

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140722.2 + phase: 0
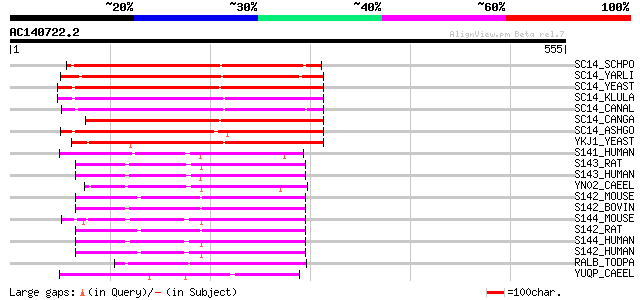
(555 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SC14_SCHPO (Q10137) Sec14 cytosolic factor (Phosphatidylinositol... 213 1e-54
SC14_YARLI (P45816) SEC14 cytosolic factor (Phosphatidylinositol... 206 1e-52
SC14_YEAST (P24280) SEC14 cytosolic factor (Phosphatidylinositol... 204 4e-52
SC14_KLULA (P24859) SEC14 cytosolic factor (Phosphatidylinositol... 203 1e-51
SC14_CANAL (P46250) SEC14 cytosolic factor (Phosphatidylinositol... 199 2e-50
SC14_CANGA (P53989) SEC14 cytosolic factor (Phosphatidylinositol... 195 3e-49
SC14_ASHGO (Q75DK1) SEC14 cytosolic factor (Phosphatidylinositol... 193 9e-49
YKJ1_YEAST (P33324) 36.1 kDa protein in BUD2-MIF2 intergenic region 175 2e-43
S141_HUMAN (Q92503) SEC14-like protein 1 126 1e-28
S143_RAT (Q9Z1J8) SEC14-like protein 3 (45 kDa secretory protein... 116 2e-25
S143_HUMAN (Q9UDX4) SEC14-like protein 3 (Tocopherol-associated ... 115 3e-25
YN02_CAEEL (Q03606) Hypothetical protein T23G5.2 in chromosome III 111 4e-24
S142_MOUSE (Q99J08) SEC14-like protein 2 (Alpha-tocopherol assoc... 110 1e-23
S142_BOVIN (P58875) SEC14-like protein 2 (Alpha-tocopherol assoc... 110 1e-23
S144_MOUSE (Q8R0F9) SEC14-like protein 4 109 2e-23
S142_RAT (Q99MS0) SEC14-like protein 2 (Alpha-tocopherol associa... 109 2e-23
S144_HUMAN (Q9UDX3) SEC14-like protein 4 (Tocopherol-associated ... 109 2e-23
S142_HUMAN (O76054) SEC14-like protein 2 (Alpha-tocopherol assoc... 107 6e-23
RALB_TODPA (P49193) Retinal-binding protein (RALBP) 100 1e-20
YUQP_CAEEL (Q19895) Hypothetical protein F28H7.8 in chromosome V 72 3e-12
>SC14_SCHPO (Q10137) Sec14 cytosolic factor
(Phosphatidylinositol/phosphatidyl-choline transfer
protein) (PI/PC TP) (Sporulation-specific protein 20)
Length = 286
Score = 213 bits (542), Expect = 1e-54
Identities = 109/255 (42%), Positives = 165/255 (63%), Gaps = 5/255 (1%)
Query: 57 VDALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMED 116
+D++R L L++L ++ D +LRFLRARK++++++ +M+ KWRKEFG D ++++
Sbjct: 32 LDSMR--LELQKLGYTERLDDATLLRFLRARKFNLQQSLEMFIKCEKWRKEFGVDDLIKN 89
Query: 117 FEFEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERA 176
F ++E + V K YPQ +H D DGRPVY+E+LG +D KL Q+T+ ER ++ V E+E
Sbjct: 90 FHYDEKEAVSKYYPQFYHKTDIDGRPVYVEQLGNIDLKKLYQITTPERMMQNLVYEYEML 149
Query: 177 FAVKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMF 236
+ PACS A I+ S TI+D++GVG+ S++ + +++ I D YPE + + +
Sbjct: 150 ALKRFPACSRKAGGLIETSCTIMDLKGVGITSIH-SVYSYIRQASSISQDYYPERMGKFY 208
Query: 237 IINAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADK 296
+INA GF +N +K FLD T KIH+LG+ Y+S LLE I A LP LGG C C
Sbjct: 209 VINAPWGFSSAFNLIKGFLDEATVKKIHILGSNYKSALLEQIPADNLPAKLGGNCQC--P 266
Query: 297 GGCMLSDKGPWNDPE 311
GGC LSD GPW++ +
Sbjct: 267 GGCELSDAGPWHEEQ 281
>SC14_YARLI (P45816) SEC14 cytosolic factor
(Phosphatidylinositol/phosphatidylcholine transfer
protein) (PI/PC TP)
Length = 492
Score = 206 bits (525), Expect = 1e-52
