

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
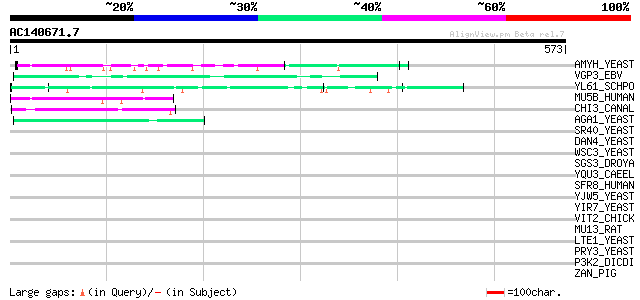
Query= AC140671.7 - phase: 0
(573 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3) (G... 53 2e-06
VGP3_EBV (P03200) Envelope glycoprotein GP340/GP220 (Membrane an... 48 8e-05
YL61_SCHPO (Q8TFG9) Hypothetical serine/threonine-rich protein P... 46 2e-04
MU5B_HUMAN (Q9HC84) Mucin 5B precursor (Mucin 5 subtype B, trach... 46 3e-04
CHI3_CANAL (P40954) Chitinase 3 precursor (EC 3.2.1.14) 45 4e-04
AGA1_YEAST (P32323) A-agglutinin attachment subunit precursor 45 5e-04
SR40_YEAST (P32583) Suppressor protein SRP40 44 0.001
DAN4_YEAST (P47179) Cell wall protein DAN4 precursor 44 0.001
WSC3_YEAST (Q12215) Cell wall integrity and stress response comp... 43 0.002
SGS3_DROYA (P13728) Salivary glue protein Sgs-3 precursor 43 0.003
YQU3_CAEEL (Q09550) Hypothetical protein F26C11.3 in chromosome II 42 0.003
SFR8_HUMAN (Q12872) Splicing factor, arginine/serine-rich 8 (Sup... 42 0.003
YJW5_YEAST (P40889) Hypothetical 197.6 kDa protein in FSP2 5'region 42 0.004
YIR7_YEAST (P40434) Hypothetical 197.5 kDa protein in SDL1 5'region 42 0.004
VIT2_CHICK (P02845) Vitellogenin II precursor (Major vitellogeni... 42 0.004
MU13_RAT (P97881) Mucin 13 precursor 42 0.004
LTE1_YEAST (P07866) Low temperature essential protein 41 0.007
PRY3_YEAST (P47033) PRY3 protein (Pathogen related in Sc 3) 41 0.010
P3K2_DICDI (P54674) Phosphatidylinositol 3-kinase 2 (EC 2.7.1.13... 41 0.010
ZAN_PIG (Q28983) Zonadhesin precursor 40 0.013
>AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3)
(Glucan 1,4-alpha-glucosidase) (1,4-alpha-D-glucan
glucohydrolase)
Length = 1367
Score = 53.1 bits (126), Expect = 2e-06
Identities = 92/421 (21%), Positives = 147/421 (34%), Gaps = 51/421 (12%)
Query: 9 TTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPP------R 62
TTT TK PTP P K ++ T+ SSS++ SS P
Sbjct: 276 TTTSCTKEKPTPPHHDTTPCTKKKTTTSKTCTKKTTTPVPTPSSSTTESSSAPVPTPSSS 335
Query: 63 RTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQS---TERQRQRQGTPRPNGETPVAQKML 119
T S T ++T+ S P TP++ S S T + P + T + +
Sbjct: 336 TTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPV 395
Query: 120 FTSARSLSVSFQGESFSIPV------SKAKPPSATVTQS----LRRSTPERRKVPATVTT 169
T + S + ES S PV S + P +++ T+S + ST E P T +T
Sbjct: 396 PTPSSSTT-----ESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSST 450
Query: 170 PTIGRGRVNGNSDQTENS-----ISRSGDQHRWPAKSQQNQANFMNRSLDCGVLLRSSNG 224
V S T S S + + P + + + + SS+
Sbjct: 451 TESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSA 510
Query: 225 FGSNVVRSLRNSLLDPRVSHDGATLRSESNKNGGSEPVIEPELVPSDNESVTSGSSSGVL 284
S S P + +T S S S VP+ + S T SS+ V
Sbjct: 511 PVPTPSSSTTESSSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSTPVT 570
Query: 285 DYGGGKPQRSARAIVVPARFLQEATNPSSRNGGIGNRSTVPPKLLVPKKSVFDS---PAS 341
+ S+ + P+ E+++ + VP P S +S PA
Sbjct: 571 ---SSTTESSSAPVPTPSSSTTESSS-----------APVP----TPSSSTTESSSAPAP 612
Query: 342 SPRGIVNNRLQGSPIRSAVRPASPSKLGTPSPRSPSRGVSPCRGRNGVASSLSSRFVNEP 401
+P +P+ S+ +S + + TPS + +P + + SS V P
Sbjct: 613 TPSSSTTES-SSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTP 671
Query: 402 S 402
S
Sbjct: 672 S 672
Score = 51.6 bits (122), Expect = 5e-06
Identities = 65/282 (23%), Positives = 114/282 (40%), Gaps = 46/282 (16%)
Query: 8 TTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPP------ 61
+++T + AP PT P +ES +A V P + S +SS++ SS P
Sbjct: 642 SSSTTESSSAPVPT-PSSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTT 700
Query: 62 RRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFT 121
+++P+ + ++T+ S P TP++ S S P P+ T + T
Sbjct: 701 ESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAP-------VPTPSSSTTESSSAPVT 753
Query: 122 SARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGRVNGNS 181
S+ + ES S PV PS++ T+S P P++ TT + +S
Sbjct: 754 SSTT-------ESSSAPV---PTPSSSTTESSSAPVP----TPSSSTTESSSAPVPTPSS 799
Query: 182 DQTENSISRSGDQHRWPAKSQQNQANFMNRSLDCGVLLRSSNGFGSNVVRSLRNSLLDPR 241
TE+S++ P + + +N + + SS F S+ S S+ P
Sbjct: 800 STTESSVA--------PVPTPSSSSNITSSA-------PSSTPFSSSTESS---SVPVPT 841
Query: 242 VSHDGATLRSESNKNGGSEPVIEPELVPSDNESVTSGSSSGV 283
S S + +E + P PS + ++TS + S +
Sbjct: 842 PSSSTTESSSAPVSSSTTESSVAPVPTPSSSSNITSSAPSSI 883
Score = 50.8 bits (120), Expect = 9e-06
Identities = 90/422 (21%), Positives = 150/422 (35%), Gaps = 51/422 (12%)
Query: 7 TTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNS 66
++TTT +T + T T SES ++ TS SS+SSS T +
Sbjct: 210 SSTTTSSTSESSTTTSST---SESSTTTSSTSESSTTTSSTSESSTSSS--------TTA 258
Query: 67 PLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARSL 126
P T + TK+K TP T + + + TP + TP +K TS
Sbjct: 259 PATPTTTSCTKEKPTPPTTTSC-----------TKEKPTPPHHDTTPCTKKKTTTSKTCT 307
Query: 127 S---------VSFQGESFSIPV-----SKAKPPSATVTQSLRRSTPERRKVPATVTTPTI 172
S ES S PV S + SA VT S S+ P++ TT +
Sbjct: 308 KKTTTPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESS 367
Query: 173 GRGRVNGNSDQTENSISRSGDQHRWPAKSQQNQANFMNRSLDCGVLLRSSNGFGSNVVRS 232
+ ++ + ++ S + + ++ S V ++ + V S
Sbjct: 368 SAPVTSSTTESSSAPVTSSTTES--SSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSS 425
Query: 233 LRNSLLDPRVSHDGATLRSESNKNGGSEPVIEPELVPSDNESVTSGSSSGVLDYGGGKPQ 292
S P S +T S S S VP+ + S T SS+ V +
Sbjct: 426 TTESSSAPVTS---STTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVT---SSTTE 479
Query: 293 RSARAIVVPARFLQEATNPSSRNGGIGNRSTVPPKLLVPKKSVFDS---PASSPRGIVNN 349
S+ + P+ E+++ + + S P P S +S PA +P
Sbjct: 480 SSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVP---TPSSSTTESSSAPAPTPSSSTTE 536
Query: 350 RLQGSPIRSAVRPASPSKLGTPSPRSPSRGVSPCRGRNGVASSLSSRFVNEPSVLSFAVD 409
+P+ S+ +S + + TPS + +P +SS + + S +
Sbjct: 537 S-SSAPVTSSTTESSSAPVPTPSSSTTESSSTPVTSSTTESSSAPVPTPSSSTTESSSAP 595
Query: 410 VP 411
VP
Sbjct: 596 VP 597
Score = 50.1 bits (118), Expect = 2e-05
Identities = 87/415 (20%), Positives = 150/415 (35%), Gaps = 38/415 (9%)
Query: 8 TTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSF------SSPP 61
T++T + AP PT P +ES +A V ++ S++ SSS SS
Sbjct: 345 TSSTTESSSAPVPT-PSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTT 403
Query: 62 RRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQR----QGTPRPNGETPVAQK 117
+++P+ + T S+ T S T ++ + ST T + P
Sbjct: 404 ESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSS 463
Query: 118 MLFTSARSLSVSFQGESFSIPV-----SKAKPPSATVTQSLRRSTPERRKVPATVTTPTI 172
S+ + S ES S PV S + SA VT S S+ P++ TT +
Sbjct: 464 STTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESS 523
Query: 173 GRGRVNGNSDQTENSISRSGDQHRWPAKSQQNQANFMNRSLDCGVLLRSSNGFGSNVVRS 232
+S TE+S + P S +++ SS+ + V S
