

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140033.2 - phase: 0
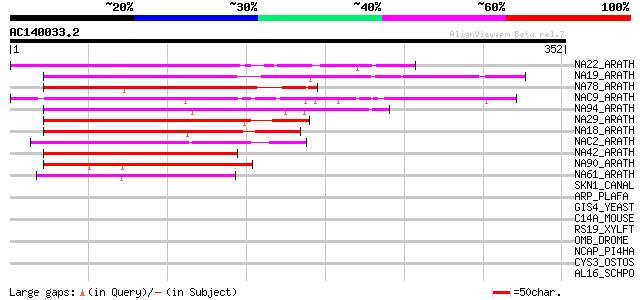
(352 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
NA22_ARATH (Q84TE6) NAC-domain containing protein 21/22 (ANAC021... 181 2e-45
NA19_ARATH (Q9C932) NAC-domain containing protein 19 (ANAC019) (... 172 9e-43
NA78_ARATH (Q84K00) NAC-domain containing protein 78 (ANAC078) 172 1e-42
NAC9_ARATH (Q9ZVH0) Putative NAC-domain containing protein 9 (AN... 170 6e-42
NA94_ARATH (Q9FIW5) Putative NAC-domain containing protein 94 (A... 169 1e-41
NA29_ARATH (O49255) NAC-domain containing protein 29 (ANAC029) (... 161 2e-39
NA18_ARATH (Q9ZNU2) NAC-domain containing protein 18 (ANAC018) (... 160 3e-39
NAC2_ARATH (Q39013) NAC-domain containing protein 2 (ANAC002) 157 4e-38
NA42_ARATH (Q9SK55) Putative NAC-domain containing protein 42 (A... 144 3e-34
NA90_ARATH (Q9FMR3) NAC-domain containing protein 90 (ANAC090) 102 1e-21
NA61_ARATH (Q9M290) Putative NAC-domain containing protein 61 (A... 96 1e-19
SKN1_CANAL (P87024) Beta-glucan synthesis-associated protein SKN1 37 0.075
ARP_PLAFA (P04931) Asparagine-rich protein (AG319) (ARP) (Fragment) 35 0.22
GIS4_YEAST (Q04233) Protein GIS4 32 1.8
C14A_MOUSE (Q6GQT0) Dual specificity protein phosphatase CDC14A ... 32 1.8
RS19_XYLFT (Q87E78) 30S ribosomal protein S19 32 2.4
OMB_DROME (Q24432) Optomotor-blind protein (Lethal(1)optomotor-b... 32 2.4
NCAP_PI4HA (P17240) Nucleocapsid protein 31 4.1
CYS3_OSTOS (Q06544) Cathepsin B-like cysteine proteinase 3 (EC 3... 31 4.1
AL16_SCHPO (P87244) Spindle pole body component alp16 (Altered p... 31 5.4
>NA22_ARATH (Q84TE6) NAC-domain containing protein 21/22 (ANAC021)
(ANAC022)
Length = 324
Score = 181 bits (460), Expect = 2e-45
Identities = 102/262 (38%), Positives = 151/262 (56%), Gaps = 23/262 (8%)
Query: 1 MGSNENVSNQKMENEKVKFDAGVRFFPTDEELINQYLVKK-VDDNSFCAIAIAEVDMNKC 59
M + E + + + K G RF P D+EL+ YL+++ + +N + + +VD+NKC
Sbjct: 1 METEEEMKESSISMVEAKLPPGFRFHPKDDELVCDYLMRRSLHNNHRPPLVLIQVDLNKC 60
Query: 60 EPWDLPEMAKMGETEWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMK 119
EPWD+P+MA +G +WYF+ RD+KY TG RTNRAT GYWKATGKD+ I + L+GM+
Sbjct: 61 EPWDIPKMACVGGKDWYFYSQRDRKYATGLRTNRATATGYWKATGKDRTILRKGKLVGMR 120
Query: 120 KTLVFYKGRAPRGEKSNWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRV 179
KTLVFY+GRAPRG K++WVMHE+RL+G S H P H+LS+ +WV+ RV
Sbjct: 121 KTLVFYQGRAPRGRKTDWVMHEFRLQG---SHHP--------PNHSLSS-PKEDWVLCRV 168
Query: 180 FEKRNCGKKMNGSKLGRSNSSREEPSNTNAASLWAPFLEF----SPYNSENKITIPDFSN 235
