

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140546.1 - phase: 0
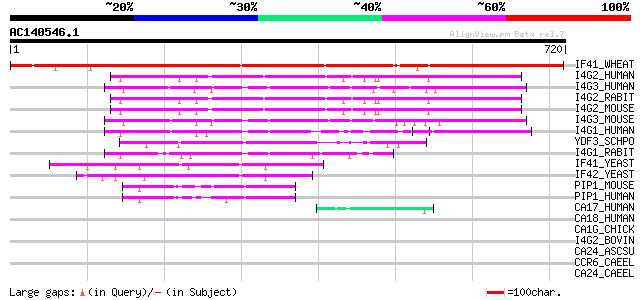
(720 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
IF41_WHEAT (Q03387) Eukaryotic initiation factor (iso)4F subunit... 874 0.0
I4G2_HUMAN (P78344) Eukaryotic translation initiation factor 4 g... 194 7e-49
I4G3_HUMAN (O43432) Eukaryotic translation initiation factor 4 g... 194 9e-49
I4G2_RABIT (P79398) Eukaryotic translation initiation factor 4 g... 193 1e-48
I4G2_MOUSE (Q62448) Eukaryotic translation initiation factor 4 g... 190 1e-47
I4G3_MOUSE (Q80XI3) Eukaryotic translation initiation factor 4 g... 188 5e-47
I4G1_HUMAN (Q04637) Eukaryotic translation initiation factor 4 g... 172 3e-42
YDF3_SCHPO (Q10475) Probable eukaryotic initiation factor C17C9.03 154 1e-36
I4G1_RABIT (P41110) Eukaryotic translation initiation factor 4 g... 134 6e-31
IF41_YEAST (P39935) Eukaryotic initiation factor 4F subunit p150... 112 3e-24
IF42_YEAST (P39936) Eukaryotic initiation factor 4F subunit p130... 100 1e-20
PIP1_MOUSE (Q8VE62) Polyadenylate-binding protein-interacting pr... 55 5e-07
PIP1_HUMAN (Q9H074) Polyadenylate-binding protein-interacting pr... 54 2e-06
CA17_HUMAN (Q02388) Collagen alpha 1(VII) chain precursor (Long-... 45 9e-04
CA18_HUMAN (P27658) Collagen alpha 1(VIII) chain precursor (Endo... 44 0.001
CA1G_CHICK (Q90584) Collagen alpha 1(XVII) chain (Bullous pemphi... 44 0.001
I4G2_BOVIN (Q95L46) Eukaryotic translation initiation factor 4 g... 42 0.004
CA24_ASCSU (P27393) Collagen alpha 2(IV) chain precursor 42 0.007
CCR6_CAEEL (P20784) Cuticle collagen rol-6 41 0.013
CA24_CAEEL (P17140) Collagen alpha 2(IV) chain precursor (Lethal... 41 0.013
>IF41_WHEAT (Q03387) Eukaryotic initiation factor (iso)4F subunit
p82-34 (eIF-(iso)4F p82-34)
Length = 788
Score = 874 bits (2257), Expect = 0.0
Identities = 453/741 (61%), Positives = 561/741 (75%), Gaps = 30/741 (4%)
Query: 1 VGDSQFESRERVRYTKEELLHIRETLEETPEDILKLRHDIDAELFGEDQSWGRVENN--- 57
+GD ESRERVRY++++LL +R+ + T E IL+L+ +I+AEL G+DQSW R ++N
Sbjct: 53 IGDLHSESRERVRYSRDQLLDLRKITDVT-EQILRLQQEIEAELNGDDQSWVRNDSNVQL 111
Query: 58 -----PPTQIQNRYSEPDNRDWRGRSAQPPANA--DERSWDNIKENREFGNTS-----QV 105
P Q QNR++E DNRDWR R+ +PPA A +E+SWDNI+E +E N S Q
Sbjct: 112 QTQAQPQVQAQNRFTETDNRDWRARTEKPPAPAVQEEKSWDNIREVKEQYNASGRQQEQF 171
Query: 106 NRQDQPRTNQGG-GPAPTLVKAEVPWSARRGTLSDKDRVLKTVKGILNKLTPEKFDLLKG 164
NRQDQ + + GP P L+KA+VPWSARRG LS+KDRVLKTVKGILNKLTPEKFDLLKG
Sbjct: 172 NRQDQSSSQKAQVGPPPALIKADVPWSARRGNLSEKDRVLKTVKGILNKLTPEKFDLLKG 231
Query: 165 QLIDSGITSADILKGVISLIFDKAVLEPTFCPMYAQLCSDLNEKLPSFPSEESGGKEITF 224
QL+DSGIT+ADILK VISLIF+KAV EPTFCPMYAQLCS+LN+ LP+FPSEE GGKEITF
Sbjct: 232 QLLDSGITTADILKDVISLIFEKAVFEPTFCPMYAQLCSELNDNLPTFPSEEPGGKEITF 291
Query: 225 KRLLLDNCQEAFEGAGKLREELAQMTSPEQETERRDKDRLVKIRTLGNIRLIGELLKQKM 284
KR+LL+NCQEAFEGA LR E+A +T P+QE E+RDK+R+ K+RTLGNIRLIGELLKQKM
Sbjct: 292 KRVLLNNCQEAFEGADSLRVEIASLTGPDQEMEKRDKERIFKLRTLGNIRLIGELLKQKM 351
Query: 285 VPERIVHHIVQELLGAADSNVCPAEENVEAICHFFNTIGKQLDESPKSRRINDMYFGRLK 344
VPE+IVHHIV+ELLG +D CP EE+VEAIC FFNTIGKQLDE+PKSRRIND YF +++
Sbjct: 352 VPEKIVHHIVKELLG-SDKKACPDEEHVEAICQFFNTIGKQLDENPKSRRINDTYFVQIR 410
Query: 345 ELSTNPQLAPRMKFMVRDVIDLRASNWVPRREEIKAKTISEIHDEAEKNLGLRPGATAGM 404
EL NPQL PR KFMVRD+IDLR++NWVPRR EIKAKTISEIH EAEKNLGLRPGATA M
Sbjct: 411 ELVANPQLTPRSKFMVRDLIDLRSNNWVPRRAEIKAKTISEIHTEAEKNLGLRPGATANM 470
Query: 405 RNTRVTGVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGIDNDNWEVPRTRSMPRGD 464
RN R G GGF + RPGTGG+MPGMPG+RKMPGMPG+DNDNWEV R+RSMPRGD
Sbjct: 471 RNGR-NAPGGPLSPGGFSVNRPGTGGMMPGMPGSRKMPGMPGLDNDNWEVQRSRSMPRGD 529
Query: 465 MSGAQTGGRGQSPYLSKPSVINSKLLPQGSSGLISGKNSALVLGGGTPSALPSNIVSGTE 524
Q + P ++KPS IN +LLPQG+ LI GK++ +LG G P + PS++ +
Sbjct: 530 PLRNQGPLINKVPSINKPSPINPRLLPQGTGALI-GKSA--LLGTGGPPSRPSSLTASPT 586
Query: 525 PAP------QIPSPVKPVSAASPEKPQAPAVKLNIDDLHRKTVSLLEEYFNVRLLDEALQ 578
P P PS P S P+K A + K+ L +KT SLLEEYF +R+LDEA Q
Sbjct: 587 PLPAQTTASPKPSSATPASVPIPDK-AASSAKVIPAGLQKKTASLLEEYFGIRILDEAQQ 645
Query: 579 CVEELKAPTYHPEVVKEAISLGLDKSPPRVEPVANLIEYLFTKKILTARDIGTGCLLFAS 638
C+EEL++P YHPE+VKEAI+L LDK V+P+ L+E+L+TKK D+ GCLL+ S
Sbjct: 646 CIEELQSPDYHPEIVKEAINLALDKGASFVDPLVKLLEHLYTKKTFKTEDLENGCLLYGS 705
Query: 639 LLDDIGIDLPKAPNNFGEIIGKLVLSAGLDFKVVKEILKKVGDDYFQKAIFNSAVQVI-S 697
LL+DIGIDLPKAP FGE++ +L+LS GL F+ + ILK + D +F+KAIF S + + +
Sbjct: 706 LLEDIGIDLPKAPTQFGEVVARLILSCGLRFEAAEGILKAMEDTFFRKAIFTSVTKTLGA 765
Query: 698 SASGQAVLDSQASDIEACQAL 718
+GQA+L S A+ ++AC +L
Sbjct: 766 DPAGQAILSSHAAVVDACNSL 786
>I4G2_HUMAN (P78344) Eukaryotic translation initiation factor 4
gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97)
(Death associated protein 5) (DAP-5)
Length = 907
Score = 194 bits (493), Expect = 7e-49
Identities = 173/600 (28%), Positives = 285/600 (46%), Gaps = 74/600 (12%)
Query: 131 SARRGTLSDKDR---VLKTVKGILNKLTPEKFDLLKGQLIDSGITSADILKGVISLIFDK 187
SA + ++K+R + + V+GILNKLTPEKFD L +L++ G+ S ILKGVI LI DK
Sbjct: 62 SAANNSANEKERHDAIFRKVRGILNKLTPEKFDKLCLELLNVGVESKLILKGVILLIVDK 121
Query: 188 AVLEPTFCPMYAQLCSDLNEKLPSFPSEESGG-----KEITFKRLLLDNCQEAFEGAGK- 241
A+ EP + +YAQLC L E P+F + G + TF+RLL+ Q+ FE +
Sbjct: 122 ALEEPKYSSLYAQLCLRLAEDAPNFDGPAAEGQPGQKQSTTFRRLLISKLQDEFENRTRN 181
Query: 242 --LREELAQMTSPEQETERRDKDRLVKIRTLGNIRLIGELLKQKMVPERIVHHIVQELLG 299
+ ++ PE+E +R + KI+ LGNI+ IGEL K ++ E I+H ++ LL
Sbjct: 182 VDVYDKRENPLLPEEEEQR----AIAKIKMLGNIKFIGELGKLDLIHESILHKCIKTLLE 237
Query: 300 AADS-NVCPAEENVEAICHFFNTIGKQLDESPKSRRINDMYFGRLKELSTNPQLAPRMKF 358
+ E++E +C T+G +LD +++ + D YF R+ L + +L R++F
Sbjct: 238 KKKRVQLKDMGEDLECLCQIMRTVGPRLDHE-RAKSLMDQYFARMCSLMLSKELPARIRF 296
Query: 359 MVRDVIDLRASNWVPRREEIK--AKTISEIHDEAEKNLGLRPGA--TAGMRNTRVTGVQG 414
+++D ++LR +WVPR+ + KTI++I +A K+LG+ A GMR+ ++G
Sbjct: 297 LLQDTVELREHHWVPRKAFLDNGPKTINQIRQDAVKDLGVFIPAPMAQGMRSDFF--LEG 354
Query: 415 NTGAGGFPIARPGTGGL---MPGMPGARKMPGMPGIDNDNWE--VPRTRS---------- 459
+ R GGL MPG+ G PG+ D + + R RS
Sbjct: 355 PFMPPRMKMDRDPLGGLADMFGQMPGSGIGTG-PGVIQDRFSPTMGRHRSNQLFNGHGGH 413
Query: 460 -MPRGDMSGAQTGGR-----------------------GQS----PYLSKPSVINS-KLL 490