Sbjct: 524 SAPAPTPSSSTTESSSA--------PVTSSTTESSSAPVPTPSSSTTESSS---TPVTSS 572
Query: 233 LRNSLLDPRVSHDGATLRSESN-----KNGGSEPVIEPELVPSDNESVTSGSSSGVLDYG 287
S P + +T S S + +E P P+ + S T SS+ V
Sbjct: 573 TTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAP--APTPSSSTTESSSAPVTS-- 628
Query: 288 GGKPQRSARAIVVPARFLQEATNPSSRNGGIGNRSTVPPKLLVPKKSVFDSPASSPRGIV 347
+ S+ + P+ E+++ + + P S +S +S+P
Sbjct: 629 -STTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTES-SSAPVTSS 686
Query: 348 NNRLQGSPIRSAVRPASPSKLGTPSPRSPSRGVSPCRGRNGVASSLSSRFVNEPS 402
+P+ S+ +S + + TPS + +P + + SS V PS
Sbjct: 687 TTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPS 741
Score = 43.1 bits (100), Expect = 0.002
Identities = 50/194 (25%), Positives = 80/194 (40%), Gaps = 29/194 (14%)
Query: 8 TTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSF---------S 58
T++T + AP PT P +ES +A V P + S + + SSS S
Sbjct: 570 TSSTTESSSAPVPT-PSSSTTESSSAPVPTPSSSTTESSSAPAPTPSSSTTESSSAPVTS 628
Query: 59 SPPRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKM 118
S +++P+ + ++T+ S P TP++ S S P P+ T +
Sbjct: 629 STTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAP-------VPTPSSSTTESSSA 681
Query: 119 LFTSARSLS-----VSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIG 173
TS+ + S S ES S PV PS++ T+S P P++ TT +
Sbjct: 682 PVTSSTTESSSAPVTSSTTESSSAPV---PTPSSSTTESSSAPVP----TPSSSTTESSS 734
Query: 174 RGRVNGNSDQTENS 187
+S TE+S
Sbjct: 735 APVPTPSSSTTESS 748
Score = 41.2 bits (95), Expect = 0.007
Identities = 57/249 (22%), Positives = 95/249 (37%), Gaps = 37/249 (14%)
Query: 8 TTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSP 67
+++T + AP PT P +ES +A V P + SSS+ SS +++P
Sbjct: 711 SSSTTESSSAPVPT-PSSSTTESSSAPVPTPSSSTT------ESSSAPVTSSTTESSSAP 763
Query: 68 LVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPN---GETPVAQKMLFTSA- 123
+ + ++T+ S P TP++ S S T + TP + + +SA
Sbjct: 764 VPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSVAPVPTPSSSSNITSSAP 823
Query: 124 RSLSVSFQGESFSIPV---------SKAKPPSATVTQSLRRSTP-----------ERRKV 163
S S ES S+PV S + P S++ T+S P +
Sbjct: 824 SSTPFSSSTESSSVPVPTPSSSTTESSSAPVSSSTTESSVAPVPTPSSSSNITSSAPSSI 883
Query: 164 PATVTTPTIGRGRVNGNS------DQTENSISRSGDQHRWPAKSQQNQANFMNRSLDCGV 217
P + TT + G S QTE S+S + + P K+ + ++ V
Sbjct: 884 PFSSTTESFSTGTTVTPSSSKYPGSQTETSVSSTTETTIVPTKTTTSVTTPSTTTITTTV 943
Query: 218 LLRSSNGFG 226
+N G
Sbjct: 944 CSTGTNSAG 952
>VGP3_EBV (P03200) Envelope glycoprotein GP340/GP220 (Membrane
antigen) (MA)
Length = 907
Score = 47.8 bits (112), Expect = 8e-05
Identities = 86/380 (22%), Positives = 133/380 (34%), Gaps = 76/380 (20%)
Query: 5 ISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRT 64
+ST T T T P+ PS S + KA ++TS ++ + + +SP
Sbjct: 468 VSTADVTSPTPAGTTSGASPVTPSPSPWDNGTESKAPDMTSSTSPVTTPTPNATSPTPAV 527
Query: 65 NSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSAR 124
+P N T TP+VT TP PN +P K TS
Sbjct: 528 TTPTPNAT------SPTPAVT-------------------TPTPNATSPTLGK---TSPT 559
Query: 125 SLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPAT--VTTPTIGR-GRVNGNS 181
S + + S + K P SA T + ++P K T VTTPT G G +
Sbjct: 560 SAVTTPTPNATSPTLGKTSPTSAVTTPTPNATSPTLGKTSPTSAVTTPTPNATGPTVGET 619
Query: 182 DQTENSISRS--GDQHRWPAKSQQNQANFMNRSLDCGVLLRSSNGFGSNVVRSLRNSLLD 239
N+ + + G SQ A S+ G + + S S +
Sbjct: 620 SPQANATNHTLGGTSPTPVVTSQPKNAT-------------SAVTTGQHNITSSSTSSMS 666
Query: 240 PRVSHDGATLRSESNKNGGSEPVIEPELVPSDNESVTSGSSSGVLDYGGGKPQRSARAIV 299
R S + TL ++ N S + P+ E++T + + + + + R
Sbjct: 667 LRPSSNPETLSPSTSDNSTSHMPLLTSAHPTGGENITQVTPASISTHHVSTSSPAPRP-- 724
Query: 300 VPARFLQEATNPSSRNGGIGNRSTVPPKLLVPKKSVFDSPASSPRGIVNNRLQGSPIRSA 359
+S+ G GN ST S+ G VN +G+P ++A
Sbjct: 725 ----------GTTSQASGPGNSST-----------------STKPGEVNVT-KGTPPQNA 756
Query: 360 VRPASPSKLGTPSPRSPSRG 379
P +PS T P S G
Sbjct: 757 TSPQAPSGQKTAVPTVTSTG 776
>YL61_SCHPO (Q8TFG9) Hypothetical serine/threonine-rich protein
PB15E9.01c in chromosome I precursor
Length = 943
Score = 46.2 bits (108), Expect = 2e-04
Identities = 87/424 (20%), Positives = 149/424 (34%), Gaps = 34/424 (8%)
Query: 3 TAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPR 62
+++++++T T +PT + + S + + + S + SSS +SS +
Sbjct: 89 SSLASSSTNSTTSASPTSSSLTSSSATSSSLASSSTTSSSLASSSITSSSLASSSITSSS 148
Query: 63 RTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTS 122
+S + ++ S+ ST S TP + S + T + + TS
Sbjct: 149 LASSSTTSSSLASSSTNSTTSATPTSSATSSSLSSTAASNSATSSSLASSSLNST---TS 205
Query: 123 ARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGRVNGNSD 182
A + S S S S + S+ + SL +T AT T+ +I +
Sbjct: 206 ATATSSSLS----STAASNSATSSSLASSSLNSTT------SATATSSSISSTVSSSTPL 255
Query: 183 QTENSISRSGDQHRWPAKSQQNQANFMNRSLDCGVLLRSSNGFGSNVVRS--LRNSLLDP 240
+ NS + + + +Q N ++ + S L S+N + S L +
Sbjct: 256 TSSNSTTAATSASATSSSAQYNTSSLLPSSTPSSTPLSSANSTTATSASSTPLTSVNSTT 315
Query: 241 RVSHDGATLRSESNKNGGSEPVIEPELVPSDNESVTSGSSSGVLDYGGGKPQRSARAIVV 300
S L S S+ N + + S N + + +SS L SA + +
Sbjct: 316 TTSASSTPLSSVSSANSTTATSTSSTPLSSVNSTTATSASSTPLTSVNSTTATSASSTPL 375
Query: 301 PARFLQEATNPSSRNGGIGN-------RSTVP---PKLLVPKKSVFDSPASSPRGIVNNR 350
+ AT+ SS N ST P ++P SV +P SS
Sbjct: 376 TSVNSTSATSASSTPLTSANSTTSTSVSSTAPSYNTSSVLPTSSVSSTPLSSANSTTATS 435
Query: 351 LQGSPIRSAVRPASPSKLGTP-----SPRSPSRGVSPCRGRNGV----ASSLSSRFVNEP 401
+P+ S + S TP S + S +P N ASS VN
Sbjct: 436 ASSTPLSSVNSTTATSASSTPLSSVNSTTATSASSTPLTSVNSTTATSASSTPLTSVNST 495
Query: 402 SVLS 405
S S
Sbjct: 496 SATS 499
Score = 45.4 bits (106), Expect = 4e-04
Identities = 90/438 (20%), Positives = 157/438 (35%), Gaps = 31/438 (7%)
Query: 41 REVTSRYMYSSSSSSSFS-------SPPRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRS 93
+ T+ Y Y++SS+SS S S ++S L + + NST S S + + S
Sbjct: 57 KRATTSYNYNTSSASSSSLTSSSAASSSLTSSSSLASSSTNSTTSASPTSSSLTSSSATS 116
Query: 94 QSTERQRQRQGTPRPNGETPVAQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSL 153
S + + T + ++ SL+ S S S SAT T S
Sbjct: 117 SSLASSSTTSSSLASSSITSSSLASSSITSSSLASSSTTSSSLASSSTNSTTSATPTSSA 176
Query: 154 RRSTPERRKVPATVTTPTIGRGRVNGNSDQTENSISRSGDQHRWPAKSQQNQANFMNRSL 213
S+ + T+ ++ +N + T S S S A S ++ +N +
Sbjct: 177 TSSSLSSTAASNSATSSSLASSSLNSTTSATATSSSLSSTAASNSATSSSLASSSLNST- 235
Query: 214 DCGVLLRSSNGFGSNVVRSLRNSLLDPRVSHDGATLRSESNKNGGSEPVIEPELVPSDNE 273
S+ S++ ++ +S P S + T + ++ S L+PS
Sbjct: 236 ------TSATATSSSISSTVSSS--TPLTSSNSTTAATSASATSSSAQYNTSSLLPSSTP 287
Query: 274 SVT--SGSSSGVLDYGGGKPQRSARAIVVPARFLQEATNPSSRNGGIGNRSTVPPKLLVP 331
S T S ++S P S + + ++ SS N ++ P L
Sbjct: 288 SSTPLSSANSTTATSASSTPLTSVNSTTTTSASSTPLSSVSSANSTTATSTSSTP-LSSV 346
Query: 332 KKSVFDSPASSPRGIVNNRLQGSPIRSAVRPASPSKLGTPSPRSPSRGVSPCRGRNGVAS 391
+ S +S+P VN+ +A +S S + S +P N S