Identities = 111/263 (42%), Positives = 161/263 (61%), Gaps = 5/263 (1%)
Query: 51 AEELQAVDALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGA 110
AE+ Q + L+ L+ + D +LRFLRARK+D+ ++MW + KWRKEFG
Sbjct: 30 AEQEQKLGELKMILLTKGY--EDRTDDATLLRFLRARKFDVPLAQEMWENCEKWRKEFGT 87
Query: 111 DTIMEDFEFEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHV 170
+TI+EDF ++E EV K YPQ +H DKDGRPVY+E +G+V+ +++ ++T+ ER L+ V
Sbjct: 88 NTILEDFWYKEKKEVAKLYPQYYHKTDKDGRPVYVENVGKVNIHEMYKITTQERMLRNLV 147
Query: 171 REFERAFAVKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPE 230
E+E +LPACS I+ S TILD++GV L S ++ L+ I + YPE
Sbjct: 148 WEYESFVRHRLPACSRVVGHLIETSCTILDLKGVSLSSASQ-VYGFLKDASNIGQNYYPE 206
Query: 231 SLNRMFIINAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGT 290
+ + ++INA GF +++ +K FLDP T SKIHV G+ Y+ KLL + A LP GG
Sbjct: 207 RMGKFYLINAPFGFSTVFSVIKRFLDPVTVSKIHVYGSNYKEKLLAQVPAYNLPIKFGG- 265
Query: 291 CTCADKGGCMLSDKGPWNDPEIL 313
+ K G LSD GPW DP+ +
Sbjct: 266 -QSSSKIGVELSDDGPWRDPQFV 287
>SC14_YEAST (P24280) SEC14 cytosolic factor
(Phosphatidylinositol/phosphatidylcholine transfer
protein) (PI/PC TP)
Length = 303
Score = 204 bits (520), Expect = 4e-52
Identities = 111/267 (41%), Positives = 164/267 (60%), Gaps = 4/267 (1%)
Query: 48 SFDAEELQAVDALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKE 107
+ D+ + +A+ LR+ +LE+ + D +LRFLRARK+D++ K+M+ + KWRK+
Sbjct: 27 NLDSAQEKALAELRK--LLEDAGFIERLDDSTLLRFLRARKFDVQLAKEMFENCEKWRKD 84
Query: 108 FGADTIMEDFEFEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLK 167
+G DTI++DF ++E + K YPQ +H DKDGRPVY E LG V+ +++ +VTS ER LK
Sbjct: 85 YGTDTILQDFHYDEKPLIAKFYPQYYHKTDKDGRPVYFEELGAVNLHEMNKVTSEERMLK 144
Query: 168 YHVREFERAFAVKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDN 227
V E+E +LPACS AA ++ S TI+D++G+ + S + ++ I +
Sbjct: 145 NLVWEYESVVQYRLPACSRAAGHLVETSCTIMDLKGISISSA-YSVMSYVREASYISQNY 203
Query: 228 YPESLNRMFIINAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFL 287
YPE + + +IINA GF + K FLDP T SKI +LG+ YQ +LL+ I A LP
Sbjct: 204 YPERMGKFYIINAPFGFSTAFRLFKPFLDPVTVSKIFILGSSYQKELLKQIPAENLPVKF 263
Query: 288 GGTCTCAD-KGGCMLSDKGPWNDPEIL 313
GG + KGG LSD GPW DP+ +
Sbjct: 264 GGKSEVDESKGGLYLSDIGPWRDPKYI 290
>SC14_KLULA (P24859) SEC14 cytosolic factor
(Phosphatidylinositol/phosphatidylcholine transfer
protein) (PI/PC TP)
Length = 301
Score = 203 bits (516), Expect = 1e-51
Identities = 112/267 (41%), Positives = 159/267 (58%), Gaps = 4/267 (1%)
Query: 48 SFDAEELQAVDALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKE 107
+ D+E+ + R+ +LE L + D +LRFLRARK+D+E +K M+ + KWRKE
Sbjct: 26 NLDSEQEAKLKEFRE--LLESLGYKERLDDSTLLRFLRARKFDLEASKIMYENCEKWRKE 83
Query: 108 FGADTIMEDFEFEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLK 167
FG DTI EDF +EE V K YPQ +H D DGRPVYIE LG V+ ++ ++T+ ER LK
Sbjct: 84 FGVDTIFEDFHYEEKPLVAKYYPQYYHKTDNDGRPVYIEELGSVNLTQMYKITTQERMLK 143
Query: 168 YHVREFERAFAVKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDN 227
V E+E +LPACS A ++ S TILD++G+ + S + ++ I +
