

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140670.16 + phase: 0
(235 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
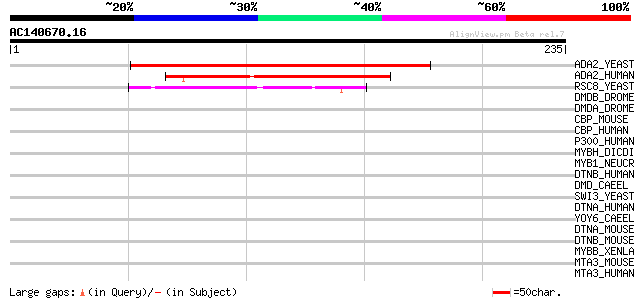
Score E
Sequences producing significant alignments: (bits) Value
ADA2_YEAST (Q02336) Transcriptional adapter 2 131 2e-30
ADA2_HUMAN (O75478) Transcriptional adapter 2-like (ADA2-like pr... 99 9e-21
RSC8_YEAST (P43609) Chromatin structure remodeling complex prote... 44 3e-04
DMDB_DROME (Q9VDW3) Dystrophin, isoform B 39 0.014
DMDA_DROME (Q9VDW6) Dystrophin, isoforms A/C/DLP1/DLP3 39 0.014
CBP_MOUSE (P45481) CREB-binding protein (EC 2.3.1.48) 38 0.024
CBP_HUMAN (Q92793) CREB-binding protein (EC 2.3.1.48) 38 0.024
P300_HUMAN (Q09472) E1A-associated protein p300 (EC 2.3.1.48) 37 0.031
MYBH_DICDI (P34127) Myb-like protein (Fragment) 36 0.069
MYB1_NEUCR (O13493) Myb-like DNA binding protein myb-1 36 0.090
DTNB_HUMAN (O60941) Dystrobrevin beta (Beta-dystrobrevin) (DTN-B) 36 0.090
DMD_CAEEL (Q9TW65) Dystrophin-1 36 0.090
SWI3_YEAST (P32591) Transcription regulatory protein SWI3 (SWI/S... 35 0.12
DTNA_HUMAN (Q9Y4J8) Dystrobrevin alpha (Dystrobrevin-alpha) 35 0.15
YOY6_CAEEL (P34664) Hypothetical protein ZK652.6 in chromosome III 35 0.20
DTNA_MOUSE (Q9D2N4) Dystrobrevin alpha (Alpha-dystrobrevin) 35 0.20
DTNB_MOUSE (O70585) Dystrobrevin beta (Beta-dystrobrevin) (DTN-B... 34 0.34
MYBB_XENLA (P52551) Myb-related protein B (B-Myb) (Myb-related p... 33 0.76
MTA3_MOUSE (Q924K8) Metastasis associated protein MTA3 33 0.76
MTA3_HUMAN (Q9BTC8) Metastasis associated protein MTA3 33 0.76
>ADA2_YEAST (Q02336) Transcriptional adapter 2
Length = 434
Score = 131 bits (329), Expect = 2e-30
Identities = 48/127 (37%), Positives = 83/127 (64%)
Query: 52 YHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPD 111
+HC+ C+ D + ++R+ CA+C ++DLC+ CFS G H+ H YR+++ S+P++CPD
Sbjct: 5 FHCDVCSADCTNRVRVSCAICPEYDLCVPCFSQGSYTGKHRPYHDYRIIETNSYPILCPD 64
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSG 171
W A+EE+ L++ G GNW D+AD+IG++ K + +HY Y+ S +P+PD++
Sbjct: 65 WGADEELQLIKGAQTLGLGNWQDIADHIGSRGKEEVKEHYLKYYLESKYYPIPDITQNIH 124
Query: 172 KNKEELL 178
++E L
Sbjct: 125 VPQDEFL 131
>ADA2_HUMAN (O75478) Transcriptional adapter 2-like (ADA2-like
protein) (KL04P)
Length = 443
Score = 99.0 bits (245), Expect = 9e-21
Identities = 46/97 (47%), Positives = 65/97 (66%), Gaps = 3/97 (3%)
Query: 67 IKCAVC--QDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPDWSAEEEMLLLEAL 124
IKCA C F LC++CF+ G E H+S+H Y +M + FP++ P W+A+EEM LLEA+
Sbjct: 29 IKCAECGPPPFFLCLQCFTRGFEYKKHQSDHTYEIMTS-DFPVLDPSWTAQEEMALLEAV 87
Query: 125 DMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCF 161
GFGNW DVA+ + TK+K +C HY ++N+P F
Sbjct: 88 MDCGFGNWQDVANQMCTKTKEECEKHYMKHFINNPLF 124
>RSC8_YEAST (P43609) Chromatin structure remodeling complex protein
RSC8 (Remodel the structure of chromatin complex subunit
8) (SWI3 homolog)
Length = 557
Score = 44.3 bits (103), Expect = 3e-04
Identities = 30/102 (29%), Positives = 52/102 (50%), Gaps = 5/102 (4%)
Query: 51 LYHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICP 110
+Y C+ C + S +R +D +LC CF G +S+ R+ +N + +
Sbjct: 257 VYICHTCGNE-SINVRYHNLRARDTNLCSRCFQEGHFGANFQSSDFIRLENNGNS--VKK 313
Query: 111 DWSAEEEMLLLEALDMYGFGNWNDVADNI-GTKSKSQCIDHY 151
