

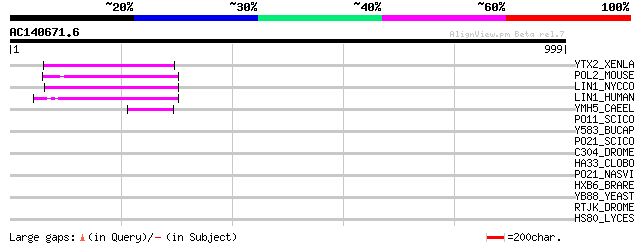
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140671.6 + phase: 0 /pseudo
(999 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 109 4e-23
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 95 1e-18
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 78 1e-13
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 76 4e-13
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 48 1e-04
PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type... 39 0.053
Y583_BUCAP (O51880) Hypothetical UPF0070 protein BUsg583 37 0.26
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 35 0.77
C304_DROME (Q9VG17) Probable cytochrome P450 304a1 (EC 1.14.-.-)... 34 1.7
HA33_CLOBO (P46084) Main hemagglutinin component (HA 33 kDa subu... 34 2.2
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 33 3.8
HXB6_BRARE (P15861) Homeobox protein Hox-B6 (ZF-22) 32 6.5
YB88_YEAST (P38330) Hypothetical 83.7 kDa protein in PRP5-THI2 i... 32 8.5
RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile elem... 32 8.5
HS80_LYCES (P36181) Heat shock cognate protein 80 32 8.5
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 109 bits (272), Expect = 4e-23
Identities = 63/237 (26%), Positives = 119/237 (49%), Gaps = 1/237 (0%)
Query: 61 FQKSRSKWIADGDRNTKYYHSKTIIRRRKNKILTLCDDSSDWIDDPNILKNLVRNFYTDL 120
F +SR + + D DR ++++++ + + +I L + ++DP +++ R+FY +L
Sbjct: 357 FVRSRMQLLCDMDRGSRFFYALEKKKGNRKQITCLFAEDGTPLEDPEAIRDRARSFYQNL 416
Query: 121 FKEDTPSRDSIVS-WTTYPNVVGKYHNKLISNVQLNECKRALFDMGPHKASGEDGYPAIF 179
F D S D+ W P V + +L + + L+E +AL M +K+ G DG F
Sbjct: 417 FSPDPISPDACEELWDGLPVVSERRKERLETPITLDELSQALRLMPHNKSPGLDGLTIEF 476
Query: 180 FQHCWDKVADSLFNFVKQVWVNHSLISFINNILIVMIPKIEKPEFVSQFRPISLCNVIYK 239
FQ WD + + + + L ++ ++PK + +RP+SL + YK
Sbjct: 477 FQFFWDTLGPDFHRVLTEAFKKGELPLSCRRAVLSLLPKKGDLRLIKNWRPVSLLSTDYK 536
Query: 240 IISKVIVNRIKPLLDRIISPYQSSFIPGRNIHHNIIVAQEMVHAMSKMKGKKDFMSI 296
I++K I R+K +L +I P QS +PGR I N+ + ++++H + F+S+
Sbjct: 537 IVAKAISLRLKSVLAEVIHPDQSYTVPGRTIFDNVFLIRDLLHFARRTGLSLAFLSL 593
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 94.7 bits (234), Expect = 1e-18
Identities = 70/248 (28%), Positives = 118/248 (47%), Gaps = 12/248 (4%)
Query: 60 WFQKSRSKWIADGDRNTKYYHSKTIIRRRKNKILTLCDDSSDWIDDPNILKNLVRNFYTD 119
WF + +K R TK + K +I + +N + D DP ++N +R+FY
Sbjct: 391 WFFEKINKIDKPLARLTKGHRDKILINKIRN-------EKGDITTDPEEIQNTIRSFYKR 443
