

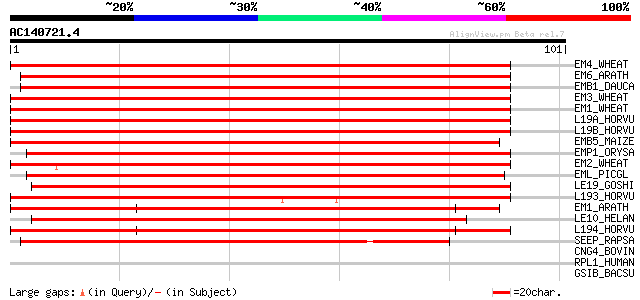
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140721.4 + phase: 0
(101 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
EM4_WHEAT (P42755) Em protein H5 153 8e-38
EM6_ARATH (Q02973) Em-like protein GEA6 149 9e-37
EMB1_DAUCA (P17639) EMB-1 protein 149 2e-36
EM3_WHEAT (Q08000) Em protein H2 147 4e-36
EM1_WHEAT (P04568) Em protein 147 6e-36
L19A_HORVU (Q05190) Late embryogenesis abundant protein B19.1A (... 146 8e-36
L19B_HORVU (P46532) Late embryogenesis abundant protein B19.1B 146 1e-35
EMB5_MAIZE (P46517) Late embryogenesis abundant protein EMB564 143 6e-35
EMP1_ORYSA (P46520) Embryonic abundant protein 1 143 8e-35
EM2_WHEAT (P22701) Em protein CS41 139 1e-33
EML_PICGL (Q40864) Em-like protein 136 1e-32
LE19_GOSHI (P09443) Late embryogenesis abundant protein D-19 (LE... 135 2e-32
L193_HORVU (Q02400) Late embryogenesis abundant protein B19.3 (L... 132 2e-31
EM1_ARATH (Q07187) Em-like protein GEA1 (EM1) 123 7e-29
LE10_HELAN (P46514) 10 kDa late embryogenesis abundant protein (... 117 4e-27
L194_HORVU (Q05191) Late embryogenesis abundant protein B19.4 115 2e-26
SEEP_RAPSA (P11573) Late seed maturation protein P8B6 115 2e-26
CNG4_BOVIN (Q28181) 240 kDa protein of rod photoreceptor CNG-cha... 37 0.008
RPL1_HUMAN (Q8IWN7) Retinitis pigmentosa 1-like 1 protein 36 0.014
GSIB_BACSU (P26907) Glucose starvation-inducible protein B (Gene... 36 0.019
>EM4_WHEAT (P42755) Em protein H5
Length = 93
Score = 153 bits (386), Expect = 8e-38
Identities = 74/91 (81%), Positives = 83/91 (90%)
Query: 1 MASKQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEM 60
MAS QQ R EL+ A++GETVVPGGTGGKSLEAQEHLA+GRS+GG+TRKEQLG EGY+EM
Sbjct: 1 MASGQQERSELDRMAREGETVVPGGTGGKSLEAQEHLADGRSRGGETRKEQLGEEGYREM 60
Query: 61 GRKGGLSTMEKSGGERAEEEGIDIDESKFKT 91
GRKGGLSTME+SGGERA EGI+IDESKFKT
Sbjct: 61 GRKGGLSTMEESGGERAAREGIEIDESKFKT 91
>EM6_ARATH (Q02973) Em-like protein GEA6
Length = 92
Score = 149 bits (377), Expect = 9e-37
Identities = 69/89 (77%), Positives = 83/89 (92%)
Query: 3 SKQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGR 62
+ QQ +++L+E+AK+GETVVPGGTGGKS EAQ+HLAEGRS+GGQTRKEQLGTEGYQ+MGR
Sbjct: 2 ASQQEKKQLDERAKKGETVVPGGTGGKSFEAQQHLAEGRSRGGQTRKEQLGTEGYQQMGR 61
Query: 63 KGGLSTMEKSGGERAEEEGIDIDESKFKT 91
KGGLST +K GGE AEEEG++IDESKF+T
Sbjct: 62 KGGLSTGDKPGGEHAEEEGVEIDESKFRT 90
>EMB1_DAUCA (P17639) EMB-1 protein
Length = 92
Score = 149 bits (375), Expect = 2e-36
Identities = 71/89 (79%), Positives = 81/89 (90%)
Query: 3 SKQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGR 62
+ QQ ++EL+ +A+QGETVVPGGTGGKSLEAQ+HLAEGRSKGGQTRKEQLG EGY EMGR