F K G + S +E ++ + L P++ F S Y S++ I +
Sbjct: 169 FHKNTEGVICRDN----MGSCFDETASASLPPLMDPYINFDQEPSSYLSDDHHYI--INE 222
Query: 236 EFNSFTNPNQSEKPKTQYDNIV 257
F+N +Q++ + N V
Sbjct: 223 HVPCFSNLSQNQTLNSNLTNSV 244
>NA19_ARATH (Q9C932) NAC-domain containing protein 19 (ANAC019)
(ANAC) (Abscicic-acid-responsive NAC)
Length = 317
Score = 172 bits (437), Expect = 9e-43
Identities = 105/310 (33%), Positives = 155/310 (49%), Gaps = 25/310 (8%)
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGETEWYFFCVR 81
G RF+PTDEEL+ QYL +K F IAE+D+ K +PW LP A GE EWYFF R
Sbjct: 17 GFRFYPTDEELMVQYLCRKAAGYDFSLQLIAEIDLYKFDPWVLPNKALFGEKEWYFFSPR 76
Query: 82 DKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRGEKSNWVMHE 141
D+KYP G R NR +GYWKATG DK I +G+KK LVFY G+AP+G K+NW+MHE
Sbjct: 77 DRKYPNGSRPNRVAGSGYWKATGTDKIISTEGQRVGIKKALVFYIGKAPKGTKTNWIMHE 136
Query: 142 YRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKKM---NGSKLGRSN 198
YRL + P + + +WV+ R+++K++ +K NG R
Sbjct: 137 YRL---------------IEPSRRNGSTKLDDWVLCRIYKKQSSAQKQVYDNGIANAREF 181
Query: 199 SSREEPSNTNAASLWAPFLEFSPYNSENKITIPDFSNEFN-SFTNPNQSEKPKTQYDNIV 257
S+ S T+++S + L+ +N+ FSN S P+ +E+ KT + +
Sbjct: 182 SNNGTSSTTSSSSHFEDVLDSFHQEIDNRNF--QFSNPNRISSLRPDLTEQ-KTGFHGLA 238
Query: 258 HNNETSILNISSSSKQMDVYPLAGATVADPNLTSMAGNSSNFFFSQEFSFGREFDADADI 317
+ + + + + + P G + PNL G +E F+ ++
Sbjct: 239 DTSNFDWASFAGNVEHNNSVPELGMSHVVPNLEYNCGYLKT---EEEVESSHGFNNSGEL 295
Query: 318 SSVVYGNDMF 327
+ YG D F
Sbjct: 296 AQKGYGVDSF 305
>NA78_ARATH (Q84K00) NAC-domain containing protein 78 (ANAC078)
Length = 567
Score = 172 bits (435), Expect = 1e-42
Identities = 87/176 (49%), Positives = 112/176 (63%), Gaps = 18/176 (10%)
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMG--ETEWYFFC 79
G RF PTDEEL+ YL +KV + F AI+ D+ K EPWDLP+ +K+ + EWYFF
Sbjct: 12 GFRFHPTDEELVRYYLKRKVCNKPFKFDAISVTDIYKSEPWDLPDKSKLKSRDLEWYFFS 71
Query: 80 VRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRGEKSNWVM 139
+ DKKY G +TNRAT GYWK TGKD+EI G+ ++GMKKTLV++KGRAPRGE++NWVM
Sbjct: 72 MLDKKYSNGSKTNRATEKGYWKTTGKDREIRNGSRVVGMKKTLVYHKGRAPRGERTNWVM 131
Query: 140 HEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKKMNGSKLG 195
HEYRL L K + E +V+ R+F+K G K NG + G
Sbjct: 132 HEYRLSDEDLKKAGVPQE---------------AYVLCRIFQKSGTGPK-NGEQYG 171
>NAC9_ARATH (Q9ZVH0) Putative NAC-domain containing protein 9
(ANAC009)
Length = 418
Score = 170 bits (430), Expect = 6e-42
Identities = 121/340 (35%), Positives = 179/340 (52%), Gaps = 33/340 (9%)
Query: 1 MGSNENVSNQKMENEKVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCE 60
MG N +QKME+ + G RF PTDEEL++ YL +KV N I ++D+ K +
Sbjct: 1 MGDRNNDGDQKMEDVLLP---GFRFHPTDEELVSFYLKRKVQHNPLSIELIRQLDIYKYD 57
Query: 61 PWDLPEMAKMGETEWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIY--KGNSLIGM 118
PWDLP+ A GE EWYF+C RD+KY R NR T AG+WKATG D+ IY +GN IG+
Sbjct: 58 PWDLPKFAMTGEKEWYFYCPRDRKYRNSSRPNRVTGAGFWKATGTDRPIYSSEGNKCIGL 117
Query: 119 KKTLVFYKGRAPRGEKSNWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTR 178
KK+LVFYKGRA +G K++W+MHE+RL SLS+ + P F + +S + W + R
Sbjct: 118 KKSLVFYKGRAAKGVKTDWMMHEFRLP--SLSEPS--PPSKRFFDSPVSPN--DSWAICR 171
Query: 179 VFEKRNCG--KKMNGS-------KLGRSNSSREEPSNT---NAASLWAPFLEFSPYNSEN 226
+F+K N + ++ S + S ++ SNT ++ + P F ++ EN
Sbjct: 172 IFKKTNTTTLRALSHSFVSSLPPETSTDTMSNQKQSNTYHFSSDKILKPSSHFQ-FHHEN 230
Query: 227 KITIPDFSNEFNSFTNPNQSEKPKTQYDNIVHNNETSILNISSSSKQMDVYPLAGAT-VA 285
T P S NS T + P + D ++ T++ N S Q + L AT
Sbjct: 231 MNT-PKTS---NSTTPSVPTISPFSYLDFTSYDKPTNVFNPVSCLDQQYLTNLFLATQET 286
Query: 286 DPNLTSMAGNSSNFFF----SQEFSFGREFDADADISSVV 321
P + ++ F S + +F EF + D+S+V+
Sbjct: 287 QPQFPRLPSSNEIPSFLLNTSSDSTFLGEFTSHIDLSAVL 326
>NA94_ARATH (Q9FIW5) Putative NAC-domain containing protein 94
(ANAC094)
Length = 337
Score = 169 bits (427), Expect = 1e-41
Identities = 95/231 (41%), Positives = 134/231 (57%), Gaps = 12/231 (5%)
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGETEWYFFCVR 81
G RF PTDEEL++ YL +KV S I +VD+ K +PWDLP++A MGE EWYF+C R
Sbjct: 23 GFRFHPTDEELVSFYLKRKVLHKSLPFDLIKKVDIYKYDPWDLPKLAAMGEKEWYFYCPR 82
Query: 82 DKKYPTGQRTNRATNAGYWKATGKDKEIYKGNS--LIGMKKTLVFYKGRAPRGEKSNWVM 139
D+KY R NR T G+WKATG D+ IY +S IG+KK+LVFY+GRA +G K++W+M
Sbjct: 83 DRKYRNSTRPNRVTGGGFWKATGTDRPIYSLDSTRCIGLKKSLVFYRGRAAKGVKTDWMM 142
Query: 140 HEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSE------WVVTRVFEKRNC---GKKMN 190
HE+RL S S H+ +P +N +H + + E W + R+F+K N + +
Sbjct: 143 HEFRLPSLSDSHHSSYPNYNNKKQHLNNNNNSKELPSNDAWAICRIFKKTNAVSSQRSIP 202
Query: 191 GSKLGRSNSSREEPSNTNAASLWAPFLEFSPYNSENKITIPDFSNEFNSFT 241
S + + + S+ N A+L A S ++ IP NE SFT
Sbjct: 203 QSWVYPTIPDNNQQSHNNTATLLASSDVLSHISTRQNF-IPSPVNEPASFT 252
>NA29_ARATH (O49255) NAC-domain containing protein 29 (ANAC029)
(NAC2) (NAC-LIKE, ACTIVATED BY AP3/PI protein) (NAP)
Length = 268
Score = 161 bits (408), Expect = 2e-39
Identities = 80/171 (46%), Positives = 105/171 (60%), Gaps = 15/171 (8%)
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGETEWYFFCVR 81
G RF PTDEELI YL + I EVD+ K +PW LPE + GE EWYFF R