MP + GG+ GQS P SK +N+ ++
Sbjct: 414 IMPPTQSQFGEMGGKFMKSQGLSQLYHNQSQGLLSQLQGQSKDMPPRFSKKGQLNADEIS 473
Query: 491 PQGSSGLISGKNSALVLGGGTPSALPSNIVSGTEPAPQIPSPVKPVSAASP---EKPQAP 547
+ + + KN L PS T+ P +P + P EKP
Sbjct: 474 LRPAQSFLMNKNQVPKLQPQITMIPPSAQPPRTQTPPLGQTPQLGLKTNPPLIQEKPAKT 533
Query: 548 AVK--LNIDDLHRKTVSLLEEYFNVRLLDEALQCVEELKAPTYH-PEVVKEAISLGLDKS 604
+ K + ++L + T +++ EY N +EA+ V E++AP + PE++ + I L LD+S
Sbjct: 534 SKKPPPSKEELLKLTETVVTEYLNSGNANEAVNGVREMRAPKHFLPEMLSKVIILSLDRS 593
Query: 605 PPRVEPVANLIEYLFTKKILTARDIGTGCLLFASLLDDIGIDLPKAPNNFGEIIGKLVLS 664
E ++LI L + I T+ + L + +D+P + + + ++S
Sbjct: 594 DEDKEKASSLISLLKQEGIATSDNFMQAFLNVLDQCPKLEVDIPLVKSYLAQFAARAIIS 653
>I4G3_HUMAN (O43432) Eukaryotic translation initiation factor 4 gamma
3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II)
(eIF4GII)
Length = 1585
Score = 194 bits (492), Expect = 9e-49
Identities = 179/628 (28%), Positives = 285/628 (44%), Gaps = 97/628 (15%)
Query: 123 LVKAEVPW--SARRGTLSDKDRVLKT------VKGILNKLTPEKFDLLKGQLIDSGITSA 174
L KAE W S +R + +D +KT V+ ILNKLTP+ F+ L Q+ + +
Sbjct: 726 LKKAENAWKPSQKRDSQADDPENIKTQELFRKVRSILNKLTPQMFNQLMKQVSGLTVDTE 785
Query: 175 DILKGVISLIFDKAVLEPTFCPMYAQLCSDLNEKLPSFPSEESGGKEITFKRLLLDNCQE 234
+ LKGVI L+F+KA+ EP+F YA +C L P + G + F++LLL+ CQ+
Sbjct: 786 ERLKGVIDLVFEKAIDEPSFSVAYANMCRCL--VTLKVPMADKPGNTVNFRKLLLNRCQK 843
Query: 235 AFEG-------AGKLREELAQMTSPEQETERRD-----KDRLVKIRTLGNIRLIGELLKQ 282
FE K ++EL ++PE+ T D KD+ + R++GNI+ IGEL K
Sbjct: 844 EFEKDKADDDVFEKKQKELEAASAPEERTRLHDELEEAKDK-ARRRSIGNIKFIGELFKL 902
Query: 283 KMVPERIVHHIVQELLGAADSNVCPAEENVEAICHFFNTIGKQLD-ESPKSRRINDMYFG 341
KM+ E I+H V +LL D EE++E +C TIGK LD E K R D YF
Sbjct: 903 KMLTEAIMHDCVVKLLKNHD------EESLECLCRLLTTIGKDLDFEKAKPRM--DQYFN 954
Query: 342 RLKELSTNPQLAPRMKFMVRDVIDLRASNWVPRREEIKAKTISEIHDEA--EKNLGLRPG 399
+++++ + + R++FM++DVIDLR NWV RR + KTI +IH EA E+ R
Sbjct: 955 QMEKIVKERKTSSRIRFMLQDVIDLRLCNWVSRRADQGPKTIEQIHKEAKIEEQEEQRKV 1014
Query: 400 ATAGMRNTRVTGVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGIDNDNWEVPRTR- 458
+ R GVQ GG+ + + K+ P ID VP+ +
Sbjct: 1015 QQLMTKEKRRPGVQ-RVDEGGWNTVQGAKNSRVLDPSKFLKIT-KPTIDEKIQLVPKAQL 1072
Query: 459 -SMPRGDMSGAQT----------------------GGRGQSPYLSKPSVINSK--LLPQG 493
S +G GA+ G +P S P +S+ L +G
Sbjct: 1073 GSWGKGSSGGAKASETDALRSSASSLNRFSALQPPAPSGSTP--STPVEFDSRRTLTSRG 1130
Query: 494 SSG---------LISGKNSALVLGGGTPSALPSNIVSGTEPAPQIPSPVKPVSAA----- 539
S G + + + + GG + L + S E ++ VK ++
Sbjct: 1131 SMGREKNDKPLPSATARPNTFMRGGSSKDLLDNQ--SQEEQRREMLETVKQLTGGVDVER 1188
Query: 540 -----------SPEKPQAPAVK------LNIDDLHRKTVSLLEEYFNVRLLDEALQCVEE 582
KP+ A+ L+ ++L RK+ S+++E+ ++ EA+QCVEE
Sbjct: 1189 NSTEAERNKTRESAKPEISAMSAHDKAALSEEELERKSKSIIDEFLHINDFKEAMQCVEE 1248
Query: 583 LKAPTYHPEVVKEAISLGLDKSPPRVEPVANLIEYLFTKKILTARDIGTGCLLFASLLDD 642
L A V+ + L++S + + L+ L + L+ +D G L DD
Sbjct: 1249 LNAQGLLHVFVRVGVESTLERSQITRDHMGQLLYQLVQSEKLSKQDFFKGFSETLELADD 1308
Query: 643 IGIDLPKAPNNFGEIIGKLVLSAGLDFK 670
+ ID+P E++ ++ G+ +
Sbjct: 1309 MAIDIPHIWLYLAELVTPMLKEGGISMR 1336
>I4G2_RABIT (P79398) Eukaryotic translation initiation factor 4
gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97)
(Novel APOBEC-1 target 1) (Translation repressor NAT1)
Length = 907
Score = 193 bits (491), Expect = 1e-48
Identities = 173/600 (28%), Positives = 286/600 (46%), Gaps = 74/600 (12%)
Query: 131 SARRGTLSDKDR---VLKTVKGILNKLTPEKFDLLKGQLIDSGITSADILKGVISLIFDK 187
SA + ++K+R + + V+GILNKLTPEKFD L +L++ G+ S ILKGVI LI DK
Sbjct: 62 SAANNSANEKERHDAIFRKVRGILNKLTPEKFDKLCLELLNVGVESKLILKGVILLIVDK 121
Query: 188 AVLEPTFCPMYAQLCSDLNEKLPSFPSEESGG-----KEITFKRLLLDNCQEAFEGAGK- 241
A+ EP + +YAQLC L E P+F + G + TF+RLL+ Q+ FE +
Sbjct: 122 ALEEPKYSSLYAQLCLRLAEDAPNFDGPAAEGQPGQKQSTTFRRLLISKLQDEFENRTRN 181
Query: 242 --LREELAQMTSPEQETERRDKDRLVKIRTLGNIRLIGELLKQKMVPERIVHHIVQELLG 299
+ ++ PE+E +R + KI+ LGNI+ IGEL K ++ E I+H ++ LL
Sbjct: 182 VDVYDKRENPLLPEEEEQR----AIAKIKMLGNIKFIGELGKLDLIHESILHKCIKTLLE 237
Query: 300 AADS-NVCPAEENVEAICHFFNTIGKQLDESPKSRRINDMYFGRLKELSTNPQLAPRMKF 358
+ E++E +C T+G +LD +++ + D YF R+ L + +L R++F
Sbjct: 238 KKKRVQLQDMGEDLECLCQIMRTVGPRLDHE-RAKSLMDQYFARMCSLMLSKELPARIRF 296
Query: 359 MVRDVIDLRASNWVPRREEIK--AKTISEIHDEAEKNLGLRPGA--TAGMRNTRVTGVQG 414
+++D ++LR +WVPR+ + KTI++I +A K+LG+ A GMR+ ++G
Sbjct: 297 LLQDTVELREHHWVPRKAFLDNGPKTINQIRQDAVKDLGVFIPAPMAQGMRSDFF--LEG 354
Query: 415 NTGAGGFPIARPGTGGL---MPGMPGARKMPGMPGIDNDNWE--VPRTRS---------- 459
+ R GGL MPG+ G PG+ D + + R RS
Sbjct: 355 PFMPPRMKMDRDPLGGLADMFGQMPGSGIGTG-PGVIQDRFSPTMGRHRSNQLFNGHGGH 413
Query: 460 -MPRGDMSGAQTGGR-----------------------GQS----PYLSKPSVINS-KLL 490
MP + GG+ GQS P SK +N+ ++
Sbjct: 414 IMPPTQSQFGEMGGKFMKSQGLSQLYHNQSQGLLSQLQGQSKDMPPRFSKKGQLNADEIS 473
Query: 491 PQGSSGLISGKNSALVLGGGTPSALPSNIVSGTEPAPQIPSPVKPVSAASP---EKPQAP 547
+ + + KN L PS T+ P +P + P EKP
Sbjct: 474 LRPAQSFLMNKNQVPKLQPQITMIPPSAQPPRTQTPPLGQTPQLGLKTNPPLIQEKPAKT 533
Query: 548 AVK--LNIDDLHRKTVSLLEEYFNVRLLDEALQCVEELKAPTYH-PEVVKEAISLGLDKS 604
+ K + ++L + T +++ EY N +EA+ V+E++AP + PE++ + I L LD+S
Sbjct: 534 SKKPPPSKEELLKLTETVVTEYLNSGNANEAVNGVKEMRAPKHFLPEMLSKVIILSLDRS 593
Query: 605 PPRVEPVANLIEYLFTKKILTARDIGTGCLLFASLLDDIGIDLPKAPNNFGEIIGKLVLS 664
E ++LI L + I T+ + L + +D+P + + + ++S
Sbjct: 594 DEDKEKASSLISLLKQEGIGTSDNFMQAFLNVLDQCPKLEVDIPLVKSYLAQFAARAIIS 653
>I4G2_MOUSE (Q62448) Eukaryotic translation initiation factor 4
gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97)
(Novel APOBEC-1 target 1) (Translation repressor NAT1)
Length = 906
Score = 190 bits (483), Expect = 1e-47
Identities = 170/599 (28%), Positives = 284/599 (47%), Gaps = 73/599 (12%)
Query: 131 SARRGTLSDKDR---VLKTVKGILNKLTPEKFDLLKGQLIDSGITSADILKGVISLIFDK 187
SA + ++K+R + + V+GILNKLTPEKFD L +L++ G+ S ILKGVI LI DK
Sbjct: 62 SAANNSANEKERHDAIFRKVRGILNKLTPEKFDKLCLELLNVGVESKLILKGVILLIVDK 121
Query: 188 AVLEPTFCPMYAQLCSDLNEKLPSFPSEESGG-----KEITFKRLLLDNCQEAFEGAGK- 241
A+ EP + +YAQLC L E P+F + G + TF+RLL+ Q+ FE +
Sbjct: 122 ALEEPKYSSLYAQLCLRLAEDAPNFDGPAAEGQPGQKQSTTFRRLLISKLQDEFENRTRN 181
Query: 242 --LREELAQMTSPEQETERRDKDRLVKIRTLGNIRLIGELLKQKMVPERIVHHIVQELLG 299
