

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140774.7 + phase: 0 /pseudo
(998 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
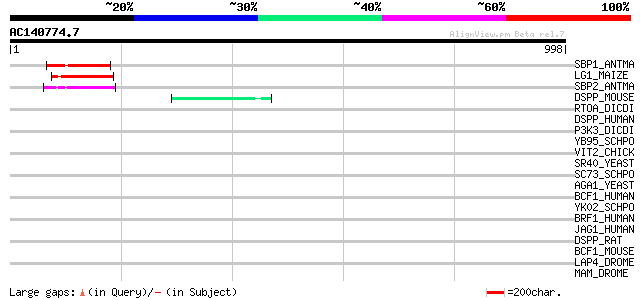
SBP1_ANTMA (Q38741) Squamosa-promoter binding protein 1 121 1e-26
LG1_MAIZE (O04003) LIGULELESS1 protein 120 1e-26
SBP2_ANTMA (Q38740) Squamosa-promoter binding protein 2 119 3e-26
DSPP_MOUSE (P97399) Dentin sialophosphoprotein precursor (Dentin... 45 7e-04
RTOA_DICDI (P54681) Protein rtoA (Ratio-A) 45 0.001
DSPP_HUMAN (Q9NZW4) Dentin sialophosphoprotein precursor [Contai... 44 0.002
P3K3_DICDI (P54675) Phosphatidylinositol 3-kinase 3 (EC 2.7.1.13... 42 0.006
YB95_SCHPO (O42970) Hypothetical serine-rich protein C1E8.05 in ... 41 0.018
VIT2_CHICK (P02845) Vitellogenin II precursor (Major vitellogeni... 40 0.041
SR40_YEAST (P32583) Suppressor protein SRP40 40 0.041
SC73_SCHPO (O13817) Protein transport protein sec73 39 0.053
AGA1_YEAST (P32323) A-agglutinin attachment subunit precursor 39 0.053
BCF1_HUMAN (Q9NYF8) Bcl-2-associated transcription factor 1 (Btf) 39 0.069
YK02_SCHPO (Q9HDY9) Hypothetical serine/threonine-rich protein P... 39 0.091
BRF1_HUMAN (P55201) Peregrin (Bromodomain and PHD finger-contain... 38 0.12
JAG1_HUMAN (P78504) Jagged 1 precursor (Jagged1) (hJ1) 38 0.15
DSPP_RAT (Q62598) Dentin sialophosphoprotein precursor [Contains... 38 0.15
BCF1_MOUSE (Q8K019) Bcl-2-associated transcription factor 1 (Btf) 38 0.15
LAP4_DROME (Q7KRY7) LAP4 protein (Scribble protein) (Smell-impai... 37 0.20
MAM_DROME (P21519) Neurogenic protein mastermind 37 0.26
>SBP1_ANTMA (Q38741) Squamosa-promoter binding protein 1
Length = 131
Score = 121 bits (303), Expect = 1e-26
Identities = 60/115 (52%), Positives = 74/115 (64%), Gaps = 4/115 (3%)
Query: 67 EPQQVEESLNLNLGSTGLVRPNKRIRSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVC 126
E + E+ + + T + P+ + SGS + CQV+NC +++ AK YHRRHKVC
Sbjct: 19 EQGEEEDDIGEDSKKTRALTPSGKRASGSTQRS----CQVENCAAEMTNAKPYHRRHKVC 74
Query: 127 EAHSKASKALLGNQMQRFCQQCSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPD 181
E H+KA L QRFCQQCSRFH L EFDE KRSCRRRLAGHN RRRK+ D
Sbjct: 75 EFHAKAPVVLHSGLQQRFCQQCSRFHELSEFDEAKRSCRRRLAGHNERRRKSSHD 129
>LG1_MAIZE (O04003) LIGULELESS1 protein
Length = 399
Score = 120 bits (302), Expect = 1e-26
Identities = 63/113 (55%), Positives = 71/113 (62%), Gaps = 5/113 (4%)
Query: 75 LNLNLGSTGLVRPNKRIRSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKASK 134
L LNLG P R G P CQ + CK DLS AK YHRRHKVCE HSKA
Sbjct: 160 LGLNLGYRTYFPP----RGGYTYGHHPPRCQAEGCKADLSSAKRYHRRHKVCEHHSKAPV 215
Query: 135 ALL-GNQMQRFCQQCSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPDEVAVG 186
+ G QRFCQQCSRFH L EFD+ K+SCR+RLA HNRRRRK++P + G
Sbjct: 216 VVTAGGLHQRFCQQCSRFHLLDEFDDAKKSCRKRLADHNRRRRKSKPSDADAG 268
>SBP2_ANTMA (Q38740) Squamosa-promoter binding protein 2
Length = 171
Score = 119 bits (299), Expect = 3e-26
Identities = 65/128 (50%), Positives = 76/128 (58%), Gaps = 2/128 (1%)
Query: 62 KSTTVEPQQVEESLNLNLGSTGLVRPNKRIRSGSPTSASYPMCQVDNCKEDLSKAKDYHR 121
K E Q ++ LNL GS G K SG A P C V+NC DL K Y++
Sbjct: 45 KVNFAESQLKKKKLNLGEGSGGK-SGEKHTASGGGVVAQ-PCCLVENCGADLRNCKKYYQ 102
Query: 122 RHKVCEAHSKASKALLGNQMQRFCQQCSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPD 181
RH+VCE H+KA + MQRFCQQCSRFH L EFD+ KRSCRRRLAGHN RRRK+ +
Sbjct: 103 RHRVCEVHAKAPVVSVEGLMQRFCQQCSRFHDLSEFDQTKRSCRRRLAGHNERRRKSSLE 162
Query: 182 EVAVGGSP 189
G SP
Sbjct: 163 SHKEGRSP 170