Sbjct: 2 ASQQEKKELDARARQGETVVPGGTGGKSLEAQQHLAEGRSKGGQTRKEQLGGEGYHEMGR 61
Query: 63 KGGLSTMEKSGGERAEEEGIDIDESKFKT 91
KGGLS + SGGERAE+EGIDIDESKF+T
Sbjct: 62 KGGLSNNDMSGGERAEQEGIDIDESKFRT 90
>EM3_WHEAT (Q08000) Em protein H2
Length = 93
Score = 147 bits (371), Expect = 4e-36
Identities = 71/91 (78%), Positives = 81/91 (88%)
Query: 1 MASKQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEM 60
MAS QQ R +L+ KA++GETVVPGGTGGKSLEA E+LAEGRS+GGQTR+EQ+G EGY EM
Sbjct: 1 MASGQQERSQLDRKAREGETVVPGGTGGKSLEAHENLAEGRSRGGQTRREQMGEEGYSEM 60
Query: 61 GRKGGLSTMEKSGGERAEEEGIDIDESKFKT 91
GRKGGLST ++SGGERA EGIDIDESKFKT
Sbjct: 61 GRKGGLSTNDESGGERAAREGIDIDESKFKT 91
>EM1_WHEAT (P04568) Em protein
Length = 93
Score = 147 bits (370), Expect = 6e-36
Identities = 70/91 (76%), Positives = 82/91 (89%)
Query: 1 MASKQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEM 60
MAS QQ R +L+ KA++GETVVPGGTGGKSLEAQE+LAEGRS+GGQTR+EQ+G EGY +M
Sbjct: 1 MASGQQERSQLDRKAREGETVVPGGTGGKSLEAQENLAEGRSRGGQTRREQMGEEGYSQM 60
Query: 61 GRKGGLSTMEKSGGERAEEEGIDIDESKFKT 91
GRKGGLST ++SGG+RA EGIDIDESKFKT
Sbjct: 61 GRKGGLSTNDESGGDRAAREGIDIDESKFKT 91
>L19A_HORVU (Q05190) Late embryogenesis abundant protein B19.1A
(B19.1)
Length = 93
Score = 146 bits (369), Expect = 8e-36
Identities = 70/91 (76%), Positives = 82/91 (89%)
Query: 1 MASKQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEM 60
MAS QQ R +L+ KA++GETVVPGGTGGKSLEAQ++LAEGRS+GGQTR+EQ+G EGY EM
Sbjct: 1 MASGQQERSQLDRKAREGETVVPGGTGGKSLEAQQNLAEGRSRGGQTRREQMGQEGYSEM 60
Query: 61 GRKGGLSTMEKSGGERAEEEGIDIDESKFKT 91
GRKGGLS+ ++SGGERA EGIDIDESKFKT
Sbjct: 61 GRKGGLSSNDESGGERAAREGIDIDESKFKT 91
>L19B_HORVU (P46532) Late embryogenesis abundant protein B19.1B
Length = 93
Score = 146 bits (368), Expect = 1e-35
Identities = 70/91 (76%), Positives = 81/91 (88%)
Query: 1 MASKQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEM 60
MAS QQ R +L+ KA++GETVVPGGTGGKSLEA ++LAEGRS+GGQTR+EQ+G EGY EM
Sbjct: 1 MASGQQERSQLDRKAREGETVVPGGTGGKSLEAHDNLAEGRSRGGQTRREQMGEEGYSEM 60
Query: 61 GRKGGLSTMEKSGGERAEEEGIDIDESKFKT 91
GRKGGLST ++SGGERA EGIDIDESKFKT
Sbjct: 61 GRKGGLSTNDESGGERAAREGIDIDESKFKT 91
>EMB5_MAIZE (P46517) Late embryogenesis abundant protein EMB564
Length = 91
Score = 143 bits (361), Expect = 6e-35
Identities = 67/89 (75%), Positives = 81/89 (90%)
Query: 1 MASKQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEM 60
MAS Q++R+EL+ KA++GETVVPGGTGGKS+EAQEHLAEGRS+GGQTR+EQLG +GY EM
Sbjct: 1 MASGQESRKELDRKAREGETVVPGGTGGKSVEAQEHLAEGRSRGGQTRREQLGQQGYSEM 60
Query: 61 GRKGGLSTMEKSGGERAEEEGIDIDESKF 89
G+KGGLST ++SGGERA EG+ IDESKF
Sbjct: 61 GKKGGLSTTDESGGERAAREGVTIDESKF 89
>EMP1_ORYSA (P46520) Embryonic abundant protein 1
Length = 95
