect = 2.6
Identities = 36/145 (24%), Positives = 62/145 (41%), Gaps = 1/145 (0%)
Query: 45 SSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSS 104
SSS+S SS S AS+ S +++ S S STS S ++S+ SSS
Sbjct: 134 SSSSSTIISSSSSLPTFTVASTSSTVASSTLSTSSSLVISTSSSTFTFSSESSSSLISSS 193
Query: 105 RPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTETLGITQ 164
+S S +S TS P S+ SS+ S + S + + + ++ + +
Sbjct: 194 ISTSVSTSSVYVPSSSTSSPPSSSSELTSSSYSSSSSSST-LFSYSSSFSSSSSSSSSSS 252
Query: 165 RKNDDFSLSTGDFPTLGSEKDKSVH 189
+ S S+ + TL + S++
Sbjct: 253 SSSSSSSSSSSSYFTLSTSSSSSIY 277
Score = 30.4 bits (67), Expect = 4.4
Identities = 31/113 (27%), Positives = 55/113 (48%), Gaps = 5/113 (4%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S SSS+ + + SS S +++ ++S S S S + + PS+S SELT++S
Sbjct: 173 STSSSTFTFSSESSSSLISSSISTSVSTSSVYVPSSSTSSPPSSS-----SELTSSSYSS 227
Query: 102 SSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGA 154
SSS + S + + +S +S S+ + SS S F +S ++ +
Sbjct: 228 SSSSSTLFSYSSSFSSSSSSSSSSSSSSSSSSSSSSSYFTLSTSSSSSIYSSS 280
>VGLX_EHV1B (P28968) Glycoprotein X precursor
Length = 797
Score = 43.9 bits (102), Expect = 4e-04
Identities = 41/143 (28%), Positives = 62/143 (42%), Gaps = 18/143 (12%)
Query: 33 SQRGTLGWGSKSSSSASNAWGS--------SLSPNANAGASSPSHLSARPSSGG--SGTR 82
S T G G +SS +N+ S S SP + SSPS S + SS S +
Sbjct: 31 SSSSTSGSGQSTSSGTTNSSSSPTTSPPTTSSSPPTSTHTSSPSSTSTQSSSTAATSSSA 90
Query: 83 PSTSGS------DMASELTTTSAWGSSSRPSSASGPPT--SNQTSQTSLRPRSAETRPGS 134
PST+ S ++E TTT+ S++ P++ + PT + T+ T+ SAET +
Sbjct: 91 PSTASSTTSIPTSTSTETTTTTPTASTTTPTTTTAAPTTAATTTAVTTAASTSAETTTAT 150
Query: 135 SHLSRFAEHVGENSVAWNGARTT 157
+ + S A TT
Sbjct: 151 ATATSTPTTTTPTSTTTTTATTT 173
Score = 43.1 bits (100), Expect = 7e-04
Identities = 31/92 (33%), Positives = 46/92 (49%), Gaps = 4/92 (4%)
Query: 44 SSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSS 103
+SSS+++ G S S +SSP+ S +S T TS S ++++A SS
Sbjct: 30 TSSSSTSGSGQSTSSGTTNSSSSPT-TSPPTTSSSPPTSTHTSSPSSTSTQSSSTAATSS 88
Query: 104 SRPSSASGP---PTSNQTSQTSLRPRSAETRP 132
S PS+AS PTS T T+ P ++ T P
Sbjct: 89 SAPSTASSTTSIPTSTSTETTTTTPTASTTTP 120
Score = 35.0 bits (79), Expect = 0.18
Identities = 29/97 (29%), Positives = 48/97 (48%), Gaps = 5/97 (5%)
Query: 44 SSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSS 103
++++A+ GS S + + +S S SA ++ + T STS + S T TSA S+
Sbjct: 252 ATTTAATTTGSPTSGSTSTTGASTSTPSASTATSATPTSTSTSAAATTSTPTPTSAATSA 311
Query: 104 SRPSSASGPPTSNQTSQTSL--RPRSAETRPGSSHLS 138
+ A PTS T+ T+ +A T P S+ +S
Sbjct: 312 ESTTEA---PTSTPTTDTTTPSEATTATTSPESTTVS 345
Score = 34.7 bits (78), Expect = 0.23
Identities = 36/183 (19%), Positives = 72/183 (38%), Gaps = 4/183 (2%)
Query: 32 PSQRGTLGWGSKSSSSASNAWGSSLSPN-ANAGASSPSHLSARPSSG---GSGTRPSTSG 87
