

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140547.14 - phase: 0
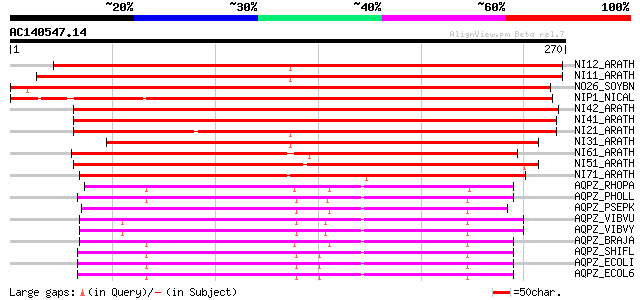
(270 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NI12_ARATH (Q8LFP7) Aquaporin NIP1.2 (NOD26-like intrinsic prote... 361 e-100
NI11_ARATH (Q8VZW1) Aquaporin NIP1.1 (NOD26-like intrinsic prote... 348 6e-96
NO26_SOYBN (P08995) Nodulin-26 (N-26) 341 1e-93
NIP1_NICAL (P49173) Probable aquaporin NIP-type (Pollen-specific... 291 1e-78
NI42_ARATH (Q8W036) Probable aquaporin NIP4.2 (NOD26-like intrin... 288 7e-78
NI41_ARATH (Q9FIZ9) Putative aquaporin NIP4.1 (NOD26-like intrin... 286 4e-77
NI21_ARATH (Q8W037) Aquaporin NIP2.1 (NOD26-like intrinsic prote... 259 6e-69
NI31_ARATH (Q9C6T0) Putative aquaporin NIP3.1 (NOD26-like intrin... 258 1e-68
NI61_ARATH (Q9SAI4) Probable aquaporin NIP6.1 (NOD26-like intrin... 223 3e-58
NI51_ARATH (Q9SV84) Probable aquaporin NIP5.1 (NOD26-like intrin... 213 4e-55
NI71_ARATH (Q8LAI1) Probable aquaporin NIP7.1 (NOD26-like intrin... 174 3e-43
AQPZ_RHOPA (P60925) Aquaporin Z 152 6e-37
AQPZ_PHOLL (Q7N5C1) Aquaporin Z 151 1e-36
AQPZ_PSEPK (Q88F17) Aquaporin Z 151 2e-36
AQPZ_VIBVU (Q8DB17) Aquaporin Z 149 9e-36
AQPZ_VIBVY (Q7MIV9) Aquaporin Z 147 3e-35
AQPZ_BRAJA (Q89EG9) Aquaporin Z 146 5e-35
AQPZ_SHIFL (O68874) Aquaporin Z 143 5e-34
AQPZ_ECOLI (P60844) Aquaporin Z (Bacterial nodulin-like intrinsi... 143 5e-34
AQPZ_ECOL6 (P60845) Aquaporin Z 143 5e-34
>NI12_ARATH (Q8LFP7) Aquaporin NIP1.2 (NOD26-like intrinsic protein
1.2) (Nodulin-26-like major intrinsic protein 2)
(AtNLM2) (NLM2 protein) (NodLikeMip2)
Length = 294
Score = 361 bits (927), Expect = e-100
Identities = 176/255 (69%), Positives = 215/255 (84%), Gaps = 7/255 (2%)
Query: 22 KCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMV 81
K D+ + VP LQKL+AEV+GT+FLIFAGCAAV VN +DK VTLPGI+IVWGL VMV
Sbjct: 38 KKQDSLLSISVPFLQKLMAEVLGTYFLIFAGCAAVAVNTQHDKAVTLPGIAIVWGLTVMV 97
Query: 82 LVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIF------ 135
LVYS+GHISGAHFNPAVTIA + GRFPLKQ+PAY+I+QV+GSTLA+ L+L+F
Sbjct: 98 LVYSLGHISGAHFNPAVTIAFASCGRFPLKQVPAYVISQVIGSTLAAATLRLLFGLDQDV 157
Query: 136 -SGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILN 194
SGK + F GTLP+GS+LQ+FV+EFIITF+LMF+ISGVATDNRAIGELAGLAVGSTV+LN
Sbjct: 158 CSGKHDVFVGTLPSGSNLQSFVIEFIITFYLMFVISGVATDNRAIGELAGLAVGSTVLLN 217
Query: 195 VLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITNKPVRE 254
V+ AGP++GASMNP RSLGPA+V+ YRG+WIY+VSPI+GA++G W Y +R T+KP+RE
Sbjct: 218 VIIAGPVSGASMNPGRSLGPAMVYSCYRGLWIYIVSPIVGAVSGAWVYNMVRYTDKPLRE 277
Query: 255 LTKSSSFLKAVSKGA 269
+TKS SFLK V G+
Sbjct: 278 ITKSGSFLKTVRNGS 292
>NI11_ARATH (Q8VZW1) Aquaporin NIP1.1 (NOD26-like intrinsic protein
1.1) (Nodulin-26-like major intrinsic protein 1)
(AtNLM1) (NLM1 protein) (NodLikeMip1)
Length = 296
Score = 348 bits (894), Expect = 6e-96
Identities = 169/263 (64%), Positives = 212/263 (80%), Gaps = 7/263 (2%)