Score = 143 bits (360), Expect = 8e-35
Identities = 68/88 (77%), Positives = 80/88 (90%)
Query: 4 KQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGRK 63
+QQ R EL+ A++G+TVVPGGTGGKSLEAQE+LAEGRS+GGQTRKEQ+G EGY+EMGRK
Sbjct: 6 QQQGRSELDRMAREGQTVVPGGTGGKSLEAQENLAEGRSRGGQTRKEQMGEEGYREMGRK 65
Query: 64 GGLSTMEKSGGERAEEEGIDIDESKFKT 91
GGLST ++SGGERA EGIDIDESK+KT
Sbjct: 66 GGLSTGDESGGERAAREGIDIDESKYKT 93
>EM2_WHEAT (P22701) Em protein CS41
Length = 94
Score = 139 bits (350), Expect = 1e-33
Identities = 70/92 (76%), Positives = 79/92 (85%), Gaps = 1/92 (1%)
Query: 1 MASKQQN-RQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQE 59
MAS Q+ R EL+ A++G+TVVPGGTGGKS EAQE LAEGRS+GGQTRKEQ+G EGY E
Sbjct: 1 MASGQEKGRSELDSLAREGQTVVPGGTGGKSYEAQEKLAEGRSRGGQTRKEQMGEEGYSE 60
Query: 60 MGRKGGLSTMEKSGGERAEEEGIDIDESKFKT 91
MGRKGGLST ++SGGERA EGIDIDESKFKT
Sbjct: 61 MGRKGGLSTNDESGGERAAREGIDIDESKFKT 92
>EML_PICGL (Q40864) Em-like protein
Length = 91
Score = 136 bits (342), Expect = 1e-32
Identities = 65/87 (74%), Positives = 78/87 (88%)
Query: 4 KQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGRK 63
+QQ+R+EL+ KA++GETVVPGGTGGKSL+AQE LAEGRS+GGQTRKEQ+G+EGYQEMGRK
Sbjct: 3 QQQDRRELDAKAREGETVVPGGTGGKSLDAQERLAEGRSRGGQTRKEQIGSEGYQEMGRK 62
Query: 64 GGLSTMEKSGGERAEEEGIDIDESKFK 90
GGLS+ GGERA EEG IDESK++
Sbjct: 63 GGLSSAGGPGGERASEEGRPIDESKYR 89
>LE19_GOSHI (P09443) Late embryogenesis abundant protein D-19 (LEA
D-19)
Length = 102
Score = 135 bits (340), Expect = 2e-32
Identities = 64/87 (73%), Positives = 77/87 (87%)
Query: 5 QQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGRKG 64
Q+ ++EL+ +AKQGETVVPGGT GKSL+AQ +LAEGR KGG+TRK+QLGTEGYQEMGRKG
Sbjct: 14 QEEKEELDARAKQGETVVPGGTRGKSLDAQINLAEGRHKGGETRKQQLGTEGYQEMGRKG 73
Query: 65 GLSTMEKSGGERAEEEGIDIDESKFKT 91
GLS + SGGERA +EG+ IDESKF+T
Sbjct: 74 GLSNSDMSGGERAADEGVTIDESKFRT 100
>L193_HORVU (Q02400) Late embryogenesis abundant protein B19.3 (LEA
B19.3)
Length = 133
Score = 132 bits (331), Expect = 2e-31
Identities = 76/131 (58%), Positives = 83/131 (63%), Gaps = 40/131 (30%)
Query: 1 MASKQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTR------------ 48
MAS QQ R EL+ A++GETVVPGGTGGK+LEAQEHLAEGRS+GGQTR
Sbjct: 1 MASGQQERSELDRMAREGETVVPGGTGGKTLEAQEHLAEGRSRGGQTRKDQLGEEGYREM 60
Query: 49 --------KEQLGTEGYQ--------------------EMGRKGGLSTMEKSGGERAEEE 80
KEQLG EGY+ EMGRKGGLSTME+SGGERA E
Sbjct: 61 GHKGGETRKEQLGEEGYREMGHKGGETRKEQMGEEGYHEMGRKGGLSTMEESGGERAARE 120
Query: 81 GIDIDESKFKT 91
GIDIDESKFKT
Sbjct: 121 GIDIDESKFKT 131
>EM1_ARATH (Q07187) Em-like protein GEA1 (EM1)
Length = 152
Score = 123 bits (309), Expect = 7e-29
Identities = 62/81 (76%), Positives = 69/81 (84%)