>DSPP_MOUSE (P97399) Dentin sialophosphoprotein precursor (Dentin
matrix protein-3) (DMP-3) [Contains: Dentin
phosphoprotein (Dentin phosphophoryn) (DPP); Dentin
sialoprotein (DSP)]
Length = 934
Score = 45.4 bits (106), Expect = 7e-04
Identities = 44/180 (24%), Positives = 69/180 (37%), Gaps = 8/180 (4%)
Query: 291 STTPSTPARNGGNGSTSSADHMRERSSGSSQSPNDDSDCQEDVRVKLPLQLFGSSPENDS 350
S + S+ + + + S+ S+D S S +D SD + GSS +DS
Sbjct: 544 SKSDSSDSSDDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSNSSSDSSDSSGSSDSSDS 603
Query: 351 PSKLPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKE 410
SS SS+SS+ D S S + + +S+ S +++
Sbjct: 604 SDTCDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSSSSDSSDSSSCSDSSDSSDS 663
Query: 411 TSQSHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPPSLNSDTQDRTG 470
+ S S + S S+N S S S SS SD S S +SD+ D +G
Sbjct: 664 SDSSDSSDSSDSSSSDSSSSSNSSDSSDSSDS--------SSSSDSSDSSDSSDSSDSSG 715
Score = 38.9 bits (89), Expect = 0.069
Identities = 43/177 (24%), Positives = 68/177 (38%), Gaps = 19/177 (10%)
Query: 291 STTPSTPARNGGNGSTSSADHMRERSSGSSQSPNDDSDCQEDVRVKLPLQLFGSSPENDS 350
S+ S+ + + S+ S+D SS S +D SD + GSS +DS
Sbjct: 672 SSDSSSSDSSSSSNSSDSSDSSDSSSSSDSSDSSDSSDSSDSS---------GSSDSSDS 722
Query: 351 PSKLPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKE 410
+ SS SS+SS+ D S S + + NS+ S + + +
Sbjct: 723 SASSDSSSSSDSSDSSSSSDSSDSSDSSDSSDSSESSDSS----NSSDSSDSSDSSDSSD 778
Query: 411 TSQSHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPPSLNSDTQD 467
+S S + SN+ SS S + +S SD S S +SD+ D
Sbjct: 779 SSDSSDSSD------SSDSSNSSDSSDSSDSSDSSDSSNSSDSSDSSDSSDSSDSSD 829
>RTOA_DICDI (P54681) Protein rtoA (Ratio-A)
Length = 400
Score = 44.7 bits (104), Expect = 0.001
Identities = 50/180 (27%), Positives = 72/180 (39%), Gaps = 19/180 (10%)
Query: 291 STTPSTPARNGGNGSTSSADHMRERSSGSSQSPNDDSDCQEDVRVKLPLQLFGSSPENDS 350
S + S + + +GS SS D S GS+ S N S D SS
Sbjct: 118 SNSGSEASGSSNSGSQSSTDSSNSGSQGSTGSSNSGSQSSTDSSNSGSQSSTDSSNSGSQ 177
Query: 351 PSKLPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKE 410
S S+ SS SSN E + SS N G + NS S+ N+ E
Sbjct: 178 GSTGSSNSGSESSGSSNSGSESSGSS-------NSGSESSSGSSNSGSESSSGSSNSGSE 230
Query: 411 TSQSHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPPSLNSDTQDRTG 470
+S S + GS+S++ S S S +G ++SGS+ S S NS ++ +G
Sbjct: 231 SSSGSSNS--------GSESSSGSSNSGSESS----SGSSNSGSESSSGSSNSGSESSSG 278
Score = 42.0 bits (97), Expect = 0.008
Identities = 49/181 (27%), Positives = 81/181 (44%), Gaps = 38/181 (20%)
Query: 291 STTPSTPARNGGNGSTSSADHMRERSSGS-SQSPNDDSDCQEDVRVKLPLQLFGSSPEND 349
S + ++ + N G+ STS++ SS S SQS D S+ GS+ ++
Sbjct: 102 SGSEASGSSNSGSQSTSNSGSEASGSSNSGSQSSTDSSNSGSQ----------GSTGSSN 151
Query: 350 SPSKLPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANK 409
S S+ S++SSN + + SS N G QG NS S+G + ++
Sbjct: 152 SGSQ-------SSTDSSNSGSQSSTDSS------NSGSQGSTGSSNSGSESSGSSNSGSE 198
Query: 410 ETSQSHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPPSLNSDTQDRT 469
+ S+S GS+S++ S S S +G ++SGS+ S S NS ++ +
Sbjct: 199 SSGSSNS----------GSESSSGSSNSGSESS----SGSSNSGSESSSGSSNSGSESSS 244
Query: 470 G 470
G
Sbjct: 245 G 245
Score = 38.5 bits (88), Expect = 0.091
Identities = 46/180 (25%), Positives = 71/180 (38%), Gaps = 34/180 (18%)
Query: 295 STPARNGGNGSTSSADHMRERSSGSSQSPNDDSDCQEDVRVKLPLQLFGSSPENDSPSKL 354
ST + N G+ ++ + + SSGSS S ++ S S+ ++S S
Sbjct: 168 STDSSNSGSQGSTGSSNSGSESSGSSNSGSESSG--------------SSNSGSESSSGS 213