+ ++ PE+E +R + KI+ LGNI+ IGEL K ++ E I+H ++ LL
Sbjct: 182 VDVYDKRENPLLPEEEEQR----AIAKIKMLGNIKFIGELGKLDLIHESILHKCIKTLLE 237
Query: 300 AADS-NVCPAEENVEAICHFFNTIGKQLDESPKSRRINDMYFGRLKELSTNPQLAPRMKF 358
+ E++E +C T+G +LD +++ + D YF R+ L + +L R++F
Sbjct: 238 KKKRVQLKDMGEDLECLCQIMRTVGPRLDHE-RAKSLMDQYFARMCSLMLSKELPARIRF 296
Query: 359 MVRDVIDLRASNWVPRREEIK--AKTISEIHDEAEKNLGL-RPGATAGMRNTRVTGVQGN 415
+++D ++LR +WVPR+ + KTI++I +A K+LG+ P A R+ ++G
Sbjct: 297 LLQDTVELREHHWVPRKAFLDNGPKTINQIRQDAVKDLGVFIPAPMAQGRSDFF--LEGP 354
Query: 416 TGAGGFPIARPGTGGL---MPGMPGARKMPGMPGIDNDNWE--VPRTRS----------- 459
+ R GGL MPG+ G PG+ D + + R RS
Sbjct: 355 FMPPRMKMDRDPLGGLADMFGQMPGSGIGTG-PGVIQDRFSPTMGRHRSNQLFNGHGGHI 413
Query: 460 MPRGDMSGAQTGGR-----------------------GQS----PYLSKPSVINS-KLLP 491
MP + GG+ GQS P SK +N+ ++
Sbjct: 414 MPPTQSQFGEMGGKFMKSQGLSQLYHNQSQGLLSQLQGQSKDMPPRFSKKGQLNADEISL 473
Query: 492 QGSSGLISGKNSALVLGGGTPSALPSNIVSGTEPAPQIPSPVKPVSAASP---EKPQAPA 548
+ + + KN L PS T+ P +P + P EKP +
Sbjct: 474 RPAQSFLMNKNQVPKLQPQITMIPPSAQPPRTQTPPLGQTPQLGLKTNPPLIQEKPAKTS 533
Query: 549 VK--LNIDDLHRKTVSLLEEYFNVRLLDEALQCVEELKAPTYH-PEVVKEAISLGLDKSP 605
K + ++L + T +++ +Y N ++A+ V E++AP + PE++ + I L LD+S
Sbjct: 534 KKPPPSKEELLKLTEAVVTDYLNSGNANDAVSGVREMRAPKHFLPEMLSKVIILSLDRSD 593
Query: 606 PRVEPVANLIEYLFTKKILTARDIGTGCLLFASLLDDIGIDLPKAPNNFGEIIGKLVLS 664
E ++LI L + I T+ + L + +D+P + + + ++S
Sbjct: 594 EDKEKASSLISLLKQEGIATSDNFMQAFLNVLEQCPKLEVDIPLVKSYLAQFAARAIIS 652
>I4G3_MOUSE (Q80XI3) Eukaryotic translation initiation factor 4 gamma
3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II)
(eIF4GII)
Length = 1579
Score = 188 bits (477), Expect = 5e-47
Identities = 178/624 (28%), Positives = 280/624 (44%), Gaps = 90/624 (14%)
Query: 123 LVKAEVPW--SARRGTLSDKDRVLKT------VKGILNKLTPEKFDLLKGQLIDSGITSA 174
L KAE W S +R + +D +KT V+ ILNKLTP+ F+ L Q+ + +
Sbjct: 721 LRKAENAWKPSQKRDSHADDPESIKTQELFRKVRSILNKLTPQMFNQLMKQVSALTVDTE 780
Query: 175 DILKGVISLIFDKAVLEPTFCPMYAQLCSDLNEKLPSFPSEESGGKEITFKRLLLDNCQE 234
+ LKGVI L+F+KA+ EP+F YA +C L P + G + F++LLL+ CQ+
Sbjct: 781 ERLKGVIDLVFEKAIDEPSFSVAYANMCRCLVTL--KVPMADKPGNTVNFRKLLLNRCQK 838
Query: 235 AFEGAG-------KLREELAQMTSPEQETERRD-----KDRLVKIRTLGNIRLIGELLKQ 282
FE K ++EL ++PE+ T D KD+ + R++GNI+ IGEL K
Sbjct: 839 EFEKDKADDDVFEKKQKELEAASAPEERTRLHDELEEAKDK-ARRRSIGNIKFIGELFKL 897
Query: 283 KMVPERIVHHIVQELLGAADSNVCPAEENVEAICHFFNTIGKQLDESPKSRRINDMYFGR 342
KM+ E I+H V +LL D EE++E +C TIGK LD R+ D YF +
Sbjct: 898 KMLTEAIMHDCVVKLLKNHD------EESLECLCRLLTTIGKDLDFEKAKPRM-DQYFNQ 950
Query: 343 LKELSTNPQLAPRMKFMVRDVIDLRASNWVPRREEIKAKTISEIHDEA--EKNLGLRPGA 400
++++ + + R++FM++DVIDLR NWV RR + KTI +IH EA E+ R
Sbjct: 951 MEKIVKERKTSSRIRFMLQDVIDLRLCNWVSRRADQGPKTIEQIHKEAKIEEQEEQRKVQ 1010
Query: 401 TAGMRNTRVTGVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGIDNDNWEVPRTR-- 458
+ R GVQ GG+ + + K+ P ID VP+ +
Sbjct: 1011 QLMTKEKRRPGVQ-RVDEGGWNTVQGAKNSRVLDPSKFLKIT-KPTIDEKIQLVPKAQLG 1068
Query: 459 SMPRGDMSGAQTGGRGQSPYLSKPSVIN--SKLLPQGSSGLISG------KNSALVLGGG 510
S +G GA+ S S +N S L P SG S AL G
Sbjct: 1069 SWGKGSSGGAKAS--ESDALRSSASSLNRFSPLQPPAPSGSPSATPLEFDSRRALTSRGS 1126
Query: 511 -----------TPSALPSNIVSGT-----------EPAPQIPSPVKPVSA------ASPE 542
+A P+ + G+ E ++ VK ++ AS E
Sbjct: 1127 MGREKSDKPIPAGTARPNTFLRGSSKDLLDNQSQEEQRREMLETVKQLTGGLDAERASTE 1186
Query: 543 KPQAPAVKLNIDDL----------------HRKTVSLLEEYFNVRLLDEALQCVEELKAP 586
++ +L ++ RK+ S+++E+ ++ EA QC+EEL A
Sbjct: 1187 ADRSKTRELAKSEMCAVPAPDKPALSEEEVERKSKSIIDEFLHINDFKEATQCIEELSAQ 1246
Query: 587 TYHPEVVKEAISLGLDKSPPRVEPVANLIEYLFTKKILTARDIGTGCLLFASLLDDIGID 646
VK + L++S + + +L+ L + L+ +D G L DD+ ID
Sbjct: 1247 GPLHVFVKVGVEFTLERSQITRDHMGHLLYQLVQSEKLSKQDFFKGFSETLELADDMAID 1306
Query: 647 LPKAPNNFGEIIGKLVLSAGLDFK 670
+P E++ ++ G+ +
Sbjct: 1307 IPHIWLYLAELVTPMLKEGGISMR 1330
>I4G1_HUMAN (Q04637) Eukaryotic translation initiation factor 4 gamma
1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G) (p220)
Length = 1600
Score = 172 bits (436), Expect = 3e-42
Identities = 146/448 (32%), Positives = 214/448 (47%), Gaps = 69/448 (15%)
Query: 123 LVKAEVPWS-ARRGTLSDKDR------------VLKTVKGILNKLTPEKFDLLKGQLIDS 169
L KAE W + + T +DKDR + + V+ ILNKLTP+ F L Q+
Sbjct: 728 LNKAEKAWKPSSKRTAADKDRGEEDADGSKTQDLFRRVRSILNKLTPQMFQQLMKQVTQL 787
Query: 170 GITSADILKGVISLIFDKAVLEPTFCPMYAQLCSDLNEKLPSFPSEESGGKEITFKRLLL 229
I + + LKGVI LIF+KA+ EP F YA +C L P+ E + F++LLL
Sbjct: 788 AIDTEERLKGVIDLIFEKAISEPNFSVAYANMCRCLMAL--KVPTTEKPTVTVNFRKLLL 845
Query: 230 DNCQEAFEGAG-------KLREELAQMTSPEQ----ETERRDKDRLVKIRTLGNIRLIGE 278
+ CQ+ FE K ++E+ + + E+ + E + + + R+LGNI+ IGE
Sbjct: 846 NRCQKEFEKDKDDDEVFEKKQKEMDEAATAEERGRLKEELEEARDIARRRSLGNIKFIGE 905
Query: 279 LLKQKMVPERIVHHIVQELLGAADSNVCPAEENVEAICHFFNTIGKQLDESPKSRRINDM 338
L K KM+ E I+H V +LL D EE++E +C TIGK LD R+ D
Sbjct: 906 LFKLKMLTEAIMHDCVVKLLKNHD------EESLECLCRLLTTIGKDLDFEKAKPRM-DQ 958
Query: 339 YFGRLKELSTNPQLAPRMKFMVRDVIDLRASNWVPRREEIKAKTISEIHDEAEKNLGLRP 398
YF +++++ + + R++FM++DV+DLR SNWVPRR + KTI +IH EAE
Sbjct: 959 YFNQMEKIIKEKKTSSRIRFMLQDVLDLRGSNWVPRRGDQGPKTIDQIHKEAE------- 1011
Query: 399 GATAGMRNTRVTGVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGIDNDNWE-VPRT 457
M R A G R G PG P +R G+P +D+ W VP +
Sbjct: 1012 -----MEEHREHIKVQQLMAKGSDKRRGGP----PGPPISR---GLPLVDDGGWNTVPIS 1059
Query: 458 RSMPRGDMSGAQTGGRGQSPYLSKPSVINSKLLPQGSSGLIS-GKNSALVLGGGTPSALP 516
+ G++ + ++KP I+S G +S GK S+ G A P
Sbjct: 1060 K--------GSRPIDTSRLTKITKPGSIDSNNQLFAPGGRLSWGKGSS-----GGSGAKP 1106
Query: 517 SNIVSGTEPAPQIPSPVKPVSAASPEKP 544
S+ S E A S + SA P
Sbjct: 1107 SDAAS--EAARPATSTLNRFSALQQAVP 1132
Score = 55.1 bits (131), Expect = 6e-07
Identities = 40/156 (25%), Positives = 75/156 (47%), Gaps = 3/156 (1%)
Query: 523 TEPAPQIPSPVKPVSAASPEKPQAPAVKLNIDDLHRKTVSLLEEYFNVRLLDEALQCVEE 582
TE + VK +A P P A L+ ++L +K+ +++EEY ++ + EA+QCV+E
Sbjct: 1212 TEDRDRGRDAVKREAALPPVSPLKAA--LSEEELEKKSKAIIEEYLHLNDMKEAVQCVQE 1269
Query: 583 LKAPTYHPEVVKEAISLGLDKSPPRVEPVANLIEYLFTKKILTARDIGTGCLLFASLLDD 642
L +P+ V+ + L++S E + L+ L L+ G L +D