Query: 120 LFK---EDTPSRDSIVSWTTYPNVVGKYHNKLISNVQLNECKRALFDMGPHKASGEDGYP 176
L+ E+ D + P + + L S + E + + + K+ G DG+
Sbjct: 444 LYSTKLENLDEMDKFLDRYQVPKLNQDQVDHLNSPISPKEIEAVINSLPTKKSPGPDGFS 503
Query: 177 AIFFQHCWDKVADSLFNFVKQVWVNHSLISFINNILIVMIPKIEK-PEFVSQFRPISLCN 235
A F+Q + + L ++ V +L + I +IPK +K P + FRPISL N
Sbjct: 504 AEFYQTFKEDLIPILHKLFHKIEVEGTLPNSFYEATITLIPKPQKDPTKIENFRPISLMN 563
Query: 236 VIYKIISKVIVNRIKPLLDRIISPYQSSFIPGRNIHHNIIVAQEMVHAMSKMKGKKDFMS 295
+ KI++K++ NRI+ + II P Q FIPG NI + ++H ++K+K K+ M
Sbjct: 564 IDAKILNKILANRIQEHIKAIIHPDQVGFIPGMQGWFNIRKSINVIHYINKLK-DKNHMI 622
Query: 296 IKIDL*KS 303
I +D K+
Sbjct: 623 ISLDAEKA 630
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 78.2 bits (191), Expect = 1e-13
Identities = 62/245 (25%), Positives = 117/245 (47%), Gaps = 5/245 (2%)
Query: 63 KSRSKWIADGDRNTKYYHSKTIIRRRKNKILTLCDDSSDWIDDPNILKNLVRNFYTDLFK 122
KS+S + ++ K + T +R K+ I ++ + + + DP+ ++ ++ +Y L+
Sbjct: 360 KSKSWFFEKINKIDKPLANLTRKKRVKSLISSIRNGNDEITTDPSEIQKILNEYYKKLYS 419
Query: 123 ---EDTPSRDSIVSWTTYPNVVGKYHNKLISNVQLNECKRALFDMGPHKASGEDGYPAIF 179
E+ D + P + K L + +E + ++ K+ G DG+ + F
Sbjct: 420 HKYENLKEIDQYLEACHLPRLSQKEVEMLNRPISSSEIASTIQNLPKKKSPGPDGFTSEF 479
Query: 180 FQHCWDKVADSLFNFVKQVWVNHSLISFINNILIVMIPKIEK-PEFVSQFRPISLCNVIY 238
+Q +++ L N + + L + I +IPK K P +RPISL N+
Sbjct: 480 YQTFKEELVPILLNLFQNIEKEGILPNTFYEANITLIPKPGKDPTRKENYRPISLMNIDA 539
Query: 239 KIISKVIVNRIKPLLDRIISPYQSSFIPGRNIHHNIIVAQEMVHAMSKMKGKKDFMSIKI 298
KI++K++ NRI+ + +II Q FIPG NI + ++ ++K+K KD M + I
Sbjct: 540 KILNKILTNRIQQHIKKIIHHDQVGFIPGSQGWFNIRKSINVIQHINKLK-NKDHMILSI 598
Query: 299 DL*KS 303
D K+
Sbjct: 599 DAEKA 603
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 76.3 bits (186), Expect = 4e-13
Identities = 65/265 (24%), Positives = 119/265 (44%), Gaps = 12/265 (4%)
Query: 43 KELHEQLAITLYQEECLWFQKSRSKWIADGDRNTKYYHSKTIIRRRKNKILTLCDDSSDW 102
KE+ Q + E WF + +K DR K +R KN+I T+ +D D
Sbjct: 347 KEIETQKTLQKINESRSWFFEKINKI----DRPLARLIKK---KREKNQIDTIKNDRGDI 399
Query: 103 IDDPNILKNLVRNFYTDLFK---EDTPSRDSIVSWTTYPNVVGKYHNKLISNVQLNECKR 159
DP ++ +R +Y L+ E+ D + T P + + L + +E +
Sbjct: 400 TTDPTEIQTTIREYYKHLYANKLENLEEMDKFLDTYTLPRLNQEEVESLNRPITSSEIEA 459
Query: 160 ALFDMGPHKASGEDGYPAIFFQHCWDKVADSLFNFVKQVWVNHSLISFINNILIVMIPKI 219
+ + K+ G +G+ A F+Q +++ L + + L + I++IPK
Sbjct: 460 IINSLPNKKSPGPEGFTAEFYQRYKEELVPFLLKLFQSIEKEGILPNSFYEASIILIPKP 519