+WS +E +LLLE ++MY W +AD++ G K CI+ +
Sbjct: 314 NWSDQEMLLLLEGIEMYE-DQWEKIADHVGGHKRVEDCIEKF 354
>DMDB_DROME (Q9VDW3) Dystrophin, isoform B
Length = 1669
Score = 38.5 bits (88), Expect = 0.014
Identities = 16/43 (37%), Positives = 21/43 (48%)
Query: 54 CNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHP 96
CN C + R +C C +FD+C +CF G HK HP
Sbjct: 1284 CNICKEYPIVGFRYRCLKCFNFDMCQKCFFFGRNAKNHKLTHP 1326
>DMDA_DROME (Q9VDW6) Dystrophin, isoforms A/C/DLP1/DLP3
Length = 3497
Score = 38.5 bits (88), Expect = 0.014
Identities = 16/43 (37%), Positives = 21/43 (48%)
Query: 54 CNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHP 96
CN C + R +C C +FD+C +CF G HK HP
Sbjct: 3112 CNICKEYPIVGFRYRCLKCFNFDMCQKCFFFGRNAKNHKLTHP 3154
>CBP_MOUSE (P45481) CREB-binding protein (EC 2.3.1.48)
Length = 2441
Score = 37.7 bits (86), Expect = 0.024
Identities = 14/33 (42%), Positives = 21/33 (63%), Gaps = 2/33 (6%)
Query: 51 LYHCNYCNKDISGKIRIKCAVCQDFDLCIECFS 83
+Y CN C + + R C VC+D+DLCI C++
Sbjct: 1705 VYTCNECKHHV--ETRWHCTVCEDYDLCINCYN 1735
>CBP_HUMAN (Q92793) CREB-binding protein (EC 2.3.1.48)
Length = 2442
Score = 37.7 bits (86), Expect = 0.024
Identities = 14/33 (42%), Positives = 21/33 (63%), Gaps = 2/33 (6%)
Query: 51 LYHCNYCNKDISGKIRIKCAVCQDFDLCIECFS 83
+Y CN C + + R C VC+D+DLCI C++
Sbjct: 1704 VYTCNECKHHV--ETRWHCTVCEDYDLCINCYN 1734
>P300_HUMAN (Q09472) E1A-associated protein p300 (EC 2.3.1.48)
Length = 2414
Score = 37.4 bits (85), Expect = 0.031
Identities = 14/33 (42%), Positives = 21/33 (63%), Gaps = 2/33 (6%)
Query: 51 LYHCNYCNKDISGKIRIKCAVCQDFDLCIECFS 83
+Y CN C + + R C VC+D+DLCI C++
Sbjct: 1667 VYTCNECKHHV--ETRWHCTVCEDYDLCITCYN 1697
>MYBH_DICDI (P34127) Myb-like protein (Fragment)
Length = 451
Score = 36.2 bits (82), Expect = 0.069
Identities = 12/43 (27%), Positives = 27/43 (61%)
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTV 154
W++EE+ +L++A++++ NW +A++ ++ QC Y V
Sbjct: 152 WTSEEDQILIKAVNLHNQKNWKKIAEHFPDRTDVQCHHRYQKV 194
>MYB1_NEUCR (O13493) Myb-like DNA binding protein myb-1
Length = 229
Score = 35.8 bits (81), Expect = 0.090
Identities = 15/41 (36%), Positives = 26/41 (62%)
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYN 152
WSA E+ L++ + G GNW +VA +GT++ QC + ++
Sbjct: 12 WSAGEDQRLIKLVKDLGPGNWVNVARILGTRTPKQCRERWH 52
>DTNB_HUMAN (O60941) Dystrobrevin beta (Beta-dystrobrevin) (DTN-B)
Length = 627
Score = 35.8 bits (81), Expect = 0.090
Identities = 14/42 (33%), Positives = 20/42 (47%)
Query: 54 CNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNH 95
C+YC + R +C C ++ LC CF G PH + H
Sbjct: 243 CSYCRCESMMGFRYRCQQCHNYQLCQNCFWRGHAGGPHSNQH 284
>DMD_CAEEL (Q9TW65) Dystrophin-1
Length = 3674
Score = 35.8 bits (81), Expect = 0.090
Identities = 16/43 (37%), Positives = 20/43 (46%)
Query: 54 CNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHP 96
CN C IR +C C + DLC CF H++NHP
Sbjct: 3306 CNVCKMFPIIGIRYRCLTCFNCDLCQNCFFSQRTAKSHRTNHP 3348
>SWI3_YEAST (P32591) Transcription regulatory protein SWI3 (SWI/SNF
complex component SWI3) (Transcription factor TYE2)
Length = 825
Score = 35.4 bits (80), Expect = 0.12
Identities = 16/38 (42%), Positives = 23/38 (60%), Gaps = 1/38 (2%)
Query: 111 DWSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCI 148
+WS E+ LL+ + +G +W VA N+G KS QCI