Query: 355 PSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKETSQS 414
+S SS SSN E + SS N G + NS S+ N+ E+S
Sbjct: 214 SNSGSESSSGSSNSGSESSSGSS------NSGSESSSGSSNSGSESSSGSSNSGSESS-- 265
Query: 415 HSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPPSLNSDTQDRTGRIMF 474
GS ++ S S S +G ++SGS+ S S S + +T I F
Sbjct: 266 -----------SGSSNSGSESSSGSSNSGSESSGSSNSGSESSSDS-GSSSDGKTTCISF 313
Score = 37.4 bits (85), Expect = 0.20
Identities = 36/126 (28%), Positives = 55/126 (43%), Gaps = 8/126 (6%)
Query: 291 STTPSTPARNGGNGSTSSADHMRERSSGSSQSPNDDSDCQEDVRVKLPLQLFGSSPENDS 350
S + S+ + N G+ S+ S++ E SSGSS S ++ S + + S E+ S
Sbjct: 185 SGSESSGSSNSGSESSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSSGSSNSGSESSS 244
Query: 351 PSKLPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKE 410
S S SS SSN E + SS N G + NS S+G + ++
Sbjct: 245 GSSNSGSES--SSGSSNSGSESSSGSS------NSGSESSSGSSNSGSESSGSSNSGSES 296
Query: 411 TSQSHS 416
+S S S
Sbjct: 297 SSDSGS 302
>DSPP_HUMAN (Q9NZW4) Dentin sialophosphoprotein precursor [Contains:
Dentin phosphoprotein (Dentin phosphophoryn) (DPP);
Dentin sialoprotein (DSP)]
Length = 1253
Score = 44.3 bits (103), Expect = 0.002
Identities = 48/215 (22%), Positives = 86/215 (39%), Gaps = 13/215 (6%)
Query: 253 NAKNDNVQMETSPSYHHRDDQLNNAPPAPLTKDFLAVLSTTPSTPARNGGNGSTSSADHM 312
++ N + ++S S D +N + + D ++ S+ + + + S S+
Sbjct: 834 DSSNSSDSSDSSDSSDSSDSDSSNRSDSSNSSDSSDSSDSSNSSDSSDSSDSSDSNESSN 893
Query: 313 RERSSGSSQSPNDDSDCQEDVRVKLPLQLFGSSPENDSPSKLPSSRKYFSSESSNPVDER 372
SS SS S + DS + SS +DS + SS SS++SN D
Sbjct: 894 SSDSSDSSNSSDSDSSDSSN-----------SSDSSDSSNSSDSSESSNSSDNSNSSDSS 942
Query: 373 TPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKETSQSHSCTTIPLDLFKGSKSNN 432
S S + + G +SN + +++ S + S ++ D S S++
Sbjct: 943 NSSDSSDSSDSSNSSDSSNSGDSSNSSDSSDSNSSDSSDSSNSSDSSDSSDSSDSSDSSD 1002
Query: 433 MIQQSSSVQSVPFKAGYASSGSDYSPPSLNSDTQD 467
S S S + +S+ SD S S +SD+ D
Sbjct: 1003 SSNSSDSSDS--SDSSDSSNSSDSSNSSDSSDSSD 1035
Score = 43.5 bits (101), Expect = 0.003
Identities = 49/221 (22%), Positives = 88/221 (39%), Gaps = 10/221 (4%)
Query: 250 NNLNAKNDNVQMETSPSYHHRDDQLNNAPPAPLTKDFLAVLSTTPSTPARNGGNGS---T 306
+N ++ +D+ ++S S +N+ + + D ++ S+ +++ + S +
Sbjct: 574 SNSSSDSDSSDSDSSDSSDSDSSDSSNSSDSSDSSDSSDSSDSSDSSDSKSDSSKSESDS 633
Query: 307 SSADHMRERSSGSSQSPNDDSDCQEDVRVKLPLQLFGSSPENDSPSKLPSSRKYFSSESS 366
S +D + S +S +D+SD + SS +DS S SS SS+SS
Sbjct: 634 SDSDSKSDSSDSNSSDSSDNSDSSDSSNSSNSSDSSDSSDSSDSSSSSDSSS---SSDSS 690
Query: 367 NPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKETSQSHSCTTIPLDLFK 426
N D S S E + +S+ + N+N S S + +
Sbjct: 691 NSSDSSDSSDSSNSSESSDSSDSS----DSDSSDSSDSSNSNSSDSDSSNSSDSSDSSDS 746
Query: 427 GSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPPSLNSDTQD 467
SN+ SS S + +S SD S S +SD+ D
Sbjct: 747 SDSSNSSDSSDSSDSSNSSDSSDSSDSSDSSDSSNSSDSND 787
Score = 42.7 bits (99), Expect = 0.005
Identities = 55/219 (25%), Positives = 84/219 (38%), Gaps = 22/219 (10%)
Query: 250 NNLNAKNDNVQMETSPSYHHRDDQLNNAPPAPLTKDFLAVLSTTPSTPARNGGNGSTSSA 309
+N + +D+ S S D +N+ + + ++ S+ + + N S SS
Sbjct: 702 SNSSESSDSSDSSDSDSSDSSDSSNSNSSDSDSSNSS----DSSDSSDSSDSSNSSDSSD 757
Query: 310 DHMRERSSGSSQSPNDDSDCQEDVRVKLPLQLFGSSPENDSPSKLPSSRKYFSSESSNPV 369
SS SS S +D SD + SS NDS + SS SS+SSN
Sbjct: 758 SSDSSNSSDSSDS-SDSSDSSDSSN---------SSDSNDSSNSSDSSDSSNSSDSSNSS 807
Query: 370 DERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKETSQSHSCTTIPLDLFKGSK 429
D S S N +SN + N++ + S S + D S