Sbjct: 1270 LASPSLLFIFVRHGVESTLERSAIAREHMGQLLHQLLCAGHLSTAQYYQGLYEILELAED 1329
Query: 643 IGIDLPKAPNNFGEIIGKLVLSAGLDF-KVVKEILK 677
+ ID+P E++ ++ G+ ++ +EI K
Sbjct: 1330 MEIDIPHVWLYLAELVTPILQEGGVPMGELFREITK 1365
>YDF3_SCHPO (Q10475) Probable eukaryotic initiation factor C17C9.03
Length = 1403
Score = 154 bits (388), Expect = 1e-36
Identities = 128/425 (30%), Positives = 185/425 (43%), Gaps = 83/425 (19%)
Query: 143 VLKTVKGILNKLTPEKFDLLKGQLIDSGITSAD-----ILKGVISLIFDKAVLEPTFCPM 197
V + VKG LNK+T EKFD + Q+++ + S LK VI L F+KA EP F M
Sbjct: 1008 VQRKVKGSLNKMTLEKFDKISDQILEIAMQSRKENDGRTLKQVIQLTFEKATDEPNFSNM 1067
Query: 198 YAQLCSDLNEKLPSFPSEES----------GGKEITFKRLLLDNCQEAFEGAGKLREELA 247
YA+ + + + +E GG + F++ LL CQE FE K
Sbjct: 1068 YARFARKMMDSIDDSIRDEGVLDKNNQPVRGG--LLFRKYLLSRCQEDFERGWKANLPSG 1125
Query: 248 QMTSPEQETERRDKDRLVKIRTLGNIRLIGELLKQKMVPERIVHHIVQELLGAADSNVCP 307
+ E ++ +K R LG +R IGEL K M+ E+I+H ++ LLG P
Sbjct: 1126 KAGEAEIMSDEYYVAAAIKRRGLGLVRFIGELFKLSMLSEKIMHECIKRLLGNVTD---P 1182
Query: 308 AEENVEAICHFFNTIGKQLDESPKSRRINDMYFGRLKELSTNPQLAPRMKFMVRDVIDLR 367
EE +E++C T+G +D + K D+Y R++ ++ P L R+KFM+ DV+D R
Sbjct: 1183 EEEEIESLCRLLMTVGVNIDATEKGHAAMDVYVLRMETITKIPNLPSRIKFMLMDVMDSR 1242
Query: 368 ASNWVPRREEIKA-KTISEIHDEAEKNLGLRPGATAGMRNTRVTGVQGNTGAGGFPIARP 426
+ W + E K KTI+EIH+EAE+ L RP
Sbjct: 1243 KNGWAVKNEVEKGPKTIAEIHEEAERKKALAES------------------------QRP 1278
Query: 427 GTGGLMPGMPGARKMPGMPGIDNDNWEVPRTRSMPRGDMSGAQTGGRGQSPYLSKPSVIN 486
+G +M G R M RGD ++ GGRG +P S N
Sbjct: 1279 SSG----------RMHG--------------RDMNRGD---SRMGGRGSNPPFSSSDWSN 1311
Query: 487 SK----LLPQGSSGLISGKNS-------ALVLGGGTPSALPSNIVSGTEPAPQIPSPVKP 535
+K L QG GL SG + +L GG+ S PS S + +P
Sbjct: 1312 NKDGYARLGQGIRGLKSGTQGSHGPTSLSSMLKGGSVSRTPSRQNSALRREQSVRAPPSN 1371
Query: 536 VSAAS 540
V+ S
Sbjct: 1372 VAVTS 1376
>I4G1_RABIT (P41110) Eukaryotic translation initiation factor 4
gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G) (p220)
Length = 1402
Score = 134 bits (338), Expect = 6e-31
Identities = 128/417 (30%), Positives = 194/417 (45%), Gaps = 68/417 (16%)
Query: 123 LVKAEVPWS-ARRGTLSDKDR------------VLKTVKGILNKLTPEKFDLLKGQLIDS 169
L KAE W + + T +DKDR + + V+ ILNKLTP+ F L Q+
Sbjct: 533 LNKAEKAWKPSSKRTAADKDRGEEDADGSKTQDLFRRVRSILNKLTPQMFQQLMKQVTQL 592
Query: 170 GITSADILKGVISLIFDKAVLEPTFCPMYAQLCSDLNEKLPSFPSEESGGKE-----ITF 224
I + KG ++ + PT +Q S + +P ES E + F
Sbjct: 593 AIDTEGASKGSLTSSLRRPFQNPT-----SQWPS---QHVPLPHGAESATTEKPTVTVNF 644
Query: 225 KRLLLDNCQ----------EAFEGAGKLREELAQMTSPEQETERRDKDRLVKIR-TLGNI 273
++LLL+ CQ E FE K +E A E+ E ++ R + R +LGNI
Sbjct: 645 RKLLLNRCQKEFEKDKDDDEVFEKKQKEMDEAATAEERERLKEELEEARDIARRCSLGNI 704
Query: 274 RLIGELLKQKMVPERIVHHIVQELLGAADSNVCPAEENVEAICHFFNTIGKQLDESPKSR 333
+ IGEL K KM+ E I+H V +LL EE++E +C TIGK LD
Sbjct: 705 KFIGELFKLKMLTEAIMHDCVVKLLRHD-------EESLEFLCRLLTTIGKDLDFEKAKP 757
Query: 334 RINDMYFGRLKELSTNPQLAPRMKFMVRDVIDLRASNWVPRREEIKAKTISEIHDEAE-- 391
R+ D YF +++++ + + R++FM++DV+DLR SNWVPRR + KTI +IH EAE
Sbjct: 758 RM-DQYFNQMEKIIKEKKTSSRIRFMLQDVLDLRQSNWVPRRGDQGPKTIDQIHKEAEME 816
Query: 392 ---KNLGLRPGATAGMRNTRVTGVQGNTGAGGFPIARPGTGGLMPGMPGAR--------K 440
+++ ++ G + R G G + G P+ G +P G+R K
Sbjct: 817 EHREHIKVQQLMAKG-SDKRRGGPPGPPISRGLPLVDDGGWNTVPISKGSRPIDTSRLTK 875
Query: 441 MPGMPGIDNDNWEVPRTRSMPRGDMSGAQTGGRGQSPYLSKPSVINSKLLPQGSSGL 497
+ ID++N P G +S G+G S +KPS S++ +S L
Sbjct: 876 ITKPGSIDSNN-----QLFAPGGRLS----WGKGSSGSGAKPSDAASEVSRPATSTL 923
>IF41_YEAST (P39935) Eukaryotic initiation factor 4F subunit p150
(eIF4F p150) (eIF-4F p150) (mRNA cap-binding protein
complex subunit p150)
Length = 952
Score = 112 bits (280), Expect = 3e-24
Identities = 106/397 (26%), Positives = 171/397 (42%), Gaps = 45/397 (11%)
Query: 52 GRVENNPPTQIQNRYSEPDNRDWRGRSAQPPANADERSWDNIKENREF--------GNTS 103
GR NN R + N D R S +R D+ + NR + G+
Sbjct: 496 GRFGNNSSRGHDFRNTSVRNMDDRANSRTSSKRRSKRMNDDRRSNRSYTSRRDRERGSYR 555
Query: 104 QVNRQDQPRTNQGGGPAPTLVKAEVPWSARRGT-----------LSDKDRVLKTVKGILN 152
+++ + + P VP + T L DKD V + +K +LN
Sbjct: 556 NEEKREDDKPKEEVAPLVPSANRWVPKFKSKKTEKKLAPDGKTELLDKDEVERKMKSLLN 615
Query: 153 KLTPEKFDLLKGQL-----IDSGITSADILKGVISLIFDKAVLEPTFCPMYAQLCSDLNE 207
KLT E FD + ++ I T+ + LK VI IF KA EP + MYAQLC + +
Sbjct: 616 KLTLEMFDAISSEILAIANISVWETNGETLKAVIEQIFLKACDEPHWSSMYAQLCGKVVK 675
Query: 208 KL-PSFPSEESGGKEITFKRLLLD----NCQEAFE--GAGKL-REELAQMTSPEQETERR 259
+L P E + GK T +L+L C F+ KL E PE +E
Sbjct: 676 ELNPDITDETNEGK--TGPKLVLHYLVARCHAEFDKGWTDKLPTNEDGTPLEPEMMSEEY 733
Query: 260 DKDRLVKIRTLGNIRLIGELLKQKMVPERIVHHIVQELLGAADSNVCPAEENVEAICHFF 319
K R LG +R IG L + ++ +++ + L+ D P+EE +E++
Sbjct: 734 YAAASAKRRGLGLVRFIGFLYRLNLLTGKMMFECFRRLM--KDLTDSPSEETLESVVELL 791
Query: 320 NTIGKQLDESP--------KSRRINDMYFGRLKELSTNPQLAPRMKFMVRDVIDLR-ASN 370
NT+G+Q + + ++ D FG L + +++ R+KF + D+ +LR N
Sbjct: 792 NTVGEQFETDSFRTGQATLEGSQLLDSLFGILDNIIQTAKISSRIKFKLIDIKELRHDKN 851
Query: 371 WVPRREEIKAKTISEIHDEAEKNLGLRPGATAGMRNT 407
W +++ KTI +IH+E E+ L+ + + R T
Sbjct: 852 WNSDKKDNGPKTIQQIHEEEERQRQLKNNSRSNSRRT 888
>IF42_YEAST (P39936) Eukaryotic initiation factor 4F subunit p130
(eIF4F p130) (eIF-4F p130) (mRNA cap-binding protein
complex subunit p130)
Length = 914
Score = 100 bits (250), Expect = 1e-20
Identities = 99/343 (28%), Positives = 157/343 (44%), Gaps = 41/343 (11%)
Query: 87 ERSWDNIKENREFGNTSQVNRQDQPRTNQGGGP----APTLVKAE--VPWSARRGT---- 136
+R D+ + NR G TS+ +R+ + P AP + A +P S + T
Sbjct: 495 KRMGDDRRSNR--GYTSRKDREKAAEKAEEQAPKEEIAPLVPSANRWIPKSRVKKTEKKL 552
Query: 137 -------LSDKDRVLKTVKGILNKLTPEKFDLLKGQLIDSGITS-----ADILKGVISLI 184
L DK+ V + +K +LNKLT E FD + +++D S + LK VI I
Sbjct: 553 APDGKTELFDKEEVERKMKSLLNKLTLEMFDSISSEILDIANQSKWEDDGETLKIVIEQI 612
Query: 185 FDKAVLEPTFCPMYAQLCSDLNEKL-PSFPSEESGGKE--ITFKRLLLDNCQEAFE--GA 239
F KA EP + MYAQLC + + L P+ +E+ GK L+ C E FE A
Sbjct: 613 FHKACDEPHWSSMYAQLCGKVVKDLDPNIKDKENEGKNGPKLVLHYLVARCHEEFEKGWA 672
Query: 240 GKL-REELAQMTSPEQETERRDKDRLVKIRTLGNIRLIGELLKQKMVPERIVHHIVQELL 298
KL E PE ++ K R LG +R IG L ++ +++ + L+
Sbjct: 673 DKLPAGEDGNPLEPEMMSDEYYIAAAAKRRGLGLVRFIGYLYCLNLLTGKMMFECFRRLM 732
Query: 299 GAADSNVCPAEENVEAICHFFNTIGKQLDESP--------KSRRINDMYFGRLKELSTNP 350