Sbjct: 144 NLVWEYEAFVRYRLPACSRKAGYLVETSCTILDLKGISISSAAQVL-SYVREASNIGQNY 202
Query: 228 YPESLNRMFIINAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFL 287
YPE + + ++INA GF + K FLDP T SKI +LG+ YQ LL+ I A LP+
Sbjct: 203 YPERMGKFYLINAPFGFSTAFRLFKPFLDPVTVSKIFILGSSYQKDLLKQIPAENLPKKF 262
Query: 288 GGTCTCAD-KGGCMLSDKGPWNDPEIL 313
GG ++ +GG LSD GPW + E +
Sbjct: 263 GGQSEVSEAEGGLYLSDIGPWREEEYI 289
>SC14_CANAL (P46250) SEC14 cytosolic factor
(Phosphatidylinositol/phosphatidylcholine transfer
protein) (PI/PC TP)
Length = 301
Score = 199 bits (506), Expect = 2e-50
Identities = 111/262 (42%), Positives = 151/262 (57%), Gaps = 4/262 (1%)
Query: 52 EELQAVDALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGAD 111
E+ +D RQ L EL D +LRFLRARK+DI+K M+ KWR++FG +
Sbjct: 32 EQKTTLDIFRQQLT--ELGYKDRLDDASLLRFLRARKFDIQKAIDMFVACEKWREDFGVN 89
Query: 112 TIMEDFEFEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVR 171
TI++DF +EE V K YP +H DKDGRPVY E LG+VD K+L++T+ ER LK V
Sbjct: 90 TILKDFHYEEKPIVAKMYPTYYHKTDKDGRPVYFEELGKVDLVKMLKITTQERMLKNLVW 149
Query: 172 EFERAFAVKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPES 231
E+E +LPACS A ++ S T+LD+ G+ + S ++ KI D YPE
Sbjct: 150 EYEAMCQYRLPACSRKAGYLVETSCTVLDLSGISVTSAYNVI-GYVREASKIGQDYYPER 208
Query: 232 LNRMFIINAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTC 291
+ + ++INA GF + K FLDP T SKIH+LG Y+ +LL+ I LP GG
Sbjct: 209 MGKFYLINAPFGFSTAFKLFKPFLDPVTVSKIHILGYSYKKELLKQIPPQNLPVKFGGMS 268
Query: 292 TCADKGGCMLSDKGPWNDPEIL 313
+D +L D GPW DPE +
Sbjct: 269 DVSD-DDLLLKDVGPWRDPEFI 289
>SC14_CANGA (P53989) SEC14 cytosolic factor
(Phosphatidylinositol/phosphatidylcholine transfer
protein) (PI/PC TP)
Length = 302
Score = 195 bits (495), Expect = 3e-49
Identities = 102/239 (42%), Positives = 146/239 (60%), Gaps = 2/239 (0%)
Query: 76 DPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDEVLKCYPQGHHG 135
D +LRFLRARK+D+ K+M+ + KWRKE+G +TIM+DF ++E V K YPQ +H
Sbjct: 52 DDSTLLRFLRARKFDVALAKEMFENCEKWRKEYGTNTIMQDFHYDEKPLVAKYYPQYYHK 111
Query: 136 VDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPACSIAAKKHIDQS 195
DKDGRPVY E LG V+ ++ ++T+ ER LK V E+E +LPACS AA ++ S
Sbjct: 112 TDKDGRPVYFEELGAVNLTEMEKITTQERMLKNLVWEYESVVNYRLPACSRAAGYLVETS 171
Query: 196 TTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGFRLLWNTVKSFL 255
T++D++G+ + S + ++ I + YPE + + ++INA GF + K FL
Sbjct: 172 CTVMDLKGISISSA-YSVLSYVREASYISQNYYPERMGKFYLINAPFGFSTAFRLFKPFL 230
Query: 256 DPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCAD-KGGCMLSDKGPWNDPEIL 313
DP T SKI +LG+ YQS+LL+ I A LP GG + GG LSD GPW D + +
Sbjct: 231 DPVTVSKIFILGSSYQSELLKQIPAENLPSKFGGKSEVDEAAGGLYLSDIGPWRDAKYI 289
>SC14_ASHGO (Q75DK1) SEC14 cytosolic factor
(Phosphatidylinositol/phosphatidylcholine transfer
protein) (PI/PC TP)
Length = 311
Score = 193 bits (491), Expect = 9e-49
Identities = 111/266 (41%), Positives = 162/266 (60%), Gaps = 8/266 (3%)
Query: 51 AEELQAVDALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGA 110
