

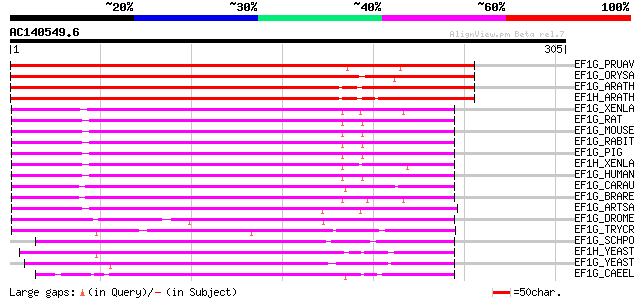
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140549.6 - phase: 0 /pseudo
(305 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
EF1G_PRUAV (Q9FUM1) Elongation factor 1-gamma (EF-1-gamma) (eEF-... 394 e-109
EF1G_ORYSA (Q9ZRI7) Elongation factor 1-gamma (EF-1-gamma) (eEF-... 360 3e-99
EF1G_ARATH (O04487) Probable elongation factor 1-gamma 1 (EF-1-g... 360 3e-99
EF1H_ARATH (Q9FVT2) Probable elongation factor 1-gamma 2 (EF-1-g... 359 4e-99
EF1G_XENLA (P26642) Elongation factor 1-gamma type 1 (EF-1-gamma... 172 9e-43
EF1G_RAT (Q68FR6) Elongation factor 1-gamma (EF-1-gamma) (eEF-1B... 159 6e-39
EF1G_MOUSE (Q9D8N0) Elongation factor 1-gamma (EF-1-gamma) (eEF-... 159 6e-39
EF1G_RABIT (P29694) Elongation factor 1-gamma (EF-1-gamma) (eEF-... 159 8e-39
EF1G_PIG (Q29387) Elongation factor 1-gamma (EF-1-gamma) (eEF-1B... 159 1e-38
EF1H_XENLA (Q91375) Elongation factor 1-gamma type 2 (EF-1-gamma... 156 7e-38
EF1G_HUMAN (P26641) Elongation factor 1-gamma (EF-1-gamma) (eEF-... 155 1e-37
EF1G_CARAU (Q90YC0) Elongation factor 1-gamma (EF-1-gamma) (eEF-... 150 3e-36
EF1G_BRARE (Q6PE25) Elongation factor 1-gamma (EF-1-gamma) (eEF-... 149 8e-36
EF1G_ARTSA (P12261) Elongation factor 1-gamma (EF-1-gamma) (eEF-... 147 4e-35
EF1G_DROME (Q9NJH0) Elongation factor 1-gamma (EF-1-gamma) (eEF-... 123 6e-28
EF1G_TRYCR (P34715) Elongation factor 1-gamma (EF-1-gamma) (eEF-... 120 5e-27
EF1G_SCHPO (P40921) Elongation factor 1-gamma (EF-1-gamma) (eEF-... 111 2e-24
EF1H_YEAST (P36008) Elongation factor 1-gamma 2 (EF-1-gamma 2) 97 5e-20
EF1G_YEAST (P29547) Elongation factor 1-gamma 1 (EF-1-gamma 1) 96 8e-20
EF1G_CAEEL (P54412) Probable elongation factor 1-gamma (EF-1-gam... 91 3e-18
>EF1G_PRUAV (Q9FUM1) Elongation factor 1-gamma (EF-1-gamma) (eEF-1B
gamma)
Length = 422
Score = 394 bits (1011), Expect = e-109
Identities = 194/261 (74%), Positives = 220/261 (83%), Gaps = 6/261 (2%)
Query: 1 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNN-LFGSSLIHQAQID 59
MGV+NKTP+++ +NPIGKVP+LETPDGP+FESNAIARYVARL +N L GSSLI A I+
Sbjct: 36 MGVTNKTPEYLKLNPIGKVPLLETPDGPIFESNAIARYVARLKADNPLIGSSLIDYAHIE 95
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
QWIDF SLEIDANI+ + PR G+A Y PP EE AIS+LKRAL ALN+HLA NTYLVGH
Sbjct: 96 QWIDFGSLEIDANIISWFRPRFGYAVYLPPAEEAAISALKRALGALNTHLASNTYLVGHF 155
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPI 179
VTLADII T NL+ GF KL++KSFTSEFPHVERYFWTLVNQP F+K+LG VKQTE++PP+
Sbjct: 156 VTLADIIVTCNLFFGFTKLMIKSFTSEFPHVERYFWTLVNQPKFKKVLGDVKQTESVPPV 215
Query: 180 PSAKQ--QPKESKPKTKDEPKKVAKSEPEKPKVEVE---EEAPKPKPKNPLDLLPPSKMI 234
PSAK+ QPKE+K K K+EPKK AK EP KPK E EEAPKPKPKNPLDLLPPS M+
Sbjct: 216 PSAKKPSQPKETKSKAKEEPKKEAKKEPAKPKAEAAEEVEEAPKPKPKNPLDLLPPSNMV 275
Query: 235 LDDWKRLYSNTKRNFREVAIK 255
LDDWKRLYSNTK NFREVAIK
Sbjct: 276 LDDWKRLYSNTKTNFREVAIK 296
>EF1G_ORYSA (Q9ZRI7) Elongation factor 1-gamma (EF-1-gamma) (eEF-1B
gamma)
Length = 418
Score = 360 bits (923), Expect = 3e-99