Query: 220 EKPEFVSQ-FRPISLCNVIYKIISKVIVNRIKPLLDRIISPYQSSFIPGRNIHHNIIVAQ 278
+ + FRPISL N+ KI++K++ N+I+ + ++I Q FIP NI +
Sbjct: 520 GRDTTKKENFRPISLMNIDAKILNKILANQIQQHIKKLIHHDQVGFIPAMQGWFNIRKSI 579
Query: 279 EMVHAMSKMKGKKDFMSIKIDL*KS 303
++ +++ K + M I ID K+
Sbjct: 580 NIIQHINRTK-DTNHMIISIDAEKA 603
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 48.1 bits (113), Expect = 1e-04
Identities = 23/83 (27%), Positives = 47/83 (55%)
Query: 212 LIVMIPKIEKPEFVSQFRPISLCNVIYKIISKVIVNRIKPLLDRIISPYQSSFIPGRNIH 271
+I+ IPK P S +RPISL + +I+ ++I +RI+ ++SP+Q F+ R+
Sbjct: 669 VIIPIPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSHLLSPHQHGFLNFRSCP 728
Query: 272 HNIIVAQEMVHAMSKMKGKKDFM 294
+++ + + H++ K + D +
Sbjct: 729 SSLVRSISLYHSILKNEKSLDIL 751
>PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1004
Score = 39.3 bits (90), Expect = 0.053
Identities = 30/129 (23%), Positives = 61/129 (47%), Gaps = 17/129 (13%)
Query: 151 NVQLNECKRALFDMGPHKASGEDGYPAIFFQHCWDKVADSLFNFVKQV---------WVN 201
+++++E ++ K+ G DG + W + + +F KQ W
Sbjct: 404 DLEMDEVSDSVRRCKVRKSPGPDGIVGEMVRAVWGAIPEYMFCLYKQCLLESYFPQKWKI 463
Query: 202 HSLISFINNILIVMIPKIEKPEFVSQFRPISLCNVIYKIISKVIVNRI-KPLLDRIISPY 260
SL+ IL+ ++ +I +RPI L + + K++ ++V R+ + L+D +SPY
Sbjct: 464 ASLV-----ILLKLLDRIRSDP--GSYRPICLLDNLGKVLEGIMVKRLDQKLMDVEVSPY 516
Query: 261 QSSFIPGRN 269
Q +F G++
Sbjct: 517 QFAFTYGKS 525
>Y583_BUCAP (O51880) Hypothetical UPF0070 protein BUsg583
Length = 194
Score = 37.0 bits (84), Expect = 0.26
Identities = 35/153 (22%), Positives = 69/153 (44%), Gaps = 10/153 (6%)
Query: 15 RKNELLARLNGIQNSHNYGYSNFLENLEKELHEQLAITLYQEECLWFQKSRSKWIADGDR 74
R E++ ++N N + NF+ K ++ L+ +L+ + K+ K + +
Sbjct: 44 RYEEVIKKINAKNNQNLKSVENFITE-NKNIYGTLS-SLFLAKKYILDKNLDKALIQLNN 101
Query: 75 NTKYYHSKT---IIRRRKNKILTLCDDSSDWID-----DPNILKNLVRNFYTDLFKEDTP 126
+ KY + I++ R KI + + D I N KN+V N D+F ++
Sbjct: 102 SLKYTKEENLQNILKIRIAKIKIQQNKNQDAIKILEEIKDNSWKNIVENMKGDIFMKNKE 161
Query: 127 SRDSIVSWTTYPNVVGKYHNKLISNVQLNECKR 159
+ +I++W + +K I N+++NE KR
Sbjct: 162 IKKAILAWKKSKYLEKSNASKEIINMKINEIKR 194
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 35.4 bits (80), Expect = 0.77
Identities = 25/91 (27%), Positives = 43/91 (46%), Gaps = 2/91 (2%)
Query: 160 ALFDMGPHKASGEDGY-PAIFFQHCWDKVADSLFNFVKQVWVNHSLI-SFINNILIVMIP 217
+L ++ +AS E G P W+ + D + ++V + + S I V P
Sbjct: 160 SLIEIKSARASNEKGAGPDGVTPRSWNALDDRYKRLLYNIFVFYGRVPSPIKGSRTVFTP 219