P+ T S S+ S+S A SS +P+ A++ S P+ S ++ S T P+T+
Sbjct: 65 PTSTHTSSPSSTSTQSSSTAATSSSAPSTASSTTSIPTSTSTETTTTTPTASTTTPTTTT 124
Query: 88 SDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGEN 147
+ + TTT+ ++S + + + ++ T+ P S T ++ + A +
Sbjct: 125 AAPTTAATTTAVTTAASTSAETTTATATATSTPTTTTPTSTTTTTATTTVPTTASTTTDT 184
Query: 148 SVAWNGARTTETLGITQRKNDDFSLSTGDFPTLGSEKDKSVHDVELQGSLLCITIISIDS 207
+ A T T T + +T T + + + T + +
Sbjct: 185 TTAATTTAATTTAATTTAATTTAATTTAATTTAATTTAATTSSATTAATTTAATTTAATT 244
Query: 208 SAA 210
+AA
Sbjct: 245 TAA 247
Score = 32.0 bits (71), Expect = 1.5
Identities = 40/187 (21%), Positives = 71/187 (37%), Gaps = 21/187 (11%)
Query: 9 TSSTRRGGMTVLGKVAVPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPS 68
T++T T P+ T G+ S+S+ S + +S +P + + +++ +
Sbjct: 240 TAATTTAATTTAATTTAATTTGSPTSGSTSTTGA-STSTPSASTATSATPTSTSTSAAAT 298
Query: 69 HLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSA 128
+ P+S + +T E T++ ++ PS A+ TS +++ S SA
Sbjct: 299 TSTPTPTSAATSAESTT-------EAPTSTPTTDTTTPSEATTATTSPESTTVSASTTSA 351
Query: 129 ETR--PGSSHLSRFAEHVGENSVAWNGARTTETLGITQRKNDDF----------SLSTGD 176
T SH S + G S A + T T D F S +
Sbjct: 352 TTTAFTTESHTSPDSS-TGSTSTAEPSSTFTLTPSTATPSTDQFTGSSASTESDSTDSST 410
Query: 177 FPTLGSE 183
PT G+E
Sbjct: 411 VPTTGTE 417
Score = 32.0 bits (71), Expect = 1.5
Identities = 22/95 (23%), Positives = 46/95 (48%), Gaps = 5/95 (5%)
Query: 44 SSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSS 103
++++A+ ++ + A ++ + +A ++ + T +T+GS + +TT A S+
Sbjct: 218 ATTTAATTSSATTAATTTAATTTAATTTAATTTAATTTAATTTGSPTSGSTSTTGA--ST 275
Query: 104 SRPSSA---SGPPTSNQTSQTSLRPRSAETRPGSS 135
S PS++ S PTS TS + T +S
Sbjct: 276 STPSASTATSATPTSTSTSAAATTSTPTPTSAATS 310
Score = 31.2 bits (69), Expect = 2.6
Identities = 28/99 (28%), Positives = 49/99 (49%), Gaps = 6/99 (6%)
Query: 37 TLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTT 96
T+ + S+++ + S SP+++ G++S +A PSS + T PST+ + + T
Sbjct: 343 TVSASTTSATTTAFTTESHTSPDSSTGSTS----TAEPSSTFTLT-PSTA-TPSTDQFTG 396
Query: 97 TSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSS 135
+SA S S++ P T ++ S A T GSS
Sbjct: 397 SSASTESDSTDSSTVPTTGTESITESSSTTEASTNLGSS 435
Score = 30.4 bits (67), Expect = 4.4
Identities = 28/144 (19%), Positives = 61/144 (41%), Gaps = 11/144 (7%)
Query: 44 SSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASEL-------TT 96
++++A+ ++ + A ++ + + P+SG + T +++ + AS T+
Sbjct: 233 TTTAATTTAATTTAATTTAATTTAATTTGSPTSGSTSTTGASTSTPSASTATSATPTSTS 292
Query: 97 TSAWGSSSRPSSASGPPTSNQTSQT-SLRPRSAETRPGSSHLSRFAEHVGENSVAWNGAR 155
TSA ++S P+ S ++ T++ + P + T P + + + S + A
Sbjct: 293 TSAAATTSTPTPTSAATSAESTTEAPTSTPTTDTTTPSEATTATTSPESTTVSASTTSAT 352
Query: 156 TTETLGITQRKNDDFSLSTGDFPT 179
TT T + STG T
Sbjct: 353 TT---AFTTESHTSPDSSTGSTST 373