Sbjct: 808 DSSDSSDSSDSDSSN-------SSDSSNSSDSSDSSNSSDSSDSSDSSDSSDSDSSNRSD 860
Query: 430 SNNMIQQS-SSVQSVPFKAGYASSGSDYSPPSLNSDTQD 467
S+N S SS S + +S SD + S +SD+ D
Sbjct: 861 SSNSSDSSDSSDSSNSSDSSDSSDSSDSNESSNSSDSSD 899
Score = 42.7 bits (99), Expect = 0.005
Identities = 54/238 (22%), Positives = 93/238 (38%), Gaps = 19/238 (7%)
Query: 239 ADLTAKLLDVGNNLNAKNDNVQMETSPSYHHRDDQLNNAPPAPLTKDFLAVLSTTPSTPA 298
+D +K +N + +DN S + + D +++ + D + ++ S+ +
Sbjct: 634 SDSDSKSDSSDSNSSDSSDNSDSSDSSNSSNSSDSSDSSD----SSDSSSSSDSSSSSDS 689
Query: 299 RNGGNGSTSSADHMRERSSGSSQSPNDDSDCQEDVRVKLPLQLFGSSPENDSPSKLPSSR 358
N + S SS SS SS S + DS D +S ++DS + SS
Sbjct: 690 SNSSDSSDSSDSSNSSESSDSSDSSDSDSSDSSD-------SSNSNSSDSDSSNSSDSSD 742
Query: 359 KYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNA-------NKET 411
SS+SSN D S S + + +SN + N+ N
Sbjct: 743 SSDSSDSSNSSDSSDSSDSSNSSDSSDSSDSSDSSDSSNSSDSNDSSNSSDSSDSSNSSD 802
Query: 412 SQSHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPPSLNSDTQDRT 469
S + S ++ D SN+ +SS S + +S SD S S +SD+ +R+
Sbjct: 803 SSNSSDSSDSSDSSDSDSSNSSDSSNSSDSSDSSNSSDSSDSSDSSDSS-DSDSSNRS 859
Score = 40.4 bits (93), Expect = 0.024
Identities = 49/229 (21%), Positives = 90/229 (38%), Gaps = 14/229 (6%)
Query: 249 GNNLNAKNDNVQMETSPSYHHRDDQLNNAPPAPLTKDFLAVLSTTPSTPARNGGNGSTSS 308
GN+ N K+D+ + ++ S D N++ + + + +++ + + + + +S
Sbjct: 533 GNDDNDKSDSGKGKSDSSDSDSSDSSNSSDSSDSSDSDSSDSNSSSDSDSSDSDSSDSSD 592
Query: 309 ADHMRERSSGSSQSPNDDSDCQE--DVRVKLPLQLFGSSPENDSPSKLPSSRKYF----- 361
+D +S S +D SD + D S +DS SK SS
Sbjct: 593 SDSSDSSNSSDSSDSSDSSDSSDSSDSSDSKSDSSKSESDSSDSDSKSDSSDSNSSDSSD 652
Query: 362 ---SSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKETSQSHSCT 418
SS+SSN + S S + + +SN + +++ + S S
Sbjct: 653 NSDSSDSSNSSNSSDSSDSSDSSDSSSSSDSSSSSDSSNSSDSSDSSDSSNSSESSDSSD 712
Query: 419 TIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPPSLNSDTQD 467
+ D S S+N S+S S + +S SD S S +SD+ D
Sbjct: 713 SSDSDSSDSSDSSN----SNSSDSDSSNSSDSSDSSDSSDSSNSSDSSD 757
Score = 39.7 bits (91), Expect = 0.041
Identities = 48/225 (21%), Positives = 89/225 (39%), Gaps = 14/225 (6%)
Query: 247 DVGNNLNAKNDNVQMETSPSYHHRDDQLNNAPPAPLTKDFLAVLSTTPSTPARNGGNGST 306
D N ++ N + ++S S + D ++ + D +++ S+ + N + +
Sbjct: 854 DSSNRSDSSNSSDSSDSSDSSNSSDSSDSSD-----SSDSNESSNSSDSSDSSNSSDSDS 908
Query: 307 SSADHMRERSSGSSQSPNDDSDCQEDVRVKLPLQLFGSSPENDSPSKLPSSRKYFSSESS 366
S + + + S S+ S + +S D SS ++S SS SS+SS
Sbjct: 909 SDSSNSSDSSDSSNSSDSSESSNSSDNS--------NSSDSSNSSDSSDSSDSSNSSDSS 960
Query: 367 NPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKETSQSHSCTTIPLDLFK 426
N D S S + + + S+ +++ S S ++ D
Sbjct: 961 NSGDSSNSSDSSDSNSSDSSDSSNSSDSSDSSDSSDSSDSSDSSNSSDSSDSSDSSDSSN 1020
Query: 427 GSKSNNMIQQS-SSVQSVPFKAGYASSGSDYSPPSLNSDTQDRTG 470
S S+N S SS S + +S+ SD S S +SD+ D +G
Sbjct: 1021 SSDSSNSSDSSDSSDSSDSSDSSDSSNSSDSSDSSDSSDSSDSSG 1065
Score = 35.4 bits (80), Expect = 0.77
Identities = 46/177 (25%), Positives = 68/177 (37%), Gaps = 10/177 (5%)
Query: 292 TTPSTPARNGGNGSTSSADHMRERSSGSSQSPNDDSDCQEDVRVKLPLQLFGSSPENDSP 351
++ S+ + + N S SS SS SS S N SD + GSS +DS
Sbjct: 1015 SSDSSNSSDSSNSSDSSDSSDSSDSSDSSDSSNS-SDSSDSSDSSDSSDSSGSSDSSDSS 1073
Query: 352 SKLPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCI-STGFGGNANKE 410
SS SS+SS+ D S S + + +S+ S+ ++N
Sbjct: 1074 DSSDSSDSSDSSDSSDSSDSSESSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSNSS 1133
Query: 411 TSQSHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPPSLNSDTQD 467
S S ++ D S S++ S S S S SD S S +SD+ D
