

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140721.1 - phase: 0
(313 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
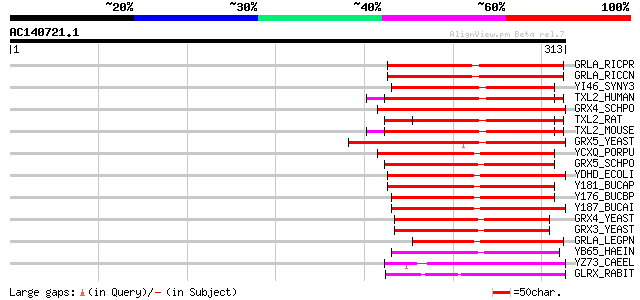
Score E
Sequences producing significant alignments: (bits) Value
GRLA_RICPR (O05957) Probable monothiol glutaredoxin grlA 105 1e-22
GRLA_RICCN (Q92GH5) Probable monothiol glutaredoxin grlA 105 2e-22
YI46_SYNY3 (P73056) Hypothetical UPF0055 protein slr1846 95 3e-19
TXL2_HUMAN (O76003) Thioredoxin-like protein 2 (PKC-interacting ... 94 3e-19
GRX4_SCHPO (Q9HDW8) Monothiol glutaredoxin 4 93 7e-19
TXL2_RAT (Q9JLZ1) Thioredoxin-like 2 protein (PKC-interacting co... 92 2e-18
TXL2_MOUSE (Q9CQM9) Thioredoxin-like protein 2 (PKC-interacting ... 92 2e-18
GRX5_YEAST (Q02784) Monothiol glutaredoxin 5, mitochondrial prec... 91 3e-18
YCXQ_PORPU (P51384) Hypothetical monothiol glutaredoxin in trpA-... 91 4e-18
GRX5_SCHPO (O74790) Monothiol glutaredoxin 5 91 4e-18
YDHD_ECOLI (P37010) Probable monothiol glutaredoxin ydhD 90 8e-18
Y181_BUCAP (Q8K9V6) Hypothetical monothiol glutaredoxin BUsg181 87 7e-17
Y176_BUCBP (Q89AR8) Hypothetical monothiol glutaredoxin bbp176 86 2e-16
Y187_BUCAI (P57284) Hypothetical monothiol glutaredoxin BU187 85 2e-16
GRX4_YEAST (P32642) Monothiol glutaredoxin 4 83 8e-16
GRX3_YEAST (Q03835) Monothiol glutaredoxin 3 82 1e-15
GRLA_LEGPN (Q48833) Probable monothiol glutaredoxin grlA 79 2e-14
YB65_HAEIN (P45085) Hypothetical monothiol glutaredoxin HI1165 77 4e-14
YZ73_CAEEL (Q19297) Hypothetical protein F10D7.3 in chromosome X 55 2e-07
GLRX_RABIT (P12864) Glutaredoxin (Thioltransferase) (TTase) 52 1e-06
>GRLA_RICPR (O05957) Probable monothiol glutaredoxin grlA
Length = 107
Score = 105 bits (262), Expect = 1e-22
Identities = 47/99 (47%), Positives = 69/99 (69%), Gaps = 3/99 (3%)
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETL 273
E + +K+NKVV F+KG++ P CGFS V+ IL K GV++ ++VL ++ LRE L
Sbjct: 8 EFIQNAIKKNKVVLFMKGTKEMPACGFSGTVVAILNKLGVEFSDINVL---FDTALREDL 64
Query: 274 KKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK 312
KK+S+WPTFPQ+++NG LVGGCDI + + GE+ + K
Sbjct: 65 KKFSDWPTFPQLYINGVLVGGCDIAKELYQNGELEKMLK 103
>GRLA_RICCN (Q92GH5) Probable monothiol glutaredoxin grlA
Length = 107
Score = 105 bits (261), Expect = 2e-22
Identities = 47/99 (47%), Positives = 70/99 (70%), Gaps = 3/99 (3%)
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETL 273
+ ++ +K NKVV F+KG + +P CGFS V+ IL K GV++ ++VL ++ LRE L
Sbjct: 8 KFIENEIKNNKVVLFMKGIKKSPACGFSGTVVAILNKLGVEFRDINVL---FDAELREDL 64
Query: 274 KKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK 312
KK+S+WPTFPQ+++NGELVGGCDI + + GE+ + K
Sbjct: 65 KKFSDWPTFPQLYINGELVGGCDIARELYQSGELEKMLK 103
>YI46_SYNY3 (P73056) Hypothetical UPF0055 protein slr1846
Length = 107
Score = 94.7 bits (234), Expect = 3e-19
Identities = 42/92 (45%), Positives = 65/92 (70%), Gaps = 3/92 (3%)
Query: 216 VDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKK 275