+E A++ LR+ +L++ +K D +LRFLRARK+D+ + M+ + KWRKE G
Sbjct: 32 SEHEAALEELRK--VLKQAGFTKRLDDSTLLRFLRARKFDVAAARAMFENCEKWRKENGV 89
Query: 111 DTIMEDFEFEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHV 170
DTI EDF +EE V K YPQ +H DKDGRPVYIE LG V+ ++ ++T+ ER LK +
Sbjct: 90 DTIFEDFHYEEKPLVAKFYPQYYHKTDKDGRPVYIEELGAVNLTEMYKITTQERMLKNLI 149
Query: 171 REFERAFAVKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDL--LQRLQKIDGDNY 228
E+E +LPA S A ++ S TILD++G+ S++ AA+ L ++ I + Y
Sbjct: 150 WEYESFSRYRLPASSRQADCLVETSCTILDLKGI---SISAAAQVLSYVREASNIGQNYY 206
Query: 229 PESLNRMFIINAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLG 288
PE + + ++INA GF + K FLDP T SKI +LG+ YQ +LL+ I A LP G
Sbjct: 207 PERMGKFYMINAPFGFSAAFRLFKPFLDPVTVSKIFILGSSYQKELLKQIPAENLPVKFG 266
Query: 289 GTCTCAD-KGGCMLSDKGPWNDPEIL 313
G ++ +GG LSD GPW +P+ +
Sbjct: 267 GQSDVSEAEGGLYLSDIGPWRNPKYI 292
>YKJ1_YEAST (P33324) 36.1 kDa protein in BUD2-MIF2 intergenic region
Length = 310
Score = 175 bits (444), Expect = 2e-43
Identities = 96/259 (37%), Positives = 156/259 (60%), Gaps = 9/259 (3%)
Query: 62 QTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEF-- 119
++++LE+ + DD +LRFLRARK+DI + +M+ + +WR+E+GA+TI+ED+E
Sbjct: 37 RSILLEKNYKERLDDS-TLLRFLRARKFDINASVEMFVETERWREEYGANTIIEDYENNK 95
Query: 120 ----EELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFER 175
+E ++ K YPQ +H VDKDGRP+Y E LG ++ K+ ++T+ ++ L+ V+E+E
Sbjct: 96 EAEDKERIKLAKMYPQYYHHVDKDGRPLYFEELGGINLKKMYKITTEKQMLRNLVKEYEL 155
Query: 176 AFAVKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRM 235
++PACS A I+ S T+LD++G+ L + ++ + I + YPE + +
Sbjct: 156 FATYRVPACSRRAGYLIETSCTVLDLKGISLSNAYHVL-SYIKDVADISQNYYPERMGKF 214
Query: 236 FIINAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCAD 295
+II++ GF ++ VK FLDP T SKI +LG+ Y+ +LL+ I LP GGT +
Sbjct: 215 YIIHSPFGFSTMFKMVKPFLDPVTVSKIFILGSSYKKELLKQIPIENLPVKYGGTSVLHN 274
Query: 296 KGG-CMLSDKGPWNDPEIL 313
SD GPW DP +
Sbjct: 275 PNDKFYYSDIGPWRDPRYI 293
>S141_HUMAN (Q92503) SEC14-like protein 1
Length = 715
Score = 126 bits (317), Expect = 1e-28
Identities = 83/250 (33%), Positives = 127/250 (50%), Gaps = 12/250 (4%)
Query: 50 DAEELQAVDALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFG 109
D LQ +R L+E K +LRFLRAR ++I+K +++ L WRK+
Sbjct: 250 DLTPLQESCLIRLRQWLQETHKGKIPKDEHILRFLRARDFNIDKAREIMCQSLTWRKQHQ 309
Query: 110 ADTIMEDFEFEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYH 169
D I+E + ++ + Y G H DKDGRP+Y+ RLGQ+D L++ E L+Y
Sbjct: 310 VDYILETWTPPQV--LQDYYAGGWHHHDKDGRPLYVLRLGQMDTKGLVRALGEEALLRYV 367
Query: 170 VREFERAFAVKLPACSIAAK---KHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGD 226
+ E +L C K + I T ++D++G+ +R + + L R+ ++
Sbjct: 368 LSVNEE----RLRRCEENTKVFGRPISSWTCLVDLEGLNMRHLWRPGVKALLRIIEVVEA 423
Query: 227 NYPESLNRMFIINAGSGFRLLWNTVKSFLDPKTTSKIHV-LGNKYQSK--LLEVIDASEL 283
NYPE+L R+ I+ A F +LW V F+D T K + GN YQ LL+ ID +
Sbjct: 424 NYPETLGRLLILRAPRVFPVLWTLVSPFIDDNTRRKFLIYAGNDYQGPGGLLDYIDKEII 483
Query: 284 PEFLGGTCTC 293
P+FL G C C