Sbjct: 1134 DSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDS--------SDSSDSSDSSDSSDSSD 1182
Score = 35.0 bits (79), Expect = 1.0
Identities = 41/188 (21%), Positives = 76/188 (39%), Gaps = 15/188 (7%)
Query: 295 STPARNGGNGSTS----------SADHMRERSSGSSQSPNDDSDCQEDVRVKLPLQLFGS 344
S +++ GNGS S ++D S+G+ + NDD+D + + K S
Sbjct: 495 SDESKDNGNGSDSKGAEDDDSDSTSDTNNSDSNGNGNNGNDDNDKSDSGKGKSDSSDSDS 554
Query: 345 SPENDS-----PSKLPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCI 399
S ++S S SS SS+S + + + SS + + +S+
Sbjct: 555 SDSSNSSDSSDSSDSDSSDSNSSSDSDSSDSDSSDSSDSDSSDSSNSSDSSDSSDSSDSS 614
Query: 400 STGFGGNANKETSQSHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPP 459
+ ++ ++S+S S ++ S SN+ +S S + +S SD S
Sbjct: 615 DSSDSSDSKSDSSKSESDSSDSDSKSDSSDSNSSDSSDNSDSSDSSNSSNSSDSSDSSDS 674
Query: 460 SLNSDTQD 467
S +S + D
Sbjct: 675 SDSSSSSD 682
>P3K3_DICDI (P54675) Phosphatidylinositol 3-kinase 3 (EC 2.7.1.137)
(PI3-kinase) (PtdIns-3-kinase) (PI3K) (Fragment)
Length = 1585
Score = 42.4 bits (98), Expect = 0.006
Identities = 41/187 (21%), Positives = 73/187 (38%), Gaps = 18/187 (9%)
Query: 250 NNLNAKNDN--VQMETSPSYHHRDDQLNNAPPAPLTKDFLAVLSTTPSTPARNGGNGSTS 307
NN N+ N+N + +SP H + NN + + + + + S S
Sbjct: 217 NNSNSNNNNNGISPSSSPPSHLNGNNNNNNSNNTNSNNTTNATTNSVGFSITMTNSNSLS 276
Query: 308 SADHMRERSSGSSQSPNDDSDCQEDVRVKLPLQLFGSSPENDS-PSKLPSSRKYFSSESS 366
+ M + S +S P S F SSP+N+ P + S ++F+S
Sbjct: 277 VSKRMNKFKSWTSSKPTSSSIG------------FASSPQNNGKPLNISGSSRFFTSRQD 324
Query: 367 NPVD-ERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKETSQSHSCTTIPLDLF 425
+ +D ++PSSSPP F N+N + N N + +++ +L
Sbjct: 325 SKIDLLKSPSSSPPTQSDIFNENNNNNNNNNN--NNNNNNNNNNNNNNNNNNNNNNEELI 382
Query: 426 KGSKSNN 432
+ +NN
Sbjct: 383 NNNNNNN 389
>YB95_SCHPO (O42970) Hypothetical serine-rich protein C1E8.05 in
chromosome II precursor
Length = 317
Score = 40.8 bits (94), Expect = 0.018
Identities = 41/158 (25%), Positives = 59/158 (36%), Gaps = 13/158 (8%)
Query: 286 FLAVLSTTPSTPARNGGNGSTSSADHMRERS-SGSSQSPNDDSDCQEDVRVKLPLQLFGS 344
F+ S+TPS+ + + + +SS+ S S SS S + S K S
Sbjct: 146 FITSSSSTPSSSSSSSSSSPSSSSSKSSSSSKSSSSSSSSSKSSSSSSSSSKSSSSSSSS 205
Query: 345 SPENDSPSKLPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFG 404
S + SPS SS K+ SS + SSS +V N+ ST
Sbjct: 206 SKSSASPSSSKSSSKFSSSSFITSTTPASSSSSGAIVS------------NAKTASTDDS 253
Query: 405 GNANKETSQSHSCTTIPLDLFKGSKSNNMIQQSSSVQS 442
+A+ TS S + S S+ SSS S
Sbjct: 254 SSASSATSSVSSVVSSASSALSASASSASASVSSSASS 291
>VIT2_CHICK (P02845) Vitellogenin II precursor (Major vitellogenin)
[Contains: Lipovitellin I (LVI); Phosvitin (PV);
Lipovitellin II (LVII); YGP40]
Length = 1850
Score = 39.7 bits (91), Expect = 0.041
Identities = 40/166 (24%), Positives = 66/166 (39%), Gaps = 9/166 (5%)
Query: 289 VLSTTPSTPARNGGNGSTSSADHMRERSSGSSQSPNDDSDCQEDVRVKLPLQLFGSSPEN 348
VL+ + P + S+S++ SS S+ SPN E+ ++ + +
Sbjct: 1110 VLAEFGTEPDAKTSSSSSSASSTATSSSSSSASSPNRKKPMDEEENDQVKQARNKDASSS 1169
Query: 349 DSPSKLPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNAN 408
SK +S K SS+SSN + SSS NS S+ +++
Sbjct: 1170 SRSSKSSNSSKRSSSKSSNSSKRSSSSSSSSSSSSRSSSSSSSSSSNSKSSSSSSKSSSS 1229
Query: 409 KETSQSHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGS 454
S+S S K S S++ SSS +S ++ +SS S
Sbjct: 1230 SSRSRSSS---------KSSSSSSSSSSSSSSKSSSSRSSSSSSKS 1266
>SR40_YEAST (P32583) Suppressor protein SRP40
Length = 406
Score = 39.7 bits (91), Expect = 0.041
Identities = 41/185 (22%), Positives = 73/185 (39%), Gaps = 10/185 (5%)