Query: 14 NINDDATKKCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISI 73
+I++ K D+ + VP LQKL+AE +GT+FL+F GCA+VVVN+ ND VVTLPGI+I
Sbjct: 33 DIHNPRPLKKQDSLLSVSVPFLQKLIAEFLGTYFLVFTGCASVVVNMQNDNVVTLPGIAI 92
Query: 74 VWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKL 133
VWGL +MVL+YS+GHISGAH NPAVTIA + GRFPLKQ+PAY+I+QV+GSTLA+ L+L
Sbjct: 93 VWGLTIMVLIYSLGHISGAHINPAVTIAFASCGRFPLKQVPAYVISQVIGSTLAAATLRL 152
Query: 134 IF-------SGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNRAIGELAGLA 186
+F SGK + F G+ P GSDLQAF +EFI+TF+LMFIISGVATDNRAIGELAGLA
Sbjct: 153 LFGLDHDVCSGKHDVFIGSSPVGSDLQAFTMEFIVTFYLMFIISGVATDNRAIGELAGLA 212
Query: 187 VGSTVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLR 246
+GSTV+LNVL A P++ ASMNP RSLGPA+V+ Y+GIWIY+V+P LGA+AG W Y +R
Sbjct: 213 IGSTVLLNVLIAAPVSSASMNPGRSLGPALVYGCYKGIWIYLVAPTLGAIAGAWVYNTVR 272
Query: 247 ITNKPVRELTKSSSFLKAVSKGA 269
T+KP+RE+TKS SFLK V G+
Sbjct: 273 YTDKPLREITKSGSFLKTVRIGS 295
>NO26_SOYBN (P08995) Nodulin-26 (N-26)
Length = 271
Score = 341 bits (875), Expect = 1e-93
Identities = 163/265 (61%), Positives = 212/265 (79%), Gaps = 2/265 (0%)
Query: 1 MGDISNG--NLDVVMNINDDATKKCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVV 58
M D S G + +VV+N+ + ++ + VP LQKLVAE VGT+FLIFAGCA++VV
Sbjct: 1 MADYSAGTESQEVVVNVTKNTSETIQRSDSLVSVPFLQKLVAEAVGTYFLIFAGCASLVV 60
Query: 59 NLNNDKVVTLPGISIVWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYII 118
N N ++T PGI+IVWGL + VLVY++GHISG HFNPAVTIA +T RFPL Q+PAY++
Sbjct: 61 NENYYNMITFPGIAIVWGLVLTVLVYTVGHISGGHFNPAVTIAFASTRRFPLIQVPAYVV 120
Query: 119 AQVVGSTLASGVLKLIFSGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNRA 178
AQ++GS LASG L+L+F G +QF+GT+P G++LQAFV EFI+TFFLMF+I GVATDNRA
Sbjct: 121 AQLLGSILASGTLRLLFMGNHDQFSGTVPNGTNLQAFVFEFIMTFFLMFVICGVATDNRA 180
Query: 179 IGELAGLAVGSTVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAG 238
+GE AG+A+GST++LNV+ GP+TGASMNPARSLGPA VH EY GIWIY+++P++GA+AG
Sbjct: 181 VGEFAGIAIGSTLLLNVIIGGPVTGASMNPARSLGPAFVHGEYEGIWIYLLAPVVGAIAG 240
Query: 239 TWTYTFLRITNKPVRELTKSSSFLK 263
W Y +R T+KP+ E TKS+SFLK
Sbjct: 241 AWVYNIVRYTDKPLSETTKSASFLK 265
>NIP1_NICAL (P49173) Probable aquaporin NIP-type (Pollen-specific
membrane integral protein)
Length = 270
Score = 291 bits (745), Expect = 1e-78
Identities = 145/264 (54%), Positives = 192/264 (71%), Gaps = 5/264 (1%)
Query: 1 MGDISNGNLDVVMNINDDATKKCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVVNL 60
+ + GN+ N +D C ++ V +LQKL+AE +GT+F+IFAGC +V VN
Sbjct: 12 ISQMEEGNIHSASN-SDSNVGFCSSVSV---VVILQKLIAEAIGTYFVIFAGCGSVAVNK 67
Query: 61 NNDKVVTLPGISIVWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQ 120
V T PGI + WGL VMV+VY++G+ISGAHFNPAVTI + GRFP KQ+P YIIAQ
Sbjct: 68 IYGSV-TFPGICVTWGLIVMVMVYTVGYISGAHFNPAVTITFSIFGRFPWKQVPLYIIAQ 126
Query: 121 VVGSTLASGVLKLIFSGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNRAIG 180