Query: 1 MASKQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEM 60
MASKQ +R+EL+EKAKQGETVVPGGTGG SLEAQEHLAEGRSKGGQTRKEQLG EGYQE+
Sbjct: 1 MASKQLSREELDEKAKQGETVVPGGTGGHSLEAQEHLAEGRSKGGQTRKEQLGHEGYQEI 60
Query: 61 GRKGGLSTMEKSGGERAEEEG 81
G KGG + E+ G E +E G
Sbjct: 61 GHKGGEARKEQLGHEGYQEMG 81
Score = 92.4 bits (228), Expect = 2e-19
Identities = 47/66 (71%), Positives = 50/66 (75%)
Query: 24 GGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGRKGGLSTMEKSGGERAEEEGID 83
GG K E E KGG+ RKEQLG EGY+EMGRKGGLSTMEKSGGERAEEEGI+
Sbjct: 84 GGEARKEQLGHEGYQEMGHKGGEARKEQLGHEGYKEMGRKGGLSTMEKSGGERAEEEGIE 143
Query: 84 IDESKF 89
IDESKF
Sbjct: 144 IDESKF 149
>LE10_HELAN (P46514) 10 kDa late embryogenesis abundant protein
(DS10)
Length = 92
Score = 117 bits (294), Expect = 4e-27
Identities = 56/79 (70%), Positives = 69/79 (86%)
Query: 5 QQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGRKG 64
+Q +++L+++A +GETVVPGGT GKSLEAQE LAEGRSKGGQTRK+QLGTEGY+EMG+KG
Sbjct: 14 EQEKKDLDQRAAKGETVVPGGTRGKSLEAQERLAEGRSKGGQTRKDQLGTEGYKEMGKKG 73
Query: 65 GLSTMEKSGGERAEEEGID 83
G +T +KS GER EEE D
Sbjct: 74 GQTTGDKSAGEREEEEEED 92
>L194_HORVU (Q05191) Late embryogenesis abundant protein B19.4
Length = 153
Score = 115 bits (288), Expect = 2e-26
Identities = 57/81 (70%), Positives = 65/81 (79%)
Query: 1 MASKQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEM 60
MAS QQ R EL+ A++GETVVPGGTGGK+LEAQEHLAEGRS+GGQTRKEQLG EGY+EM
Sbjct: 1 MASGQQERSELDRMAREGETVVPGGTGGKTLEAQEHLAEGRSRGGQTRKEQLGEEGYREM 60
Query: 61 GRKGGLSTMEKSGGERAEEEG 81
G KGG + E+ G E E G
Sbjct: 61 GHKGGETRKEQLGEEGYREMG 81
Score = 91.7 bits (226), Expect = 3e-19
Identities = 46/68 (67%), Positives = 51/68 (74%)
Query: 24 GGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGRKGGLSTMEKSGGERAEEEGID 83
GG K +E E KGG+TRKEQ+G EGY+EMGRKGGLSTM +SGGERA EGID
Sbjct: 84 GGETRKEQLGEEGYREMGHKGGETRKEQMGEEGYREMGRKGGLSTMNESGGERAAREGID 143
Query: 84 IDESKFKT 91
IDESKFKT
Sbjct: 144 IDESKFKT 151
>SEEP_RAPSA (P11573) Late seed maturation protein P8B6
Length = 83
Score = 115 bits (287), Expect = 2e-26
Identities = 55/78 (70%), Positives = 69/78 (87%), Gaps = 1/78 (1%)
Query: 3 SKQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGR 62
+ QQ +++L+E+AK+GETVVPGGTGGKS EAQ+HLAEGRS+GG TRKEQLG+EGYQ+MGR
Sbjct: 2 ASQQEKKQLDERAKKGETVVPGGTGGKSFEAQQHLAEGRSRGGNTRKEQLGSEGYQQMGR 61
Query: 63 KGGLSTMEKSGGERAEEE 80
KGG ST +K+ E AE+E
Sbjct: 62 KGG-STPDKTDKEDAEDE 78
>CNG4_BOVIN (Q28181) 240 kDa protein of rod photoreceptor
CNG-channel [Contains: Glutamic acid-rich protein
(GARP); Cyclic-nucleotide-gated cation channel 4 (CNG
channel 4) (CNG-4) (Cyclic nucleotide-gated cation
channel modulatory subunit)]