Sbjct: 484 PDFLSGECMC 493
>S143_RAT (Q9Z1J8) SEC14-like protein 3 (45 kDa secretory protein)
(rsec45)
Length = 400
Score = 116 bits (290), Expect = 2e-25
Identities = 75/234 (32%), Positives = 127/234 (54%), Gaps = 11/234 (4%)
Query: 66 LEELLPS-KHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDE 124
++++LP+ + D + +LR+LRAR +D++K++ M +++RK D I++ ++ +
Sbjct: 23 VQDVLPALPNPDDYFLLRWLRARNFDLQKSEAMLRKYMEFRKTMDIDHILD---WQPPEV 79
Query: 125 VLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPAC 184
+ K P G G D+DG PV+ + +G +D LL + + LK +R+ ER L C
Sbjct: 80 IQKYMPGGLCGYDRDGCPVWYDIIGPLDPKGLLFSVTKQDLLKTKMRDCERI----LHEC 135
Query: 185 SIAAKK---HIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAG 241
+ ++ I+ I D +G+GL+ K ++ Q + +NYPE+L M I+ A
Sbjct: 136 DLQTERLGRKIETIVMIFDCEGLGLKHFWKPLVEVYQEFFGLLEENYPETLKFMLIVKAT 195
Query: 242 SGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCAD 295
F + +N +K FL T KI VLGN ++ LL++I ELP GGT T D
Sbjct: 196 KLFPVGYNLMKPFLSEDTRRKIVVLGNSWKEGLLKLISPEELPAHFGGTLTDPD 249
>S143_HUMAN (Q9UDX4) SEC14-like protein 3 (Tocopherol-associated
protein 2)
Length = 400
Score = 115 bits (288), Expect = 3e-25
Identities = 75/234 (32%), Positives = 127/234 (54%), Gaps = 11/234 (4%)
Query: 66 LEELLPS-KHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDE 124
++++LP+ + D + +LR+LRAR +D++K++ + +++RK D I++ ++ +
Sbjct: 23 VQDVLPALPNPDDYFLLRWLRARNFDLQKSEALLRKYMEFRKTMDIDHILD---WQPPEV 79
Query: 125 VLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPAC 184
+ K P G G D+DG PV+ + +G +D LL + + LK +R+ ER L C
Sbjct: 80 IQKYMPGGLCGYDRDGCPVWYDIIGPLDPKGLLFSVTKQDLLKTKMRDCERI----LHEC 135
Query: 185 SIAAK---KHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAG 241
+ + K I+ I D +G+GL+ K ++ Q + +NYPE+L M I+ A
Sbjct: 136 DLQTERLGKKIETIVMIFDCEGLGLKHFWKPLVEVYQEFFGLLEENYPETLKFMLIVKAT 195
Query: 242 SGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCAD 295
F + +N +K FL T KI VLGN ++ LL++I ELP GGT T D
Sbjct: 196 KLFPVGYNLMKPFLSEDTRRKIIVLGNNWKEGLLKLISPEELPAQFGGTLTDPD 249
>YN02_CAEEL (Q03606) Hypothetical protein T23G5.2 in chromosome III
Length = 743
Score = 111 bits (278), Expect = 4e-24
Identities = 71/229 (31%), Positives = 118/229 (51%), Gaps = 13/229 (5%)
Query: 75 DDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDEVLKCYPQGHH 134
+D H+ LRFLRAR +D+ K K M + WRK+ D I+E E+ + + +P H
Sbjct: 300 NDAHL-LRFLRARDFDVAKAKDMVHASIIWRKQHNVDKILE--EWTRPTVIKQYFPGCWH 356
Query: 135 GVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPACSIAAKK---H 191
DK GRP+YI R GQ+D +L+ VE +K + E L + A +K
Sbjct: 357 NSDKAGRPMYILRFGQLDTKGMLRSCGVENLVKLTLSICEDG----LQRAAEATRKLGTP 412
Query: 192 IDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGFRLLWNTV 251
I + ++D+ G+ +R + + L ++ +I NYPE++ ++ ++ A F +LW +
Sbjct: 413 ISSWSLVVDLDGLSMRHLWRPGVQCLLKIIEIVEANYPETMGQVLVVRAPRVFPVLWTLI 472
Query: 252 KSFLDPKTTSKIHVLGNK---YQSKLLEVIDASELPEFLGGTCTCADKG 297
F+D KT K V G + +L + I+ +P+FLGG+C + G
Sbjct: 473 SPFIDEKTRKKFMVSGGSGGDLKEELRKHIEEKFIPDFLGGSCLTTNCG 521