++GS LASG L L+F + GT+P GS+ Q+ +E II+F LMF+ISGVATD+RAIG
Sbjct: 127 LMGSILASGTLALLFDVTPQAYFGTVPVGSNGQSLAIEIIISFLLMFVISGVATDDRAIG 186
Query: 181 ELAGLAVGSTVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAGTW 240
++AG+AVG T+ LNV AGPI+GASMNPARS+GPAIV H Y G+W+Y+V PI+G LAG +
Sbjct: 187 QVAGIAVGMTITLNVFVAGPISGASMNPARSIGPAIVKHVYTGLWVYVVGPIIGTLAGAF 246
Query: 241 TYTFLRITNKPVRELTKSSSFLKA 264
Y +R T+KP+REL KS+S L++
Sbjct: 247 VYNLIRSTDKPLRELAKSASSLRS 270
>NI42_ARATH (Q8W036) Probable aquaporin NIP4.2 (NOD26-like intrinsic
protein 4.2) (Nodulin-26-like major intrinsic protein 5)
(AtNLM5) (NLM5 protein) (NodLikeMip5)
Length = 283
Score = 288 bits (738), Expect = 7e-78
Identities = 142/236 (60%), Positives = 178/236 (75%)
Query: 32 VPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISG 91
V L QKL+AE++GT+F+IF+GC VVVN+ +T PGI + WGL VMV++YS GHISG
Sbjct: 39 VCLTQKLIAEMIGTYFIIFSGCGVVVVNVLYGGTITFPGICVTWGLIVMVMIYSTGHISG 98
Query: 92 AHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGKENQFAGTLPAGSD 151
AHFNPAVT+ RFP Q+P YI AQ+ GS LAS L+L+F+ F GT P S
Sbjct: 99 AHFNPAVTVTFAVFRRFPWYQVPLYIGAQLTGSLLASLTLRLMFNVTPKAFFGTTPTDSS 158
Query: 152 LQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGASMNPARS 211
QA V E II+F LMF+ISGVATD+RA GELAG+AVG T+ILNV AGPI+GASMNPARS
Sbjct: 159 GQALVAEIIISFLLMFVISGVATDSRATGELAGIAVGMTIILNVFVAGPISGASMNPARS 218
Query: 212 LGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITNKPVRELTKSSSFLKAVSK 267
LGPAIV Y+GIW+Y+V P +G AG + Y F+R T+KP+RELTKS+SFL++V++
Sbjct: 219 LGPAIVMGRYKGIWVYIVGPFVGIFAGGFVYNFMRFTDKPLRELTKSASFLRSVAQ 274
>NI41_ARATH (Q9FIZ9) Putative aquaporin NIP4.1 (NOD26-like intrinsic
protein 4.1)
Length = 283
Score = 286 bits (732), Expect = 4e-77
Identities = 139/235 (59%), Positives = 177/235 (75%)
Query: 32 VPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISG 91
V L QKL+AE++GT+F++F+GC VVVN+ +T PGI + WGL VMV++YS GHISG
Sbjct: 39 VCLTQKLIAEMIGTYFIVFSGCGVVVVNVLYGGTITFPGICVTWGLIVMVMIYSTGHISG 98
Query: 92 AHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGKENQFAGTLPAGSD 151
AHFNPAVT+ RFP Q+P YI AQ GS LAS L+L+F F GT PA S
Sbjct: 99 AHFNPAVTVTFAIFRRFPWHQVPLYIGAQFAGSLLASLTLRLMFKVTPEAFFGTTPADSP 158
Query: 152 LQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGASMNPARS 211
+A V E II+F LMF+ISGVATDNRA+GELAG+AVG T+++NV AGPI+GASMNPARS
Sbjct: 159 ARALVAEIIISFLLMFVISGVATDNRAVGELAGIAVGMTIMVNVFVAGPISGASMNPARS 218
Query: 212 LGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITNKPVRELTKSSSFLKAVS 266
LGPA+V Y+ IW+Y+V P+LG ++G + Y +R T+KP+RELTKS+SFL+AVS
Sbjct: 219 LGPALVMGVYKHIWVYIVGPVLGVISGGFVYNLIRFTDKPLRELTKSASFLRAVS 273
>NI21_ARATH (Q8W037) Aquaporin NIP2.1 (NOD26-like intrinsic protein
2.1) (Nodulin-26-like major intrinsic protein 4)
(AtNLM4) (NLM4 protein) (NodLikeMip4)
Length = 288
Score = 259 bits (661), Expect = 6e-69
Identities = 133/242 (54%), Positives = 176/242 (71%), Gaps = 8/242 (3%)