Sbjct: 347 NSTTATSASSTPLTSVNS-------TTATSASSTPLTSVNSTSATSASSTPLTSANSTTS 399
Query: 392 -SLSSRFVNEPSVLSFAVDVPRGKIGENRVIDAHSLRLLHNRLMQWRFVNARADASLSVQ 450
S+SS PS + +V +P + + A+S VN+ S S
Sbjct: 400 TSVSS---TAPSYNTSSV-LPTSSVSSTPLSSANSTTATSASSTPLSSVNSTTATSASST 455
Query: 451 TLNSEKSLYAAWVATSKL 468
L+S S A +++ L
Sbjct: 456 PLSSVNSTTATSASSTPL 473
Score = 44.7 bits (104), Expect = 7e-04
Identities = 76/341 (22%), Positives = 133/341 (38%), Gaps = 36/341 (10%)
Query: 2 VTAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPP 61
+T+ ++TT+T + AP+ +LP+ S ++ + T+ SS+ SS +S
Sbjct: 391 LTSANSTTSTSVSSTAPSYNTSSVLPTSSVSSTPLS--SANSTTATSASSTPLSSVNSTT 448
Query: 62 RRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFT 121
+ S +VNST S S TP V + +T + T + L +
Sbjct: 449 ATSASSTPLSSVNSTTATSASS-TPLTSVNSTTATSASSTPLTSVNSTSATSASSTPLTS 507
Query: 122 --SARSLSVSFQGESF------------SIPVSKAKPPSATVTQSLRRSTPERRKVPATV 167
S S SVS S+ S P+S A +AT S ++ +
Sbjct: 508 ANSTTSTSVSSTAPSYNTSSVLPTSSVSSTPLSSANSTTATSASSTPLTSVNSTTATSAS 567
Query: 168 TTPTIGRGRV----NGNSDQTENSISRSGDQHRWPAKSQQNQANFMNRSLDCGVLLRSSN 223
+TP G + +G++ + N+ S +GD + ++ S +S+
Sbjct: 568 STP-FGNSTITSSASGSTGEFTNTNSGNGDVS---GSVTTPTSTPLSNSTVAPTSTFTSS 623
Query: 224 GFGSNVVRSLRNSLLDPRVSHDGATLRSESNKNGGSEPVIEPELVPSDNESVTSGSSSGV 283
GF N L S + +S+ A S S+ + ++P++ TSGS++G
Sbjct: 624 GF--NTTSGLPTSSVSTPLSNSSAYPSSGSSTFSRLSSTLTSSIIPTETFGSTSGSATGT 681
Query: 284 LDYGGGKPQRSARAIVVPARFLQEATNPSSRNGGIGNRSTV 324
G S++ VVP +T S + G G + V
Sbjct: 682 RPTGS-----SSQGSVVPT----TSTGSSVTSTGTGTTTGV 713
>MU5B_HUMAN (Q9HC84) Mucin 5B precursor (Mucin 5 subtype B,
tracheobronchial) (High molecular weight salivary mucin
MG1) (Sublingual gland mucin)
Length = 5703
Score = 45.8 bits (107), Expect = 3e-04
Identities = 47/179 (26%), Positives = 73/179 (40%), Gaps = 13/179 (7%)
Query: 2 VTAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPP 61
++ +T TTT T T T P +P + A V V + M + SSS+ S P
Sbjct: 4560 ISTTTTPTTTTPTTSGSTVT-PSSIPGTTHTARVLTTTTTTVATGSMATPSSSTQTSGTP 4618
Query: 62 RR--TNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQ-----STERQRQRQGTPRPNGETPV 114
T + + T ++T STP TP V S +T + +PR PV
Sbjct: 4619 PSLTTTATTITATGSTTNPSSTPGTTPIPPVLTSMATTPAATSSKATSSSSPRTATTLPV 4678
Query: 115 ----AQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTT 169
A K TS + S ++++P ++ P +T++ STPE +TT
Sbjct: 4679 LTSTATKSTATSFTPIPSSTLWTTWTVP-AQTTTPMSTMSTIHTSSTPETTHTSTVLTT 4736
Score = 39.7 bits (91), Expect = 0.022
Identities = 56/192 (29%), Positives = 77/192 (39%), Gaps = 38/192 (19%)
Query: 5 ISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRR- 63
ISTTTT T R T T P +P + A V V + M + SSS+ S P
Sbjct: 3304 ISTTTTP--TTRGSTVT-PSSIPGTTHTATVLTTTTTTVATGSMATPSSSTQTSGTPPSL 3360
Query: 64 -TNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTS 122
T + + T ++T STP TP V + +T TP A T
Sbjct: 3361 TTTATTITATGSTTNPSSTPGTTPIPPVLTTTAT---------------TPAATSSTVTP 3405
Query: 123 ARSLSVSFQGESFSIPVSKAKPPSATVT---QSLRRSTPERRKVPATVTTPTIGRGRVNG 179
+ +L G + + PV P+ T T +SL S+P VP T+ T G + G
Sbjct: 3406 SSAL-----GTTHTPPV-----PNTTATTHGRSLPPSSP--HTVPTAWTSATSG---ILG 3450
Query: 180 NSDQTENSISRS 191
+ TE S S
Sbjct: 3451 TTHITEPSTGTS 3462
Score = 38.5 bits (88), Expect = 0.048
Identities = 56/211 (26%), Positives = 86/211 (40%), Gaps = 36/211 (17%)
Query: 5 ISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYS-SSSSSSFSSPPR- 62
ISTTTT T R T T P +P + A V V + M + SSS+ + +PP
Sbjct: 2606 ISTTTTP--TTRGSTVT-PSSIPGTTHTATVLTTTTTTVATGSMATPSSSTQTSGTPPSL 2662
Query: 63 RTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTS 122
T + + T ++T STP P V + +T TP A T
Sbjct: 2663 TTTATTITATGSTTNPSSTPGTRPIPPVLTTTAT---------------TPAATSSTVTP 2707
Query: 123 ARSLSVSFQGESFSIPVSKAKPPSATVT---QSLRRSTPER-RKVPATVTTPTIGRGRVN 178
+ +L G + + PV P+ T T +SL S+P R + T+ T+G +
Sbjct: 2708 SSAL-----GTTHTPPV-----PNTTATTHGRSLSPSSPHTVRTAWTSATSGTLGTTHIT 2757
Query: 179 GNSDQTEN--SISRSGDQHRWPAKSQQNQAN 207
S T + + + QH PA S + ++
Sbjct: 2758 EPSTGTSHTPAATTGTTQHSTPALSSPHPSS 2788
Score = 38.5 bits (88), Expect = 0.048
Identities = 94/428 (21%), Positives = 152/428 (34%), Gaps = 47/428 (10%)
Query: 6 STTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRR-- 63
+TTTTT+ P PT P L + P E + Y+S+ ++ + PR+
Sbjct: 1620 ATTTTTQALFSTPQPTSSPGLTRAPPASTTAVPTLSEGLTSPRYTSTLGTATTGGPRQSA 1679
Query: 64 -TNSPLVNRTVNST--KQKSTPSVT----------PAAFVKRSQSTERQRQRQGTPRPNG 110
+ P V ST + + P T P + +T R R GT
Sbjct: 1680 GSTEPTVPGVATSTLPTRSALPGTTGSLGTWRPSQPPTLAPTTMATSRARP-TGTASTAS 1738
Query: 111 ETPVAQKMLFTSARSLSVSFQGESFSIPVSKAKPP--SATVTQSLRRSTPERRKVP-ATV 167
+ P+ + T LS S Q E+ + P + T +Q R P+ V
Sbjct: 1739 KEPLTTSLAPTLTSELSTS-QAETSTPRTETTMSPLTNTTTSQGTTRCQPKCEWTEWFDV 1797
Query: 168 TTPTIGRGRVNGNSDQTENSISRSGDQHRWPAKSQQNQA------------NFMNRSLDC 215
PT G V +T +I +G + W KS + +A + SL+
Sbjct: 1798 DFPTSG---VASGDMETFENIRAAGGKMCWAPKSIECRAENYPEVSIDQVGQVLTCSLET 1854
Query: 216 GVLLRSSNGFGS-NVVRSLR-NSLLDPRVSHDGATLRSESNKNGGSEP-----VIEPELV 268
G+ ++ + G N+ + L SH +TL + S S P + +P
Sbjct: 1855 GLTCKNEDQTGRFNMCFNYNVRVLCCDDYSHCPSTLATSSTATPSSTPGTTWILTKPTTT 1914
Query: 269 PSDNESVTSGSSSGVLDYGGGKPQRSARAIVVPARFLQEATNPSSRNGGIGNRSTVPPKL 328
+ S S +++ G P S A P +AT P S G +
Sbjct: 1915 ATTTASTGSTATASSTQATAGTPHVSTTA-TTPTVTSSKAT-PFSSPGTATALPALRSTA 1972
Query: 329 LVPKKSVFDSPASSPRGIVNNRL-QGSPIRSAVRPASPSKLGTPSPRSPSRGVSPCRGRN 387
P + F + SS G RL Q + + + A+PS TP S ++
Sbjct: 1973 TTPTATSFTAIPSSSLGTTWTRLSQTTTPMATMSTATPS--STPETVHTSTVLTTTATTT 2030
Query: 388 GVASSLSS 395
G S+++
Sbjct: 2031 GATGSVAT 2038
Score = 37.7 bits (86), Expect = 0.082
Identities = 79/409 (19%), Positives = 140/409 (33%), Gaps = 37/409 (9%)
Query: 10 TTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLV 69
+T T TP+ P I+ K T+ S+++ SS PP T P V
Sbjct: 3643 STPATSSTATPSSTP-----GTTWILTKLTTTATTTESTGSTATPSSTQGPPAGT--PHV 3695
Query: 70 NRTVNS---TKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARSL 126
+ T + T K+TP +P + + R TP T + L T+ L
Sbjct: 3696 STTATTPTVTSSKATPFSSPGT----ATALPALRSTATTPTATSFTAIPSSSLGTTWTRL 3751
Query: 127 SVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGRVNGNSDQTEN 186
S + + + + P T ++ +T +V TP+ G + T
Sbjct: 3752 SQTTTPMATMSTATPSSTPETVHTSTVLTTTATTTGATGSVATPSSTPGTAHTTKVPTTT 3811
Query: 187 SISRSGDQHRWP--AKSQQNQANFMNRSLDCGVLLRSSNGFGSNVVRSLRNSLLDPRVSH 244
+ + P A++ + G + S+ G+ ++ + P +
Sbjct: 3812 TTGFTVTPSSSPGTARTPPVWISTTTTPTTSGSTVTPSSIPGTTHTPTVLTTTTQPVATG 3871
Query: 245 DGATLRSESNKNGGSEPVIEPELVPSDNESVTSGSSSGVLDYGGGKPQRSARAIVVPARF 304
AT S + +G P L+ + +GS++ G P +P
Sbjct: 3872 SMATPSSSTQTSG-----TPPSLITTATTITATGSTTNPSSTPGTTP--------IPPEL 3918
Query: 305 LQEATNPSSRNGGIGNRSTVPPKLLVPKKSVFDSPASSPRGIVNNRLQGSP--IRSAVRP 362