Sbjct: 12 GFRFHPTDEELIVYYLRNQTMSKPCPVSIIPEVDIYKFDPWQLPEKTEFGENEWYFFSPR 71
Query: 82 DKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRGEKSNWVMHE 141
++KYP G R NRA +GYWKATG DK I+ G+S +G+KK LVFYKGR P+G K++W+MHE
Sbjct: 72 ERKYPNGVRPNRAAVSGYWKATGTDKAIHSGSSNVGVKKALVFYKGRPPKGIKTDWIMHE 131
Query: 142 YRLEGN--SLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKKMN 190
YRL + + +K N + + EWV+ R+++KR K +N
Sbjct: 132 YRLHDSRKASTKRN-------------GSMRLDEWVLCRIYKKRGASKLLN 169
>NA18_ARATH (Q9ZNU2) NAC-domain containing protein 18 (ANAC018) (NO
APICAL MERISTEM protein) (AtNAM)
Length = 320
Score = 160 bits (406), Expect = 3e-39
Identities = 77/167 (46%), Positives = 102/167 (60%), Gaps = 11/167 (6%)
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGETEWYFFCVR 81
G RF PTDEEL+ YL +K D IA+VD+ K +PW+LP A GE EWYFF R
Sbjct: 20 GFRFHPTDEELVIHYLKRKADSVPLPVAIIADVDLYKFDPWELPAKASFGEQEWYFFSPR 79
Query: 82 DKKYPTGQRTNRATNAGYWKATGKDKEIYK----GNSLIGMKKTLVFYKGRAPRGEKSNW 137
D+KYP G R NRA +GYWKATG DK + G+ +G+KK LVFY G+ P+G KS+W
Sbjct: 80 DRKYPNGARPNRAATSGYWKATGTDKPVISTGGGGSKKVGVKKALVFYSGKPPKGVKSDW 139
Query: 138 VMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRN 184
+MHEYRL N P H + ++ + +WV+ R+++K N
Sbjct: 140 IMHEYRLTDNK-------PTHICDFGNKKNSLRLDDWVLCRIYKKNN 179
>NAC2_ARATH (Q39013) NAC-domain containing protein 2 (ANAC002)
Length = 289
Score = 157 bits (397), Expect = 4e-38
Identities = 76/175 (43%), Positives = 105/175 (59%), Gaps = 12/175 (6%)
Query: 14 NEKVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGET 73
+E ++ G RF PTDEEL+ YL +K S IAE+D+ K +PW+LP +A GE
Sbjct: 2 SELLQLPPGFRFHPTDEELVMHYLCRKCASQSIAVPIIAEIDLYKYDPWELPGLALYGEK 61
Query: 74 EWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRGE 133
EWYFF RD+KYP G R NR+ +GYWKATG DK I +G+KK LVFY G+AP+GE
Sbjct: 62 EWYFFSPRDRKYPNGSRPNRSAGSGYWKATGADKPIGLPKP-VGIKKALVFYAGKAPKGE 120
Query: 134 KSNWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKK 188
K+NW+MHEYRL S ++ + +WV+ R++ K+ ++
Sbjct: 121 KTNWIMHEYRLADVDRSVRK-----------KKNSLRLDDWVLCRIYNKKGATER 164
>NA42_ARATH (Q9SK55) Putative NAC-domain containing protein 42
(ANAC042)
Length = 275
Score = 144 bits (363), Expect = 3e-34
Identities = 61/123 (49%), Positives = 87/123 (70%)
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGETEWYFFCVR 81
G RF PTDEEL+ YL +KV++ + I ++D+ K +PWDLP ++ +GE EWYFFC+R
Sbjct: 21 GFRFHPTDEELLGYYLRRKVENKTIKLELIKQIDIYKYDPWDLPRVSSVGEKEWYFFCMR 80
Query: 82 DKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRGEKSNWVMHE 141
+KY R NR T +G+WKATG DK +Y +G+KK+LV+Y G A +G K++W+MHE