+D+LV NKV+ F+KG++ P CGFS V+ IL G+ +E++DVL + +R+ +K+
Sbjct: 9 IDQLVTANKVMVFMKGTKLMPQCGFSNNVVQILNMLGIPFETLDVLADAE---IRQGIKE 65
Query: 276 YSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
YSNWPT PQ+++NGE VGG DI+ + + GE+
Sbjct: 66 YSNWPTIPQVYVNGEFVGGSDIMIELYQNGEL 97
>TXL2_HUMAN (O76003) Thioredoxin-like protein 2 (PKC-interacting
cousin of thioredoxin) (PKC-theta-interacting protein)
(PKCq-interacting protein) (HUSSY-22)
Length = 335
Score = 94.4 bits (233), Expect = 3e-19
Identities = 42/101 (41%), Positives = 69/101 (67%), Gaps = 3/101 (2%)
Query: 212 LEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRE 271
LEE + L + V+ F+KG++ CGFS++++ IL GV+YE+ D+L+++ +R+
Sbjct: 235 LEERLKVLTNKASVMLFMKGNKQEAKCGFSKQILEILNSTGVEYETFDILEDEE---VRQ 291
Query: 272 TLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK 312
LK YSNWPT+PQ+++ GELVGG DI+ + E GE+ + +
Sbjct: 292 GLKAYSNWPTYPQLYVKGELVGGLDIVKELKENGELLPILR 332
Score = 78.6 bits (192), Expect = 2e-14
Identities = 36/106 (33%), Positives = 62/106 (57%), Gaps = 3/106 (2%)
Query: 202 PGRHVQLTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVL 261
P + L L + KL + F+KG+ P CGFS++++ IL K + + S D+
Sbjct: 123 PSANEHLKEDLNLRLKKLTHAAPCMLFMKGTPQEPRCGFSKQMVEILHKHNIQFSSFDIF 182
Query: 262 DEDYNYGLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
++ +R+ LK YS+WPT+PQ++++GEL+GG DI+ + E+
Sbjct: 183 SDEE---VRQGLKAYSSWPTYPQLYVSGELIGGLDIIKELEASEEL 225
>GRX4_SCHPO (Q9HDW8) Monothiol glutaredoxin 4
Length = 146
Score = 93.2 bits (230), Expect = 7e-19
Identities = 44/106 (41%), Positives = 70/106 (65%)
Query: 208 LTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNY 267
L+ + +++ VKE+ +V F+KG+ + P+CGFS K I IL E V + + + N
Sbjct: 21 LSTQTRQALEQAVKEDPIVLFMKGTPTRPMCGFSLKAIQILSLENVASDKLVTYNVLSND 80
Query: 268 GLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
LRE +K++S+WPT PQ+++NGE VGG DIL SM++ GE+ + K+
Sbjct: 81 ELREGIKEFSDWPTIPQLYINGEFVGGSDILASMHKSGELHKILKE 126
>TXL2_RAT (Q9JLZ1) Thioredoxin-like 2 protein (PKC-interacting
cousin of thioredoxin) (PKCq-interacting protein)
(PKC-theta-interacting protein)
Length = 279
Score = 91.7 bits (226), Expect = 2e-18
Identities = 40/101 (39%), Positives = 69/101 (67%), Gaps = 3/101 (2%)
Query: 212 LEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRE 271
LEE + L + V+ F+KG++ CGFS++++ IL GV+YE+ D+L+++ +R+
Sbjct: 179 LEERLKVLTNKASVMLFMKGNKQEAKCGFSKQILEILNSTGVEYETFDILEDEE---VRQ 235
Query: 272 TLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK 312
LK +SNWPT+PQ+++ G+LVGG DI+ + + GE+ + K
Sbjct: 236 GLKTFSNWPTYPQLYVRGDLVGGLDIVKELKDNGELLPILK 276
Score = 77.8 bits (190), Expect = 3e-14
Identities = 32/80 (40%), Positives = 54/80 (67%), Gaps = 3/80 (3%)
Query: 228 FIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKKYSNWPTFPQIFL 287
F+KG+ P CGFS++++ IL K + + S D+ ++ +R+ LK YSNWPT+PQ+++
Sbjct: 93 FMKGTPQEPRCGFSKQMVEILHKHNIQFSSFDIFSDEE---VRQGLKTYSNWPTYPQLYV 149
Query: 288 NGELVGGCDILTSMNEKGEV 307
+GEL+GG DI+ + E+