AT P++ + STV P + P ++ + SP +R+A
Sbjct: 3919 TTTATTPAATS------STVTPSSALGTTHTPPVPNTTATTHGRSLSPSSPHTVRTAWTS 3972
Query: 363 ASPSKLGTPSPRSPSRGVSPCRGRNGVASSLSSRFVNEPSVLSFAVDVP 411
A+ LGT PS G S ++ S+ ++ P S + P
Sbjct: 3973 ATSGTLGTTHITEPSTGTSHTPAATTGTTTTSTPALSSPHPSSRTTESP 4021
Score = 37.4 bits (85), Expect = 0.11
Identities = 85/431 (19%), Positives = 146/431 (33%), Gaps = 68/431 (15%)
Query: 3 TAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPR 62
T I T TT T A T + L + I+ +P T+ + S+++ SS
Sbjct: 4359 TWILTELTTTATTTASTGSTATLSSTPGTTWILTEPST---TATVTVPTGSTATASSTQA 4415
Query: 63 RTNSPLVNRTVNS---TKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKML 119
+P V+ T + T K+TPS +P + + R TP T + L
Sbjct: 4416 TAGTPHVSTTATTPTVTSSKATPSSSPGT----ATALPALRSTATTPTATSFTAIPSSSL 4471
Query: 120 FTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGRVNG 179
T+ LS + P+AT++ + STPE +TT G
Sbjct: 4472 GTTWTRLS-------------QTTTPTATMSTATPSSTPETVHTSTVLTT----TATTTG 4514
Query: 180 NSDQTENSISRSGDQHRWPAKSQQNQANFMNRSLDCGVLLRSSNGFGSNVVRSLRNSLLD 239
+ S G H V ++ GF + S +L
Sbjct: 4515 ATGSVATPSSTPGTAH------------------TTKVPTTTTTGFTATPSSSPGTALTP 4556
Query: 240 PRVSHDGATLRSESNKNGGSEPVIEPELVPSDNES--VTSGSSSGVLDYGGGKPQRSARA 297
P T + + GS + P +P + V + +++ V P S +
Sbjct: 4557 PVWISTTTTPTTTTPTTSGS--TVTPSSIPGTTHTARVLTTTTTTVATGSMATPSSSTQT 4614
Query: 298 IVVPARFLQEATNPSSRNGGIGNRSTVPPKLLVPKKSVFDSPASSPRGIVNNRLQGS--- 354
P AT ++ G N S+ P +P V S A++P + S
Sbjct: 4615 SGTPPSLTTTATTITA-TGSTTNPSSTPGTTPIP--PVLTSMATTPAATSSKATSSSSPR 4671
Query: 355 -----PIRSAVRPASPSKLGTPSPRS--------PSRGVSPCRGRNGVASSLSSRFVNEP 401
P+ ++ S + TP P S P++ +P + + +S + +
Sbjct: 4672 TATTLPVLTSTATKSTATSFTPIPSSTLWTTWTVPAQTTTPMSTMSTIHTSSTPETTHTS 4731
Query: 402 SVLSFAVDVPR 412
+VL+ + R
Sbjct: 4732 TVLTTTATMTR 4742
Score = 37.4 bits (85), Expect = 0.11
Identities = 58/214 (27%), Positives = 80/214 (37%), Gaps = 42/214 (19%)
Query: 5 ISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRR- 63
ISTTTT T R T T P +P + V V + M + SSS+ S P
Sbjct: 2077 ISTTTTP--TTRGSTVT-PSSIPGTTHTPTVLTTTTTTVATGSMATPSSSTQTSGTPPSL 2133
Query: 64 -TNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTS 122
T + + T ++T STP TP V + +T TP A T
Sbjct: 2134 TTTATTITATGSTTNPSSTPGTTPIPPVLTTTAT---------------TPAATSSTVTP 2178
Query: 123 ARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTP-TIGRGRVNGNS 181
+ +L G + + PV P+ T T R +P P TV T T + G +
Sbjct: 2179 SSAL-----GTTHTPPV-----PNTTATTHGRSLSPSS---PHTVCTAWTSATSGILGTT 2225
Query: 182 DQTENSISRS--------GDQHRWPAKSQQNQAN 207
TE S S QH PA S + ++
Sbjct: 2226 HITEPSTGTSHTPAATTGTTQHSTPALSSPHPSS 2259
Score = 37.0 bits (84), Expect = 0.14
Identities = 49/186 (26%), Positives = 68/186 (36%), Gaps = 21/186 (11%)
Query: 3 TAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPR 62
T ++TT TT T TP+ P A K T SSS + +PP
Sbjct: 3249 TVLTTTATTTRTGSVATPSSTP------GTAHTTKVPTTTTTGFTATPSSSPGTALTPPV 3302
Query: 63 RTNSPLVNRTVNST-KQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFT 121
++ T ST S P T A V + +T TP + +T L T
Sbjct: 3303 WISTTTTPTTRGSTVTPSSIPGTTHTATVLTTTTTTVATGSMATPSSSTQTSGTPPSLTT 3362
Query: 122 SARSLSVSFQGES-FSIPVSKAKPP------------SATVTQSLRRSTPERRKVPATVT 168
+A +++ + + S P + PP S+TVT S T VP T
Sbjct: 3363 TATTITATGSTTNPSSTPGTTPIPPVLTTTATTPAATSSTVTPSSALGTTHTPPVPNTTA 3422
Query: 169 TPTIGR 174
T T GR
Sbjct: 3423 T-THGR 3427
Score = 36.6 bits (83), Expect = 0.18
Identities = 72/352 (20%), Positives = 116/352 (32%), Gaps = 55/352 (15%)
Query: 50 SSSSSSSFSSPPRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPN 109
S+ S+++ +S R P V T +T ++ TP++ + + R TP
Sbjct: 2448 STGSTATPTSTLRTAPPPKVLTTTATTPTVTSSKATPSSSPGTATALPALRSTATTPTAT 2507
Query: 110 GETPVAQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTT 169
TP+ L T+ L S+ P+AT++ + STPE +TV T
Sbjct: 2508 SVTPIPSSSLGTTWTRL-------------SQTTTPTATMSTATPSSTPETAHT-STVLT 2553
Query: 170 PTIGRGRVNGNSDQTENSISRSGDQHRWPAKSQQNQANFMNRSLDCGVLLRSSNGFGSNV 229
T G + S G H + S G L +
Sbjct: 2554 AT---ATTTGATGSVATPSSTPGTAHTTKVPTTTTTGFTATPSSSPGTALTPPVWISTTT 2610
Query: 230 VRSLRNSLLDPR----VSHDGATLR--------------SESNKNGGSEPVIEPELVPSD 271
+ R S + P +H L S S + G+ P L +
Sbjct: 2611 TPTTRGSTVTPSSIPGTTHTATVLTTTTTTVATGSMATPSSSTQTSGT----PPSLTTTA 2666
Query: 272 NESVTSGSSSGVLDYGGGKPQRSARAIVVPARFLQEATNPSSRNGGIGNRSTVPPKLLVP 331
+GS++ G +P +P AT P++ + STV P +
Sbjct: 2667 TTITATGSTTNPSSTPGTRP--------IPPVLTTTATTPAATS------STVTPSSALG 2712
Query: 332 KKSVFDSPASSPRGIVNNRLQGSP--IRSAVRPASPSKLGTPSPRSPSRGVS 381
P ++ + SP +R+A A+ LGT PS G S
Sbjct: 2713 TTHTPPVPNTTATTHGRSLSPSSPHTVRTAWTSATSGTLGTTHITEPSTGTS 2764
Score = 35.0 bits (79), Expect = 0.53
Identities = 82/417 (19%), Positives = 143/417 (33%), Gaps = 54/417 (12%)
Query: 3 TAIST-TTTTKNTKRAPTPTRPP--------LLPSESDNAIVRK--PKAREVTSRYMYSS 51
T ++T TTT+ T TP+ P L + + A + P + T+ +
Sbjct: 3079 TVLTTKATTTRATSSMSTPSSTPGTTWILTELTTAATTTAALPHGTPSSTPGTTWILTEP 3138
Query: 52 SSSSSFSSPPRRTNSPLVNR-TVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTP--RP 108
S++++ + P T + R T + K ++ + TP R+ + P R
Sbjct: 3139 STTATVTVPTGSTATASSTRATAGTLKVLTSTATTPTVISSRATPSSSPGTATALPALRS 3198
Query: 109 NGETPVAQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVT 168
TP A TS ++ S G +++ +S+ P+AT++ + STPE +T
Sbjct: 3199 TATTPTA-----TSVTAIPSSSLGTAWT-RLSQTTTPTATMSTATPSSTPETVHTSTVLT 3252
Query: 169 TPTIGRGRVNGNSDQTENSISRSGDQHRWPAKSQQNQANFMNRSLDCGVLLRSSNGFGSN 228
T + S G H + S G L +
Sbjct: 3253 TTA-----TTTRTGSVATPSSTPGTAHTTKVPTTTTTGFTATPSSSPGTALTPPVWISTT 3307
Query: 229 VVRSLRNSLLDPRVSHDGATLRSESNKNGGSEPVIEPELVPSDNESVTSGSSSGVLDYGG 288
+ R S + P S T + + + V + + + TSG+
Sbjct: 3308 TTPTTRGSTVTP--SSIPGTTHTATVLTTTTTTVATGSMATPSSSTQTSGT--------- 3356
Query: 289 GKPQRSARAIVVPARFLQEATNPSSRNGGIGNRSTVPPKLLVPKKSVFDSPASSPRGIVN 348
P + A + A TNPSS G + +PP V + A++P +
Sbjct: 3357 -PPSLTTTATTITA--TGSTTNPSSTPG----TTPIPP--------VLTTTATTPAATSS 3401
Query: 349 NRLQGSPIRSAVRPASPSKLGTPSPRS---PSRGVSPCRGRNGVASSLSSRFVNEPS 402
S + + P P+ T RS S P + + L + + EPS
Sbjct: 3402 TVTPSSALGTTHTPPVPNTTATTHGRSLPPSSPHTVPTAWTSATSGILGTTHITEPS 3458
Score = 35.0 bits (79), Expect = 0.53
Identities = 38/135 (28%), Positives = 51/135 (37%), Gaps = 16/135 (11%)
Query: 5 ISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRR- 63
ISTTTT + TP+ +P + V + V + M + SSS+ S P
Sbjct: 3833 ISTTTTPTTSGSTVTPSS---IPGTTHTPTVLTTTTQPVATGSMATPSSSTQTSGTPPSL 3889
Query: 64 -TNSPLVNRTVNSTKQKSTPSVTP--------AAFVKRSQSTERQRQRQGTPRPNGETPV 114
T + + T ++T STP TP A + ST GT PV
Sbjct: 3890 ITTATTITATGSTTNPSSTPGTTPIPPELTTTATTPAATSSTVTPSSALGTTH---TPPV 3946
Query: 115 AQKMLFTSARSLSVS 129
T RSLS S
Sbjct: 3947 PNTTATTHGRSLSPS 3961