Identities = 180/260 (69%), Positives = 208/260 (79%), Gaps = 8/260 (3%)
Query: 1 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNN-LFGSSLIHQAQID 59
MGVSNKTP+++ MNPIGKVP+LETPDGPVFESNAIARYV R +N L+GSSLI A I+
Sbjct: 36 MGVSNKTPEYLKMNPIGKVPILETPDGPVFESNAIARYVTRSKSDNPLYGSSLIEYAHIE 95
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
QWIDFS+ E+DAN K PRLGFA Y EE AI++LKR+L ALN+HLA NTYLVGHS
Sbjct: 96 QWIDFSATEVDANTGKWLFPRLGFAPYVAVSEEAAIAALKRSLGALNTHLASNTYLVGHS 155
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPI 179
VTLADI+ T NLY+GFA+++ K+FTSEFPHVERYFWT+VNQPNF+K++G VKQ +++P +
Sbjct: 156 VTLADIVMTCNLYMGFARIMTKNFTSEFPHVERYFWTMVNQPNFKKVMGDVKQADSVPQV 215
Query: 180 PSAKQQPKESKPKTKDEPKKVAKSEPEKPKV----EVEEEAPKPKPKNPLDLLPPSKMIL 235
PKE KPK E KK A E KPK E EEEAPKPKPKNPLDLLPPSKMIL
Sbjct: 216 QKKAAAPKEQKPK---EAKKEAPKEAPKPKAAEKPEEEEEAPKPKPKNPLDLLPPSKMIL 272
Query: 236 DDWKRLYSNTKRNFREVAIK 255
D+WKRLYSNTK NFREVAIK
Sbjct: 273 DEWKRLYSNTKTNFREVAIK 292
>EF1G_ARATH (O04487) Probable elongation factor 1-gamma 1
(EF-1-gamma) (eEF-1B gamma)
Length = 414
Score = 360 bits (923), Expect = 3e-99
Identities = 183/256 (71%), Positives = 210/256 (81%), Gaps = 4/256 (1%)
Query: 1 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARL-GHNNLFGSSLIHQAQID 59
MGV+NKTP F+ MNPIGKVPVLETP+G VFESNAIARYV+RL G N+L GSSLI AQI+
Sbjct: 36 MGVTNKTPAFLKMNPIGKVPVLETPEGSVFESNAIARYVSRLNGDNSLNGSSLIEYAQIE 95
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
QWIDFSSLEI A+I++ + PR+GF Y P EE AIS+LKRAL+ALN+HL NTYLVGHS
Sbjct: 96 QWIDFSSLEIYASILRWFGPRMGFMPYSAPAEEGAISTLKRALDALNTHLTSNTYLVGHS 155
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPI 179
+TLADIIT NL +GFA ++ K FTSEFPHVERYFWT+VNQPNF K+LG VKQTEA+PPI
Sbjct: 156 ITLADIITVCNLNLGFATVMTKKFTSEFPHVERYFWTVVNQPNFTKVLGDVKQTEAVPPI 215
Query: 180 PSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPKPKNPLDLLPPSKMILDDWK 239
S K+ + +KP K+EPKK E PK+ EEEAPKPK KNPLDLLPPS M+LDDWK
Sbjct: 216 AS-KKAAQPAKP--KEEPKKKEAPVAEAPKLAEEEEAPKPKAKNPLDLLPPSPMVLDDWK 272
Query: 240 RLYSNTKRNFREVAIK 255
RLYSNTK NFREVAIK
Sbjct: 273 RLYSNTKSNFREVAIK 288
>EF1H_ARATH (Q9FVT2) Probable elongation factor 1-gamma 2
(EF-1-gamma) (eEF-1B gamma)
Length = 413
Score = 359 bits (922), Expect = 4e-99
Identities = 179/256 (69%), Positives = 212/256 (81%), Gaps = 5/256 (1%)
Query: 1 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARL-GHNNLFGSSLIHQAQID 59
MGV+NK+P+F+ MNPIGKVPVLETP+GP+FESNAIARYV+R G N+L GSSLI A I+
Sbjct: 36 MGVTNKSPEFLKMNPIGKVPVLETPEGPIFESNAIARYVSRKNGDNSLNGSSLIEYAHIE 95
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
QWIDFSSLEIDAN++K + PR+G+A + P EE AIS+LKR LEALN+HLA NT+LVGHS
Sbjct: 96 QWIDFSSLEIDANMLKWFAPRMGYAPFSAPAEEAAISALKRGLEALNTHLASNTFLVGHS 155
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPI 179
VTLADI+T NL +GFA ++ K FTS FPHVERYFWT+VNQP F+K+LG KQTEA+PP+
Sbjct: 156 VTLADIVTICNLNLGFATVMTKKFTSAFPHVERYFWTMVNQPEFKKVLGDAKQTEAVPPV 215
Query: 180 PSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPKPKNPLDLLPPSKMILDDWK 239