Sbjct: 150 SGELIGGLDIIKELEASEEL 169
>TXL2_MOUSE (Q9CQM9) Thioredoxin-like protein 2 (PKC-interacting
cousin of thioredoxin) (PKC-theta-interacting protein)
(PKCq-interacting protein)
Length = 337
Score = 91.7 bits (226), Expect = 2e-18
Identities = 40/101 (39%), Positives = 69/101 (67%), Gaps = 3/101 (2%)
Query: 212 LEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRE 271
LEE + L + V+ F+KG++ CGFS++++ IL GV+YE+ D+L+++ +R+
Sbjct: 237 LEERLKVLTNKASVMLFMKGNKQEAKCGFSKQILEILNSTGVEYETFDILEDEE---VRQ 293
Query: 272 TLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK 312
LK +SNWPT+PQ+++ G+LVGG DI+ + + GE+ + K
Sbjct: 294 GLKTFSNWPTYPQLYVRGDLVGGLDIVKELKDNGELLPILK 334
Score = 80.5 bits (197), Expect = 5e-15
Identities = 37/106 (34%), Positives = 62/106 (57%), Gaps = 3/106 (2%)
Query: 202 PGRHVQLTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVL 261
P + L L + KL + F+KG+ P CGFS++++ IL K + + S D+
Sbjct: 125 PSTNEHLKEDLSLRLKKLTHAAPCMLFMKGTPQEPRCGFSKQMVEILHKHNIQFSSFDIF 184
Query: 262 DEDYNYGLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
++ +R+ LK YSNWPT+PQ++++GEL+GG DI+ + E+
Sbjct: 185 SDEE---VRQGLKTYSNWPTYPQLYVSGELIGGLDIIKELEASEEL 227
>GRX5_YEAST (Q02784) Monothiol glutaredoxin 5, mitochondrial
precursor
Length = 150
Score = 91.3 bits (225), Expect = 3e-18
Identities = 45/125 (36%), Positives = 76/125 (60%), Gaps = 6/125 (4%)
Query: 192 PKKKADLRLTPGRHVQLTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKE 251
P +A L + L+ + + ++ ++ VV F+KG+ P CGFS+ IG+L +
Sbjct: 14 PILRAKTLLRYQNRMYLSTEIRKAIEDAIESAPVVLFMKGTPEFPKCGFSRATIGLLGNQ 73
Query: 252 GVD---YESVDVLDEDYNYGLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVA 308
GVD + + +VL++ LRE +K++S WPT PQ+++N E +GGCD++TSM GE+A
Sbjct: 74 GVDPAKFAAYNVLEDPE---LREGIKEFSEWPTIPQLYVNKEFIGGCDVITSMARSGELA 130
Query: 309 GLFKK 313
L ++
Sbjct: 131 DLLEE 135
>YCXQ_PORPU (P51384) Hypothetical monothiol glutaredoxin in
trpA-ycf12 intergenic region (ORF107)
Length = 107
Score = 90.9 bits (224), Expect = 4e-18
Identities = 38/100 (38%), Positives = 68/100 (68%), Gaps = 3/100 (3%)
Query: 208 LTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNY 267
+ + ++++++++ NK+V F+KGS+ P+CGFS I IL DY + D+L+ N
Sbjct: 1 MDIETKKVIEQILDNNKIVLFMKGSKLMPMCGFSNTAIQILNTLNTDYFTYDILE---NE 57
Query: 268 GLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
+R+ +K++S+WPT PQ+++N E VGG DI+ + E+GE+
Sbjct: 58 NIRQAIKEHSSWPTIPQLYINREFVGGADIMLELFEQGEL 97
>GRX5_SCHPO (O74790) Monothiol glutaredoxin 5
Length = 244
Score = 90.9 bits (224), Expect = 4e-18
Identities = 41/96 (42%), Positives = 65/96 (67%), Gaps = 3/96 (3%)
Query: 212 LEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRE 271
L E + L + V+ F+KG+ S P CGFS+K++G+L ++ V Y ++L +D +R+
Sbjct: 146 LNERLSTLTNAHNVMLFLKGTPSEPACGFSRKLVGLLREQNVQYGFFNILADD---SVRQ 202
Query: 272 TLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
LK +S+WPTFPQ+++ GE VGG DI++ M E GE+
Sbjct: 203 GLKVFSDWPTFPQLYIKGEFVGGLDIVSEMIENGEL 238