Score = 35.0 bits (79), Expect = 0.53
Identities = 43/176 (24%), Positives = 62/176 (34%), Gaps = 18/176 (10%)
Query: 5 ISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRT 64
+STT TT PT T P S P R + +S ++ SS
Sbjct: 3695 VSTTATT------PTVTSSKATPFSSPGTATALPALRSTATTPTATSFTAIPSSSLGTTW 3748
Query: 65 NSPLVNRTVNSTKQKSTPSVTP-----AAFVKRSQSTERQRQRQGTPRPNGETPVAQKML 119
T +T +TPS TP + + + +T TP T K+
Sbjct: 3749 TRLSQTTTPMATMSTATPSSTPETVHTSTVLTTTATTTGATGSVATPSSTPGTAHTTKVP 3808
Query: 120 FTSARSLSVSFQGESFSIPVSKAKPP---SATVTQSLRRSTPERRKVPATVTTPTI 172
T+ +V+ S P + PP S T T + ST +P T TPT+
Sbjct: 3809 TTTTTGFTVT----PSSSPGTARTPPVWISTTTTPTTSGSTVTPSSIPGTTHTPTV 3860
Score = 33.1 bits (74), Expect = 2.0
Identities = 36/157 (22%), Positives = 65/157 (40%), Gaps = 10/157 (6%)
Query: 3 TAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPR 62
T ++TTTTT T TP+ S P + T+ + ++ S+++ SS P
Sbjct: 2103 TVLTTTTTTVATGSMATPS--------SSTQTSGTPPSLTTTATTITATGSTTNPSSTPG 2154
Query: 63 RTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTS 122
T P V T +T ++ +VTP++ + + T T +P + + T+
Sbjct: 2155 TTPIPPVLTTTATTPAATSSTVTPSSALGTTH-TPPVPNTTATTHGRSLSPSSPHTVCTA 2213
Query: 123 ARSLSVSFQGES-FSIPVSKAKPPSATVTQSLRRSTP 158
S + G + + P + A T + + STP
Sbjct: 2214 WTSATSGILGTTHITEPSTGTSHTPAATTGTTQHSTP 2250
Score = 31.6 bits (70), Expect = 5.9
Identities = 33/121 (27%), Positives = 53/121 (43%), Gaps = 12/121 (9%)
Query: 3 TAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPR 62
TA S+ T+ ++ R T T P+L S + K A VT SS+ + + P
Sbjct: 4232 TATSSKATSSSSPR--TATTLPVLTSTAT-----KSTATSVTP---IPSSTLGTTGTLPE 4281
Query: 63 RTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTS 122
+T +P+ T+++ STP T + V +++T R TP T L T+
Sbjct: 4282 QTTTPVA--TMSTIHPSSTPETTHTSTVLTTKATTRATSSTSTPSSTPGTTWILTELTTA 4339
Query: 123 A 123
A
Sbjct: 4340 A 4340
>CHI3_CANAL (P40954) Chitinase 3 precursor (EC 3.2.1.14)
Length = 567
Score = 45.4 bits (106), Expect = 4e-04
Identities = 48/178 (26%), Positives = 78/178 (42%), Gaps = 20/178 (11%)
Query: 3 TAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPR 62
T+ STTTTT T + S S ++ K TS + S++SS++ S+
Sbjct: 320 TSSSTTTTTTTTSTT--------ISSSSSSSKTSKTSTTSTTSSSISSTTSSTTSSTSSS 371
Query: 63 RTNSPLVNRTVNST--KQKSTPSVTPAAFVKRSQST--ERQRQRQGTPRPNGETPVAQKM 118
T+S + T +ST Q ST S P + S ST T + ET +
Sbjct: 372 STSSSTSSTTSSSTTSSQISTTSTAPTSSTSLSSSTISTSASTSDTTSVTSSET---TPV 428
Query: 119 LFTSARSLSVSFQGESFSIPVSKAK--PPSATVTQSLRRSTPERRKVP---ATVTTPT 171
+ S+ S +++ G+S + +SK+ P+ + T +L ST +P + TPT
Sbjct: 429 VTPSSLSSAITIPGDSTTTGISKSSSTKPATSTTSALSSSTTTVATIPDDKEIINTPT 486
>AGA1_YEAST (P32323) A-agglutinin attachment subunit precursor
Length = 725
Score = 45.1 bits (105), Expect = 5e-04
Identities = 44/197 (22%), Positives = 77/197 (38%), Gaps = 7/197 (3%)
Query: 5 ISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRT 64
IS T+T ++ + PT L + + + + TS S+SSSS+ +S +
Sbjct: 166 ISPVTSTLSSTTSSNPTTTSLSSTSTSPSSTSTSPSSTSTSSSSTSTSSSSTSTSSSSTS 225
Query: 65 NSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSAR 124
SP T +S S+ S + + + S+ T + T + TSA
Sbjct: 226 TSPSSTSTSSSLTSTSSSSTSTSQSSTSTSSSSTSTSPSSTSTSSSSTSTSPSSKSTSAS 285
Query: 125 SLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGRVNGNSDQT 184
S S S S S ++ + P +L ++P + +T T T G +S +
Sbjct: 286 STSTSSYSTSTSPSLTSSSP-------TLASTSPSSTSISSTFTDSTSSLGSSIASSSTS 338
Query: 185 ENSISRSGDQHRWPAKS 201
+ S S + P+ S
Sbjct: 339 VSLYSPSTPVYSVPSTS 355
>SR40_YEAST (P32583) Suppressor protein SRP40
Length = 406
Score = 43.9 bits (102), Expect = 0.001
Identities = 58/243 (23%), Positives = 97/243 (39%), Gaps = 20/243 (8%)
Query: 50 SSSSSSSFSSPPRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPN 109
SSSSSSS SS +S + + + + S+ S ++ S+ ++R R+
Sbjct: 79 SSSSSSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSESESEDETKKRARESDNEDA 138
Query: 110 GETPVAQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTT 169
ET A+ +S+ S S S S S S ++ S + + S S E + ++
Sbjct: 139 KETKKAKTEPESSSSSESSSSGSSSSSESESGSESDSDSSSSSSSSSDSESDSESDSQSS 198
Query: 170 PTIGRGRVNGNSDQTENSISRSGDQHRWPAKSQQNQANFMNRSLDCGVLLRSSNGFGSNV 229
+ + +SD + + S D + S + + + S D S+ GS+
Sbjct: 199 SSSSSSDSSSDSDSSSSDSSSDSDSSSSSSSSSSDSDSDSDSSSD-------SDSSGSSD 251
Query: 230 VRSLRNSLLDPRVSHDGATLRSESNKNGGSEPVIE-----------PELVPSDNESVTSG 278
S +S D S D + S+S+ + GS +E E S NES S
Sbjct: 252 SSSSSDSSSDESTSSDSSD--SDSDSDSGSSSELETKEATADESKAEETPASSNESTPSA 309
Query: 279 SSS 281
SSS
Sbjct: 310 SSS 312
Score = 39.7 bits (91), Expect = 0.022
Identities = 68/336 (20%), Positives = 127/336 (37%), Gaps = 30/336 (8%)
Query: 37 KPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQST 96
K K E S SSSSSSS SS ++S + +S+ S+ S + ++ S+S+
Sbjct: 17 KEKEIEEKSSSSSSSSSSSSSSSSSSSSSSSSSGESSSSSSSSSSSSSSDSSDSSDSESS 76
Query: 97 ERQRQRQGTPRPNGETPVAQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRS 156
+ + ++ + + S+ S S S S S+++ T ++
Sbjct: 77 SSSSSSSSSSSSSSDSESSSES--DSSSSGSSSSSSSSSDESSSESESEDETKKRARESD 134
Query: 157 TPERRKVPATVTTPTIGRGRVNGNSDQTENSISRSGDQHRWPAKSQQNQANFMNRSLDCG 216
+ ++ T P + +S + +S S SG ++S + ++ + S D
Sbjct: 135 NEDAKETKKAKTEPESSSSSESSSSGSSSSSESESG------SESDSDSSSSSSSSSD-- 186
Query: 217 VLLRSSNGFGSNVVRSLRNSLLDPRVSHDGATLRSESNKNGGSEPVIEPELVPSDNESVT 276
S + S+ S +S D D ++ S S+ + S SD++S +
Sbjct: 187 ----SESDSESDSQSSSSSSSSDSSSDSDSSSSDSSSDSDSSSSSSSSSSDSDSDSDSSS 242
Query: 277 SGSSSGVLDYGGGKPQRSARAIVVPARFLQEATNPSSRNGGIGNRSTVPPKLLVPKKSVF 336
SSG D S E+T+ S + + S +L + +
Sbjct: 243 DSDSSGSSDSSSSSDSSS-----------DESTSSDSSDSDSDSDSGSSSELETKEATAD 291
Query: 337 DSPASSPRGIVNNRLQGSPIRSAVRPASPSKLGTPS 372
+S A N + +P SA +S +KL P+
Sbjct: 292 ESKAEETPASSN---ESTP--SASSSSSANKLNIPA 322
>DAN4_YEAST (P47179) Cell wall protein DAN4 precursor
Length = 1161
Score = 43.5 bits (101), Expect = 0.001
Identities = 47/193 (24%), Positives = 70/193 (35%), Gaps = 9/193 (4%)
Query: 3 TAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPR 62
TAI T+T+T TK + T T P + + + P TS + ++S++ ++P
Sbjct: 119 TAIPTSTSTTTTKSS-TSTTPTTTITSTTSTTSTTP----TTSTTSTTPTTSTTSTTPTT 173
Query: 63 RTNSPL-VNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFT 121
T S T ++T ST S TP + T T TP T
Sbjct: 174 STTSTTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTTSTTPTT 233
Query: 122 SARSLSVSFQGESFSIPVSKAK---PPSATVTQSLRRSTPERRKVPATVTTPTIGRGRVN 178
S S + +S + S S T T S + P T TT TI
Sbjct: 234 STTSTTSQTSTKSTTPTTSSTSTTPTTSTTPTTSTTSTAPTTSTTSTTSTTSTISTAPTT 293
Query: 179 GNSDQTENSISRS 191
+ T ++ S S
Sbjct: 294 STTSSTFSTSSAS 306
Score = 43.5 bits (101), Expect = 0.001
Identities = 40/169 (23%), Positives = 58/169 (33%), Gaps = 26/169 (15%)
Query: 3 TAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPR 62