Sbjct: 526 NWSKEDLQKLLKGIQEFG-ADWYKVAKNVGNKSPEQCI 562
>DTNA_HUMAN (Q9Y4J8) Dystrobrevin alpha (Dystrobrevin-alpha)
Length = 743
Score = 35.0 bits (79), Expect = 0.15
Identities = 13/42 (30%), Positives = 21/42 (49%)
Query: 54 CNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNH 95
C+YC+ + R +C C ++ LC +CF G H + H
Sbjct: 243 CSYCHSESMMGFRYRCQQCHNYQLCQDCFWRGHAGGSHSNQH 284
>YOY6_CAEEL (P34664) Hypothetical protein ZK652.6 in chromosome III
Length = 575
Score = 34.7 bits (78), Expect = 0.20
Identities = 17/64 (26%), Positives = 29/64 (44%), Gaps = 9/64 (14%)
Query: 54 CNYCNKDISGKIRIKCAVCQDFDLCIECFSVG--------VELTPHKSNHPYR-VMDNLS 104
C+ C R KC C D+DLC CF+ ++ H +HP + ++ ++
Sbjct: 14 CDGCAFTAFAGNRYKCLRCSDYDLCFSCFTTKNYGDQQTIADIPIHDESHPMQLILSSVD 73
Query: 105 FPLI 108
F L+
Sbjct: 74 FDLV 77
>DTNA_MOUSE (Q9D2N4) Dystrobrevin alpha (Alpha-dystrobrevin)
Length = 746
Score = 34.7 bits (78), Expect = 0.20
Identities = 13/42 (30%), Positives = 21/42 (49%)
Query: 54 CNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNH 95
C+YC+ + R +C C ++ LC +CF G H + H
Sbjct: 243 CSYCHSESIMGFRYRCQQCHNYQLCQDCFWRGHAGGSHSNQH 284
>DTNB_MOUSE (O70585) Dystrobrevin beta (Beta-dystrobrevin) (DTN-B)
(MDTN-B)
Length = 700
Score = 33.9 bits (76), Expect = 0.34
Identities = 13/42 (30%), Positives = 20/42 (46%)
Query: 54 CNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNH 95
C+YC+ + R +C C ++ LC CF G H + H
Sbjct: 243 CSYCHCESMMGFRYRCQQCHNYQLCQNCFWRGHASGAHSNQH 284
>MYBB_XENLA (P52551) Myb-related protein B (B-Myb) (Myb-related
protein 1) (XMYB1)
Length = 743
Score = 32.7 bits (73), Expect = 0.76
Identities = 11/36 (30%), Positives = 20/36 (55%)
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQC 147
W+ EE+ L + +G G W +A N+ +++ QC
Sbjct: 34 WTPEEDETLKALVKKHGQGEWKTIASNLNNRTEQQC 69
>MTA3_MOUSE (Q924K8) Metastasis associated protein MTA3
Length = 591
Score = 32.7 bits (73), Expect = 0.76
Identities = 22/58 (37%), Positives = 33/58 (55%), Gaps = 6/58 (10%)
Query: 99 VMDNLSFPLICPD----WSAEEEMLLLEALDMYGFGNWNDV-ADNIGTKSKSQCIDHY 151
V+ L P++C D WSA E L EAL+ YG ++ND+ D + KS + I++Y
Sbjct: 253 VLVPLGGPVLCRDEMEEWSASEACLFEEALEKYG-KDFNDIRQDFLPWKSLTSIIEYY 309
>MTA3_HUMAN (Q9BTC8) Metastasis associated protein MTA3
Length = 594
Score = 32.7 bits (73), Expect = 0.76
Identities = 22/58 (37%), Positives = 33/58 (55%), Gaps = 6/58 (10%)
Query: 99 VMDNLSFPLICPD----WSAEEEMLLLEALDMYGFGNWNDV-ADNIGTKSKSQCIDHY 151
V+ L P++C D WSA E L EAL+ YG ++ND+ D + KS + I++Y
Sbjct: 254 VLVPLGGPVLCRDEMEEWSASEASLFEEALEKYG-KDFNDIRQDFLPWKSLTSIIEYY 310
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.135 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,359,769
Number of Sequences: 164201
Number of extensions: 1170675
Number of successful extensions: 2763
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 36
Number of HSP's successfully gapped in prelim test: 27
Number of HSP's that attempted gapping in prelim test: 2708
Number of HSP's gapped (non-prelim): 80
length of query: 235
length of database: 59,974,054
effective HSP length: 107
effective length of query: 128
effective length of database: 42,404,547
effective search space: 5427782016
effective search space used: 5427782016
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC140670.16