P+ K+ P+ +KP K+EPKK A E PK EEEAPKPK KNPLDLLPPS M+LDDWK
Sbjct: 216 PT-KKAPQPAKP--KEEPKKAA-PVAEAPKPAEEEEAPKPKAKNPLDLLPPSPMVLDDWK 271
Query: 240 RLYSNTKRNFREVAIK 255
RLYSNTK NFREVAIK
Sbjct: 272 RLYSNTKSNFREVAIK 287
>EF1G_XENLA (P26642) Elongation factor 1-gamma type 1 (EF-1-gamma)
(eEF-1B gamma) (p47)
Length = 436
Score = 172 bits (436), Expect = 9e-43
Identities = 105/261 (40%), Positives = 145/261 (55%), Gaps = 21/261 (8%)
Query: 2 GVSNKTPQFINMNPIGKVPVLETPDG-PVFESNAIARYVARLGHNNLFGSSLIHQAQIDQ 60
GV+NKTP+F+ P+GKVP E DG +FES+AIA YV G++ L G++ +HQAQ+ Q
Sbjct: 41 GVTNKTPEFLKKFPLGKVPAFEGKDGFCLFESSAIAHYV---GNDELRGTTRLHQAQVIQ 97
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 120
W+ FS I P LG Y E A +K L L+SHL T+LVG +
Sbjct: 98 WVSFSDSHIVPPASAWVFPTLGIMQYNKQATEQAKEGIKTVLGVLDSHLQTRTFLVGERI 157
Query: 121 TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIP 180
TLADI T +L + ++L SF F +V R+F T VNQP FR +LG+VK + M
Sbjct: 158 TLADITVTCSLLWLYKQVLEPSFRQPFGNVTRWFVTCVNQPEFRAVLGEVKLCDKMAQFD 217
Query: 181 S--------AKQQPKESKP-----KTKDEPKKVAKSEPEKPKVEVEEE----APKPKPKN 223
+ K+ PK+ KP K K+E KK A + P+ +++E A +PK K+
Sbjct: 218 AKKFAEMQPKKETPKKEKPAKEPKKEKEEKKKAAPTPAPAPEDDLDESEKALAAEPKSKD 277
Query: 224 PLDLLPPSKMILDDWKRLYSN 244
P LP S I+D++KR YSN
Sbjct: 278 PYAHLPKSSFIMDEFKRKYSN 298
>EF1G_RAT (Q68FR6) Elongation factor 1-gamma (EF-1-gamma) (eEF-1B
gamma)
Length = 436
Score = 159 bits (403), Expect = 6e-39
Identities = 103/262 (39%), Positives = 138/262 (52%), Gaps = 22/262 (8%)
Query: 2 GVSNKTPQFINMNPIGKVPVLETPDG-PVFESNAIARYVARLGHNNLFGSSLIHQAQIDQ 60
G +N+TP+F+ P GKVP E DG VFESNAIA YV+ + L GS+ AQ+ Q
Sbjct: 40 GQTNRTPEFLRKFPAGKVPAFEGDDGFCVFESNAIAYYVS---NEELRGSTPEAAAQVVQ 96
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 120
W+ F+ +I P LG + ENA +KR L L++HL T+LVG V
Sbjct: 97 WVSFADSDIVPPASTWVFPTLGIMHHNKQATENAKEEVKRILGLLDTHLKTRTFLVGERV 156
Query: 121 TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIP 180
TLADI L + ++L SF FP+ R+F T +NQP FR ILG+VK E M
Sbjct: 157 TLADITVVCTLLWLYKQVLEPSFRQAFPNTNRWFLTCINQPQFRAILGEVKLCEKMAQFD 216
Query: 181 S---AKQQPKESKPK--------------TKDEPKKVAKSEPEKPKVEVEEE-APKPKPK 222
+ A+ QPK+ P+ + E KK A PE+ E E+ A +PK K
Sbjct: 217 AKKFAESQPKKDTPRKEKGSREEKQKPQTERKEEKKAAAPAPEEEMDECEQALAAEPKAK 276
Query: 223 NPLDLLPPSKMILDDWKRLYSN 244
+P LP S +LD++KR YSN
Sbjct: 277 DPFAHLPKSTFVLDEFKRKYSN 298
>EF1G_MOUSE (Q9D8N0) Elongation factor 1-gamma (EF-1-gamma) (eEF-1B
gamma)
Length = 436
Score = 159 bits (403), Expect = 6e-39
Identities = 103/262 (39%), Positives = 138/262 (52%), Gaps = 22/262 (8%)
Query: 2 GVSNKTPQFINMNPIGKVPVLETPDG-PVFESNAIARYVARLGHNNLFGSSLIHQAQIDQ 60
G +N+TP+F+ P GKVP E DG VFESNAIA YV+ + L GS+ AQ+ Q
Sbjct: 40 GQTNRTPEFLRKFPAGKVPAFEGDDGFCVFESNAIAYYVS---NEELRGSTPEAAAQVVQ 96
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 120
W+ F+ +I P LG + ENA +KR L L++HL T+LVG V
Sbjct: 97 WVSFADSDIVPPASTWVFPTLGIMHHNKQATENAKEEVKRILGLLDTHLKTRTFLVGERV 156
Query: 121 TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIP 180