Length = 1394
Score = 37.0 bits (84), Expect = 0.008
Identities = 24/87 (27%), Positives = 43/87 (48%), Gaps = 15/87 (17%)
Query: 5 QQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGRK- 63
Q+ +++ EE+ + GE E +E E + G+ ++E+ G E +E G K
Sbjct: 354 QEEKEDEEEEKEDGE------------EEEEEGREKEEEEGEEKEEEEGREKEEEEGEKK 401
Query: 64 --GGLSTMEKSGGERAEEEGIDIDESK 88
G E+ GGE+ +EEG + +E +
Sbjct: 402 EEEGREKEEEEGGEKEDEEGREKEEEE 428
Score = 35.4 bits (80), Expect = 0.025
Identities = 24/82 (29%), Positives = 39/82 (47%), Gaps = 4/82 (4%)
Query: 4 KQQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGRK 63
K+ +E EE ++ E + E +E E + + G+ ++E+ G E E GR+
Sbjct: 364 KEDGEEEEEEGREKEEEEGEEKEEEEGREKEEEEGEKKEEEGREKEEEEGGEKEDEEGRE 423
Query: 64 G----GLSTMEKSGGERAEEEG 81
G E+ GGE+ EEEG
Sbjct: 424 KEEEEGRGKEEEEGGEKEEEEG 445
>RPL1_HUMAN (Q8IWN7) Retinitis pigmentosa 1-like 1 protein
Length = 2480
Score = 36.2 bits (82), Expect = 0.014
Identities = 24/84 (28%), Positives = 40/84 (47%), Gaps = 1/84 (1%)
Query: 12 EEKAKQGETVVPGGTGGKSLEAQE-HLAEGRSKGGQTRKEQLGTEGYQEMGRKGGLSTME 70
EE + ET G + ++ +E EG + G + TEG + + L +E
Sbjct: 1381 EEGVQLEETKTEEGLQEEGVQLEETKTEEGLQEEGVQLEGTKETEGEGQQEEEAQLEEIE 1440
Query: 71 KSGGERAEEEGIDIDESKFKTGGG 94
++GGE +EEG+ ++E K GG
Sbjct: 1441 ETGGEGLQEEGVQLEEVKEGPEGG 1464
Score = 29.3 bits (64), Expect = 1.8
Identities = 23/82 (28%), Positives = 37/82 (45%), Gaps = 5/82 (6%)
Query: 12 EEKAKQGETVVPGGTGGKSLEAQE-HLAEGRSKGGQTRKEQLGTEGYQEMGRKGGLSTME 70
EE + ET G + ++ +E EG + G +E EG QE G + E
Sbjct: 1333 EEGVQLEETKTEEGLQEEGVQLEETKTEEGLQEEGVQLEETKTEEGLQEEG----VQLEE 1388
Query: 71 KSGGERAEEEGIDIDESKFKTG 92
E +EEG+ ++E+K + G
Sbjct: 1389 TKTEEGLQEEGVQLEETKTEEG 1410
Score = 29.3 bits (64), Expect = 1.8
Identities = 26/95 (27%), Positives = 41/95 (42%), Gaps = 13/95 (13%)
Query: 7 NRQELEEKAKQGETVVPG---GTGGKSLEAQ------EHLAEGRSKGGQTRKEQLGTEGY 57
N ++L E Q E + GT G+ L+ + EG + G +E EG
Sbjct: 1288 NLEQLAENTVQEEVQLEETKEGTEGEGLQEEGVQLEETKTEEGLQEEGVQLEETKTEEGL 1347
Query: 58 QEMGRKGGLSTMEKSGGERAEEEGIDIDESKFKTG 92
QE G + E E +EEG+ ++E+K + G
Sbjct: 1348 QEEG----VQLEETKTEEGLQEEGVQLEETKTEEG 1378
Score = 29.3 bits (64), Expect = 1.8
Identities = 23/82 (28%), Positives = 37/82 (45%), Gaps = 5/82 (6%)
Query: 12 EEKAKQGETVVPGGTGGKSLEAQE-HLAEGRSKGGQTRKEQLGTEGYQEMGRKGGLSTME 70
EE + ET G + ++ +E EG + G +E EG QE G + E
Sbjct: 1317 EEGVQLEETKTEEGLQEEGVQLEETKTEEGLQEEGVQLEETKTEEGLQEEG----VQLEE 1372
Query: 71 KSGGERAEEEGIDIDESKFKTG 92
E +EEG+ ++E+K + G
Sbjct: 1373 TKTEEGLQEEGVQLEETKTEEG 1394
Score = 28.5 bits (62), Expect = 3.0
Identities = 22/82 (26%), Positives = 33/82 (39%), Gaps = 2/82 (2%)
Query: 9 QELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEG--YQEMGRKGGL 66