>YDHD_ECOLI (P37010) Probable monothiol glutaredoxin ydhD
Length = 115
Score = 89.7 bits (221), Expect = 8e-18
Identities = 42/100 (42%), Positives = 64/100 (64%), Gaps = 3/100 (3%)
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETL 273
E + + + EN ++ ++KGS P CGFS + + L G + VD+L N +R L
Sbjct: 6 EKIQRQIAENPILLYMKGSPKLPSCGFSAQAVQALAACGERFAYVDILQ---NPDIRAEL 62
Query: 274 KKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
KY+NWPTFPQ++++GELVGGCDI+ M ++GE+ L K+
Sbjct: 63 PKYANWPTFPQLWVDGELVGGCDIVIEMYQRGELQQLIKE 102
>Y181_BUCAP (Q8K9V6) Hypothetical monothiol glutaredoxin BUsg181
Length = 107
Score = 86.7 bits (213), Expect = 7e-17
Identities = 37/94 (39%), Positives = 63/94 (66%), Gaps = 3/94 (3%)
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETL 273
E +++ +K+N ++ ++KG+ +P CGFS + + L G + VD+L+ N +R+ L
Sbjct: 5 EKIERQIKDNIILIYMKGTPQSPSCGFSAQAVQALSICGEKFAYVDILE---NLDIRKEL 61
Query: 274 KKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
+Y+NWPTFPQ+++ GEL+GGC I+ M E GE+
Sbjct: 62 PRYANWPTFPQLWIKGELIGGCSIILEMLENGEL 95
>Y176_BUCBP (Q89AR8) Hypothetical monothiol glutaredoxin bbp176
Length = 108
Score = 85.5 bits (210), Expect = 2e-16
Identities = 37/92 (40%), Positives = 60/92 (65%), Gaps = 3/92 (3%)
Query: 216 VDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKK 275
+ ++ N ++ ++KGS AP CGFS + + + G + +DVL N +R L K
Sbjct: 8 IQNQIQNNPIIIYMKGSPDAPSCGFSAQAVHAISSCGKKFAYIDVLK---NPDIRLELPK 64
Query: 276 YSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
Y+NWPTFPQ+++NGEL+GGC+I+ + +KGE+
Sbjct: 65 YANWPTFPQLWVNGELIGGCNIILELFQKGEL 96
>Y187_BUCAI (P57284) Hypothetical monothiol glutaredoxin BU187
Length = 108
Score = 85.1 bits (209), Expect = 2e-16
Identities = 39/98 (39%), Positives = 63/98 (63%), Gaps = 3/98 (3%)
Query: 216 VDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKK 275
+ K +++N ++ ++KG+ AP CGFS + + L G + V++L+ N +R L K
Sbjct: 7 IKKQIQDNIILIYMKGTPEAPSCGFSAQAVQALSFCGEKFAYVNILE---NPDIRSELPK 63
Query: 276 YSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
Y+NWPTFPQ++++GEL+GGC I+ M E GE+ L K
Sbjct: 64 YANWPTFPQLWIDGELIGGCSIILEMLENGELKKLILK 101
>GRX4_YEAST (P32642) Monothiol glutaredoxin 4
Length = 244
Score = 83.2 bits (204), Expect = 8e-16
Identities = 38/87 (43%), Positives = 61/87 (69%), Gaps = 3/87 (3%)
Query: 218 KLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKKYS 277
KLV+ V+ F+KGS S P CGFS++++GIL + + + D+L ++ +R++LKK+S
Sbjct: 151 KLVQAAPVMLFMKGSPSEPKCGFSRQLVGILREHQIRFGFFDILRDE---NVRQSLKKFS 207
Query: 278 NWPTFPQIFLNGELVGGCDILTSMNEK 304
+WPTFPQ+++NGE GG DI+ E+
Sbjct: 208 DWPTFPQLYINGEFQGGLDIIKESIEE 234
>GRX3_YEAST (Q03835) Monothiol glutaredoxin 3
Length = 285
Score = 82.4 bits (202), Expect = 1e-15
Identities = 39/87 (44%), Positives = 58/87 (65%), Gaps = 3/87 (3%)
Query: 218 KLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKKYS 277
KLV V+ F+KGS S P CGFS++++GIL + V + D+L ++ +R+ LKK+S
Sbjct: 191 KLVNAAPVMLFMKGSPSEPKCGFSRQLVGILREHQVRFGFFDILRDE---SVRQNLKKFS 247