TLADI L + ++L SF FP+ R+F T +NQP FR ILG+VK E M
Sbjct: 157 TLADITVVCTLLWLYKQVLEPSFRQAFPNTNRWFLTCINQPQFRAILGEVKLCEKMAQFD 216
Query: 181 S---AKQQPKESKPK--------------TKDEPKKVAKSEPEKPKVEVEEE-APKPKPK 222
+ A+ QPK+ P+ + E KK A PE+ E E+ A +PK K
Sbjct: 217 AKKFAESQPKKDTPRKEKGSREEKQKPQAERKEEKKAAAPAPEEEMDECEQALAAEPKAK 276
Query: 223 NPLDLLPPSKMILDDWKRLYSN 244
+P LP S +LD++KR YSN
Sbjct: 277 DPFAHLPKSTFVLDEFKRKYSN 298
>EF1G_RABIT (P29694) Elongation factor 1-gamma (EF-1-gamma) (eEF-1B
gamma)
Length = 436
Score = 159 bits (402), Expect = 8e-39
Identities = 102/262 (38%), Positives = 138/262 (51%), Gaps = 22/262 (8%)
Query: 2 GVSNKTPQFINMNPIGKVPVLETPDG-PVFESNAIARYVARLGHNNLFGSSLIHQAQIDQ 60
G +N+TP+F+ P GKVP E DG VFESNAIA YV+ + L GS+ AQ+ Q
Sbjct: 40 GQTNRTPEFLRKFPAGKVPAFEGDDGFCVFESNAIAYYVS---NEELRGSTPEAAAQVVQ 96
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 120
W+ F+ +I P LG + ENA +KR L L++HL T+LVG V
Sbjct: 97 WVSFADSDIVPPASTWVFPTLGIMHHNKQATENAKEEVKRILGLLDAHLKTRTFLVGERV 156
Query: 121 TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIP 180
TLADI L + ++L SF FP+ R+F T +NQP FR +LG+VK E M
Sbjct: 157 TLADITVVCTLLWLYKQVLEPSFRQAFPNTNRWFLTCINQPQFRAVLGEVKLCEKMAQFD 216
Query: 181 S---AKQQPKESKPK--------------TKDEPKKVAKSEPEKPKVEVEEE-APKPKPK 222
+ A+ QPK+ P+ + E KK A PE+ E E+ A +PK K
Sbjct: 217 AKKFAESQPKKDTPRKEKGSREEKQKPQAERKEEKKAAAPAPEEEMDECEQALAAEPKAK 276
Query: 223 NPLDLLPPSKMILDDWKRLYSN 244
+P LP S +LD++KR YSN
Sbjct: 277 DPFAHLPKSTFVLDEFKRKYSN 298
>EF1G_PIG (Q29387) Elongation factor 1-gamma (EF-1-gamma) (eEF-1B
gamma) (Fragment)
Length = 432
Score = 159 bits (401), Expect = 1e-38
Identities = 102/262 (38%), Positives = 137/262 (51%), Gaps = 22/262 (8%)
Query: 2 GVSNKTPQFINMNPIGKVPVLETPDG-PVFESNAIARYVARLGHNNLFGSSLIHQAQIDQ 60
G +N TP+F+ P GKVP E DG VFESNAIA YV+ + L GS+ AQ+ Q
Sbjct: 36 GQTNHTPEFLRKFPAGKVPAFEGDDGFCVFESNAIAYYVS---NEELRGSTPEAAAQVVQ 92
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 120
W+ F+ +I P LG Y ENA ++R L L++HL T+LVG V
Sbjct: 93 WVSFADSDIVPPASTWVFPTLGIMHYNKQATENAKDEVRRVLGLLDAHLKTRTFLVGERV 152
Query: 121 TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIP 180
TLADI L + ++L SF FP+ R+F T +NQP FR +LG+VK E M
Sbjct: 153 TLADITVVCTLLWLYKQVLEPSFRQAFPNTNRWFLTCINQPQFRAVLGEVKLCEKMAQFD 212
Query: 181 S---AKQQPKESKPK--------------TKDEPKKVAKSEPEKPKVEVEEE-APKPKPK 222
+ A+ QPK+ P+ + E KK A PE+ E E+ A +PK K
Sbjct: 213 AKKFAESQPKKDTPRKEKGSREEKQKPQAERKEEKKAAAPAPEEELDECEQALAAEPKAK 272
Query: 223 NPLDLLPPSKMILDDWKRLYSN 244
+P LP S +LD++KR YSN
Sbjct: 273 DPFAHLPKSTFVLDEFKRKYSN 294
>EF1H_XENLA (Q91375) Elongation factor 1-gamma type 2 (EF-1-gamma)
(eEF-1B gamma) (p47)
Length = 437
Score = 156 bits (394), Expect = 7e-38
Identities = 100/263 (38%), Positives = 142/263 (53%), Gaps = 24/263 (9%)
Query: 2 GVSNKTPQFINMNPIGKVPVLETPDG-PVFESNAIARYVARLGHNNLFGSS-LIHQAQID 59
G++NKTP+F+ P+GKVP E +G +FES+AIA YVA ++ L GS+ +HQAQ+
Sbjct: 41 GLTNKTPEFLKKFPLGKVPAFEGNNGFCLFESSAIAHYVA---NDELRGSNNRLHQAQVI 97