Sbjct: 81 GRKYRNSVRPNRVTGSGFWKATGIDKPVYSNLDCVGLKKSLVYYLGSAGKGTKTDWMMHE 140
Query: 142 YRL 144
+RL
Sbjct: 141 FRL 143
>NA90_ARATH (Q9FMR3) NAC-domain containing protein 90 (ANAC090)
Length = 235
Score = 102 bits (255), Expect = 1e-21
Identities = 55/140 (39%), Positives = 85/140 (60%), Gaps = 7/140 (5%)
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAI--AIAEVDMNKCEPWDLPEMAKM---GETE-W 75
G RF+PT+EEL++ YL +++ S ++ I +D+ + EP LP +A + G+ E W
Sbjct: 8 GFRFYPTEEELVSFYLRNQLEGRSDDSMHRVIPVLDVFEVEPSHLPNVAGVRCRGDAEQW 67
Query: 76 YFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIY-KGNSLIGMKKTLVFYKGRAPRGEK 134
+FF R ++ G R +R T +GYWKATG ++ K N +IG KKT+VFY G+AP G K
Sbjct: 68 FFFVPRQEREARGGRPSRTTGSGYWKATGSPGPVFSKDNKMIGAKKTMVFYTGKAPTGRK 127
Query: 135 SNWVMHEYRLEGNSLSKHNI 154
+ W M+EY +++ I
Sbjct: 128 TKWKMNEYHAVDETVNASTI 147
>NA61_ARATH (Q9M290) Putative NAC-domain containing protein 61
(ANAC061)
Length = 228
Score = 95.9 bits (237), Expect = 1e-19
Identities = 52/132 (39%), Positives = 76/132 (57%), Gaps = 6/132 (4%)
Query: 18 KFDAGVRFFPTDEELINQYL-VKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAK---MGET 73
+ G RF+PT+ EL+ YL ++ N+ I +D+ EP LP +A G+
Sbjct: 4 ELSVGFRFYPTEVELLTYYLRIQLGGGNATIHSLIPILDVFSVEPTQLPNLAGERCRGDA 63
Query: 74 E-WYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYK-GNSLIGMKKTLVFYKGRAPR 131
E W FF R ++ G R +R T +GYWKATG ++ N +IG+KKT+VFY G+AP
Sbjct: 64 EQWIFFVPRQEREARGGRPSRTTGSGYWKATGSPGPVFSPDNRVIGVKKTMVFYTGKAPT 123
Query: 132 GEKSNWVMHEYR 143
G K+ W M+EY+
Sbjct: 124 GRKTKWKMNEYK 135
>SKN1_CANAL (P87024) Beta-glucan synthesis-associated protein SKN1
Length = 737
Score = 37.0 bits (84), Expect = 0.075
Identities = 40/185 (21%), Positives = 74/185 (39%), Gaps = 17/185 (9%)
Query: 156 PEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKKMNGSKLGRSNSSREEPSNTNAASLWAP 215
P HN F +++ S + F + N S N+ + PS + S
Sbjct: 59 PPHNPFSDNSHENSTESLYQSETRFHQPLLHNDSNNSNSSIGNNRQRIPSQQHDTS---- 114
Query: 216 FLEFSPYNSENKITIPDFSNEFNSFTNPNQSEKPKTQYDNIVHNNETSILNISSSSKQMD 275
S Y++ T P SN F S+++ NQ E + +Y+ + + ++ + S +S D
Sbjct: 115 ----SLYSASPISTSPLVSN-FQSYSD-NQDEMTRGKYNQNTNRSSSNYIQHSPTSAGYD 168
Query: 276 VYPLAGATVADPNLTSMAGNSSN-------FFFSQEFSFGREFDADADISSVVYGNDMFQ 328
YPL + +++ + +SS+ + S + D+ +VY N F
Sbjct: 169 RYPLKTQSSIGGSMSRIGLSSSSPSQQQQQHMYDNNSSNRSSYSPDSATDLMVYENGEFS 228
Query: 329 RWSGY 333
+ GY
Sbjct: 229 PFGGY 233
>ARP_PLAFA (P04931) Asparagine-rich protein (AG319) (ARP) (Fragment)
Length = 537
Score = 35.4 bits (80), Expect = 0.22