D N P+EE +E++ NT+G+Q + + + D F L+ +
Sbjct: 733 --KDLNNDPSEETLESVIELLNTVGEQFEHDKFVTPQATLEGSVLLDNLFMLLQHIIDGG 790
Query: 351 QLAPRMKFMVRDVIDLR-ASNWVPRREEIKAKTISEIHDEAEK 392
++ R+KF + DV +LR +W +++ KTI +IH E E+
Sbjct: 791 TISNRIKFKLIDVKELREIKHWNSAKKDAGPKTIQQIHQEEEQ 833
>PIP1_MOUSE (Q8VE62) Polyadenylate-binding protein-interacting
protein 1 (Poly(A) binding protein-interacting protein
1) (PolyA binding protein-interacting protein 1)
(PABP-interacting protein 1) (PAIP-1)
Length = 400
Score = 55.5 bits (132), Expect = 5e-07
Identities = 60/234 (25%), Positives = 108/234 (45%), Gaps = 27/234 (11%)
Query: 147 VKGILNKLT--PEKFDLLKGQL---IDSGITSADILKGVISLIFDKAVLEPTFCPMYAQL 201
V+ LN LT P F+ Q ++ +T+ D L+ ++ LI+ +A P F M A+L
Sbjct: 83 VQDFLNHLTEQPGSFETEIEQFAETLNGWVTTDDALQELVELIYQQATSIPNFSYMGARL 142
Query: 202 CSDLNEKLPSFPSEESGGKEITFKRLLLDNCQEAFEGAGKLREELAQMTSPEQETERRDK 261
C+ L+ L P +SG F++LLL C+ +E Q ++ T +R
Sbjct: 143 CNYLSHHLTISP--QSG----NFRQLLLQRCRTEYEAKD-------QAAKGDEVTRKRFH 189
Query: 262 DRLVKIRTLGNIRLIGEL--LKQKMVPERIVHHIVQELLGAADSNVCPAEENVEAICHFF 319
+ LG + L E+ ++ I+ ++ELL A SN P ++N+
Sbjct: 190 ---AFVLFLGELYLNLEIKGTNGQVTRADILQVGLRELLNALFSN--PMDDNLICAVKLL 244
Query: 320 NTIGKQLDESPKSRRINDM--YFGRLKELSTNPQLAPRMKFMVRDVIDLRASNW 371
G L+++ K + DM R++ + + + +K M+ +++LR+SNW
Sbjct: 245 KLTGSVLEDTWKEKGKTDMEEIIQRIENVVLDANCSRDVKQMLLKLVELRSSNW 298
>PIP1_HUMAN (Q9H074) Polyadenylate-binding protein-interacting
protein 1 (Poly(A) binding protein-interacting protein
1) (PABP-interacting protein 1) (PAIP-1)
Length = 479
Score = 53.5 bits (127), Expect = 2e-06
Identities = 59/240 (24%), Positives = 110/240 (45%), Gaps = 39/240 (16%)
Query: 147 VKGILNKLT--PEKFDLLKGQL---IDSGITSADILKGVISLIFDKAVLEPTFCPMYAQL 201
V+ LN LT P F+ Q ++ +T+ D L+ ++ LI+ +A P F M A+L
Sbjct: 162 VQDFLNHLTEQPGSFETEIEQFAETLNGCVTTDDALQELVELIYQQATSIPNFSYMGARL 221
Query: 202 CSDLNEKLPSFPSEESGGKEITFKRLLLDNCQEAFEGAGKLREELAQMTSPEQETERRDK 261
C+ L+ L P +SG F++LLL C+ +E ++++ A+
Sbjct: 222 CNYLSHHLTISP--QSG----NFRQLLLQRCRTEYE----VKDQAAK------------G 259
Query: 262 DRLVKIRTLGNIRLIGEL--------LKQKMVPERIVHHIVQELLGAADSNVCPAEENVE 313
D + + R + +GEL ++ I+ ++ELL A SN P ++N+
Sbjct: 260 DEVTRKRFHAFVLFLGELYLNLEIKGTNGQVTRADILQVGLRELLNALFSN--PMDDNLI 317
Query: 314 AICHFFNTIGKQLDESPKSRRINDM--YFGRLKELSTNPQLAPRMKFMVRDVIDLRASNW 371
G L+++ K + DM R++ + + + +K M+ +++LR+SNW
Sbjct: 318 CAVKLLKLTGSVLEDAWKEKGKMDMEEIIQRIENVVLDANCSRDVKQMLLKLVELRSSNW 377
>CA17_HUMAN (Q02388) Collagen alpha 1(VII) chain precursor (Long-chain
collagen) (LC collagen)
Length = 2944
Score = 44.7 bits (104), Expect = 9e-04
Identities = 47/163 (28%), Positives = 60/163 (35%), Gaps = 16/163 (9%)
Query: 398 PGATAGMRNTRVTGVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGIDNDNWEV-PR 456
P G+ T + G +G+ G G P PG GG+ PG PG +PG+PG V P
Sbjct: 1387 PRGPPGLPGTAMKGDKGDRGERGPP--GPGEGGIAPGEPG---LPGLPGSPGPQGPVGPP 1441
Query: 457 TRSMPRGDMSGAQTGGRGQ--SPYLSKPSVINSKLLPQGSSGLISGKNSALVLG-GGTPS 513
+ +GD G GQ SP P + P+G G A G G P
Sbjct: 1442 GKKGEKGDSEDGAPGLPGQPGSPGEQGPRGPPGAIGPKGDRGFPGPLGEAGEKGERGPPG 1501
Query: 514 ALPSNIVSGTEPAPQIPSPVKPV-------SAASPEKPQAPAV 549
S + G P P P P +P PAV
Sbjct: 1502 PAGSRGLPGVAGRPGAKGPEGPPGPTGRQGEKGEPGRPGDPAV 1544
Score = 36.2 bits (82), Expect = 0.31
Identities = 34/119 (28%), Positives = 44/119 (36%), Gaps = 15/119 (12%)
Query: 389 EAEKNLGLRPGATAGMRNTRVTGVQGNTGAGGFPIAR-----PGTGGLMPGMPGARKMPG 443
+ L +PGA G G+ G G P R PG G PG+PG G
Sbjct: 1778 DGRSGLDGKPGAAGPSGPNGAAGKAGDPGRDGLPGLRGEQGLPGPSG-PPGLPGKPGEDG 1836
Query: 444 MPGIDNDNWEV----PRTRSMPRGDMSGAQTGGR-GQSPYLSKPSVINSKLLPQGSSGL 497
PG++ N E R +GD + GR G P ++ PQG GL
Sbjct: 1837 KPGLNGKNGEPGDPGEDGRKGEKGDSGASGREGRDGPKGERGAPGILG----PQGPPGL 1891
Score = 34.3 bits (77), Expect = 1.2
Identities = 28/81 (34%), Positives = 34/81 (41%), Gaps = 7/81 (8%)
Query: 398 PGATAGMRNTRVTGVQGNTGAGGFPIARPGTGGL--MPGMPGARKMPGMPGIDNDNWEVP 455
PG + G +G GA G P PG G PG PG + PG+PG D E
Sbjct: 1283 PGPQGPPGSATAKGERGFPGADGRP-GSPGRAGNPGTPGAPGLKGSPGLPGPRGDPGE-- 1339
Query: 456 RTRSMPRGD--MSGAQTGGRG 474
R P+G+ G GG G
Sbjct: 1340 RGPRGPKGEPGAPGQVIGGEG 1360
Score = 32.0 bits (71), Expect = 5.9
Identities = 27/87 (31%), Positives = 33/87 (37%), Gaps = 5/87 (5%)
Query: 398 PGATAGMRNTRVTGVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGIDNDNWEVPRT 457
PG T G V G+ G G G P G G + G PGA+ G+PG + E R
Sbjct: 2292 PGPT-GAPGQAVVGLPGAKGEKGAP---GGLAGDLVGEPGAKGDRGLPGPRGEKGEAGRA 2347
Query: 458 RSMPRGDMSGAQTGGRGQSPYLSKPSV 484
P Q G G + P V
Sbjct: 2348 -GEPGDPGEDGQKGAPGPKGFKGDPGV 2373
Score = 31.6 bits (70), Expect = 7.7
Identities = 28/95 (29%), Positives = 42/95 (43%), Gaps = 19/95 (20%)
Query: 411 GVQGNTGAGGFPIAR--------PGTGGLMPGMPGARKMPGMPGIDNDNWEV----PRTR 458
G G+ G+ G P R PG G+ PG PG+ G+PGI + +V PR
Sbjct: 2566 GEPGDKGSAGLPGLRGLLGPQGQPGAAGI-PGDPGSPGKDGVPGIRGEKGDVGFMGPRGL 2624
Query: 459 SMPRGDMSGA-----QTGGRGQSPYLSKPSVINSK 488
RG + GA + G +G++ +P + K
Sbjct: 2625 KGERG-VKGACGLDGEKGDKGEAGPPGRPGLAGHK 2658
Score = 31.2 bits (69), Expect = 10.0
Identities = 29/101 (28%), Positives = 35/101 (33%), Gaps = 6/101 (5%)
Query: 398 PGATAGMRNTRVTGVQGNTGAGGFPIARPGTGGLM--PGMPGARKMPGMPGIDNDNWEVP 455
PG T G G G G P R G G + PG PG+ PG G+ D +
Sbjct: 2406 PGQTGPRGEMGQPGPSGERGLAG-PPGREGIPGPLGPPGPPGSVGPPGASGLKGDKGDPG 2464
Query: 456 RTRSMPRGDMSGAQTGGRGQSPYLSKPSVINSKLLPQGSSG 496
PRG+ G P P + P GS G
Sbjct: 2465 VGLPGPRGERGEPGIRGEDGRPGQEGPRGLTG---PPGSRG 2502
>CA18_HUMAN (P27658) Collagen alpha 1(VIII) chain precursor
(Endothelial collagen)
Length = 744
Score = 44.3 bits (103), Expect = 0.001
Identities = 45/155 (29%), Positives = 57/155 (36%), Gaps = 19/155 (12%)
Query: 398 PGATAGMRNTRVTGVQGNTGAGGFP-IARPGTGGLMPGMPGARKMPGMPGIDNDNWEV-- 454
P G+ + G++G G G+P + +PG G MPG PGA MPG G E+
Sbjct: 131 PPGPPGLPGHGIPGIKGKPGPQGYPGVGKPGMPG-MPGKPGAMGMPGAKGEIGQKGEIGP 189
Query: 455 -----PRTRSMPRGDMSGAQTGGRGQSPYLSKPSVINSKLLPQGSSGLISGKNSALVLGG 509
P+ P G + GG G K LP G GL K G
Sbjct: 190 MGIPGPQGPPGPHGLPGIGKPGGPGLPGQPGPKGDRGPKGLP-GPQGLRGPKGDK---GF 245
Query: 510 GTPSALPSNIVSGTEPAPQIPSPVKPVSAASPEKP 544