Query: 218 KIEKPEFVSQFRPISLCNVIYKIISKVIVNR 248
KIE FRP+S+C+VI + +K++ R
Sbjct: 220 KIEGGPDPGVFRPLSICSVILREFNKILARR 250
>C304_DROME (Q9VG17) Probable cytochrome P450 304a1 (EC 1.14.-.-)
(CYPCCCIVA1)
Length = 510
Score = 34.3 bits (77), Expect = 1.7
Identities = 25/90 (27%), Positives = 37/90 (40%), Gaps = 18/90 (20%)
Query: 720 WSRHYWVGWNQYPPFHSSKCLFFATSGFSDFGR*QEKYLEG-ANGVVESLLLARVVEMKM 778
W RH W W+ Y + S F F+DF +KYL+ GV + + + EM+
Sbjct: 240 WIRHIWPEWSGYNKLNESN--LFVRQFFADF---VDKYLDSYEEGVERNFMDVYIAEMRR 294
Query: 779 KLFFMCFAIASTLPRYGFTSFHMIL*LISF 808
P YGF +I+ L+ F
Sbjct: 295 G------------PGYGFNRDQLIMGLVDF 312
>HA33_CLOBO (P46084) Main hemagglutinin component (HA 33 kDa
subunit) (HA1)
Length = 285
Score = 33.9 bits (76), Expect = 2.2
Identities = 29/119 (24%), Positives = 52/119 (43%), Gaps = 9/119 (7%)
Query: 6 TKTFGNIFRRKNELLARLNGIQNSHNYGYSNFLENLEKELHEQLAITLYQEECLWFQKSR 65
T + F+ N + LN +N N L K L+ Q+ I L+Q WF SR
Sbjct: 129 TSSSNQFFKFSNCIYEALNN-RNCKLQTQLNSDRFLSKNLNSQI-IVLWQ----WFDSSR 182
Query: 66 SKWIADGDRNTKYYHSKTIIRRRKNKILTLCDDSSDWIDDPNILKNLVRNFYTDLFKED 124
KWI + + Y T+ + N+ LT +S+++++ +L++ + + D
Sbjct: 183 QKWIIEYNETKSAY---TLKCQENNRYLTWIQNSNNYVETYQSTDSLIQYWNINYLDND 238
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 33.1 bits (74), Expect = 3.8
Identities = 37/145 (25%), Positives = 65/145 (44%), Gaps = 18/145 (12%)
Query: 149 ISNVQLNE---CKRALFDMGPHKASGEDGYPAIFFQHCWDKVADSLFNFVKQVWVNHSLI 205
ISN ++ E CKR A+G DG + D+ SLFN + ++
Sbjct: 319 ISNDEIKEVEACKRT--------AAGPDGMTTTAWNSI-DECIKSLFNMI--MYHGQCPR 367
Query: 206 SFINNILIVMIPKIEKPEFVSQFRPISLCNVIYKIISKVIVNRIKPLLDRIISPYQSSFI 265
++++ V+IPK + FRP+S+ +V + +++ NRI ++ Q +FI
Sbjct: 368 RYLDS-RTVLIPKEPGTMDPACFRPLSIASVALRHFHRILANRIGE--HGLLDTRQRAFI 424
Query: 266 PGRNIHHNIIVAQEMV-HAMSKMKG 289
+ N + M+ A K+KG
Sbjct: 425 VADGVAENTSLLSAMIKEARMKIKG 449
>HXB6_BRARE (P15861) Homeobox protein Hox-B6 (ZF-22)
Length = 228
Score = 32.3 bits (72), Expect = 6.5
Identities = 18/62 (29%), Positives = 30/62 (48%), Gaps = 2/62 (3%)
Query: 8 TFGNIFRRKNELLARLNGIQNSHNYGYSNFLENLEK-ELHEQLAITLYQEECLWFQKSRS 66
TFGN RR + R ++ + ++ +L + E+ L +T Q + +WFQ R
Sbjct: 145 TFGNAGRRGRQTYTRYQTLELEKEFHFNRYLTRRRRIEIAHALCLTERQIK-IWFQNRRM 203
Query: 67 KW 68
KW
Sbjct: 204 KW 205
>YB88_YEAST (P38330) Hypothetical 83.7 kDa protein in PRP5-THI2
intergenic region
Length = 731
Score = 32.0 bits (71), Expect = 8.5