QE EE+A+ V + EAQE E + G + E G E +E ++
Sbjct: 2090 QEAEEEAQPESDGVEAQPKSEGEEAQEVEGETQKTEGDAQPESDGVEAPEAEEEAQEAEG 2149
Query: 67 STMEKSGGERAEEEGIDIDESK 88
E G E E +D E++
Sbjct: 2150 EVQEAEGEAHPESEDVDAQEAE 2171
Score = 27.3 bits (59), Expect = 6.7
Identities = 19/88 (21%), Positives = 38/88 (42%)
Query: 5 QQNRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGRKG 64
+ R E++A + T + ++ +E +G Q QL +E ++
Sbjct: 1275 EAERDSEEQRASSNLEQLAENTVQEEVQLEETKEGTEGEGLQEEGVQLEETKTEEGLQEE 1334
Query: 65 GLSTMEKSGGERAEEEGIDIDESKFKTG 92
G+ E E +EEG+ ++E+K + G
Sbjct: 1335 GVQLEETKTEEGLQEEGVQLEETKTEEG 1362
>GSIB_BACSU (P26907) Glucose starvation-inducible protein B (General
stress protein B)
Length = 122
Score = 35.8 bits (81), Expect = 0.019
Identities = 21/58 (36%), Positives = 27/58 (46%)
Query: 24 GGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGRKGGLSTMEKSGGERAEEEG 81
GG +E E KGG+ E E YQE+GRKGG +T + E +E G
Sbjct: 55 GGEATSKNHDKEFYQEIGEKGGEATSENHDKEFYQEIGRKGGEATSKNHDKEFYQEIG 112
Score = 35.0 bits (79), Expect = 0.032
Identities = 26/86 (30%), Positives = 35/86 (40%), Gaps = 9/86 (10%)
Query: 24 GGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGRKGGLSTMEKSGGERAEEEGID 83
GG +E E KGG+ + E YQE+G KGG +T E E +E G
Sbjct: 35 GGEATSKNHDKEFYQEIGEKGGEATSKNHDKEFYQEIGEKGGEATSENHDKEFYQEIGRK 94
Query: 84 IDESKFKT---------GGGGGRSQN 100
E+ K G GG ++N
Sbjct: 95 GGEATSKNHDKEFYQEIGSKGGNARN 120
Score = 33.9 bits (76), Expect = 0.072
Identities = 24/75 (32%), Positives = 36/75 (48%), Gaps = 6/75 (8%)
Query: 7 NRQELEEKAKQGETVVPGGTGGKSLEAQEHLAEGRSKGGQTRKEQLGTEGYQEMGRKGGL 66
N+ EE ++G G T K+ + +E E KGG+ + E YQE+G KGG
Sbjct: 4 NKMSREEAGRKG-----GETTSKNHD-KEFYQEIGQKGGEATSKNHDKEFYQEIGEKGGE 57
Query: 67 STMEKSGGERAEEEG 81
+T + E +E G
Sbjct: 58 ATSKNHDKEFYQEIG 72
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.299 0.126 0.331
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,846,530
Number of Sequences: 164201
Number of extensions: 621275
Number of successful extensions: 1049
Number of sequences better than 10.0: 100
Number of HSP's better than 10.0 without gapping: 33
Number of HSP's successfully gapped in prelim test: 68
Number of HSP's that attempted gapping in prelim test: 934
Number of HSP's gapped (non-prelim): 166
length of query: 101
length of database: 59,974,054
effective HSP length: 77
effective length of query: 24
effective length of database: 47,330,577
effective search space: 1135933848
effective search space used: 1135933848
T: 11
A: 40
X1: 17 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 44 (22.0 bits)
S2: 58 (26.9 bits)
Medicago: description of AC140721.4