G P A G + P + P PV KP
Sbjct: 246 GMPGA------PGVKGPPGMHGPPGPVGLPGVGKP 274
Score = 41.6 bits (96), Expect = 0.007
Identities = 39/127 (30%), Positives = 49/127 (37%), Gaps = 14/127 (11%)
Query: 411 GVQGNTGAGGFP-IARPGTGGLM--PGMPGARKMPGMPGIDNDNWEVPRTRSMPRGDMSG 467
G G G GFP I +PG GL PG PGA G PG+ P P G +
Sbjct: 519 GEPGLPGPPGFPGIGKPGVAGLHGPPGKPGALGPQGQPGLPG-----PPGPPGPPGPPAV 573
Query: 468 AQTGGRGQSPYLSKPSVINSKLLPQGSSGLISGKNSALVLGGGTPSALPSNIVSGTEPAP 527
Q YL + + P + G GKN GG +P+ T P P
Sbjct: 574 MPPTPPPQGEYLPDMGLGIDGVKPPHAYGAKKGKN------GGPAYEMPAFTAELTAPFP 627
Query: 528 QIPSPVK 534
+ +PVK
Sbjct: 628 PVGAPVK 634
Score = 31.2 bits (69), Expect = 10.0
Identities = 20/64 (31%), Positives = 29/64 (45%), Gaps = 6/64 (9%)
Query: 388 DEAEKNLGLRPGATAGMRNTRVTGVQGNTGAGGFPIARPGTGGLM-----PGMPGARKMP 442
+ +K + PG + G+ G+ G G P PG GG PG+PG + P
Sbjct: 463 EAGQKGVPGLPGVPGLLGPKGEPGIPGDQGLQG-PPGIPGIGGPSGPIGPPGIPGPKGEP 521
Query: 443 GMPG 446
G+PG
Sbjct: 522 GLPG 525
>CA1G_CHICK (Q90584) Collagen alpha 1(XVII) chain (Bullous
pemphigoid antigen 2) (180 kDa bullous pemphigoid
antigen 2) (Fragment)
Length = 1146
Score = 43.9 bits (102), Expect = 0.001
Identities = 47/167 (28%), Positives = 66/167 (39%), Gaps = 18/167 (10%)
Query: 409 VTGVQGNTGAGGFPIARPGTGGLM-----PGMPGARKMPGMPGIDNDNWEVPRTRSMPRG 463
+TG QG+ G G P PG G G PG R PG+ GI R P G
Sbjct: 303 MTGEQGSRGIPG-PPGEPGAKGPAGQAGRDGQPGERGEPGLMGIPG-----ARGPPGPSG 356
Query: 464 DM-SGAQTGGRGQSPYLSKPSVINSKLLPQGSSGLISGKNSALVLGGGTPSALPSNIVSG 522
D TG +G P +K P +IS + S+ + G P P +
Sbjct: 357 DTGEPGLTGPQGPPGLPGNPGRPGAKGEPGAPGKVISAEGSSTIALPGPPG--PPGPIGP 414
Query: 523 TEPAPQIPSPVKPVSAASPEKP---QAPAVKLNIDDLHRKTVSLLEE 566
T P P +P PV P + P + AV++ I+ + + SL +
Sbjct: 415 TGP-PGVPGPVGPAGLPGQQGPRGEKGSAVEVVIETIKTEVSSLASQ 460
Score = 31.2 bits (69), Expect = 10.0
Identities = 21/49 (42%), Positives = 23/49 (46%), Gaps = 5/49 (10%)
Query: 398 PGATAGMRNTRVTGVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPG 446
PGA + TG G TG G P PG PG PGA+ PG PG
Sbjct: 346 PGARGPPGPSGDTGEPGLTGPQG-PPGLPGN----PGRPGAKGEPGAPG 389
>I4G2_BOVIN (Q95L46) Eukaryotic translation initiation factor 4
gamma 2 (eIF-4-gamma 2) (eIF-4G 2) (eIF4G 2) (p97)
(Fragment)
Length = 136
Score = 42.4 bits (98), Expect = 0.004
Identities = 33/120 (27%), Positives = 60/120 (49%), Gaps = 6/120 (5%)
Query: 530 PSPVKPVSAASPEKPQAPAVKLNIDDLHRKTVSLLEEYFNVRLLDEALQCVEELKAPTYH 589
P ++ A + +KP P K ++L + T +++ EY N +EA+ V E++AP +
Sbjct: 10 PPLIQEKPAKTSKKP--PPSK---EELLKLTETVVTEYLNSGNANEAVNGVREMRAPKHF 64
Query: 590 -PEVVKEAISLGLDKSPPRVEPVANLIEYLFTKKILTARDIGTGCLLFASLLDDIGIDLP 648
PE++ + I L LD+S E ++LI L + I T+ + L + +D+P
Sbjct: 65 LPEMLSKVIILSLDRSDEDKEKASSLISLLKQEGIATSDNFMQAFLNVLDQCPKLEVDIP 124
>CA24_ASCSU (P27393) Collagen alpha 2(IV) chain precursor
Length = 1763
Score = 41.6 bits (96), Expect = 0.007
Identities = 49/182 (26%), Positives = 66/182 (35%), Gaps = 37/182 (20%)
Query: 389 EAEKNLGLRPGATAGMRNTRVTGVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGID 448
+ E+ L PGAT G G+ G G P RPG PG PG +PG+PG
Sbjct: 559 QGERGLPGIPGAT---------GAPGDDGLPGAP-GRPGP----PGPPGQDGLPGLPGQK 604
Query: 449 NDNWEVPRTRSMPRGDMSGAQT---GGRGQSPYLSKPSVINSKLL-----PQGSSGLIS- 499
+ ++ P +T G RGQ KP ++ + L P+G GL
Sbjct: 605 GEPTQLTLRPGPPGYPGQKGETGFPGPRGQEGLPGKPGIVGAPGLPGPPGPKGEPGLTGL 664
Query: 500 ----GKNSALVLGG----------GTPSALPSNIVSGTEPAPQIPSPVKPVSAASPEKPQ 545
GK+ L G G P +G P +P V P+ PE
Sbjct: 665 PEKPGKDGIPGLPGLKGEPGYGQPGMPGLPGMKGDAGLPGLPGLPGAVGPMGPPVPESQL 724
Query: 546 AP 547
P
Sbjct: 725 RP 726
Score = 38.1 bits (87), Expect = 0.082
Identities = 44/168 (26%), Positives = 54/168 (31%), Gaps = 28/168 (16%)
Query: 398 PGATAGMRNTR----VTGVQGNTGAGGFPIARPGTGGL--------------MPGMPGAR 439
P G++ R V G GN GA G P PG G +PG+PG
Sbjct: 83 PPGPQGIKGDRGIIGVPGFPGNDGANGRP-GEPGPPGAPGWDGCNGTDGAPGVPGLPGPP 141
Query: 440 KMPGMPGIDNDNWEVPRTRSMPRGDMSGAQTGGRGQSPYLSKPSVINSKLLPQGSSGLIS 499
MPG PG VP + P +GA G +G + P + P G G
Sbjct: 142 GMPGFPGPPG----VPGMKGEPAIGYAGA-PGEKGDAGMPGMPGLPG----PPGRDGFPG 192
Query: 500 GKNSALVLGGGTPSALPSNIVSGTEPAPQIPSPVKPVSAASPEKPQAP 547
K +G P P P P P PQ P
Sbjct: 193 EKGDRGDVGQAGPRGPPGEAGPPGNPGIGSIGPKGDPGEQGPRGPQGP 240
Score = 38.1 bits (87), Expect = 0.082
Identities = 43/176 (24%), Positives = 65/176 (36%), Gaps = 36/176 (20%)
Query: 398 PGATAGMRNTRVTGVQGNTGAGGFP------IARPGTGGLMPGMPGARKMPGMPGIDNDN 451
PGA + G++G G G P I P +PG+PG +PG+PG+ +
Sbjct: 1240 PGAPGRDGLPGLPGMKGEAGLPGLPGQPGKSITGPKGNAGLPGLPGKDGLPGLPGLKGEP 1299
Query: 452 WE-----------------VPRTRSMPRGDMSGAQTGGRGQSPYLSKPSVINSKLLPQGS 494
+ +P + P +SG G RG KP P G
Sbjct: 1300 GKPGYAGAAGIKGEPGLPGIPGAKGEP--GLSGI-PGKRGNDGIPGKPG-------PAGL 1349
Query: 495 SGLISGKNSALVLGGGTPSALPSNIVSGTEPAPQIPS-PVKPVSAASPEKPQAPAV 549
GL K + + G P+ LP + G + P +P P + P +P P +
Sbjct: 1350 PGLPGMKGESGLPGPQGPAGLPG--LPGLKGEPGLPGFPGQKGETGFPGQPGIPGL 1403
Score = 37.7 bits (86), Expect = 0.11
Identities = 40/156 (25%), Positives = 61/156 (38%), Gaps = 14/156 (8%)
Query: 398 PGATAGMRNTRVTGVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGIDNDNW--EVP 455
P AG+ + G++G G GFP + TG PG PG +PG+PG+ D+ P
Sbjct: 1364 PQGPAGLPG--LPGLKGEPGLPGFPGQKGETG--FPGQPG---IPGLPGMKGDSGYPGAP 1416
Query: 456 RTRSMPRGDMSGAQTGGRGQSPYLSKPSVINSKLLPQGSSGLISGKNSALVLGGGTPSAL 515
P G G P + + ++ P G+ G+ K + G PS
Sbjct: 1417 GRDGAPGKQGEPGPMGPPGAQPIVQRGE--KGEMGPMGAPGIRGEKGLPGLDGLPGPSGP 1474
Query: 516 PSNIVSGTEPAPQIP-SPVKPVSAASPEKPQAPAVK 550
P +G + P P P +P P P ++
Sbjct: 1475 PG--FAGAKGRDGFPGQPGMPGEKGAPGLPGFPGIE 1508
Score = 37.7 bits (86), Expect = 0.11
Identities = 38/142 (26%), Positives = 51/142 (35%), Gaps = 22/142 (15%)
Query: 411 GVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGIDNDNWEVPRTRSMP--RGDMSGA 468
G QG G G P + G +PG PG +PG+PG+ D +P +G+ A
Sbjct: 978 GEQGLAGLPGIPGMKGAPG--IPGAPGQDGLPGLPGVKGDR----GFNGLPGEKGEPGPA 1031
Query: 469 QTGGRGQSPYLSKPSVINSKLLPQGSSGLISGKNSALVLGGGTPSALPSNIVSGTEPAPQ 528
G P L + P G GL K G G P + G + P
Sbjct: 1032 ARDGEKGEPGLPGQPGLRGPQGPPGLPGLPGLKGDEGQPGYGAPGLM------GEKGLPG 1085
Query: 529 IPSPVKPVSAASPEKPQAPAVK 550
+P P +P AP K
Sbjct: 1086 LP--------GKPGRPGAPGPK 1099