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
QW+ FS + P LG + E A +K L L+ HL T+LVG
Sbjct: 98 QWVGFSDSHVVPPASAWVFPTLGIMQFNKQATEQAKEEIKTVLGVLDCHLQTRTFLVGER 157
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPI 179
+TLADI T +L + ++L SF + +V R+F T VNQP FR +LG+VK + M
Sbjct: 158 ITLADITLTCSLLWLYKQVLEPSFRQPYGNVTRWFVTCVNQPEFRAVLGEVKLCDKMAQF 217
Query: 180 PS---AKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAP---------------KPKP 221
+ A+ QPK+ PK K++P K K + +K K AP +PK
Sbjct: 218 DAKKFAEVQPKKETPK-KEKPAKEPKKKKKKKKKATPAPAPAPEDDLDESEKALAAEPKS 276
Query: 222 KNPLDLLPPSKMILDDWKRLYSN 244
K+P LP S I+D++KR YSN
Sbjct: 277 KDPYAHLPKSSFIMDEFKRKYSN 299
>EF1G_HUMAN (P26641) Elongation factor 1-gamma (EF-1-gamma) (eEF-1B
gamma) (PRO1608)
Length = 436
Score = 155 bits (392), Expect = 1e-37
Identities = 100/262 (38%), Positives = 138/262 (52%), Gaps = 22/262 (8%)
Query: 2 GVSNKTPQFINMNPIGKVPVLETPDG-PVFESNAIARYVARLGHNNLFGSSLIHQAQIDQ 60
G +N+TP+F+ P GKVP E DG VFESNAIA YV+ + L GS+ AQ+ Q
Sbjct: 40 GQTNRTPEFLRKFPAGKVPAFEGDDGFCVFESNAIAYYVS---NEELRGSTPEAAAQVVQ 96
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 120
W+ F+ +I P LG + ENA ++R L L+++L T+LVG V
Sbjct: 97 WVSFADSDIVPPASTWVFPTLGIMHHNKQATENAKEEVRRILGLLDAYLKTRTFLVGERV 156
Query: 121 TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIP 180
TLADI L + ++L SF FP+ R+F T +NQP FR +LG+VK E M
Sbjct: 157 TLADITVVCTLLWLYKQVLEPSFRQAFPNTNRWFLTCINQPQFRAVLGEVKLCEKMAQFD 216
Query: 181 S---AKQQPKESKPK--------------TKDEPKKVAKSEPEKPKVEVEEE-APKPKPK 222
+ A+ QPK+ P+ + E KK A PE+ E E+ A +PK K
Sbjct: 217 AKKFAETQPKKDTPRKEKGSREEKQKPQAERKEEKKAAAPAPEEEMDECEQALAAEPKAK 276
Query: 223 NPLDLLPPSKMILDDWKRLYSN 244
+P LP S +LD++KR YSN
Sbjct: 277 DPFAHLPKSTFVLDEFKRKYSN 298
>EF1G_CARAU (Q90YC0) Elongation factor 1-gamma (EF-1-gamma) (eEF-1B
gamma)
Length = 442
Score = 150 bits (380), Expect = 3e-36
Identities = 96/268 (35%), Positives = 138/268 (50%), Gaps = 29/268 (10%)
Query: 2 GVSNKTPQFINMNPIGKVPVLETPDG-PVFESNAIARYVARLGHNNLFGSSLIHQAQIDQ 60
G +N++P F++ P+GKVP + DG +FESNAIA + L ++ L GS+ AQ+ Q
Sbjct: 41 GQTNRSPAFLSNFPLGKVPAYQGDDGFCLFESNAIAHF---LSNDALRGSTPQASAQVLQ 97
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 120
W+ F+ EI P LG + E A +KR L LN HL T+LVG V
Sbjct: 98 WVSFADSEIIPPASAWVFPTLGIMQFNKQATEQAKEEVKRVLAVLNQHLNTRTFLVGERV 157
Query: 121 TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIP 180
+LADI +L + ++L +F +P+V R+F T VNQP F+ +LG+VK E M
Sbjct: 158 SLADITVVCSLLWLYKQVLEPAFRQPYPNVTRWFLTCVNQPQFKAVLGEVKLCEKMAQFD 217
Query: 181 SAK------------------------QQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEA 216
+ K QQP++ K + K E KK A +E E + E A
Sbjct: 218 AKKFAEMQPKKEAPAKKEKAGKEGGKQQQPQQEKKEKKKEEKKAAPAEEEMDECEA-ALA 276
Query: 217 PKPKPKNPLDLLPPSKMILDDWKRLYSN 244
+PK K+P LP S ++D++KR YSN
Sbjct: 277 SEPKAKDPYAHLPKSSFVMDEFKRKYSN 304
>EF1G_BRARE (Q6PE25) Elongation factor 1-gamma (EF-1-gamma) (eEF-1B
gamma)
Length = 442
Score = 149 bits (376), Expect = 8e-36