Query: 295 STPARNGGNGSTSSADHMRERSSGSSQSPN---------DDSDCQEDVRVKLPLQLFGSS 345
S+ + + +GS+SS+ + SS S+S + D+ D +E + K + SS
Sbjct: 95 SSESDSSSSGSSSSSSSSSDESSSESESEDETKKRARESDNEDAKETKKAKTEPES-SSS 153
Query: 346 PENDSPSKLPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGG 405
E+ S SS SES + + SSS + Q +S+ S
Sbjct: 154 SESSSSGSSSSSESESGSESDSDSSSSSSSSSDSESDSESDSQSSSSSSSSDSSSDSDSS 213
Query: 406 NANKETSQSHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPPSLNSDT 465
+++ + S ++ S S++ SS S + +SS S S +SD+
Sbjct: 214 SSDSSSDSDSSSSSSSSSSDSDSDSDSSSDSDSSGSSDSSSSSDSSSDESTSSDSSDSDS 273
Query: 466 QDRTG 470
+G
Sbjct: 274 DSDSG 278
Score = 37.7 bits (86), Expect = 0.15
Identities = 43/175 (24%), Positives = 76/175 (42%), Gaps = 14/175 (8%)
Query: 291 STTPSTPARNGGNGSTSSADHMRERSSGSSQSPNDDSDCQEDVRVKLPLQLFGSSPENDS 350
S+ S+ + + S S ++ + SS SS S + +SD + D Q SS +DS
Sbjct: 153 SSESSSSGSSSSSESESGSESDSDSSSSSSSSSDSESDSESD------SQSSSSSSSSDS 206
Query: 351 PSKLPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKE 410
S SS SS+SS+ D + SSS + + +S + +++ +
Sbjct: 207 SSDSDSS----SSDSSSDSDSSSSSSSSS-SDSDSDSDSSSDSDSSGSSDSSSSSDSSSD 261
Query: 411 TSQSHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPPSLNSDT 465
S S + D GS S +++++ +S KA + S+ S PS +S +
Sbjct: 262 ESTSSDSSDSDSDSDSGSSSELETKEATADES---KAEETPASSNESTPSASSSS 313
>SC73_SCHPO (O13817) Protein transport protein sec73
Length = 1082
Score = 39.3 bits (90), Expect = 0.053
Identities = 56/239 (23%), Positives = 95/239 (39%), Gaps = 28/239 (11%)
Query: 252 LNAKNDNVQMETSPSYHHRDDQLNNAPPAPLTKDFLAVLSTTPSTPARNGGNGSTSSADH 311
LN N ++ S + + + D L+N+P + F + S + + A+
Sbjct: 48 LNEHNSKLKSSGSFTSNSQTD-LSNSPSSKKKNIFYPIARAKHSFYFNHNWSSDLQQANS 106
Query: 312 MRERSSGSSQSPNDDSDCQEDVRVKLPLQLFGSSPENDSPS-----KLPSSRKYF----- 361
+S SSQS ++DS+ +DV + L GS+P SPS KL +SR +F
Sbjct: 107 --PVASYSSQSVSNDSNFPKDVTSPISTDLSGSNPSLKSPSSINSAKLRASRSFFRFPKW 164
Query: 362 SSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSN--------CISTGFGGNANKETSQ 413
S +S NP D + +SSP V G + N N + G ++ K +
Sbjct: 165 SRKSWNP-DVNSTTSSPSEVGSLDKAGGPLNQQNENLSAESDNTILQNGTRKSSVKMSDL 223
Query: 414 SHSCTTI------PLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPPSLNSDTQ 466
++ P+ S + +S +VP + S Y PS+ +D +
Sbjct: 224 GKRIASLPVVKRPPIQFSISSDYIKPVDKSDQTSNVPSPSSSVGSLRLYRSPSIPNDVE 282
>AGA1_YEAST (P32323) A-agglutinin attachment subunit precursor
Length = 725
Score = 39.3 bits (90), Expect = 0.053
Identities = 50/237 (21%), Positives = 92/237 (38%), Gaps = 17/237 (7%)
Query: 243 AKLLDVGNNLNAKNDNVQMETSPSYHHRDDQLNNAPPAPLTKDFLAVLSTTPSTPARNGG 302
+ L +VG + ++ ++ L++ + T L+ ST+PS+ + +
Sbjct: 142 SSLSEVGTTTVVSSSAIEPSSASIISPVTSTLSSTTSSNPTTTSLSSTSTSPSSTSTSPS 201
Query: 303 NGSTSSADHMRERSSGSS---------QSPNDDSDCQEDVRVKLPLQLFGSSPENDSPSK 353
+ STSS+ SS S+ S + S +S + S S
Sbjct: 202 STSTSSSSTSTSSSSTSTSSSSTSTSPSSTSTSSSLTSTSSSSTSTSQSSTSTSSSSTST 261
Query: 354 LPSSRKYFSSE-SSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKETS 412
PSS SS S++P + T +SS + + + ST + T
Sbjct: 262 SPSSTSTSSSSTSTSPSSKSTSASSTSTSSYSTSTSPSLTSSSPTLASTSPSSTSISSTF 321
Query: 413 QSHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPPSLNSDTQDRT 469
+ S +++ + S S ++ S+ V SVP S+ S+ + PS+ S T + T
Sbjct: 322 -TDSTSSLGSSIASSSTSVSLYSPSTPVYSVP------STSSNVATPSMTSSTVETT 371
>BCF1_HUMAN (Q9NYF8) Bcl-2-associated transcription factor 1 (Btf)
Length = 920
Score = 38.9 bits (89), Expect = 0.069