Score = 37.4 bits (85), Expect = 0.14
Identities = 40/159 (25%), Positives = 58/159 (36%), Gaps = 32/159 (20%)
Query: 398 PGATAGMRNTRVTGVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGIDNDNWEVPRT 457
PG G +G GA GFP + G +PG PG G+PG+
Sbjct: 1084 PGLPGKPGRPGAPGPKGLDGAPGFPGLKGEAG--LPGAPGLPGQDGLPGL---------- 1131
Query: 458 RSMPRGDMSGAQTGGRGQSPYLSKPSVINSKLLP--QGSSGLISGKNSALVLG----GGT 511
G +G+S + +P ++ LP G+ G+ K A + G G
Sbjct: 1132 ------------PGQKGESGFPGQPGLVGPPGLPGKMGAPGIRGEKGDAGLPGLPGERGL 1179
Query: 512 PSALPSNIVSGTEPAPQIPSPVKPVSAASPEKPQAPAVK 550
+G AP +P PV P +A P P +K
Sbjct: 1180 DGLPGQKGEAGFPGAPGLPGPVGPKGSAG--APGFPGLK 1216
Score = 35.8 bits (81), Expect = 0.41
Identities = 40/145 (27%), Positives = 57/145 (38%), Gaps = 30/145 (20%)
Query: 411 GVQGNTGAGGFPIARPGTGGLM--------PGMPGARKMPGMPGIDNDNWEVPRTRSM-- 460
G+ G G GFP A PG G + PG PG + PG+PG++ P R M
Sbjct: 1181 GLPGQKGEAGFPGA-PGLPGPVGPKGSAGAPGFPGLKGEPGLPGLEGQ----PGPRGMKG 1235
Query: 461 -------PRGDMSGAQTGGRGQSPYLSKPSVINSKLL-PQGSSGL--ISGKNSALVLGG- 509
P D G +G++ P + P+G++GL + GK+ L G
Sbjct: 1236 EAGLPGAPGRDGLPGLPGMKGEAGLPGLPGQPGKSITGPKGNAGLPGLPGKDGLPGLPGL 1295
Query: 510 ----GTPSALPSNIVSGTEPAPQIP 530
G P + + G P IP
Sbjct: 1296 KGEPGKPGYAGAAGIKGEPGLPGIP 1320
Score = 34.7 bits (78), Expect = 0.90
Identities = 18/42 (42%), Positives = 23/42 (53%), Gaps = 6/42 (14%)
Query: 411 GVQGNTGAGGFP-----IARPGTGGLMPGMPGARKMPGMPGI 447
G++G G G P I PG G PG PGA+ PG+PG+
Sbjct: 849 GLEGQRGLPGVPGQKGEIGLPGLAGA-PGFPGAKGEPGLPGL 889
Score = 34.3 bits (77), Expect = 1.2
Identities = 20/59 (33%), Positives = 28/59 (46%), Gaps = 2/59 (3%)
Query: 411 GVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGIDNDNWE--VPRTRSMPRGDMSG 467
G+ G GA G P R G +PG+PG PG+ G+ E +P R P ++G
Sbjct: 439 GLAGPPGAKGEPGPRGVDGQSIPGLPGKDGRPGLDGLPGRKGEMGLPGVRGPPGDSLNG 497
Score = 33.9 bits (76), Expect = 1.5
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 2/39 (5%)
Query: 409 VTGVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGI 447
V+G++G+TG G P G PG PG + PG PG+
Sbjct: 922 VSGMKGDTGLPGVPGLAGPPG--QPGFPGQKGQPGFPGV 958
Score = 32.0 bits (71), Expect = 5.9
Identities = 26/86 (30%), Positives = 38/86 (43%), Gaps = 7/86 (8%)
Query: 411 GVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGIDNDNWEVPRTRSMPRGDMSGAQT 470
G G GA G+P PG G G+PG +PG+PG++ +P + +GD
Sbjct: 379 GQPGPPGADGYP-GPPGPQGPQ-GLPGGPGLPGLPGLEG----LPGPKG-EKGDSGIPGA 431
Query: 471 GGRGQSPYLSKPSVINSKLLPQGSSG 496
G P L+ P + P+G G
Sbjct: 432 PGVQGPPGLAGPPGAKGEPGPRGVDG 457
Score = 31.2 bits (69), Expect = 10.0
Identities = 40/150 (26%), Positives = 49/150 (32%), Gaps = 31/150 (20%)
Query: 398 PGATAGMRNTRVTGVQGNTGAGGFPIARPGTGGLM---PGMPGARKMPGMPGIDNDNWEV 454
PG + G+ G G G P R G + PG PG R G G D +
Sbjct: 461 PGLPGKDGRPGLDGLPGRKGEMGLPGVRGPPGDSLNGLPGPPGPRGPQGPKGYDGRD-GA 519
Query: 455 PRTRSM--PRGDMSG-------AQTGGRGQSPYLSKPSVINSKLLPQGSSGLISGKNSAL 505
P + P+GD G G +G + Y P PQG GL
Sbjct: 520 PGLPGIPGPKGDRGGTCAFCAHGAKGEKGDAGYAGLPG-------PQGERGL-------- 564
Query: 506 VLGGGTPSALPSNIVSGTEPAPQIPSPVKP 535
G P A + G AP P P P
Sbjct: 565 ---PGIPGATGAPGDDGLPGAPGRPGPPGP 591
>CCR6_CAEEL (P20784) Cuticle collagen rol-6
Length = 348
Score = 40.8 bits (94), Expect = 0.013
Identities = 36/105 (34%), Positives = 41/105 (38%), Gaps = 14/105 (13%)
Query: 398 PGATAGMRNTR-VTGVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGIDNDNWEVPR 456
P G+R R G G G G P PG G PG PG+ PG PG D+ E P
Sbjct: 203 PNGAPGLRGMRGARGQPGRPGRDGNP-GMPGDCG-PPGAPGSDGKPGSPGGKGDDGERPL 260
Query: 457 TRSMPRGDMSGA----QTGGRGQSPYLSKPSVINSKLLPQGSSGL 497
R PRG A G G+ Y + PQG GL
Sbjct: 261 GRPGPRGPPGEAGPEGPQGPTGRDAYPGQSG-------PQGEPGL 298
>CA24_CAEEL (P17140) Collagen alpha 2(IV) chain precursor (Lethal
protein 2)
Length = 1758
Score = 40.8 bits (94), Expect = 0.013
Identities = 48/171 (28%), Positives = 66/171 (38%), Gaps = 18/171 (10%)
Query: 389 EAEKNLGLRPGATAGMRNTRVTGVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGID 448
+ +K G PGA + + G+ G G G+ R G +PG+PG PG+ G
Sbjct: 418 KGDKGDGGIPGAPGVSGPSGIPGLPGPKGEPGY---RGTPGQSIPGLPGKDGKPGLDGAP 474
Query: 449 NDNWE--VPRTRSMPRGDMSG-----AQTGGRGQSPYLSKPSVINSKLLPQGSSGLISGK 501
E +P R P ++G Q G G + Y + V P G+ G G
Sbjct: 475 GRKGENGLPGVRGPPGDSLNGLPGAPGQRGAPGPNGYDGRDGVNGLPGAP-GTKGDRGGT 533
Query: 502 NSALVLGGGTPSALPSNIVSGTEPAPQ----IPSPVKPVSAASPEKPQAPA 548
SA G LP SG +P PQ +P PV A + PA
Sbjct: 534 CSACAPGTKGEKGLPG--YSG-QPGPQGDRGLPGMPGPVGDAGDDGLPGPA 581
Score = 39.7 bits (91), Expect = 0.028
Identities = 43/155 (27%), Positives = 56/155 (35%), Gaps = 22/155 (14%)
Query: 388 DEAEKNLGLRPGATA--GMRNTRVTGVQGNTGAGGFPIARPGTGGL--MPGMPGARKMPG 443
D L +PG G+ + G G GFP A+ G GGL +PG PG + MPG
Sbjct: 656 DAGLPGLSGKPGQDGLPGLPGNKGEAGYGQPGQPGFPGAK-GDGGLPGLPGTPGLQGMPG 714
Query: 444 MPGIDND-NWEVPRTRSMPRGDMSGAQTGGRGQSPYLSKPSVINSKLLP-----QGSSGL 497
P +N N P +P G +G+ Y +P + P +G SGL
Sbjct: 715 EPAPENQVNPAPPGQPGLP------GLPGTKGEGGYPGRPGEVGQPGFPGLPGMKGDSGL 768
Query: 498 ISGKNSALVLGGGTPSALPSNIVSGTEPAPQIPSP 532
G P G P IP P
Sbjct: 769 PGPPGLP-----GHPGVPGDKGFGGVPGLPGIPGP 798
Score = 38.9 bits (89), Expect = 0.048
Identities = 38/145 (26%), Positives = 55/145 (37%), Gaps = 25/145 (17%)
Query: 411 GVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGIDNDNWEVPRTRSMPRGDMSGAQT 470
G +G+ GA G P R G +PG+PG PG+PG+ +G++ GA
Sbjct: 1319 GAKGDLGANGIPGKRGEDG--LPGVPGRDGQPGIPGL--------------KGEVGGA-- 1360
Query: 471 GGRGQSPYLSKPSVINSKLLP-----QGSSGLISGKNSALVLGGGTPSALPSNIVSGTEP 525
G GQ + P + LP +G +G G LP + G +
Sbjct: 1361 GLPGQPGFPGIPGLKGEGGLPGFPGAKGEAGFPGTPGVPGYAGEKGDGGLPG--LPGRDG 1418
Query: 526 APQIPSPVKPVSAASPEKPQAPAVK 550
P PV P + P+ P K
Sbjct: 1419 LPGADGPVGPPGPSGPQNLVEPGEK 1443
Score = 36.6 bits (83), Expect = 0.24
Identities = 42/149 (28%), Positives = 54/149 (36%), Gaps = 20/149 (13%)
Query: 389 EAEKNLGLRPGATAGMRNTRVTGVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGID 448
E ++ PG G G+ G +G G+P G GL PG+PG PG PG D
Sbjct: 849 EGQRGFPGAPGLKGG------DGLPGLSGQPGYP-GEKGDAGL-PGVPGREGSPGFPGQD 900
Query: 449 NDNWEVPRTRSMPRGDMSGAQTGGRGQSPYLSKPSVINSKLLPQGSSGLISGKNSALVLG 508