T+ STTTT +T PT T + S++S++S +
Sbjct: 123 TSTSTTTTKSSTSTTPTTT--------------------------ITSTTSTTSTTPTTS 156
Query: 63 RTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTS 122
T++ T ++T ST S TP + T T TP T
Sbjct: 157 TTSTTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTTSTTSTTP 216
Query: 123 ARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPT 171
S + + S + S S T T+S +T P T TTPT
Sbjct: 217 TTSTTSTTPTTSTTPTTSTTSTTSQTSTKSTTPTTSSTSTTPTTSTTPT 265
Score = 39.7 bits (91), Expect = 0.022
Identities = 40/193 (20%), Positives = 70/193 (35%), Gaps = 9/193 (4%)
Query: 6 STTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSS-SSSFSSPPRRT 64
STT TT T PT + P+ S + T +S++ ++S +S T
Sbjct: 150 STTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTT 209
Query: 65 NSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSAR 124
++ T ++T T S TP + S + T TP TS
Sbjct: 210 STTSTTPTTSTTSTTPTTSTTPTTSTTSTTSQTSTKSTTPTTSSTSTTPTTSTTPTTSTT 269
Query: 125 SLSVSFQGESFSIPVS--KAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGRV----- 177
S + + S + S P ++T + + S+ V +T T + +
Sbjct: 270 STAPTTSTTSTTSTTSTISTAPTTSTTSSTFSTSSASASSVISTTATTSTTFASLTTPAT 329
Query: 178 -NGNSDQTENSIS 189
++D T +S+S
Sbjct: 330 STASTDHTTSSVS 342
Score = 38.9 bits (89), Expect = 0.037
Identities = 43/171 (25%), Positives = 67/171 (39%), Gaps = 12/171 (7%)
Query: 6 STTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTN 65
STT TT T APT + + S I P +S + SS+S+SS S T+
Sbjct: 261 STTPTTSTTSTAPTTSTTSTTSTTS--TISTAPTTSTTSSTFSTSSASASSVISTTATTS 318
Query: 66 SPLVNRTVNSTKQKSTPSVTPA-----AFVKRSQSTERQRQRQGTPRPNGETPVAQKMLF 120
+ + T +T ST T + AF + +T + P+ T A+
Sbjct: 319 TTFASLTTPATSTASTDHTTSSVSTTNAFTTSATTTTTSDTYISSSSPSQVTSSAEP--- 375
Query: 121 TSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPT 171
T+ ++ S + S S A+P TV++ P R + PT
Sbjct: 376 TTVSEVTSSVEPTRSSQVTSSAEP--TTVSEFTSSVEPTRSSQVTSSAEPT 424
Score = 32.3 bits (72), Expect = 3.4
Identities = 30/124 (24%), Positives = 45/124 (36%), Gaps = 8/124 (6%)
Query: 48 MYSSSSSSSFSSPPRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPR 107
+Y++ +S+ ++ + + S T+ ST ST S TP + T T
Sbjct: 117 IYTAIPTSTSTTTTKSSTSTTPTTTITSTT--STTSTTPTTSTTSTTPTTSTTSTTPTTS 174
Query: 108 PNGETPVAQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATV 167
TP TS S + + S + S T T S +T P T
Sbjct: 175 TTSTTPT------TSTTSTTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTTSTTSTTPTTS 228
Query: 168 TTPT 171
TTPT
Sbjct: 229 TTPT 232
>WSC3_YEAST (Q12215) Cell wall integrity and stress response
component 3 precursor
Length = 556
Score = 43.1 bits (100), Expect = 0.002
Identities = 42/192 (21%), Positives = 78/192 (39%), Gaps = 17/192 (8%)
Query: 2 VTAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPK-----AREVTSRYMYSSSSSSS 56
V+++ ++++ + + + P+ L S ++ R+ + + + +S++SSS
Sbjct: 136 VSSVESSSSKEGSSTSYMPSTTSSLSSAQISSTTRRTSTDMKSSEMIATTVSTTSTTSSS 195
Query: 57 FSSPPRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQ 116
SS T S + T +ST ST S T + + ST T T
Sbjct: 196 TSSTTSSTTSSTTSSTTSSTTSSSTSSTTSSTTSSTTSSTTSSTTSSTTSSTTSST---- 251
Query: 117 KMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGR 176
TS+ + S S SI +S S T R S+P VP + ++ T+ +
Sbjct: 252 ----TSSTTSIFSVTSSSSSITLSS----SEHTTVDSRTSSPSSTLVPVSSSSSTLSTPK 303
Query: 177 VNGNSDQTENSI 188
V + T ++I
Sbjct: 304 VTSMTPSTSSTI 315
Score = 37.7 bits (86), Expect = 0.082
Identities = 37/150 (24%), Positives = 62/150 (40%), Gaps = 17/150 (11%)
Query: 1 MVTAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSP 60
+ T +STT+TT ++ + T + S + ++ T+ SS++SS+ SS
Sbjct: 182 IATTVSTTSTTSSSTSSTTSSTTSSTTSSTTSS----------TTSSSTSSTTSSTTSST 231
Query: 61 PRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLF 120
T S + T +ST +T S T V S S+ T + T V +
Sbjct: 232 TSSTTSSTTSSTTSSTTSSTTSSTTSIFSVTSSSSSI-------TLSSSEHTTVDSRTSS 284
Query: 121 TSARSLSVSFQGESFSIPVSKAKPPSATVT 150
S+ + VS + S P + PS + T
Sbjct: 285 PSSTLVPVSSSSSTLSTPKVTSMTPSTSST 314
>SGS3_DROYA (P13728) Salivary glue protein Sgs-3 precursor
Length = 263
Score = 42.7 bits (99), Expect = 0.003
Identities = 40/171 (23%), Positives = 62/171 (35%), Gaps = 8/171 (4%)
Query: 3 TAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPR 62
T TTTTT T AP PTRPP P ++ + T R ++ +++ +
Sbjct: 42 TCAPTTTTTTTTTCAP-PTRPPPPPCTDAPTTTKRTTEKSTTRRTTTTTRQTTTRPTTTT 100
Query: 63 RTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTS 122
T + T ST + T + T + +T +R T T +
Sbjct: 101 TTTTTTRRPTTRSTTTRHTTTTTTTTRRPTTTTTTTRRPTTTTTTTRRPTTTTTTTRLPT 160
Query: 123 ARSLSVSFQGESFSIPVSKAKPPSATVTQSLR---RSTPERRKVPATVTTP 170
RS + +S + +P T T S R +T R+ TTP
Sbjct: 161 TRSTTTRHTTKS----TTSKRPTHETTTTSKRPTQETTTTTRRATQATTTP 207
Score = 34.7 bits (78), Expect = 0.69
Identities = 25/109 (22%), Positives = 44/109 (39%), Gaps = 10/109 (9%)
Query: 3 TAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPR 62
T TTTT T R PT T + R+P T+R + S+++ ++
Sbjct: 114 TTTRHTTTTTTTTRRPTTTTTTTRRPTTTTTTTRRPTTTTTTTRLPTTRSTTTRHTTKST 173
Query: 63 RTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGE 111
+ P + T ++K+ + + T +T R Q TP+P +
Sbjct: 174 TSKRP-THETTTTSKRPTQETTT---------TTRRATQATTTPKPTNK 212
>YQU3_CAEEL (Q09550) Hypothetical protein F26C11.3 in chromosome II
Length = 1240
Score = 42.4 bits (98), Expect = 0.003
Identities = 48/198 (24%), Positives = 79/198 (39%), Gaps = 23/198 (11%)
Query: 6 STTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTN 65
ST TTT +T+ + T T P E + + K R + S++ +S S+ T+
Sbjct: 239 STQTTTPSTEPSTTITTP----MEQSSTVSSVQKTRTSEDKPSSSTTVPTSASTSESSTS 294
Query: 66 SPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARS 125
SP+ + +ST +S+P+ S ST + G+ G T L T+ +S
Sbjct: 295 SPMAETSSSSTTSQSSPA---------STSTVPESSTVGSTPTTGLT-----TLSTNEQS 340
Query: 126 LSVSFQGESFSI--PVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGRVNGN--- 180
S S G S S S+ S T + S+ + + T TI G N
Sbjct: 341 TSTSSGGHSTSTFGTTSETPETSTDFTATSTSSSSDSSTQSSNAQTSTIENGSTTTNFTS 400
Query: 181 SDQTENSISRSGDQHRWP 198
+ T ++ + + WP
Sbjct: 401 APSTSSTPATPTTTYNWP 418
>SFR8_HUMAN (Q12872) Splicing factor, arginine/serine-rich 8
(Suppressor of white apricot protein homolog)
Length = 951
Score = 42.4 bits (98), Expect = 0.003
Identities = 50/183 (27%), Positives = 71/183 (38%), Gaps = 30/183 (16%)
Query: 12 KNTKRAPTPTRPPLLPSESDNAIVRKPKARE-VTSRYMYSSSSSSSFSSPPRRTNSPLVN 70
K+ KR+ T +R P S S + KA+ + S Y S S SP RR +SP
Sbjct: 760 KHKKRSRTRSRSPKYHSSSKSRSRSHSKAKHSLPSAYRTVRRSRSRSRSPRRRAHSP--- 816
Query: 71 RTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARSLSVSF 130
+++ SV A V RS R+R R +P + ++ + ARS SVS
Sbjct: 817 -----ERRREERSVPTAYRVSRSPGASRKRTRSRSPHEKKKKRRSRSRTKSKARSQSVS- 870