Identities = 95/267 (35%), Positives = 138/267 (51%), Gaps = 27/267 (10%)
Query: 2 GVSNKTPQFINMNPIGKVPVLETPDG-PVFESNAIARYVARLGHNNLFGSSLIHQAQIDQ 60
G +N++P F+ P+GKVP + DG +FESNAIA Y L ++ L GS+ AQ+ Q
Sbjct: 41 GQTNRSPAFLGNFPLGKVPAYQGDDGFCLFESNAIAHY---LSNDVLRGSTPQASAQVLQ 97
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 120
W+ F+ E+ P LG + E A +KR L LN HL T+LVG +
Sbjct: 98 WVSFADSEVIPPASAWVFPTLGIMQFNKQATEQAKEEVKRVLAVLNQHLNTRTFLVGERI 157
Query: 121 TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIP 180
+LADI +L + ++L +F +P+V R+F T VNQP F+ +LG+VK E M
Sbjct: 158 SLADITVVCSLLWLYKQVLEPAFRQPYPNVTRWFVTCVNQPQFKTVLGEVKLCEKMAQFD 217
Query: 181 S---AKQQPKESKPKTKD------------EPKKVAKSEPEKPKVEVEEE--------AP 217
+ A+ QPK+ P K+ + +K K + EK EEE A
Sbjct: 218 AKKFAEMQPKKEAPIKKEKGGKEGGKQQPQQQEKKEKKKEEKKAAPAEEEMDECEAALAS 277
Query: 218 KPKPKNPLDLLPPSKMILDDWKRLYSN 244
+PK K+P LP S ++D++KR YSN
Sbjct: 278 EPKAKDPFAHLPKSSFVMDEFKRKYSN 304
>EF1G_ARTSA (P12261) Elongation factor 1-gamma (EF-1-gamma) (eEF-1B
gamma)
Length = 429
Score = 147 bits (370), Expect = 4e-35
Identities = 95/262 (36%), Positives = 135/262 (51%), Gaps = 20/262 (7%)
Query: 2 GVSNKTPQFINMNPIGKVPVLETPDGP-VFESNAIARYVARLGHNNLFGSSLIHQAQIDQ 60
G +NK+ F+ P+GKVP E+ DG + ESNAIA YVA + L GSS + +AQI Q
Sbjct: 37 GETNKSDAFLKSFPLGKVPAFESADGHCIAESNAIAYYVA---NETLRGSSDLEKAQIIQ 93
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 120
W+ F+ EI P LG + A + +AL+AL+ HL TYLVG +
Sbjct: 94 WMTFADTEILPASCTWVFPVLGIMQFNKQATARAKEDIDKALQALDDHLLTRTYLVGERI 153
Query: 121 TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQV---------- 170
TLADI+ T L + +L ++F + + R+F TL+NQ + ++G
Sbjct: 154 TLADIVVTCTLLHLYQHVLDEAFRKSYVNTNRWFITLINQKQVKAVIGDFKLCEKAGEFD 213
Query: 171 --KQTEAMPPIPSAKQQPKESKP---KTKDEPKKVAKSEPEKPKVEVEEE-APKPKPKNP 224
K E I S +++ E P K K E K+V K E E+P EE A +PK K+P
Sbjct: 214 PKKYAEFQAAIGSGEKKKTEKAPKAVKAKPEKKEVPKKEQEEPADAAEEALAAEPKSKDP 273
Query: 225 LDLLPPSKMILDDWKRLYSNTK 246
D +P +DD+KR YSN +
Sbjct: 274 FDEMPKGTFNMDDFKRFYSNNE 295
>EF1G_DROME (Q9NJH0) Elongation factor 1-gamma (EF-1-gamma) (eEF-1B
gamma)
Length = 431
Score = 123 bits (308), Expect = 6e-28
Identities = 90/264 (34%), Positives = 125/264 (47%), Gaps = 27/264 (10%)
Query: 2 GVSNKTPQFINMNPIGKVPVLETPDGPVF-ESNAIARYVARLGHNNLFGSSLIHQAQIDQ 60
G +NK+ +F+ P GKVP ET +G ESNAIA +A G QAQ+ Q
Sbjct: 38 GETNKSAEFLKKFPGGKVPAFETAEGQYLSESNAIAYLLANEQLRG--GKCPFVQAQVQQ 95
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISS--LKRALEALNSHLAHNTYLVGH 118
WI F+ EI P LG P ++N+ + + L+ LN L T+L G
Sbjct: 96 WISFADNEIVPASCAWVFPLLGIL----PQQKNSTAKQEAEAVLQQLNQKLQDATFLAGE 151
Query: 119 SVTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVK------- 171
+TLADI+ S+L + +L S S F +V R+F T++NQ + ++ K
Sbjct: 152 RITLADIVVFSSLLHLYEYVLEPSVRSAFGNVNRWFVTILNQKQVQAVVKDYKLCEKALV 211
Query: 172 -----------QTEAMPPIPSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPK 220
+T A P A+QQ +E KPK K E K A E+ E A +PK
Sbjct: 212 FDPKKYAEFQAKTGAAKPQQQAQQQKQEKKPKEKKEAPKKAAEPAEELDAADEALAAEPK 271