+P M D G G L P + LP G+ G K +A + G
Sbjct: 901 G----LPGVPGMKGEDGLPGLPGVTGLKGDLGAPGQSGAPGLP-GAPGYPGMKGNAGIPG 955
Query: 509 -------GGTPSALPSNIVSGTEPAPQIP 530
GG P N G P +P
Sbjct: 956 VPGFKGDGGLPGLPGLNGPKGEPGVPGMP 984
Score = 36.2 bits (82), Expect = 0.31
Identities = 34/122 (27%), Positives = 49/122 (39%), Gaps = 30/122 (24%)
Query: 398 PGATAGMRNTRVTGVQGNTGAGGFP--------------IARPGTGGL-----MPGMPGA 438
PG N + GV G G GG P PGT G+ +PG+PG
Sbjct: 942 PGYPGMKGNAGIPGVPGFKGDGGLPGLPGLNGPKGEPGVPGMPGTPGMKGNGGLPGLPGR 1001
Query: 439 RKMPGMPGIDNDNWEVPRTRSMP--RGDMS-GAQTGGRGQSPYLSKPSVINSKLLPQGSS 495
+ G+PG+ D +P +G+ A+ G +G + +P + PQG S
Sbjct: 1002 DGLSGVPGMKGDR----GFNGLPGEKGEAGPAARDGQKGDAGLPGQPGLRG----PQGPS 1053
Query: 496 GL 497
GL
Sbjct: 1054 GL 1055
Score = 35.0 bits (79), Expect = 0.69
Identities = 44/158 (27%), Positives = 55/158 (33%), Gaps = 10/158 (6%)
Query: 397 RPGATAGMRNTRVTGVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGIDNDNWEVPR 456
RPG+ G+ G G RPG G PG+ G PG PG+ D P
Sbjct: 583 RPGSPGPPGQDGFPGLPGQKGEPTQLTLRPGPPGY-PGLKGENGFPGQPGV--DGLPGPS 639
Query: 457 TRSMPRGDMSGAQTGGRGQSPYLS-KPSVINSKLLP--QGSSGLISGKNSAL--VLGGGT 511
P G G P LS KP LP +G +G G G
Sbjct: 640 GPVGPPGAPGYPGEKGDAGLPGLSGKPGQDGLPGLPGNKGEAGYGQPGQPGFPGAKGDGG 699
Query: 512 PSALPSNIVSGTEPAPQIPSPVKPVSAASPEKPQAPAV 549
LP G + P P+P V+ A P +P P +
Sbjct: 700 LPGLPG--TPGLQGMPGEPAPENQVNPAPPGQPGLPGL 735
Score = 33.9 bits (76), Expect = 1.5
Identities = 32/95 (33%), Positives = 40/95 (41%), Gaps = 14/95 (14%)
Query: 393 NLGLRPGATAGMRNTRVTGVQGNTGAGGFPIAR-----PGTGGLMPGMPGARKMPGMPGI 447
N GL PG G + GV G G+ GFP + PG G PG+ G R PG PG+
Sbjct: 804 NPGL-PGLN-GQKGEPGVGVPGQPGSPGFPGLKGDAGLPGLPGT-PGLEGQRGFPGAPGL 860
Query: 448 DNDNWEVPRTRSMP-----RGDMSGAQTGGRGQSP 477
+ +P P +GD GR SP
Sbjct: 861 KGGD-GLPGLSGQPGYPGEKGDAGLPGVPGREGSP 894
Score = 33.9 bits (76), Expect = 1.5
Identities = 43/153 (28%), Positives = 58/153 (37%), Gaps = 22/153 (14%)
Query: 411 GVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGIDNDNWE-------VPRTRSMPRG 463
G+ G G GFP +PG GL PG+ G + M G+PG+ + P +
Sbjct: 1227 GIPGLKGDSGFP-GQPGQEGL-PGLSGEKGMGGLPGMPGQPGQSIAGPVGPPGAPGLQGK 1284
Query: 464 DMSGAQTGGRGQSPYLSKPSVINSKLLPQGSSGLISGKNSALVLG-GGTP-----SALPS 517
D G +G+S P K G SG+ + LG G P LP
Sbjct: 1285 DGFPGLPGQKGESGLSGLPGAPGLK----GESGMPGFPGAKGDLGANGIPGKRGEDGLPG 1340
Query: 518 NIVSGTEPAPQIPSPVKPVSAAS-PEKPQAPAV 549
V G + P IP V A P +P P +
Sbjct: 1341 --VPGRDGQPGIPGLKGEVGGAGLPGQPGFPGI 1371
Score = 33.9 bits (76), Expect = 1.5
Identities = 27/87 (31%), Positives = 32/87 (36%), Gaps = 19/87 (21%)
Query: 411 GVQGNTGAGGFP-------IARPGTGGL--MPGMPGARKMPGMPGIDNDNWEVPRTRSMP 461
G+ G G G P + PG GL +PG PG R GMPG+D P
Sbjct: 1418 GLPGADGPVGPPGPSGPQNLVEPGEKGLPGLPGAPGLRGEKGMPGLDGP----------P 1467
Query: 462 RGDMSGAQTGGRGQSPYLSKPSVINSK 488
D G RG Y P + K
Sbjct: 1468 GNDGPPGLPGQRGNDGYPGAPGLSGEK 1494
Score = 33.9 bits (76), Expect = 1.5
Identities = 18/36 (50%), Positives = 20/36 (55%), Gaps = 2/36 (5%)
Query: 411 GVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPG 446
G+ G G GG P PG G PG PGA +PG PG
Sbjct: 1489 GLSGEKGMGGLP-GFPGLDG-QPGGPGAPGLPGAPG 1522
Score = 33.5 bits (75), Expect = 2.0
Identities = 43/148 (29%), Positives = 47/148 (31%), Gaps = 26/148 (17%)
Query: 388 DEAEKNLGLRPGATAGMRNTRVTGVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGI 447
D LG PG G G G G P PG G G PG +PG PG
Sbjct: 320 DRGLDGLGGIPGLPGQKGEAGYPGRDGPKGNSG-PPGPPGGGTFNDGAPGPPGLPGRPG- 377
Query: 448 DNDNWEVPRTRSMPRGDMSG---AQTGGRGQSPYLSKPSVINSKLLPQGSSGLISGKNSA 504
N P T P TGG G Y + P+G G
Sbjct: 378 ---NPGPPGTDGYPGAPGPAGPIGNTGGPGLPGYPGNEGLPG----PKGDKG-------- 422
Query: 505 LVLGGGTPSALPSNIVSGTEPAPQIPSP 532
GG P A P VSG P +P P
Sbjct: 423 ---DGGIPGA-PG--VSGPSGIPGLPGP 444
Score = 32.7 bits (73), Expect = 3.4
Identities = 17/50 (34%), Positives = 26/50 (52%), Gaps = 2/50 (4%)
Query: 398 PGATAGMRNTRVTGVQGNTGAGGFPI-ARPGTGGLMPGMPGARKMPGMPG 446
PG + N + G+ G G G + +PG+ G PG+ G +PG+PG
Sbjct: 796 PGPKGDVGNPGLPGLNGQKGEPGVGVPGQPGSPGF-PGLKGDAGLPGLPG 844
Score = 32.7 bits (73), Expect = 3.4
Identities = 20/45 (44%), Positives = 24/45 (52%), Gaps = 3/45 (6%)
Query: 409 VTGVQGNTGAGGFPIAR--PGTGGLMPGMPGARKMPGMPGIDNDN 451
++G G G G P R G GGL PG+PG R M G PG +N
Sbjct: 1144 LSGAPGLDGQPGVPGIRGDKGQGGL-PGIPGDRGMDGYPGQKGEN 1187
Score = 32.0 bits (71), Expect = 5.9
Identities = 21/54 (38%), Positives = 23/54 (41%), Gaps = 5/54 (9%)
Query: 397 RPGATAGMRNTRVTGVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGIDND 450
RPG G G GA G P RPG PGMPG PGM G+ +
Sbjct: 109 RPGEPGPPGAPGWDGCNGTDGAPGIP-GRPGP----PGMPGFPGPPGMDGLKGE 157
Score = 31.6 bits (70), Expect = 7.7
Identities = 17/41 (41%), Positives = 22/41 (53%), Gaps = 2/41 (4%)
Query: 411 GVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGIDNDN 451
G+ G G G P PG G PG PG +PG+PG+ D+
Sbjct: 1197 GLGGEKGFAGTP-GFPGLKG-SPGYPGQDGLPGIPGLKGDS 1235
Score = 31.2 bits (69), Expect = 10.0
Identities = 30/97 (30%), Positives = 41/97 (41%), Gaps = 17/97 (17%)
Query: 395 GLR-PGATAGMRNTRVTGVQGNTGAGGF-PIARPGTGGL-----------MPGMPGARKM 441
GLR P +G+ V G +G TG G+ +PG GL PG PG +
Sbjct: 1045 GLRGPQGPSGLPG--VPGFKGETGLPGYGQPGQPGEKGLPGIPGKAGRQGAPGSPGQDGL 1102
Query: 442 PGMPGIDNDNWEVPRTRSMPRGDMSG--AQTGGRGQS 476
PG PG+ ++ + R + G Q G GQS
Sbjct: 1103 PGFPGMKGESGYPGQDGLPGRDGLPGVPGQKGDLGQS 1139
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.135 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 91,095,365
Number of Sequences: 164201
Number of extensions: 4440610
Number of successful extensions: 14194
Number of sequences better than 10.0: 233
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 217
Number of HSP's that attempted gapping in prelim test: 11932
Number of HSP's gapped (non-prelim): 1321
length of query: 720
length of database: 59,974,054
effective HSP length: 118
effective length of query: 602
effective length of database: 40,598,336
effective search space: 24440198272
effective search space used: 24440198272
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 69 (31.2 bits)
Medicago: description of AC140546.1