Score = 30.4 bits (67), Expect = 4.4
Identities = 18/52 (34%), Positives = 29/52 (55%), Gaps = 1/52 (1%)
Query: 87 GSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLS 138
GS +E TTTS+ +S S S T++ +S T+ P ++ + P S+H S
Sbjct: 21 GSTTTTE-TTTSSSSTSGSGQSTSSGTTNSSSSPTTSPPTTSSSPPTSTHTS 71
Score = 29.3 bits (64), Expect = 9.7
Identities = 30/118 (25%), Positives = 50/118 (41%), Gaps = 11/118 (9%)
Query: 63 GASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTS 122
G+++ + + SS + ++SG+ +S TTS +S S PPTS TS
Sbjct: 21 GSTTTTETTTSSSSTSGSGQSTSSGTTNSSSSPTTSPPTTS------SSPPTSTHTSS-- 72
Query: 123 LRPRSAETRPGS-SHLSRFAEHVGENSVAWNGARTTETLGITQRKNDDFSLSTGDFPT 179
P S T+ S + S A ++ + + +TET T + +T PT
Sbjct: 73 --PSSTSTQSSSTAATSSSAPSTASSTTSIPTSTSTETTTTTPTASTTTPTTTTAAPT 128
>MID2_YEAST (P36027) Mating process protein MID2 (Serine-rich
protein SMS1) (Protein kinase A interference protein)
Length = 376
Score = 43.9 bits (102), Expect = 4e-04
Identities = 35/107 (32%), Positives = 55/107 (50%), Gaps = 5/107 (4%)
Query: 33 SQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMAS 92
S R + S S+S AS ++ +S S ++++ SS S S+ SS S + STS + +S
Sbjct: 76 SSRSLVSHTSSSTSIASISF-TSFSFSSDSSTSSSSSASSDSSSSSSFSISSTSATSESS 134
Query: 93 ----ELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSS 135
+ +T+S+ SS PSS+S P T TS P + GS+
Sbjct: 135 TSSTQTSTSSSSSLSSTPSSSSSPSTITSAPSTSSTPSTTAYNQGST 181
Score = 38.5 bits (88), Expect = 0.016
Identities = 33/90 (36%), Positives = 50/90 (54%), Gaps = 6/90 (6%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S SS S++++ S+ S ++++ + S S SA S S T+ STS S S L++T +
Sbjct: 99 SFSSDSSTSSSSSASSDSSSSSSFSISSTSATSESSTSSTQTSTSSS---SSLSSTPS-- 153
Query: 102 SSSRPSS-ASGPPTSNQTSQTSLRPRSAET 130
SSS PS+ S P TS+ S T+ S T
Sbjct: 154 SSSSPSTITSAPSTSSTPSTTAYNQGSTIT 183
Score = 37.4 bits (85), Expect = 0.036
Identities = 27/78 (34%), Positives = 43/78 (54%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S SSS++S++ SS ++ A+S S S+ +S S + S++ S +S T TSA
Sbjct: 107 SSSSSASSDSSSSSSFSISSTSATSESSTSSTQTSTSSSSSLSSTPSSSSSPSTITSAPS 166
Query: 102 SSSRPSSASGPPTSNQTS 119
+SS PS+ + S TS
Sbjct: 167 TSSTPSTTAYNQGSTITS 184
Score = 31.6 bits (70), Expect = 2.0
Identities = 29/133 (21%), Positives = 60/133 (44%), Gaps = 4/133 (3%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S S+SSA + SS+S +++ + S + + S+ S STS + S +++++
Sbjct: 32 SSSNSSAVSTARSSVSRVSSSSSILSSSMVSSSSADSSSLTSSTSSRSLVSHTSSSTSIA 91
Query: 102 SSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTETLG 161
S S S + +S +S ++ S+++ SS E+S + T+ +
Sbjct: 92 SISFTSFSFSSDSSTSSSSSA----SSDSSSSSSFSISSTSATSESSTSSTQTSTSSSSS 147
Query: 162 ITQRKNDDFSLST 174
++ + S ST
Sbjct: 148 LSSTPSSSSSPST 160
>CHI3_CANAL (P40954) Chitinase 3 precursor (EC 3.2.1.14)