Identities = 33/150 (22%), Positives = 62/150 (41%), Gaps = 28/150 (18%)
Query: 151 NVQLNECKRALFDMGPHKASGEDGYPAIFFQHCWDKVADSLFNFVKQVW----------V 200
NV +AL D P+K + Y F ++ W + D FN + Q+W V
Sbjct: 395 NVAQTFFDKALNDEMPYKIDLQVSYVKSFLRNIWSQTRD--FNHIYQIWYKSSLHYGRNV 452
Query: 201 NHSLISFINNILIVMIPKIEKPEFVSQFRPISLCNVIYK--------------IISKVIV 246
NH + S +N+ + + + + F+ +L N+I I++K V
Sbjct: 453 NHGISSSLNDTFFDIFFENYAVDKMQGFQ--TLQNIIQTYNNIKHIDEPFFNIILAKCTV 510
Query: 247 NRIKPLLDRIISPYQSSFIPGRNIHHNIIV 276
+ +L+ I Y++ IP + + I++
Sbjct: 511 WHDRSILEYIDKSYEAYHIPKTIVAYRILL 540
>RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 32.0 bits (71), Expect = 8.5
Identities = 32/123 (26%), Positives = 54/123 (43%), Gaps = 11/123 (8%)
Query: 149 ISNVQLNECKRALFDMGPHKASGEDGYPAIFFQHCWDKVADSLFNFVKQVWVNHSLISFI 208
+S V L E K + + KA GED + + D F+ ++ + + +
Sbjct: 435 LSAVTLEEVKNLIAKLPLKKAPGED----LLDNRTIRLLPDQALQFLALIFNSVLDVGYF 490
Query: 209 ----NNILIVMIPKIEK-PEFVSQFRPISLCNVIYKIISKVIVNRIKPLLD--RIISPYQ 261
+ I+MI K K P V +RP SL + KI+ ++I+NR+ D + I +Q
Sbjct: 491 PKAWKSASIIMIHKTGKTPTDVDSYRPTSLLPSLGKIMERLILNRLLTCKDVTKAIPKFQ 550
Query: 262 SSF 264
F
Sbjct: 551 FGF 553
>HS80_LYCES (P36181) Heat shock cognate protein 80
Length = 699
Score = 32.0 bits (71), Expect = 8.5
Identities = 27/121 (22%), Positives = 50/121 (41%), Gaps = 5/121 (4%)
Query: 11 NIFRRKNELLARLNGIQNSHNYGYSNFLENLEKELHEQLAITLYQEECLWFQKSRSKWIA 70
N+ ++ EL + + +N Y F +NL+ +HE E L + ++S
Sbjct: 386 NLVKKCVELFFEIAENKEDYNKFYEAFSKNLKLGIHEDSQNRAKFAELLRYHSTKS---- 441
Query: 71 DGDRNTKYYHSKTIIRRRKNKILTLCDDSSDWIDDPNILKNLVRNFYTDLFKEDTPSRDS 130
GD T T ++ +N I + +S +++ L+ L + Y L+ D S
Sbjct: 442 -GDEMTSLKDYVTRMKEGQNDIYYITGESKKAVENSPFLEKLKKKGYEVLYMVDAIDEYS 500
Query: 131 I 131
I
Sbjct: 501 I 501
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.360 0.161 0.604
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 97,881,035
Number of Sequences: 164201
Number of extensions: 3557898
Number of successful extensions: 16605
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 16584
Number of HSP's gapped (non-prelim): 23
length of query: 999
length of database: 59,974,054
effective HSP length: 120
effective length of query: 879
effective length of database: 40,269,934
effective search space: 35397271986
effective search space used: 35397271986
T: 11
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.9 bits)
S2: 71 (32.0 bits)
Medicago: description of AC140671.6