Identities = 44/165 (26%), Positives = 65/165 (38%), Gaps = 21/165 (12%)
Query: 315 RSSGSSQSPNDDSDCQEDVRVKLPLQLFGSS-----------PENDSPSKLPSSRK---Y 360
RS SS+SP S K P+ S P+ +SP K S +
Sbjct: 131 RSYRSSRSPRSSSSRSSSPYSKSPVSKRRGSQEKQTKKAEGEPQEESPLKSKSQEEPKDT 190
Query: 361 FSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKETSQSHSCTTI 420
F + S +DE SS+ + + G+ ++++ S SQS SC+
Sbjct: 191 FEHDPSESIDEFNKSSAT-----SGDIWPGLSAYDNSPRSPHSPSPIATPPSQSSSCSDA 245
Query: 421 PLDLFKGSKSNNMIQQSSSVQSVPFKAGYAS--SGSDYSPPSLNS 463
P+ S N Q S S+Q P ++G S +GS PS NS
Sbjct: 246 PMLSTVHSAKNTPSQHSHSIQHSPERSGSGSVGNGSSRYSPSQNS 290
>YK02_SCHPO (Q9HDY9) Hypothetical serine/threonine-rich protein
P11E10.02c in chromosome I precursor
Length = 1082
Score = 38.5 bits (88), Expect = 0.091
Identities = 53/268 (19%), Positives = 110/268 (40%), Gaps = 16/268 (5%)
Query: 199 EIFNLLTAIADGSQGKFEERRSQVPDKEQLVQILNRIPLPADLTAKLLDVGNNLNAKNDN 258
+ FN+ ++ S E +S + + + + + L AK + + + + N +
Sbjct: 25 DTFNVSRSVGLISSSNLESCQSSPLEVGNIYNSTSASEILSTLDAKYITIIGVIGSSNSS 84
Query: 259 VQMETSPSYHHRDDQLNNAPPAPLTKDFLAVLSTTPSTPARNGGNGSTSSADHMRERSSG 318
+Q + S + ++ ++ P + +T +++ + T + G TS+ D SS
Sbjct: 85 IQ-DLIDSVGNSNNAASSNPTSTVT-EYVDRVQTVTEYVTLSCGQAFTSTVDISSSTSSS 142
Query: 319 SSQSPNDDSDCQEDVRVKL-PLQLFGSSPENDSPSKLPSSRKYFSSESSNPVDERTPSSS 377
SP + + + + P SP SK+ SS Y SSE S+ SSS
Sbjct: 143 VINSPTGTAVSSQISTLSMSPSSTPVFSPSASVSSKVASSVSYVSSEPSD------SSSS 196
Query: 378 PPVVEMNFGLQGGIRGFNSNCISTGFGGNANKETSQSHSCTTIPLDLFKGSKSNNMIQQS 437
V + + NS +S+ + TS + T+ +D+ + +++ + S
Sbjct: 197 TNTVILTTSV-------NSPAVSSSETLTSVSITSTESAYTSSSVDIAASTTASSTLPVS 249
Query: 438 SSVQSVPFKAGYASSGSDYSPPSLNSDT 465
+S +V F ++ S S P+ +S +
Sbjct: 250 TSEATVSFSTDIPATPSTLSSPASSSSS 277
>BRF1_HUMAN (P55201) Peregrin (Bromodomain and PHD finger-containing
protein 1) (BR140 protein)
Length = 1214
Score = 38.1 bits (87), Expect = 0.12
Identities = 58/271 (21%), Positives = 99/271 (36%), Gaps = 11/271 (4%)
Query: 217 ERRSQVPDKEQLVQILNRIPLPADLTAKLLDVGNNLNAKNDNVQMETSPSYHHRDDQLNN 276
E + +P +EQL +L R+ ++ A VG + AK ++ E + Q
Sbjct: 775 ENQKHLPVEEQLKLLLERLD---EVNASKQSVGRSRRAKM--IKKEMTALRRKLAHQRET 829
Query: 277 APPAPLTKDFLAVLSTTPSTPARNGGNGSTSSADHMRERSSGSSQSPNDDSDCQEDVRVK 336
P + S TP A + + S+A+ + + PN S +V +
Sbjct: 830 GRDGPERHGPSSRGSLTPHPAACDKDGQTDSAAEESSSQETSKGLGPNMSSTPAHEVGRR 889
Query: 337 LPLQLFGSSPENDSPSKLPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNS 396
+ +P+ P K P P +P PP + + L+ RG +
Sbjct: 890 TSVLFSKKNPKTAGPPKRPGRPPKNRESQMTPSHGGSP-VGPPQLPIMSSLRQRKRGRSP 948
Query: 397 NCISTGFGGNANKETSQSHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDY 456
S+ +++ + S +P + F G N +++S V +SS S+
Sbjct: 949 RPSSS---SDSDSDKSTEDPPMDLPANGFSG--GNQPVKKSFLVYRNDCSLPRSSSDSES 1003
Query: 457 SPPSLNSDTQDRTGRIMFKLFDKHPSHFPGT 487
S S +S DRT K PS GT
Sbjct: 1004 SSSSSSSAASDRTSTTPSKQGRGKPSFSRGT 1034
>JAG1_HUMAN (P78504) Jagged 1 precursor (Jagged1) (hJ1)
Length = 1218
Score = 37.7 bits (86), Expect = 0.15
Identities = 36/115 (31%), Positives = 56/115 (48%), Gaps = 10/115 (8%)
Query: 873 TPHAYAMMRN-------NHSYNMLVARKCSDRQRSEVSVRIDNE-IEHPSLGI-ELMQKR 923
T H + +RN + Y++ +A + S +E+ V I E I I E+ K
Sbjct: 976 TEHICSELRNLNILKNVSAEYSIYIACEPSPSANNEIHVAISAEDIRDDGNPIKEITDKI 1035
Query: 924 INQV-KRVGDSCSKCAIAEVRAKRRFSGSRSWLHGPFIHSMLAVAAVCVCVCVLF 977
I+ V KR G+S A+AEVR +RR +R+ P + S+L VA +C V +
Sbjct: 1036 IDLVSKRDGNSSLIAAVAEVRVQRRPLKNRTDFLVPLLSSVLTVAWICCLVTAFY 1090
>DSPP_RAT (Q62598) Dentin sialophosphoprotein precursor [Contains:
Dentin phosphoprotein (Dentin phosphophoryn) (DPP);
Dentin sialoprotein (DSP)]
Length = 687
Score = 37.7 bits (86), Expect = 0.15
Identities = 42/176 (23%), Positives = 67/176 (37%), Gaps = 9/176 (5%)
Query: 296 TPARNGGNGSTSSADHMRERSSGSSQSPNDDSDCQ--EDVRVKLPLQLFGSSPENDSPSK 353
T + NGS S + H E S S DDSD +D K + S+ +NDS S+
Sbjct: 482 TSDESSDNGSDSDS-HAGEDDSSDDTSDTDDSDSNGDDDSESKDKDESDNSNHDNDSDSE 540
Query: 354 LPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKETSQ 413
S S+SS+ D S S + + S+ + +++ +
Sbjct: 541 SKSDSSDSDSDSSDSSDSSDSSDSSETSDSSDSSD------TSDSSDSSDSSDSSNSSDT 594
Query: 414 SHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPPSLNSDTQDRT 469
S S + D G S++ S S S + +S S +SD++D T
Sbjct: 595 SDSSDSSDGDSSDGDSSDSDSSDSDSSNSSDSDSSDSSDSSSSDSSDSDSDSKDST 650
>BCF1_MOUSE (Q8K019) Bcl-2-associated transcription factor 1 (Btf)
Length = 919
Score = 37.7 bits (86), Expect = 0.15
Identities = 43/165 (26%), Positives = 65/165 (39%), Gaps = 22/165 (13%)
Query: 315 RSSGSSQSPNDDSDCQEDVRVKLPLQLFGSS-----------PENDSPSKLPSSRK---Y 360
RS SS+SP S K P+ S P+ +SP K S +
Sbjct: 131 RSYRSSRSPRSSSSRSSSPYSKSPVSKRRGSQEKQTKKAEGEPQEESPLKSKSQEEPKDT 190
Query: 361 FSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKETSQSHSCTTI 420
F + S +DE S++ + + G+ ++++ S SQS SC+
Sbjct: 191 FEHDPSESIDEFNKSAT------SGDIWPGLSAYDNSPRSPHSPSPIATPPSQSSSCSDA 244
Query: 421 PLDLFKGSKSNNMIQQSSSVQSVPFKAGYAS--SGSDYSPPSLNS 463
P+ S N Q S S+Q P ++G S +GS PS NS
Sbjct: 245 PMLSTVHSAKNTPSQHSHSIQHSPERSGSGSVGNGSSRYSPSQNS 289
>LAP4_DROME (Q7KRY7) LAP4 protein (Scribble protein) (Smell-impaired
protein)
Length = 1851
Score = 37.4 bits (85), Expect = 0.20
Identities = 30/121 (24%), Positives = 46/121 (37%), Gaps = 3/121 (2%)
Query: 264 SPSYHHRDDQLNNAPPAPLTKDFLAVLSTTPSTPARNGGNGSTSSADHMRERSSGSSQSP 323
SPS HH D++ ++K L TTP+TPA SA+ S S
Sbjct: 1605 SPSEHHEQDKIQKTTTVVISKHTLDTNPTTPTTPAAPLSIAGAESANSAGAPSPAVPAST 1664
Query: 324 NDDSDCQEDVRVKLPLQLFGSSPENDSPSKL---PSSRKYFSSESSNPVDERTPSSSPPV 380
+ V V+ Q + + + S+L P+SR + + TP+ P
Sbjct: 1665 PGSAPVLPAVAVQTQTQTTSTEKDEEEESQLQSTPASRDGAEEQQEEVRAKPTPTKVPKS 1724
Query: 381 V 381
V
Sbjct: 1725 V 1725
>MAM_DROME (P21519) Neurogenic protein mastermind
Length = 1596
Score = 37.0 bits (84), Expect = 0.26
Identities = 32/118 (27%), Positives = 53/118 (44%), Gaps = 9/118 (7%)
Query: 298 ARNGGNGSTS--SADHMRERSSGSSQSPNDDSDCQEDVRVKLPLQL-FGSSPENDSPSKL 354
A NGGNGS + + ++ ++ + S N+ S+ V++ QL F +SP N P ++
Sbjct: 275 ANNGGNGSNTGNNTNNNGNSTNNNGGSNNNGSENLTKFSVEIVQQLEFTTSPANSQPQQI 334
Query: 355 PSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKETS 412
++ + + + S P V G GG G N+N G GGN N +
Sbjct: 335 STN------VTVKALTNTSVKSEPGVGGGGGGGGGGNSGNNNNNGGGGGGGNGNNNNN 386
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.132 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 122,599,314
Number of Sequences: 164201
Number of extensions: 5420271
Number of successful extensions: 14975
Number of sequences better than 10.0: 93
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 80
Number of HSP's that attempted gapping in prelim test: 14729
Number of HSP's gapped (non-prelim): 216
length of query: 998
length of database: 59,974,054
effective HSP length: 120
effective length of query: 878
effective length of database: 40,269,934
effective search space: 35357002052
effective search space used: 35357002052
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 71 (32.0 bits)
Medicago: description of AC140774.7