Query: 32 VPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISG 91
V LQKL+AE+VGT++LIFAGCAA+ VN ++ VVTL GI++VWG+ +MVLVY +GH+S
Sbjct: 44 VHFLQKLLAELVGTYYLIFAGCAAIAVNAQHNHVVTLVGIAVVWGIVIMVLVYCLGHLS- 102
Query: 92 AHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIF-------SGKENQFAG 144
AHFNPAVT+A ++ RFPL Q+PAYI QV+GSTLAS L+L+F S K + F G
Sbjct: 103 AHFNPAVTLALASSQRFPLNQVPAYITVQVIGSTLASATLRLLFDLNNDVCSKKHDVFLG 162
Query: 145 TLPAGSDLQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGA 204
+ P+GSDLQAFV+EFIIT FLM ++ V T R EL GL +G+TV LNV+FAG ++GA
Sbjct: 163 SSPSGSDLQAFVMEFIITGFLMLVVCAVTTTKRTTEELEGLIIGATVTLNVIFAGEVSGA 222
Query: 205 SMNPARSLGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITNKPVRELTKSSSFLKA 264
SMNPARS+GPA+V Y+GIWIY+++P LGA++G + L E +K+ S K
Sbjct: 223 SMNPARSIGPALVWGCYKGIWIYLLAPTLGAVSGALIHKMLPSIQNAEPEFSKTGSSHKR 282
Query: 265 VS 266
V+
Sbjct: 283 VT 284
>NI31_ARATH (Q9C6T0) Putative aquaporin NIP3.1 (NOD26-like intrinsic
protein 3.1)
Length = 269
Score = 258 bits (658), Expect = 1e-68
Identities = 122/217 (56%), Positives = 167/217 (76%), Gaps = 7/217 (3%)
Query: 48 LIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGR 107
+IFAGC+A+VVN K VTLPGI++VWGL V V++YSIGH+SGAHFNPAV+IA ++ +
Sbjct: 1 MIFAGCSAIVVNETYGKPVTLPGIALVWGLVVTVMIYSIGHVSGAHFNPAVSIAFASSKK 60
Query: 108 FPLKQLPAYIIAQVVGSTLASGVLKLIF-------SGKENQFAGTLPAGSDLQAFVVEFI 160
FP Q+P YI AQ++GSTLA+ VL+L+F S K + + GT P+ S+ +FV+EFI
Sbjct: 61 FPFNQVPGYIAAQLLGSTLAAAVLRLVFHLDDDVCSLKGDVYVGTYPSNSNTTSFVMEFI 120
Query: 161 ITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGASMNPARSLGPAIVHHE 220
TF LMF+IS VATD RA G AG+A+G+T++L++LF+GPI+GASMNPARSLGPA++
Sbjct: 121 ATFNLMFVISAVATDKRATGSFAGIAIGATIVLDILFSGPISGASMNPARSLGPALIWGC 180
Query: 221 YRGIWIYMVSPILGALAGTWTYTFLRITNKPVRELTK 257
Y+ +W+Y+VSP++GAL+G WTY LR T K E+ +
Sbjct: 181 YKDLWLYIVSPVIGALSGAWTYGLLRSTKKSYSEIIR 217
>NI61_ARATH (Q9SAI4) Probable aquaporin NIP6.1 (NOD26-like intrinsic
protein 6.1)
Length = 305
Score = 223 bits (569), Expect = 3e-58
Identities = 119/219 (54%), Positives = 150/219 (68%), Gaps = 5/219 (2%)
Query: 31 HVPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHIS 90
+V L +KL AE VGT LIFAG A +VN D TL G + GLAVM+++ S GHIS
Sbjct: 75 NVSLYRKLGAEFVGTLILIFAGTATAIVNQKTDGAETLIGCAASAGLAVMIVILSTGHIS 134
Query: 91 GAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGKENQFAG--TLPA 148
GAH NPAVTIA FP K +P YI AQV+ S A+ LK +F E +G T+P
Sbjct: 135 GAHLNPAVTIAFAALKHFPWKHVPVYIGAQVMASVSAAFALKAVF---EPTMSGGVTVPT 191
Query: 149 GSDLQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGASMNP 208
QAF +EFII+F LMF+++ VATD RA+GELAG+AVG+TV+LN+L AGP T ASMNP
Sbjct: 192 VGLSQAFALEFIISFNLMFVVTAVATDTRAVGELAGIAVGATVMLNILIAGPATSASMNP 251
Query: 209 ARSLGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRI 247
R+LGPAI + YR IW+Y+ +PILGAL G TYT +++
Sbjct: 252 VRTLGPAIAANNYRAIWVYLTAPILGALIGAGTYTIVKL 290