>S142_MOUSE (Q99J08) SEC14-like protein 2 (Alpha-tocopherol
associated protein) (TAP)
Length = 403
Score = 110 bits (275), Expect = 1e-23
Identities = 73/231 (31%), Positives = 123/231 (52%), Gaps = 5/231 (2%)
Query: 66 LEELLPS-KHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDE 124
++++LP+ + D + +LR+LRAR +D++K++ M +++RK+ D I+ E + +
Sbjct: 23 VQDVLPTLPNPDDYFLLRWLRARSFDLQKSEAMLRKHVEFRKQKDIDKIISWQPPEVIQQ 82
Query: 125 VLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPAC 184
L G G D DG PV+ + +G +D LL S + L+ +R+ E +
Sbjct: 83 YLS---GGRCGYDLDGCPVWYDIIGPLDAKGLLFSASKQDLLRTKMRDCELLLQECIQQT 139
Query: 185 SIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGF 244
+ KK I+ T I D +G+GL+ + K A + + +NYPE+L R+F++ A F
Sbjct: 140 TKLGKK-IETITMIYDCEGLGLKHLWKPAVEAYGEFLTMFEENYPETLKRLFVVKAPKLF 198
Query: 245 RLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCAD 295
+ +N +K FL T KI VLG ++ LL+ I +LP GGT T D
Sbjct: 199 PVAYNLIKPFLSEDTRRKIMVLGANWKEVLLKHISPDQLPVEYGGTMTDPD 249
>S142_BOVIN (P58875) SEC14-like protein 2 (Alpha-tocopherol
associated protein) (TAP) (bTAP) (Fragment)
Length = 387
Score = 110 bits (275), Expect = 1e-23
Identities = 72/231 (31%), Positives = 123/231 (53%), Gaps = 5/231 (2%)
Query: 66 LEELLPS-KHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDE 124
++++LP+ + D + +LR+LRAR ++++K++ M +++RK+ D IM ++ +
Sbjct: 23 VQDVLPALPNPDDYFLLRWLRARNFNLQKSEAMLRKHVEFRKQKDIDNIMS---WQPPEV 79
Query: 125 VLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPAC 184
V + G G D +G P++ + +G +D LL S + K +R+ E +
Sbjct: 80 VQQYLSGGMCGYDLEGSPIWYDIIGPLDAKGLLLSASKQDLFKTKMRDCELLLQECVRQT 139
Query: 185 SIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGF 244
KK I+ +T I D +G+GL+ + K A + + +NYPE+L R+FI+ A F
Sbjct: 140 EKMGKK-IEATTLIYDCEGLGLKHLWKPAVEAYGEFLCMFEENYPETLKRLFIVKAPKLF 198
Query: 245 RLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCAD 295
+ +N VK FL T KI VLG ++ LL+ I +LP GGT T D
Sbjct: 199 PVAYNLVKPFLSEDTRKKIQVLGANWKEVLLKYISPDQLPVEYGGTMTDPD 249
>S144_MOUSE (Q8R0F9) SEC14-like protein 4
Length = 403
Score = 109 bits (273), Expect = 2e-23
Identities = 74/250 (29%), Positives = 137/250 (54%), Gaps = 17/250 (6%)
Query: 52 EELQAVDALRQTLILEELLPS--KHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFG 109
++ +A+ R+TL ++LLP+ K DD + +LR+LRAR +D++K++ M +++R +
Sbjct: 11 QQQEALARFRETL--QDLLPTLPKADD-YFLLRWLRARNFDLKKSEDMLRKHVEFRNQQN 67
Query: 110 ADTIMEDFEFEELDEVLKCYPQGH-HGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKY 168
D I+ + EV++ Y G G D +G PV+ + +G +D L S + ++
Sbjct: 68 LDQILT----WQAPEVIQLYDSGGLSGYDYEGCPVWFDIIGTMDPKGLFMSASKQDMIRK 123
Query: 169 HVREFERAFAVKLPACSIAAKK---HIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDG 225
++ E + L C + ++K I++ + D++G+ LR + K A ++ Q+ I
Sbjct: 124 RIKVCE----MLLHECELQSQKLGRKIERMVMVFDMEGLSLRHLWKPAVEVYQQFFAILE 179
Query: 226 DNYPESLNRMFIINAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPE 285
NYPE++ + II A F + +N VKSF+ +T KI +LG ++ +L++ + +LP