Identities = 25/117 (21%), Positives = 51/117 (43%), Gaps = 3/117 (2%)
Query: 147 NSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEK--RNCGKKMNGSKLGRSNSSREEP 204
N S +N + +N N + M+ + + N MN + N++
Sbjct: 247 NQNSNNNFYMNYNYQNRKNSMNNNMNNNMNNNMNHNMNNNMNHNMNNNMNHNMNNNMNHN 306
Query: 205 SNTNAASLWAPFLEFSP-YNSENKITIPDFSNEFNSFTNPNQSEKPKTQYDNIVHNN 260
N N ++ + + SP Y++ K+++ +++N ++ NPNQ +T DN+ N
Sbjct: 307 MNNNMNNINSLDSDMSPNYHAHVKMSMMNYNNNESNTANPNQMNFEQTNNDNMKREN 363
>GIS4_YEAST (Q04233) Protein GIS4
Length = 774
Score = 32.3 bits (72), Expect = 1.8
Identities = 39/187 (20%), Positives = 75/187 (39%), Gaps = 18/187 (9%)
Query: 150 SKHNIFPEHNLFPEHNLS--THGMSEWVVTRVFEKRNCGKKMNGSKLGRSNSSREEPSNT 207
SK N P+ +FP + T SE + F + G L + + RE+ S
Sbjct: 249 SKPNRLPK--IFPSYTNEDYTPSHSEIMSIDSFAGEDVSSTYPGQDLSLTTARREDESGQ 306
Query: 208 NAAS---------LWAPFLEFSPYNSENKITIPDFSNEFNSFT---NPNQSEKPKTQYDN 255
+ L ++ + Y+ E+ ++ +S+ NS + + + S + + +N
Sbjct: 307 DEVEDHYSRVSHDLGDESIDQASYSMESSVSYTSYSSSSNSSSAHYSLSSSSRGNPKREN 366
Query: 256 IVHNNETSILNISSSSKQMDVYPLAGATVADPNLTSMAGNSSNFFFSQEFSFGREFDADA 315
I H N T + +SS + +D + + NL ++ + ++ G E D
Sbjct: 367 IDHTNATYVSELSSITSSIDNLTTSTTPEEEDNLIHHNYDAQGYGSGED--DGEEVYDDE 424
Query: 316 DISSVVY 322
D+SS Y
Sbjct: 425 DLSSSDY 431
>C14A_MOUSE (Q6GQT0) Dual specificity protein phosphatase CDC14A (EC
3.1.3.48) (EC 3.1.3.16) (CDC14 cell division cycle 14
homolog A)
Length = 554
Score = 32.3 bits (72), Expect = 1.8
Identities = 30/117 (25%), Positives = 41/117 (34%), Gaps = 13/117 (11%)
Query: 192 SKLGRSNSSREEPSNTNAASLWAPFLEFSPYNSENKITIPDFSNEFNSFTNPNQSEKPKT 251
S LG N+ EEP SL SP+ S F N N E
Sbjct: 434 SSLGNLNAGTEEPETKKTTSLTKAAFIASPFTS--------FLNGSTQTPGRNYPELNNN 485
Query: 252 QYDNIVHNNETSIL-----NISSSSKQMDVYPLAGATVADPNLTSMAGNSSNFFFSQ 303
QY ++N +S N++SS P T+ P+ +SS F S+
Sbjct: 486 QYTRSSNSNSSSSSSGLGGNLNSSPVPQSAKPEEHTTILRPSFPGSLSSSSVRFLSR 542
>RS19_XYLFT (Q87E78) 30S ribosomal protein S19
Length = 89
Score = 32.0 bits (71), Expect = 2.4
Identities = 16/49 (32%), Positives = 29/49 (58%), Gaps = 1/49 (2%)
Query: 143 RLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKKMNG 191
++ G++++ HN H + N+ H + E+ +TRVF K +CG K +G
Sbjct: 41 QMVGHTIAIHNGKNYHPVVINENMVGHKLGEFSITRVF-KGHCGDKKSG 88
>OMB_DROME (Q24432) Optomotor-blind protein
(Lethal(1)optomotor-blind) (L(1)omb) (Bifid protein)
Length = 972
Score = 32.0 bits (71), Expect = 2.4
Identities = 28/105 (26%), Positives = 44/105 (41%), Gaps = 10/105 (9%)
Query: 175 VVTRVFEKRNCGKKMNGSKLGRSNSSREEPSNTN------AASLWAPFLEFSPYNSENKI 228