>NI51_ARATH (Q9SV84) Probable aquaporin NIP5.1 (NOD26-like intrinsic
protein 5.1) (Nodulin-26-like major intrinsic protein 6)
(AtNLM6) (NLM6 protein) (NodLikeMip6)
Length = 304
Score = 213 bits (542), Expect = 4e-55
Identities = 115/232 (49%), Positives = 152/232 (64%), Gaps = 7/232 (3%)
Query: 32 VPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISG 91
V L +KL AE VGTF LIF A +VN D TL G + GLAVM+++ S GHISG
Sbjct: 74 VSLTRKLGAEFVGTFILIFTATAGPIVNQKYDGAETLIGNAACAGLAVMIIILSTGHISG 133
Query: 92 AHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGKENQFAGTLPAGSD 151
AH NP++TIA FP +PAYI AQV S AS LK +F + T+P+ S
Sbjct: 134 AHLNPSLTIAFAALRHFPWAHVPAYIAAQVSASICASFALKGVFHPFMSGGV-TIPSVSL 192
Query: 152 LQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGASMNPARS 211
QAF +EFIITF L+F+++ VATD RA+GELAG+AVG+TV+LN+L AGP TG SMNP R+
Sbjct: 193 GQAFALEFIITFILLFVVTAVATDTRAVGELAGIAVGATVMLNILVAGPSTGGSMNPVRT 252
Query: 212 LGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITN------KPVRELTK 257
LGPA+ YR +W+Y+V+P LGA++G YT +++ + +PVR +
Sbjct: 253 LGPAVASGNYRSLWVYLVAPTLGAISGAAVYTGVKLNDSVTDPPRPVRSFRR 304
>NI71_ARATH (Q8LAI1) Probable aquaporin NIP7.1 (NOD26-like intrinsic
protein 7.1)
Length = 275
Score = 174 bits (440), Expect = 3e-43
Identities = 92/218 (42%), Positives = 136/218 (62%), Gaps = 2/218 (0%)
Query: 35 LQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISGAHF 94
L+ ++AE+VGTF L+F+ C + + V L ++ GL+V+V+VYSIGHISGAH
Sbjct: 45 LRIVMAELVGTFILMFSVCGVISSTQLSGGHVGLLEYAVTAGLSVVVVVYSIGHISGAHL 104
Query: 95 NPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGKENQFAGTLPAGSDLQA 154
NP++TIA G FP Q+P YI AQ +G+T A+ V ++ G T PA S + A
Sbjct: 105 NPSITIAFAVFGGFPWSQVPLYITAQTLGATAATLVGVSVY-GVNADIMATKPALSCVSA 163
Query: 155 FVVEFIITFFLMFIISGV-ATDNRAIGELAGLAVGSTVILNVLFAGPITGASMNPARSLG 213
F VE I T ++F+ S + ++ +G L G +G+ + L VL GPI+G SMNPARSLG
Sbjct: 164 FFVELIATSIVVFLASALHCGPHQNLGNLTGFVIGTVISLGVLITGPISGGSMNPARSLG 223
Query: 214 PAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITNKP 251
PA+V ++ +WIYM +P++GA+ G TY + + +P
Sbjct: 224 PAVVAWDFEDLWIYMTAPVIGAIIGVLTYRSISLKTRP 261
>AQPZ_RHOPA (P60925) Aquaporin Z
Length = 240
Score = 152 bits (385), Expect = 6e-37
Identities = 88/223 (39%), Positives = 134/223 (59%), Gaps = 15/223 (6%)
Query: 37 KLVAEVVGTFFLIFAGCAAVVVNLNNDKV-VTLPGISIVWGLAVMVLVYSIGHISGAHFN 95
K +AE++GTF+L FAGC + V+ +V + L G+S+ +GL+V+ + Y+IGHISG H N
Sbjct: 5 KYLAEMIGTFWLTFAGCGSAVIAAGFPQVGIGLVGVSLAFGLSVVTMAYAIGHISGCHLN 64
Query: 96 PAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSG----------KENQFAGT 145
PAVT+ GRFP+KQ+ YIIAQV+G+ A+ +L LI SG N +
Sbjct: 65 PAVTLGLAAGGRFPVKQIAPYIIAQVLGAIAAAALLYLIASGAAGFDLAKGFASNGYGAH 124
Query: 146 LPAGSDLQA-FVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGA 204
P +L A FV+E ++T +F+I G +T +A A LA+G +++ L + P+T
Sbjct: 125 SPGQYNLVACFVMEVVMTMMFLFVIMG-STHGKAPAGFAPLAIGLALVMIHLVSIPVTNT 183