Length = 567
Score = 43.9 bits (102), Expect = 4e-04
Identities = 38/129 (29%), Positives = 61/129 (46%), Gaps = 3/129 (2%)
Query: 8 WTSSTRRGGMTVLGKVAVPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSP 67
W+++ G K V K P + T + ++++ S SS S + + S+
Sbjct: 293 WSNTNSNGNFVENMKAIVKKAS--PGEETTSSSTTTTTTTTSTTISSSSSSSKTSKTSTT 350
Query: 68 SHLSARPSSGGSGTRPSTSGSDMASELT-TTSAWGSSSRPSSASGPPTSNQTSQTSLRPR 126
S S+ SS S T STS S +S + TTS+ +SS+ S+ S PTS+ + +S
Sbjct: 351 STTSSSISSTTSSTTSSTSSSSTSSSTSSTTSSSTTSSQISTTSTAPTSSTSLSSSTIST 410
Query: 127 SAETRPGSS 135
SA T +S
Sbjct: 411 SASTSDTTS 419
Score = 35.8 bits (81), Expect = 0.10
Identities = 29/97 (29%), Positives = 45/97 (45%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S SSSS S++ S+ S + + S + + S+ S + STS S + T+S
Sbjct: 367 STSSSSTSSSTSSTTSSSTTSSQISTTSTAPTSSTSLSSSTISTSASTSDTTSVTSSETT 426
Query: 102 SSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLS 138
PSS S T S T+ +S+ T+P +S S
Sbjct: 427 PVVTPSSLSSAITIPGDSTTTGISKSSSTKPATSTTS 463
Score = 31.6 bits (70), Expect = 2.0
Identities = 28/135 (20%), Positives = 48/135 (34%), Gaps = 5/135 (3%)
Query: 40 WGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSA 99
+G S S W N N+ + ++ A G ++S + + T+T+
Sbjct: 281 FGGISMWDVSAGWS-----NTNSNGNFVENMKAIVKKASPGEETTSSSTTTTTTTTSTTI 335
Query: 100 WGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTET 159
SSS ++ TS +S S S + SS S +S + TT T
Sbjct: 336 SSSSSSSKTSKTSTTSTTSSSISSTTSSTTSSTSSSSTSSSTSSTTSSSTTSSQISTTST 395
Query: 160 LGITQRKNDDFSLST 174
+ ++ST
Sbjct: 396 APTSSTSLSSSTIST 410
Score = 29.3 bits (64), Expect = 9.7
Identities = 26/90 (28%), Positives = 40/90 (43%), Gaps = 6/90 (6%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S S+SAS + +S++ + +PS LS+ + G T S S T+T++
Sbjct: 406 STISTSASTSDTTSVTSSETTPVVTPSSLSSAITIPGDSTTTGISKSSSTKPATSTTSAL 465
Query: 102 SSSRPSSASGPP------TSNQTSQTSLRP 125
SSS + A+ P T T TS P
Sbjct: 466 SSSTTTVATIPDDKEIINTPTDTETTSKPP 495
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.312 0.125 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,786,143
Number of Sequences: 164201
Number of extensions: 1229544
Number of successful extensions: 9905
Number of sequences better than 10.0: 614
Number of HSP's better than 10.0 without gapping: 178
Number of HSP's successfully gapped in prelim test: 449
Number of HSP's that attempted gapping in prelim test: 6206
Number of HSP's gapped (non-prelim): 2745
length of query: 256
length of database: 59,974,054
effective HSP length: 108
effective length of query: 148
effective length of database: 42,240,346
effective search space: 6251571208
effective search space used: 6251571208
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC140849.12