++T N G +G+ SNS+ SNTN A S + SP ++ +
Sbjct: 45 LLTAGSNNNNSGNTNSGNN--NSNSNNNTNSNTNNTNNLVAVSPTGGGAQLSPQSNHSSS 102
Query: 229 TIPDFSNEFNSFTNPNQSEKPKTQYDNIVHNNETSILNISSSSKQ 273
SN NS +N N + +N +NN + N ++S KQ
Sbjct: 103 NTTTTSNTNNSSSNNNNNNSTHNNNNNHTNNNNNN--NNNTSQKQ 145
>NCAP_PI4HA (P17240) Nucleocapsid protein
Length = 551
Score = 31.2 bits (69), Expect = 4.1
Identities = 16/59 (27%), Positives = 30/59 (50%), Gaps = 1/59 (1%)
Query: 186 GKKMNGSKLGRSNSSREEPSNTNAASLWAPFLEFSPYNSE-NKITIPDFSNEFNSFTNP 243
G +++ + G S ++E +T S F NS+ N++ +P+F N+ N F +P
Sbjct: 437 GARLSNYEQGWSGIDQDETRDTLPESTMHRFQNIDSTNSDHNELQMPEFENDINPFNHP 495
>CYS3_OSTOS (Q06544) Cathepsin B-like cysteine proteinase 3 (EC
3.4.22.-) (Fragment)
Length = 174
Score = 31.2 bits (69), Expect = 4.1
Identities = 17/53 (32%), Positives = 24/53 (45%), Gaps = 3/53 (5%)
Query: 59 CEPWDLPEMAKMGETEWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYK 111
C P++ P + G+ +Y C K P Q+T GY KA +DK K
Sbjct: 22 CRPYEFPPCGRHGKEPYYGECYDTAKTPKCQKT---CQRGYLKAYKEDKHFGK 71
>AL16_SCHPO (P87244) Spindle pole body component alp16 (Altered
polarity protein 16)
Length = 759
Score = 30.8 bits (68), Expect = 5.4
Identities = 39/182 (21%), Positives = 69/182 (37%), Gaps = 22/182 (12%)
Query: 118 MKKTLVFYKGRA----PRGEKSNWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHG--M 171
MK L+++ P S++V + EG K +F F + + +
Sbjct: 5 MKNDLLWFGNHVFDLTPNSFDSDFVEQLSKFEGKEGLKRKLFDSSEYFQNFSFQVNDDLL 64
Query: 172 SEWVVTRVFEKR------NCGKKMNGSKLGRSNSSREEPSNTNAASLWAPFLEFSP--YN 223
+ V+ + ++ C N + S E ++ + + SP +
Sbjct: 65 GKDVILDISNQQLTPAVPGCETSSNSKLISASKEITSERKRAKSSVSPSYLTDSSPSDLS 124
Query: 224 SENKITIPDFSNEFNSFTN-----PNQSEKPKTQYDNIVHNNETSILNISSSSKQMDVYP 278
ENK+ + S + +F N P SE P YD +HN + S + S+ + VYP
Sbjct: 125 VENKVLLTCPSWDGENFKNLEARSPFISEAPSRVYDYFLHNQDWSPKPLFSA---LQVYP 181
Query: 279 LA 280
LA
Sbjct: 182 LA 183
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.313 0.130 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,892,658
Number of Sequences: 164201
Number of extensions: 1892113
Number of successful extensions: 3098
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 3070
Number of HSP's gapped (non-prelim): 30
length of query: 352
length of database: 59,974,054
effective HSP length: 111
effective length of query: 241
effective length of database: 41,747,743
effective search space: 10061206063
effective search space used: 10061206063
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 66 (30.0 bits)
Medicago: description of AC140033.2