Query: 205 SMNPARSLGPAIVHHEYR--GIWIYMVSPILGALAGTWTYTFL 245
S+NPARS GPA+ + +W++ V+P+LG + G Y L
Sbjct: 184 SVNPARSTGPALFVGGWAIGQLWLFWVAPLLGGVLGGVIYRVL 226
>AQPZ_PHOLL (Q7N5C1) Aquaporin Z
Length = 231
Score = 151 bits (382), Expect = 1e-36
Identities = 91/227 (40%), Positives = 130/227 (57%), Gaps = 16/227 (7%)
Query: 34 LLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKV-VTLPGISIVWGLAVMVLVYSIGHISGA 92
+L+KL AE++GTF L+F GC +VV ++ + G+S+ +GL V+ ++Y++GHISG
Sbjct: 1 MLRKLAAELLGTFVLVFGGCGSVVFAAAFPELGIGFVGVSLAFGLTVLTMIYAVGHISGG 60
Query: 93 HFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGK-----------ENQ 141
HFNPAVTI GRF ++ YII+QV+G LA+ VL +I SG+ N
Sbjct: 61 HFNPAVTIGLWAGGRFRAVEVIPYIISQVIGGILAAAVLYVIASGQVGFDATTSGFASNG 120
Query: 142 FAGTLPAGSDLQ-AFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGP 200
F P G LQ A V E ++T + +I G ATD RA A LA+G ++L L + P
Sbjct: 121 FGEHSPGGFSLQSAIVAEIVLTAIFLIVIIG-ATDRRAPPGFAPLAIGLALVLINLISIP 179
Query: 201 ITGASMNPARSLGPAIVHHEY--RGIWIYMVSPILGALAGTWTYTFL 245
IT S+NPARS AI + + +W + V PI+G + G Y L
Sbjct: 180 ITNTSVNPARSTAVAIFQNTWALEQLWFFWVMPIIGGIVGGGIYRLL 226
>AQPZ_PSEPK (Q88F17) Aquaporin Z
Length = 230
Score = 151 bits (381), Expect = 2e-36
Identities = 85/220 (38%), Positives = 132/220 (59%), Gaps = 14/220 (6%)
Query: 36 QKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISGAHFN 95
+++ AE++GTF+L+ GC + V+ ++ + + G++ +GL V+ + ++IGHISG H N
Sbjct: 5 KRMGAELIGTFWLVLGGCGSAVLAASSPLGIGVLGVAFAFGLTVLTMAFAIGHISGCHLN 64
Query: 96 PAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGK----------ENQFAGT 145
PAV+ GRFP K+L Y+IAQV+G+ LA+GV+ LI SGK N +A
Sbjct: 65 PAVSFGLVVGGRFPAKELLPYVIAQVIGAILAAGVIYLIASGKAGFELSAGLASNGYADH 124
Query: 146 LPAGSDLQA-FVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGA 204
P G L A FV E ++T + +I G ATD RA A +A+G + L L + P+T
Sbjct: 125 SPGGYTLGAGFVSEVVMTAMFLVVIMG-ATDARAPAGFAPIAIGLALTLIHLISIPVTNT 183
Query: 205 SMNPARSLGPAIVHHEY--RGIWIYMVSPILGALAGTWTY 242
S+NPARS GPA+ + + +W++ V+P++GA G Y
Sbjct: 184 SVNPARSTGPALFVGGWALQQLWLFWVAPLIGAAIGGALY 223
>AQPZ_VIBVU (Q8DB17) Aquaporin Z
Length = 231
Score = 149 bits (375), Expect = 9e-36
Identities = 90/231 (38%), Positives = 131/231 (55%), Gaps = 16/231 (6%)
Query: 35 LQKLVAEVVGTFFLIFAGC-AAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISGAH 93
+ K +AE+ GTF+L+ GC +AV+ D + L G+S+ +GL V+ + ++IGHISG H
Sbjct: 1 MNKYLAELFGTFWLVLGGCGSAVLAAAFPDVGIGLLGVSLAFGLTVLTMAFAIGHISGCH 60
Query: 94 FNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGK-----------ENQF 142
NPAVTI GRF K++ YI+AQV+G +A GVL I SG+ N +
Sbjct: 61 LNPAVTIGLWAGGRFEAKEIVPYILAQVIGGVIAGGVLYTIASGQMGFDATSSGFASNGY 120
Query: 143 AGTLPAGSDL-QAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPI 201
P G L A V E ++T + +I G ATD RA A +A+G + L L + P+
Sbjct: 121 GEHSPGGYSLTSALVTEVVMTMMFLLVILG-ATDQRAPQGFAPIAIGLCLTLIHLISIPV 179