Sbjct: 180 ANYPETVKNLIIIRAPKLFPVAFNLVKSFMGEETQKKIVILGGNWKQELVKFVSPDQLPV 239
Query: 286 FLGGTCTCAD 295
GGT T D
Sbjct: 240 EFGGTMTDPD 249
>S142_RAT (Q99MS0) SEC14-like protein 2 (Alpha-tocopherol associated
protein) (TAP) (Supernatant protein factor) (SPF)
(Squalene transfer protein)
Length = 403
Score = 109 bits (273), Expect = 2e-23
Identities = 73/231 (31%), Positives = 122/231 (52%), Gaps = 5/231 (2%)
Query: 66 LEELLPS-KHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDE 124
++++LP+ + D + +LR+LRAR +D++K++ M +++RK+ D I+ E + +
Sbjct: 23 VQDVLPALPNPDDYFLLRWLRARSFDLQKSEAMLRKHVEFRKQKDIDKIISWQPPEVIQQ 82
Query: 125 VLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPAC 184
L G G D DG PV+ + +G +D LL S + L+ +R+ E
Sbjct: 83 YLS---GGRCGYDLDGCPVWYDIIGPLDAKGLLFSASKQDLLRTKMRDCELLLQECTQQT 139
Query: 185 SIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGF 244
+ KK I+ T I D +G+GL+ + K A + + +NYPE+L R+F++ A F
Sbjct: 140 AKLGKK-IETITMIYDCEGLGLKHLWKPAVEAYGEFLTMFEENYPETLKRLFVVKAPKLF 198
Query: 245 RLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCAD 295
+ +N +K FL T KI VLG ++ LL+ I +LP GGT T D
Sbjct: 199 PVAYNLIKPFLSEDTRKKIMVLGANWKEVLLKHISPDQLPVEYGGTMTDPD 249
>S144_HUMAN (Q9UDX3) SEC14-like protein 4 (Tocopherol-associated
protein 3)
Length = 406
Score = 109 bits (272), Expect = 2e-23
Identities = 72/235 (30%), Positives = 127/235 (53%), Gaps = 13/235 (5%)
Query: 66 LEELLPS-KHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDE 124
L++LLP + D + +LR+LRAR +D++K++ M +++RK+ D I+ + E
Sbjct: 23 LQDLLPILPNADDYFLLRWLRARNFDLQKSEDMLRRHMEFRKQQDLDNIVT----WQPPE 78
Query: 125 VLKCYPQGHH-GVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPA 183
V++ Y G G D +G PVY +G +D LL S + ++ ++ E + L
Sbjct: 79 VIQLYDSGGLCGYDYEGCPVYFNIIGSLDPKGLLLSASKQDMIRKRIKVCE----LLLHE 134
Query: 184 CSIAAKK---HIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINA 240
C + +K I+ + + D++G+ L+ + K A ++ Q+ I NYPE+L + +I A
Sbjct: 135 CELQTQKLGRKIEMALMVFDMEGLSLKHLWKPAVEVYQQFFSILEANYPETLKNLIVIRA 194
Query: 241 GSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCAD 295
F + +N VKSF+ +T KI +LG+ ++ +L + I +LP GGT T D
Sbjct: 195 PKLFPVAFNLVKSFMSEETRRKIVILGDNWKQELTKFISPDQLPVEFGGTMTDPD 249
>S142_HUMAN (O76054) SEC14-like protein 2 (Alpha-tocopherol
associated protein) (TAP) (hTAP) (Supernatant protein
factor) (SPF) (Squalene transfer protein)
Length = 403
Score = 107 bits (268), Expect = 6e-23
Identities = 73/234 (31%), Positives = 124/234 (52%), Gaps = 11/234 (4%)
Query: 66 LEELLPS-KHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDE 124
++++LP+ + D + +LR+LRAR +D++K++ M +++RK+ D I+ E + +
Sbjct: 23 VQDVLPALPNPDDYFLLRWLRARSFDLQKSEAMLRKHVEFRKQKDIDNIISWQPPEVIQQ 82
Query: 125 VLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPAC 184
L G G D DG PV+ + +G +D LL S + L+ +RE E + L C
Sbjct: 83 YLS---GGMCGYDLDGCPVWYDIIGPLDAKGLLFSASKQDLLRTKMRECE----LLLQEC 135
Query: 185 SIAAKK---HIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAG 241
+ K ++ T I D +G+GL+ + K A + + +NYPE+L R+F++ A