Query: 131 QGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRG----RVNGNSDQTEN 186
P +A P R + P + +P RG R G S + E
Sbjct: 871 -------PSKQAAP---------RPAAPAAHSAHSASVSPVESRGSSQERSRGVSQEKEA 914
Query: 187 SIS 189
IS
Sbjct: 915 QIS 917
>YJW5_YEAST (P40889) Hypothetical 197.6 kDa protein in FSP2 5'region
Length = 1758
Score = 42.0 bits (97), Expect = 0.004
Identities = 56/259 (21%), Positives = 96/259 (36%), Gaps = 22/259 (8%)
Query: 2 VTAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPP 61
+T ST T T A T + S NA + T+ ++++++S +
Sbjct: 1136 ITTASTNVRTNATTNASTNATTNASTNASTNATTNA--STNATTNSSTNATTTASTNVRT 1193
Query: 62 RRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFT 121
T + +N ++T +ST S T A + + S+ + T T A + T
Sbjct: 1194 SATTTASINVRTSATTTESTNSSTNATTTESTNSSTNATTTESTNSNTSATTTASINVRT 1253
Query: 122 SARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGRVNGNS 181
SA + + S + S SAT T+S+ ST AT T T N N+
Sbjct: 1254 SATTTESTNSSTSATTTASINVRTSATTTKSINSST------NATTTEST----NSNTNA 1303
Query: 182 DQTENSISRSGDQHRWPAKSQQNQANFMNRSLDCGVLLRSSNGFGSNVVRSLRNSLLDPR 241
TE++ S + S N S + ++ SN + S
Sbjct: 1304 TTTESTNSSTNATTTESTNSSTNATT--TESTNSNTSAATTESTNSNTSATTTES----- 1356
Query: 242 VSHDGATLRSESNKNGGSE 260
A+ + ++NK+G +E
Sbjct: 1357 ---TNASAKEDANKDGNAE 1372
>YIR7_YEAST (P40434) Hypothetical 197.5 kDa protein in SDL1 5'region
Length = 1758
Score = 42.0 bits (97), Expect = 0.004
Identities = 56/259 (21%), Positives = 96/259 (36%), Gaps = 22/259 (8%)
Query: 2 VTAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPP 61
+T ST T T A T + S NA + T+ ++++++S +
Sbjct: 1136 ITTASTNVRTNATTNASTNATTNASTNASTNATTNA--STNATTNSSTNATTTASTNVRT 1193
Query: 62 RRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFT 121
T + +N ++T +ST S T A + + S+ + T T A + T
Sbjct: 1194 SATTTASINVRTSATTTESTNSSTNATTTESTNSSTNATTTESTNSNTSATTTASINVRT 1253
Query: 122 SARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGRVNGNS 181
SA + + S + S SAT T+S+ ST AT T T N N+
Sbjct: 1254 SATTTESTNSSTSATTTASINVRTSATTTKSINSST------NATTTEST----NSNTNA 1303
Query: 182 DQTENSISRSGDQHRWPAKSQQNQANFMNRSLDCGVLLRSSNGFGSNVVRSLRNSLLDPR 241
TE++ S + S N S + ++ SN + S
Sbjct: 1304 TTTESTNSSTNATTTESTNSSTNATT--TESTNSNTSAATTESTNSNTSATTTES----- 1356
Query: 242 VSHDGATLRSESNKNGGSE 260
A+ + ++NK+G +E
Sbjct: 1357 ---TNASAKEDANKDGNAE 1372
>VIT2_CHICK (P02845) Vitellogenin II precursor (Major vitellogenin)
[Contains: Lipovitellin I (LVI); Phosvitin (PV);
Lipovitellin II (LVII); YGP40]
Length = 1850
Score = 42.0 bits (97), Expect = 0.004
Identities = 39/193 (20%), Positives = 86/193 (44%), Gaps = 22/193 (11%)
Query: 3 TAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPR 62
++ S+T T+ ++ A +P R + E ++ + ++ ++ SSS SS S+ +
Sbjct: 1127 SSASSTATSSSSSSASSPNRKKPMDEEENDQV------KQARNKDASSSSRSSKSSNSSK 1180
Query: 63 RTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTS 122
R++S ++ NS+K+ S+ S + ++ + S S+ + + ++ + +S
Sbjct: 1181 RSSS----KSSNSSKRSSSSSSSSSSSSRSSSSSSSSSSNSKSSSSSSKSSSSSSRSRSS 1236
Query: 123 ARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGRVNGNSD 182
++S S S S S S + S++ ++S + G +NG+S
Sbjct: 1237 SKSSSSSSSSSSSSSSKSSSSRSSSSSSKSSSHHSHSHHS------------GHLNGSSS 1284
Query: 183 QTENSISRSGDQH 195
+ +S S S H
Sbjct: 1285 SSSSSRSVSHHSH 1297
Score = 31.6 bits (70), Expect = 5.9
Identities = 38/203 (18%), Positives = 78/203 (37%), Gaps = 6/203 (2%)
Query: 11 TKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLVN 70
T++ K P+ +L + + S SSSSSS+ SSP R+ P+
Sbjct: 1096 TRHPKNRPSKKGNTVLAEFGTEPDAKTSSSSSSASSTATSSSSSSA-SSPNRK--KPMDE 1152
Query: 71 RTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARSLSVSF 130
+ KQ + ++ RS + +R + N + +S+ S S S
Sbjct: 1153 EENDQVKQARNKDASSSS---RSSKSSNSSKRSSSKSSNSSKRSSSSSSSSSSSSRSSSS 1209
Query: 131 QGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGRVNGNSDQTENSISR 190
S S S + ++ + S RS+ + ++ ++ + + + +S + S S
Sbjct: 1210 SSSSSSNSKSSSSSSKSSSSSSRSRSSSKSSSSSSSSSSSSSSKSSSSRSSSSSSKSSSH 1269
Query: 191 SGDQHRWPAKSQQNQANFMNRSL 213
H + + ++ +RS+
Sbjct: 1270 HSHSHHSGHLNGSSSSSSSSRSV 1292
>MU13_RAT (P97881) Mucin 13 precursor
Length = 547
Score = 42.0 bits (97), Expect = 0.004
Identities = 48/195 (24%), Positives = 72/195 (36%), Gaps = 15/195 (7%)
Query: 6 STTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTN 65
++T TT T+ T T+ P A + TS +S+ + + P T+
Sbjct: 8 TSTPTTTATQPTSTSTQTPGTTQLLSTTSTPTTTATQPTS----TSTQTPGTTQLPSTTS 63
Query: 66 SPLVNRT---VNSTKQKST---PSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKML 119
+P T ST+ T P T +Q T Q GT + T
Sbjct: 64 TPTTTATQPTXTSTQTPGTTQLPGTTSTPTTTATQPTSTSFQTPGTTQLPSSTSTPTT-- 121
Query: 120 FTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRS--TPERRKVPATVTTPTIGRGRV 177
T+ + S + Q + P A P+ TVTQ S TP + P +TPT +
Sbjct: 122 -TATQPTSTASQTPGTTQPPGGASSPTTTVTQPTGSSSQTPGTTQPPGGASTPTTTVTQP 180
Query: 178 NGNSDQTENSISRSG 192
G+S QT + G
Sbjct: 181 TGSSSQTSGTTQPPG 195
Score = 35.0 bits (79), Expect = 0.53
Identities = 36/145 (24%), Positives = 58/145 (39%), Gaps = 8/145 (5%)
Query: 51 SSSSSSFSSPPRRTNSPLVN--RTVNSTKQKSTPSV-TPAAFVKRSQSTERQRQRQGTPR 107
S SS S+P P +T +T+ ST S T A S ST+ Q
Sbjct: 2 SQSSGGTSTPTTTATQPTSTSTQTPGTTQLLSTTSTPTTTATQPTSTSTQTPGTTQLPST 61
Query: 108 PNGETPVAQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATV 167
+ T A + TS ++ + + S P + A P++T Q TP ++P++
Sbjct: 62 TSTPTTTATQPTXTSTQTPGTTQLPGTTSTPTTTATQPTSTSFQ-----TPGTTQLPSST 116
Query: 168 TTPTIGRGRVNGNSDQTENSISRSG 192
+TPT + + QT + G
Sbjct: 117 STPTTTATQPTSTASQTPGTTQPPG 141
>LTE1_YEAST (P07866) Low temperature essential protein
Length = 1435
Score = 41.2 bits (95), Expect = 0.007
Identities = 74/345 (21%), Positives = 135/345 (38%), Gaps = 27/345 (7%)
Query: 40 AREVTSRYMYSSSSSSSFSSPPRRTNSP-------LVNRTVNSTKQKSTPSVTPAAFV-K 91
+RE T YS S S SSPPR T + +VN T + T + T + F +
Sbjct: 648 SREDTESITYSYDSELSSSSPPRDTVTKKSRKVRNIVNNTDSPTLKTKTGFLNLREFTFE 707
Query: 92 RSQSTERQRQRQGTPRPNGETPVAQKMLFTSAR----SLSVSFQGESFSIPVSKAKPP-S 146
++S + ++ N + Q+ + S + SL S + ++ I K +
Sbjct: 708 DTKSLDEKKSTIDGLEKNYDNKENQESEYESTKKLDNSLDASSEANNYDITTRKKHSSCN 767
Query: 147 ATVTQSLRRSTPER---RKVPATVTTPTIGRGRVNGNSDQTENSISRSGDQHRWPAKSQQ 203
+ Q++ R R +V + TPT V+ +++ + I + P +
Sbjct: 768 HKIKQAVVRPASGRISISRVQSIAITPTKELSIVDPEQNKSNSVIEEISEIE--PLNLEY 825
Query: 204 NQANFMNRSLDCGVLLRSSNGFGSNVVRSLRNSLLDPRVSHDGATLRSESNKNGGS-EPV 262
N+ + + V+ S++ + S +P+ T SE+N+ S P
Sbjct: 826 NKKSALYSDTSSTVISISTSKLFESAQNSPLKQTQNPQREFPNGTSVSETNRIRLSIAPT 885
Query: 263 IEPELVPSDNESVTSGSSSGVLDYGGGKPQRSARAIVVPARFLQEATNPSSRNGGIGNRS 322
IE V SD S+T+GS+ + P R I + + + + S +
Sbjct: 886 IES--VVSDLNSITTGSTVETFETSRDLPVPHQRIINLREEYQRGNQDIISNTSSLHELK 943