Query: 202 TGASMNPARSLGPAIVHHEY--RGIWIYMVSPILGALAGTWTYTFLRITNK 250
T S+NPARS G A+ ++ +W++ V+PILGAL G Y + +NK
Sbjct: 180 TNTSVNPARSTGVALYVGDWATAQLWLFWVAPILGALLGAVAYKLISGSNK 230
>AQPZ_VIBVY (Q7MIV9) Aquaporin Z
Length = 231
Score = 147 bits (371), Expect = 3e-35
Identities = 89/231 (38%), Positives = 130/231 (55%), Gaps = 16/231 (6%)
Query: 35 LQKLVAEVVGTFFLIFAGC-AAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISGAH 93
+ K +AE+ GTF+L+ GC +AV+ D + L G+S+ +GL V+ + ++IGHISG H
Sbjct: 1 MNKYLAELFGTFWLVLGGCGSAVLAAAFPDVGIGLLGVSLAFGLTVLTMAFAIGHISGCH 60
Query: 94 FNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGK-----------ENQF 142
NPAVTI GRF K++ YI+AQV+G +A GVL I SG+ N +
Sbjct: 61 LNPAVTIGLWAGGRFEAKEIVPYILAQVIGGVIAGGVLYTIASGQMGFDATSSGFASNGY 120
Query: 143 AGTLPAGSDL-QAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPI 201
P G L A V E ++ + +I G ATD RA A +A+G + L L + P+
Sbjct: 121 GEHSPGGYSLTSALVTEIVMAMMFLLVILG-ATDQRAPQGFAPIAIGLCLTLIHLISIPV 179
Query: 202 TGASMNPARSLGPAIVHHEY--RGIWIYMVSPILGALAGTWTYTFLRITNK 250
T S+NPARS G A+ ++ +W++ V+PILGAL G Y + +NK
Sbjct: 180 TNTSVNPARSTGVALYVGDWATAQLWLFWVAPILGALLGAVAYKLISGSNK 230
>AQPZ_BRAJA (Q89EG9) Aquaporin Z
Length = 240
Score = 146 bits (369), Expect = 5e-35
Identities = 85/225 (37%), Positives = 130/225 (57%), Gaps = 15/225 (6%)
Query: 35 LQKLVAEVVGTFFLIFAGCAAVVVNLNNDKV-VTLPGISIVWGLAVMVLVYSIGHISGAH 93
++K AE +GTF+L FAGC + V+ +V + L G+S+ +GL+V+ + Y+IGHISG H
Sbjct: 3 MKKYAAEAIGTFWLTFAGCGSAVIAAGFPQVGIGLVGVSLAFGLSVVTMAYAIGHISGCH 62
Query: 94 FNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSG----------KENQFA 143
NPAVT+ GRFP Q+ Y+IAQV G+ +A+ +L +I SG N +
Sbjct: 63 LNPAVTVGLAAGGRFPAGQILPYVIAQVCGAIVAAELLYIIASGAPGFDVTKGFASNGYD 122
Query: 144 GTLPAGSDLQA-FVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPIT 202
P + A F+ E ++T +FII G AT RA A LA+G +++ L + P+T
Sbjct: 123 AHSPGQYSMMACFLTEVVMTMMFLFIIMG-ATHGRAPAGFAPLAIGLALVMIHLVSIPVT 181
Query: 203 GASMNPARSLGPAIVHHEY--RGIWIYMVSPILGALAGTWTYTFL 245
S+NPARS GPA+ + +W++ V+P++G G Y +L
Sbjct: 182 NTSVNPARSTGPALFVGGWAMAQLWLFWVAPLIGGALGGVIYRWL 226
>AQPZ_SHIFL (O68874) Aquaporin Z
Length = 231
Score = 143 bits (360), Expect = 5e-34
Identities = 86/227 (37%), Positives = 127/227 (55%), Gaps = 16/227 (7%)
Query: 34 LLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKV-VTLPGISIVWGLAVMVLVYSIGHISGA 92
+ +KL AE GTF+L+F GC + V+ ++ + G+++ +GL V+ + +++GHISG
Sbjct: 1 MFRKLAAECFGTFWLVFGGCGSAVLAAGFPELGIGFAGVALAFGLTVLTMAFAVGHISGG 60
Query: 93 HFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGK-----------ENQ 141
HFNPAVTI GRFP K++ Y+IAQVVG +A+ +L LI SGK N
Sbjct: 61 HFNPAVTIGLWAGGRFPAKEVVGYVIAQVVGGIVAAALLYLIASGKTGFDAAASGFASNG 120
Query: 142 FAGTLPAG-SDLQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGP 200
+ P G S L A VVE +++ + +I G ATD A A +A+G + L L + P
Sbjct: 121 YGEHSPGGYSMLSALVVELVLSAGFLLVIHG-ATDKFAPAGFAPIAIGLALTLIHLISIP 179