Sbjct: 136 AHQTTKLGRKVETITIIYDCEGLGLKHLWKPAVEAYGEFLCMFEENYPETLKRLFVVKAP 195
Query: 242 SGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCAD 295
F + +N +K FL T KI VLG ++ LL+ I ++P GGT T D
Sbjct: 196 KLFPVAYNLIKPFLSEDTRKKIMVLGANWKEVLLKHISPDQVPVEYGGTMTDPD 249
>RALB_TODPA (P49193) Retinal-binding protein (RALBP)
Length = 342
Score = 100 bits (248), Expect = 1e-20
Identities = 67/192 (34%), Positives = 98/192 (50%), Gaps = 3/192 (1%)
Query: 105 RKEFGADTIMEDFEFEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVER 164
R++ GADT++ E+ D + K G G DKDG + IE G +D ++
Sbjct: 3 REQMGADTLIA--EYTPPDVIQKFMTGGDVGHDKDGSVLRIEPWGYLDMKGIMYSCKKSD 60
Query: 165 YLKYHVREFERAFAVKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKID 224
K + + E+ L A S K T + D++ VG + M K D+ L ++
Sbjct: 61 LEKSKLLQCEKHLK-DLEAQSEKVGKPCTGLTVVFDMENVGSKHMWKPGLDMYLYLVQVL 119
Query: 225 GDNYPESLNRMFIINAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELP 284
DNYPE + R+F+INA + F +L+ VK L +KI VLG Y+ LLE IDA ELP
Sbjct: 120 EDNYPEMMKRLFVINAPTLFPVLYKLVKPLLSEDMKNKIFVLGGDYKDTLLEYIDAEELP 179
Query: 285 EFLGGTCTCADK 296
+LGGT + D+
Sbjct: 180 AYLGGTKSEGDE 191
>YUQP_CAEEL (Q19895) Hypothetical protein F28H7.8 in chromosome V
Length = 453
Score = 72.4 bits (176), Expect = 3e-12
Identities = 61/246 (24%), Positives = 112/246 (44%), Gaps = 10/246 (4%)
Query: 50 DAEELQAVDALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFG 109
D E + + L + +++ ++D MLR+L++ ++I KT + LKWRK+
Sbjct: 10 DIENMNDAAIEQVRLQVSDVIDPRYDTKWNMLRWLQSNDFNIPKTVHLLKKHLKWRKDRK 69
Query: 110 ADTIMEDFEFEELDEVLKCYPQGHHGVDK--DG-RPVYIERLGQVDCNKLLQVTSVERYL 166
D + D K P G + DG R V ++R G++D + L++ YL
Sbjct: 70 LDEPESQSLLQFSDARRKHAPIDIIGPQRKEDGDRLVVVDRAGRIDVSGLMKSVQPTEYL 129
Query: 167 KYHVREFE--RAFAVKLPA-CSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKI 223
R FE + +K+ A + H L+ L +N R Q +
Sbjct: 130 HEMFRSFEEIQRRLMKMEAETGVQCYMHYIFDLEALNFDPTLLGVVNGPFRVSWQLV--- 186
Query: 224 DGDNYPESLNRMFIINAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASEL 283
G +Y E +++ +IN+ S +LW+ + F+ ++ +I G+ ++ +LL+++D L
Sbjct: 187 -GQHYREFIDKFIVINSPSYINVLWSALSPFIPEQSKQRIVFAGSNWKEELLDIVDKECL 245
Query: 284 PEFLGG 289
PE GG
Sbjct: 246 PERYGG 251
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.133 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 64,234,533
Number of Sequences: 164201
Number of extensions: 2764553
Number of successful extensions: 9529
Number of sequences better than 10.0: 76
Number of HSP's better than 10.0 without gapping: 34
Number of HSP's successfully gapped in prelim test: 42
Number of HSP's that attempted gapping in prelim test: 9410
Number of HSP's gapped (non-prelim): 114
length of query: 555
length of database: 59,974,054
effective HSP length: 115
effective length of query: 440
effective length of database: 41,090,939
effective search space: 18080013160
effective search space used: 18080013160
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 68 (30.8 bits)
Medicago: description of AC140722.2