Query: 221 PKNPLDLLPPSKMILDDWKRLYSN 244
K+P D LP DD+KR+YSN
Sbjct: 272 SKDPFDALPKGTFNFDDFKRVYSN 295
>EF1G_TRYCR (P34715) Elongation factor 1-gamma (EF-1-gamma) (eEF-1B
gamma)
Length = 411
Score = 120 bits (300), Expect = 5e-27
Identities = 85/250 (34%), Positives = 133/250 (53%), Gaps = 14/250 (5%)
Query: 2 GVSNKTPQFI-NMNPIGKVPVLETPDGPVFESNAIARYVARLGHNN--LFGSSLIHQAQI 58
G N+T ++ N +P G+ PVL+T +G VFESNAI R++ARL + L+G + + +Q+
Sbjct: 38 GRENETAEYCRNCSPCGRYPVLQTEEGCVFESNAILRHIARLDRSGGFLYGRTPLEGSQV 97
Query: 59 DQWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGH 118
D W+DFS+ E+DA P + A P+ NA+ + L AL + L T+LVG
Sbjct: 98 DMWLDFSATELDA----ASEPFVHHAFRGEPLPANAMDRVHEVLRALEAWLETRTFLVGE 153
Query: 119 SVTLADIITTSNL--YIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAM 176
+T+AD+ L + ++ T ++ + R + T++ QP ++L T
Sbjct: 154 RMTVADVAVAFALQWHYRLNGAEGEALTKKYRNAYRMYNTVMQQPKTVEVLRSQGATFGA 213
Query: 177 PPIPSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPKPK-NPLDLLPPSKMIL 235
AK Q + ++E ++ A + E E+EAP+ K K NPLD LPPS +L
Sbjct: 214 RE-GGAKGQGRGCARPGREEAERAAAA---ADGAEEEDEAPREKKKPNPLDELPPSPFVL 269
Query: 236 DDWKRLYSNT 245
D +KR YSNT
Sbjct: 270 DAFKREYSNT 279
>EF1G_SCHPO (P40921) Elongation factor 1-gamma (EF-1-gamma) (eEF-1B
gamma)
Length = 409
Score = 111 bits (278), Expect = 2e-24
Identities = 74/232 (31%), Positives = 119/232 (50%), Gaps = 7/232 (3%)
Query: 15 PIGKVPVLETPDG-PVFESNAIARYVARLGHNN-LFGSSLIHQAQIDQWIDFSSLEIDAN 72
P+ K+PV DG P+ E+ AIA Y+A L L G++ +A++ Q+ F++ E+
Sbjct: 48 PLQKMPVFVGKDGFPLSETLAIAFYLASLNKTRALNGTTAEEKAKVLQYCSFTNSELPGA 107
Query: 73 IMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSVTLADIITTSNLY 132
+ PR+ A Y + A +++ + LA TYLVG +TLADI T L
Sbjct: 108 FRPIIAPRVFGAPYDEQAAKEAETAIALIFARFDEELASKTYLVGSRLTLADIFFTCFLK 167
Query: 133 IGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIPSAKQQPKESKPK 192
G +L KS+ +++ H+ RY+ T+ +Q I +K + P+P K + KE+ P
Sbjct: 168 FGATYVLTKSYLAKYTHIYRYYQTIYHQAKLDAITEPLKFID--QPLPIIKAENKEAAPA 225
Query: 193 TKDEPKKVAKSEPEKPKVEVEEEAPKPKPKNPLDLLPPSKMILDDWKRLYSN 244
K E K K E +K + + E P PK+PL P ++++KR+YSN
Sbjct: 226 KKAEKK---KDEKKKNAPKPQAERPAKPPKHPLASAPNGSFDIEEYKRVYSN 274
>EF1H_YEAST (P36008) Elongation factor 1-gamma 2 (EF-1-gamma 2)
Length = 412
Score = 97.1 bits (240), Expect = 5e-20
Identities = 68/246 (27%), Positives = 119/246 (47%), Gaps = 13/246 (5%)
Query: 6 KTPQFINMNPIGKVPVLETPDG-PVFESNAIARYVARLGHNN-----LFGSSLIHQAQID 59
++ +F ++ P+ + P P G + E+ AI Y+A + L GS +I ++QI
Sbjct: 35 QSSEFASLFPLKQAPAFLGPKGLKLTEALAIQFYLANQVADEKERARLLGSDVIEKSQIL 94
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
+W ++ ++ +NI + +L G Y + + ++ L T++ +
Sbjct: 95 RWASLANSDVMSNIARPFLSFKGLIPYNKKDVDACFVKIDNLAAVFDARLRDYTFVATEN 154
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTE-AMPP 178
++L D+ + G A +L + ++ PH+ R+F T+ P + +VK E A+
Sbjct: 155 ISLGDLHAAGSWAFGLATILGPEWRAKHPHLMRWFNTVAASPIVKTPFAEVKLAEKALTY 214
Query: 179 IPSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPKPKNPLDLLPPSKMILDDW 238