Query: 323 TVPPKLLVPKKSVFDSPASSPRGIVNNRLQGSPIRSAVRPASPSK 367
T+ L + +SP++ + NN+ SP ++ ASP K
Sbjct: 944 TID---LSDSNNDLESPSTHAK---NNKYFFSPDDGSIDVASPMK 982
>PRY3_YEAST (P47033) PRY3 protein (Pathogen related in Sc 3)
Length = 881
Score = 40.8 bits (94), Expect = 0.010
Identities = 57/273 (20%), Positives = 102/273 (36%), Gaps = 18/273 (6%)
Query: 50 SSSSSSSFSSPPRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPN 109
SSSSSSS +S T S + + + + Q T +V+ AA +ST + T +
Sbjct: 167 SSSSSSSSTSTTSDTVSTISSSIMPAVAQGYTTTVSSAASSSSLKSTTINPAKTATLTAS 226
Query: 110 GETPVAQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTT 169
T + T + S S S+ S A S V+ S T TT
Sbjct: 227 SSTVITSS---TESVGSSTVSSASSSSVTTSYATSSSTVVSSDATSS---------TTTT 274
Query: 170 PTIGRGRVNGNSDQTENSISRSGDQHRWPAKSQQNQANFMNRSLDCGVLLRSSNGFGSNV 229
++ +SD T ++ + S + + + A+ N + + SS S V
Sbjct: 275 SSVATSSSTTSSDPTSSTAAASSSDPASSSAAASSSASTENAASSSSAISSSS----SMV 330
Query: 230 VRSLRNSLLDPRVSHDGATLRSESNKNGGSEPVIEPELVPSDNESVTSGSSSGVLDYGGG 289
L ++L S T S ++ + V + S + S++S ++ D G
Sbjct: 331 SAPLSSTLTTSTASSRSVTSNSVNSVKFANTTVFSAQTTSSVSASLSSSVAAD--DIQGS 388
Query: 290 KPQRSARAIVVPARFLQEATNPSSRNGGIGNRS 322
+ + ++ + ATN + +G+ S
Sbjct: 389 TSKEATSSVSEHTSIVTSATNAAQYATRLGSSS 421
Score = 33.1 bits (74), Expect = 2.0
Identities = 61/296 (20%), Positives = 114/296 (37%), Gaps = 43/296 (14%)
Query: 2 VTAISTTTTTKNTKRAPTPTRPPLLPSE-------SDNAIVRKPKAREVTSRYMYSSSSS 54
+TA S+T T +T+ + T S S + +V T+ ++SSS
Sbjct: 223 LTASSSTVITSSTESVGSSTVSSASSSSVTTSYATSSSTVVSSDATSSTTTTSSVATSSS 282
Query: 55 SSFSSPPRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNG---E 111
++ S P T ++ S+ + +A S STE +
Sbjct: 283 TTSSDP-----------TSSTAAASSSDPASSSAAASSSASTENAASSSSAISSSSSMVS 331
Query: 112 TPVAQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPT 171
P++ + ++A S SV+ S V+ K + TV + S+ V A++++
Sbjct: 332 APLSSTLTTSTASSRSVT------SNSVNSVKFANTTVFSAQTTSS-----VSASLSSSV 380
Query: 172 IGRGRVNGNSDQTENSISRSGDQHRWPAKSQQNQANFMNRSLDCGVLLRSSNG--FGSNV 229
S + +S+S +H S N A + R G RSS+G S V
Sbjct: 381 AADDIQGSTSKEATSSVS----EHTSIVTSATNAAQYATR---LGSSSRSSSGAVSSSAV 433
Query: 230 VRSLRNSLL--DPRVSHDGATLRSESNKNGGSEPVIEPELVPSDNESVTSGSSSGV 283
+S+ NS++ + VS + + + K+ + V E + S+ +S + +
Sbjct: 434 SQSVLNSVIAVNTDVSVTSVSSTAHTTKDTATTSVTASESITSETAQASSSTEKNI 489
>P3K2_DICDI (P54674) Phosphatidylinositol 3-kinase 2 (EC 2.7.1.137)
(PI3-kinase) (PtdIns-3-kinase) (PI3K)
Length = 1858
Score = 40.8 bits (94), Expect = 0.010
Identities = 79/314 (25%), Positives = 124/314 (39%), Gaps = 58/314 (18%)
Query: 4 AISTTTTTKNTKRAPTPTRP------------PLLPSE--------SDNAIVRK------ 37
A +TTTTT NT TPT P +PS +A+ +K
Sbjct: 293 ATTTTTTTTNTSAPTTPTNRVQSSLDDLLFNLPTIPSNVPTVNGGPKISAVPKKVSSSKL 352
Query: 38 --PKAREVTS----RYMYSSSSSSSFSSPPRRTNSPLVNRTVNSTKQKSTPSVTPAAFVK 91
P + V+S SSSS SS SS T P+V ++S+ ++PS + +
Sbjct: 353 LIPPSSNVSSSSNITLSLSSSSPSSSSSSSTSTVVPIVQ--LSSSNSTNSPSTSLPTTPR 410
Query: 92 RSQSTERQRQRQGTPRPNGETPV-AQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVT 150
SQ T Q P + P + T+ + S S S S+ +S +P +
Sbjct: 411 LSQPTTSYTQL--IPSQQQQQPTESNSSSNTNTTTTSSSSSSSSSSLTISSPQPSN---- 464
Query: 151 QSLRRSTPERRKVPATVTTPTI--GRGRVNGNSDQTENSISRSGDQHRWPAKSQQNQANF 208
S+R S R T+++ I G+V+ ++S S + Q + N
Sbjct: 465 NSIRISAFGRSSTQFTISSNGIPSSPGQVSNKVYNNIGNLSNSSGER--VKNKQYSMLNI 522
Query: 209 MNRS-LDCGVLLRSSNGFGS-NVVRSLRNSLLDPRVSHDGATLRSESNKNGGSEPVI--- 263
++ LD + S GS N +R+ +S L P +S G GGS P +
Sbjct: 523 SKKTILDESDISSSPRSIGSPNSIRASISSQLPPSLSSIG------GGGGGGSGPNVVSN 576
Query: 264 EPELV--PSDNESV 275
+P +V PS +E +
Sbjct: 577 KPLVVKKPSTSEHI 590
>ZAN_PIG (Q28983) Zonadhesin precursor
Length = 2476
Score = 40.4 bits (93), Expect = 0.013
Identities = 42/166 (25%), Positives = 71/166 (42%), Gaps = 14/166 (8%)
Query: 5 ISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKP--KAREVTSRYMYSSSSSSSFSSPPR 62
+ T TT T+R PT P +P+E + KP E T + + ++P +
Sbjct: 534 VLTERTTTPTERTTIPTEKPTVPTEKPSVPTEKPTVPTEEPTIPTEKLTVPTERTTTPTK 593
Query: 63 RTNSPLVNRTVNSTKQKSTPSV-TPAAFVKRSQSTERQRQRQGTPRPNGETPV-AQKMLF 120
RT +P + RT T + +TP+ T ++ + TER T P +T V +K +
Sbjct: 594 RTTTPTI-RTTTPTIRTTTPTERTTTPTIRTTTPTER------TTIPTKKTTVPTEKTII 646
Query: 121 TSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPAT 166
+ R+++ + S P P+A V S +T R A+
Sbjct: 647 PTERTIAPTTPQPS---PTLVPTQPAAVVMPSTSATTVTPRTTIAS 689
Score = 36.6 bits (83), Expect = 0.18
Identities = 45/190 (23%), Positives = 75/190 (38%), Gaps = 26/190 (13%)
Query: 3 TAISTTTTTKN---TKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSS 59
T ST T K TKR TPT +P+E + KP T R + ++ +
Sbjct: 438 TGKSTIPTEKPMVPTKRTTTPTERTTIPAEKPTVPIEKPMV--PTERTTIPTERTTIPTE 495
Query: 60 PPR--------RTNSPLV--NRTVNSTKQKSTPSVTPAAFVKRSQS-TER---QRQRQGT 105
P T P+V + + T++ + P+ +R+ + TER ++
Sbjct: 496 KPTVPTEKLTVPTEKPIVPTEKPIVPTEKHTIPTEKLTVLTERTTTPTERTTIPTEKPTV 555
Query: 106 PRPNGETPVAQKMLFTSARSLSVSFQGESFSIPVSKAKPPS-ATVTQSLRRSTPERRKVP 164
P P + + T ++ E ++P + P+ T T ++R +TP R
Sbjct: 556 PTEKPSVPTEKPTVPTEEPTIPT----EKLTVPTERTTTPTKRTTTPTIRTTTPTIRTTT 611
Query: 165 AT--VTTPTI 172
T TTPTI
Sbjct: 612 PTERTTTPTI 621
Score = 35.8 bits (81), Expect = 0.31
Identities = 41/179 (22%), Positives = 66/179 (35%), Gaps = 32/179 (17%)
Query: 5 ISTTTTTKNTKRAPTPTRPPLLPSE-----SDNAIVRKPKAREVTSRYMYSSSS----SS 55
+ T TT T+R PT P +P+E ++ IV K T ++ + +
Sbjct: 478 VPTERTTIPTERTTIPTEKPTVPTEKLTVPTEKPIVPTEKPIVPTEKHTIPTEKLTVLTE 537
Query: 56 SFSSPPRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKR----SQSTERQRQRQGTPRPNGE 111
++P RT P TV T++ S P+ P + ++ +R TP
Sbjct: 538 RTTTPTERTTIPTEKPTV-PTEKPSVPTEKPTVPTEEPTIPTEKLTVPTERTTTPTKRTT 596
Query: 112 TPVAQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTP-ERRKVPATVTT 169
TP + T + P T T ++R +TP ER +P TT
Sbjct: 597 TPTIRTTTPT-----------------IRTTTPTERTTTPTIRTTTPTERTTIPTKKTT 638
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.312 0.127 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 64,454,234
Number of Sequences: 164201
Number of extensions: 2824973
Number of successful extensions: 9611
Number of sequences better than 10.0: 276
Number of HSP's better than 10.0 without gapping: 29
Number of HSP's successfully gapped in prelim test: 250
Number of HSP's that attempted gapping in prelim test: 8487
Number of HSP's gapped (non-prelim): 806
length of query: 573
length of database: 59,974,054
effective HSP length: 116
effective length of query: 457
effective length of database: 40,926,738
effective search space: 18703519266
effective search space used: 18703519266
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 68 (30.8 bits)
Medicago: description of AC140671.7