Query: 278 NWPTFPQIFLNGELVGGCDILTSMNEK 304
WPTFPQ+++NGE GG DI+ E+
Sbjct: 248 EWPTFPQLYINGEFQGGLDIIKESLEE 274
>GRLA_LEGPN (Q48833) Probable monothiol glutaredoxin grlA
Length = 89
Score = 78.6 bits (192), Expect = 2e-14
Identities = 34/85 (40%), Positives = 57/85 (67%), Gaps = 3/85 (3%)
Query: 228 FIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKKYSNWPTFPQIFL 287
++KG+ P CGFS + + +E GVD+ VD+L N +R+ L ++S+WPTFPQ+++
Sbjct: 3 YMKGTPKMPQCGFSARAVQCIEACGVDFAYVDILA---NPDIRQVLPQFSDWPTFPQLYV 59
Query: 288 NGELVGGCDILTSMNEKGEVAGLFK 312
GEL+GG DI+ M ++GE+ + +
Sbjct: 60 KGELIGGSDIIAEMFQQGELEPMLR 84
>YB65_HAEIN (P45085) Hypothetical monothiol glutaredoxin HI1165
Length = 120
Score = 77.4 bits (189), Expect = 4e-14
Identities = 37/95 (38%), Positives = 55/95 (56%), Gaps = 3/95 (3%)
Query: 216 VDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKK 275
+ K + EN ++ ++KGS P CGF + L V + VD+L +R L
Sbjct: 20 IKKQISENPILIYMKGSPKLPSCGFPARASEALMHCKVPFGYVDILQHP---DIRAELPT 76
Query: 276 YSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGL 310
Y+NWPTFPQ+++ GEL+GGCDI+ M + GE+ L
Sbjct: 77 YANWPTFPQLWVEGELIGGCDIILEMYQAGELQTL 111
>YZ73_CAEEL (Q19297) Hypothetical protein F10D7.3 in chromosome X
Length = 146
Score = 55.5 bits (132), Expect = 2e-07
Identities = 36/106 (33%), Positives = 60/106 (55%), Gaps = 9/106 (8%)
Query: 212 LEELVDKLVKE---NKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDE-DYNY 267
L++L DK+V + +KV+ + K C +S+++ IL +D + LD +
Sbjct: 30 LKDLEDKIVNDVMTHKVMVYSK-----TYCPWSKRLKAILANYEIDDMKIVELDRSNQTE 84
Query: 268 GLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
++E LKKYS T PQ+F++G+ VGG D ++ EKGE+ L +K
Sbjct: 85 EMQEILKKYSGRTTVPQLFISGKFVGGHDETKAIEEKGELRPLLEK 130
>GLRX_RABIT (P12864) Glutaredoxin (Thioltransferase) (TTase)
Length = 106
Score = 52.4 bits (124), Expect = 1e-06
Identities = 32/102 (31%), Positives = 57/102 (55%), Gaps = 4/102 (3%)
Query: 213 EELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILE-KEGVDYESVDVLDEDYNYGLRE 271
+E V+ ++ KVV FIK + P C +Q+++ L K+G+ E VD+ +++
Sbjct: 2 QEFVNSKIQPGKVVVFIKPT--CPYCRKTQEILSQLPFKQGL-LEFVDITATSDMSEIQD 58
Query: 272 TLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
L++ + T P++FL + +GGC L +M EKGE+ K+
Sbjct: 59 YLQQLTGARTVPRVFLGKDCIGGCSDLIAMQEKGELLARLKE 100
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,914,102
Number of Sequences: 164201
Number of extensions: 1866347
Number of successful extensions: 3646
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 42
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 3576
Number of HSP's gapped (non-prelim): 71
length of query: 313
length of database: 59,974,054
effective HSP length: 110
effective length of query: 203
effective length of database: 41,911,944
effective search space: 8508124632
effective search space used: 8508124632
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 66 (30.0 bits)
Medicago: description of AC140721.1