P KQ K KPK ++ K K + KP ++ AP KPK+PL+ L S +LDDW
Sbjct: 215 TPPKKQ--KAEKPKA-EKSKAEKKKDEAKP---ADDAAPAKKPKHPLEALGKSTFVLDDW 268
Query: 239 KRLYSN 244
KR YSN
Sbjct: 269 KRKYSN 274
>EF1G_YEAST (P29547) Elongation factor 1-gamma 1 (EF-1-gamma 1)
Length = 415
Score = 96.3 bits (238), Expect = 8e-20
Identities = 69/244 (28%), Positives = 112/244 (45%), Gaps = 13/244 (5%)
Query: 9 QFINMNPIGKVPVLETPDG-PVFESNAIARYVARLGHNNLFGSSLIH-------QAQIDQ 60
QF P+ KVP P G + E+ AI Y+ +L ++ + L+ QAQI +
Sbjct: 39 QFARDFPLKKVPAFVGPKGYKLTEAMAINYYLVKLSQDDKMKTQLLGADDDLNAQAQIIR 98
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 120
W ++ ++ I +P G A Y ++A+ ++ + ++ + L + TYL ++
Sbjct: 99 WQSLANSDLCIQIANTIVPLKGGAPYNKKSVDSAMDAVDKIVDIFENRLKNYTYLATENI 158
Query: 121 TLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIP 180
+LAD++ S F L + ++ P + R+F T+ P + K + P
Sbjct: 159 SLADLVAASIFTRYFESLFGTEWRAQHPAIVRWFNTVRASPFLKDEYKDFKFADK----P 214
Query: 181 SAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPKPKNPLDLLPPSKMILDDWKR 240
+ Q K+ K P K E KP E E KPK+PL+LL S +LDDWKR
Sbjct: 215 LSPPQKKKEKKAPAAAPAASKKKEEAKP-AATETETSSKKPKHPLELLGKSTFVLDDWKR 273
Query: 241 LYSN 244
YSN
Sbjct: 274 KYSN 277
>EF1G_CAEEL (P54412) Probable elongation factor 1-gamma (EF-1-gamma)
(eEF-1B gamma)
Length = 398
Score = 91.3 bits (225), Expect = 3e-18
Identities = 71/236 (30%), Positives = 115/236 (48%), Gaps = 15/236 (6%)
Query: 15 PIGKVPVLETPDGPVFESNAIARYVARLGHNNLFGSSLIHQAQIDQWIDFSSLEIDANIM 74
P K P+ TP FE +A+ +G + L G+S A+ QW+ F+ + ++
Sbjct: 36 PADKFPLGVTP---AFEGDALLFGAESIGLH-LTGTSA--NAETVQWLQFAEGYLLPAVL 89
Query: 75 KLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSVTLADIITTSNLYIG 134
LP + A + E + L L+ L+ L TYLVG ++LAD+ +L
Sbjct: 90 GYVLPSVSAANFDKKTVEQYKNELNGQLQVLDRVLVKKTYLVGERLSLADVSVALDLLPA 149
Query: 135 FAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPIPSAK-----QQPKES 189
F +L + +V R+F T+VNQP +++LG+V ++ AK + +S
Sbjct: 150 FQYVLDANARKSIVNVTRWFRTVVNQPAVKEVLGEVSLASSVAQFNQAKFTELSAKVAKS 209
Query: 190 KPKTKDEPKKVAKSEPEKPKVEVEEEAPK-PKPKNPLDLLPPSKMILDDWKRLYSN 244
PK ++PKK AK P + E++ PK K K+P +P +LD++KR YSN
Sbjct: 210 APKA-EKPKKEAK--PAAAAAQPEDDEPKEEKSKDPFQDMPKGTFVLDNFKRSYSN 262
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.134 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,253,728
Number of Sequences: 164201
Number of extensions: 1569391
Number of successful extensions: 10690
Number of sequences better than 10.0: 438
Number of HSP's better than 10.0 without gapping: 171
Number of HSP's successfully gapped in prelim test: 279
Number of HSP's that attempted gapping in prelim test: 8743
Number of HSP's gapped (non-prelim): 1457
length of query: 305
length of database: 59,974,054
effective HSP length: 110
effective length of query: 195
effective length of database: 41,911,944
effective search space: 8172829080
effective search space used: 8172829080
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC140549.6