Query: 201 ITGASMNPARSLGPAIVHHEY--RGIWIYMVSPILGALAGTWTYTFL 245
+T S+NPARS AI + +W + V PI+G + G Y L
Sbjct: 180 VTNTSVNPARSTAVAIFQGGWALEQLWFFWVVPIVGGIIGGLIYRTL 226
>AQPZ_ECOLI (P60844) Aquaporin Z (Bacterial nodulin-like intrinsic
protein)
Length = 231
Score = 143 bits (360), Expect = 5e-34
Identities = 86/227 (37%), Positives = 127/227 (55%), Gaps = 16/227 (7%)
Query: 34 LLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKV-VTLPGISIVWGLAVMVLVYSIGHISGA 92
+ +KL AE GTF+L+F GC + V+ ++ + G+++ +GL V+ + +++GHISG
Sbjct: 1 MFRKLAAECFGTFWLVFGGCGSAVLAAGFPELGIGFAGVALAFGLTVLTMAFAVGHISGG 60
Query: 93 HFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGK-----------ENQ 141
HFNPAVTI GRFP K++ Y+IAQVVG +A+ +L LI SGK N
Sbjct: 61 HFNPAVTIGLWAGGRFPAKEVVGYVIAQVVGGIVAAALLYLIASGKTGFDAAASGFASNG 120
Query: 142 FAGTLPAG-SDLQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGP 200
+ P G S L A VVE +++ + +I G ATD A A +A+G + L L + P
Sbjct: 121 YGEHSPGGYSMLSALVVELVLSAGFLLVIHG-ATDKFAPAGFAPIAIGLALTLIHLISIP 179
Query: 201 ITGASMNPARSLGPAIVHHEY--RGIWIYMVSPILGALAGTWTYTFL 245
+T S+NPARS AI + +W + V PI+G + G Y L
Sbjct: 180 VTNTSVNPARSTAVAIFQGGWALEQLWFFWVVPIVGGIIGGLIYRTL 226
>AQPZ_ECOL6 (P60845) Aquaporin Z
Length = 231
Score = 143 bits (360), Expect = 5e-34
Identities = 86/227 (37%), Positives = 127/227 (55%), Gaps = 16/227 (7%)
Query: 34 LLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKV-VTLPGISIVWGLAVMVLVYSIGHISGA 92
+ +KL AE GTF+L+F GC + V+ ++ + G+++ +GL V+ + +++GHISG
Sbjct: 1 MFRKLAAECFGTFWLVFGGCGSAVLAAGFPELGIGFAGVALAFGLTVLTMAFAVGHISGG 60
Query: 93 HFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGK-----------ENQ 141
HFNPAVTI GRFP K++ Y+IAQVVG +A+ +L LI SGK N
Sbjct: 61 HFNPAVTIGLWAGGRFPAKEVVGYVIAQVVGGIVAAALLYLIASGKTGFDAAASGFASNG 120
Query: 142 FAGTLPAG-SDLQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGP 200
+ P G S L A VVE +++ + +I G ATD A A +A+G + L L + P
Sbjct: 121 YGEHSPGGYSMLSALVVELVLSAGFLLVIHG-ATDKFAPAGFAPIAIGLALTLIHLISIP 179
Query: 201 ITGASMNPARSLGPAIVHHEY--RGIWIYMVSPILGALAGTWTYTFL 245
+T S+NPARS AI + +W + V PI+G + G Y L
Sbjct: 180 VTNTSVNPARSTAVAIFQGGWALEQLWFFWVVPIVGGIIGGLIYRTL 226
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.139 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,850,465
Number of Sequences: 164201
Number of extensions: 1224846
Number of successful extensions: 3979
Number of sequences better than 10.0: 158
Number of HSP's better than 10.0 without gapping: 153
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 3410
Number of HSP's gapped (non-prelim): 187
length of query: 270
length of database: 59,974,054
effective HSP length: 108
effective length of query: 162
effective length of database: 42,240,346
effective search space: 6842936052
effective search space used: 6842936052
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 65 (29.6 bits)
Medicago: description of AC140547.14


